Abstract
The genome of the radiation-resistant eubacterium Deinococcus radiodurans contains an ortholog of an RNA-binding protein known as the Ro 60-kD autoantigen. This protein, which was previously identified only in higher eukaryotes, is normally bound to small RNAs known as Y RNAs. We show that the Ro protein ortholog Rsr contributes to the resistance of D. radiodurans to UV irradiation. Rsr binds several small RNAs, encoded upstream of rsr, that accumulate following UV irradiation. One of these RNAs resembles a Y RNA. These results suggest that Ro RNPs could similarly contribute to the recovery of higher cells following UV irradiation.
Keywords: Ro ribonucleoproteins, D. radiodurans, UV irradiation
A major target of the immune response in patients suffering from the systemic rheumatic diseases Sjogren's syndrome and systemic lupus erythematosus is a 60-kD RNA-binding protein known as the Ro autoantigen (for review, see von Muhlen and Tan 1995). In most higher eukaryotic cells, the Ro 60 kDa protein is complexed with one of several small cytoplasmic RNAs known as Y RNAs. The number of distinct Y RNAs present in Ro RNPs varies, ranging from four in humans and Xenopus to one in Caenorhabditis elegans (Hendrick et al. 1981; O'Brien et al. 1993; Van Horn et al. 1995). Although there is little primary sequence conservation among different Y RNAs (both within and across species), all Y RNAs can be folded into a structure containing a large internal loop and a long stem formed by base-pairing the 5′ and 3′ ends (Van Horn et al. 1995). Experimental evidence that the stem and loop exist has been provided by chemical and enzymatic modification experiments (van Gelder et al. 1994; Green et al. 1998) and by oligonucleotide-directed RNase H cleavage (Matera et al. 1995). Within the stem is a highly conserved bulged helix that is the binding site for the Ro protein (Green et al. 1998). As the level of the C. elegans Y RNA is drastically decreased in worms lacking the Ro protein, binding by Ro may stabilize Y RNAs from degradation (Labbe et al. 1999).
Although Ro RNPs were first described nearly 20 years ago, their function has remained mysterious. One of the few clues has come from the finding that in Xenopus laevis oocytes, the Ro protein is complexed with a large class of variant 5S rRNA molecules (O'Brien and Wolin 1994). To synthesize the vast numbers of ribosomes required for early development, X. laevis contains ∼20,000 genes encoding the major oocyte 5S rRNA, many of which contain sequence heterogeneities (Peterson et al. 1980). The 5S rRNA variants bound by the Ro protein contain 8–10 additional nucleotides at their 3′ ends and are apparently generated by readthrough of the first termination signal for RNA polymerase III. In addition to being longer than mature 5S rRNA, all of the 5S rRNA variants bound by the Ro protein contain one or more point mutations. These mutations disrupt the normal 5S rRNA structure, causing the variant 5S rRNAs to misfold into an alternative structure that is bound by the 60-kD Ro protein (Shi et al. 1996). Because the misfolded, variant 5S rRNAs are inefficiently processed to 5S rRNA and eventually degraded, the Ro protein was proposed to function in a quality control pathway for ribosome biogenesis (O'Brien and Wolin 1994). It is not known what role the Y RNAs have in this process.
The recently sequenced genome of the eubacterium Deinococcus radiodurans contains a potential ortholog of the Ro 60-kD protein (White et al. 1999). This bacterium is remarkable for its ability to tolerate exposure to DNA-damaging agents, yet little is known of the mechanisms involved (for review, see Battista 1997). Here we show that the Rsr protein (Ro sixty related) contributes to the resistance of D. radiodurans to UV irradiation. D. radiodurans cells lacking rsr are more sensitive to UV damage than are wild-type cells. During recovery from UV irradiation, the levels of Rsr increase, consistent with a role in repairing radiation damage. Rsr binds several small RNAs, encoded upstream of rsr, that are major transcripts during recovery from UV irradiation. Remarkably, at least one of these small RNAs resembles the Y RNAs bound by the Ro autoantigen in higher eukaryotes. The finding that Ro RNPs contribute to the survival of D. radiodurans following UV irradiation suggests that this mysterious class of RNPs could similarly function in the recovery of higher eukaryotic cells following irradiation with UV light.
Results and Discussion
An ortholog of the Ro 60-kD protein in D. radiodurans
The genome sequence of D. radiodurans (White et al. 1999) predicts the existence of a protein, encoded on chromosome I, that is highly related in sequence to all previously identified Ro proteins (Fig. 1). This was unexpected, as Ro proteins have not been identified in either budding yeast (Goffeau et al. 1996) or other microbial genomes. As one explanation for the presence of a Ro protein in D. radiodurans would be the lateral transfer of the gene from a eukaryote, we used several sequence analysis methods to examine this possibility. Codon usage for nine of the most frequent amino acids present in Rsr was consistent with the codon usage of 50 housekeeping genes located on D. radiodurans chromosome I (data not shown). A phylogenetic tree constructed from the nucleotide sequences of the Ro protein coding regions from humans, X. laevis, C. elegans, and D. radiodurans placed Rsr in a clade separate from the C. elegans and vertebrate clades (not shown). Although alignment of the amino acid sequences reveals Rsr to be about as related to the vertebrate Ro proteins as the worm Ro ortholog, the C. elegans protein contains several inserts that are not present in Rsr or the vertebrate sequences (Fig. 1). These results, coupled with the location of Ro on the deinococcal chromosome rather than on the deinococcal plasmids [which may have been transferred from another species (White et al. 1999)], imply that if there was a transfer from a vertebrate species, it was not a recent one.
Figure 1.

Comparison of human and worm Ro proteins with D. radiodurans Rsr. Rsr (D.r.) is aligned with the human (H.s.; Deutscher et al. 1988) and C. elegans (C.e.; Van Horn et al. 1995) Ro 60-kD proteins. The alignment was created with MegAlign (Lasergene) using the PAM250 similarity matrix. Similar residues were defined as D = E, H = K = R, A = G, I = L = V, F = W = Y, S = T. The RNA recognition motif (Birney et al. 1993) is boxed. Two conserved sequences within the motif, RNP1 and RNP2, are indicated by overlines. The human and D. radiodurans sequences are 31.8% identical and 45% similar, whereas the C. elegans and D. radiodurans proteins are 29.8% identical and 50% similar.
Rsr contributes to the resistance of D. radiodurans to UV irradiation
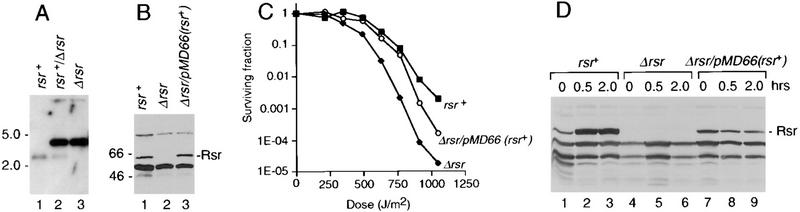
To determine the phenotype of cells lacking rsr, we constructed an allele of this gene in which most of the coding sequence was replaced by the Escherichia coli cat gene fused to a deinococcal promoter (Gutman et al. 1993). Introduction of this allele into a wild-type strain and selection on medium containing chloramphenicol resulted in isolates in which 1 or more of the 4–10 independently segregating copies of chromosome I were replaced with Δrsr chromosomes. Southern blotting confirmed that, at least for some isolates, all of the wild-type chromosomes were replaced by chromosomes carrying the mutant allele (Fig. 2A, lane 3). We verified that Rsr was undetectable in these strains by subjecting extracts from wild-type and Δrsr strains to protein immunoblotting using an antibody against Rsr (Fig. 2B, lane 2). The growth of Δrsr strains on agar plates was indistinguishable from that of wild-type strains.
Figure 2.
Rsr contributes to the survival of D. radiodurans following UV irradiation. (A) Genomic DNA was isolated from wild-type cells (lane 1) and cells that were converted to Δrsr as described in Materials and methods (lanes 2,3). Following digestion with StuI and BsrGI, the DNA was subjected to Southern blotting and probed with a random-primed HindIII–NruI fragment containing 226 bp of 3′ coding sequence of rsr and 341 bp of 3′ flanking sequence. In wild-type cells, the probe detects a 2.8-kb StuI–BsrGI fragment that contains most of the rsr coding region and 3′ flanking sequences. In Δrsr cells, this fragment is 4.7 kb, due to replacement of rsr with the cat gene. The increased intensity of the Δrsr band, relative to the wild-type band, is most likely due to gene amplification that occurs during the drug selection step (Agostini et al. 1996). (B) Extracts from wild-type cells (lane 1), Δrsr cells (lane 2), and Δrsr/pMD66(rsr+) cells (lane 3) were subjected to Western blot analysis. (C) Wild-type cells (█), Δrsr cells (♦), and Δrsr/pMD66(rsr+) cells (○) were subjected to irradiation with the indicated doses of UV light. After irradiation, aliquots were removed, plated on TGY agar, and counted to determine the fraction of surviving cells. Shown are the means for three separate experiments. For each radiation dose, the t-test was used to evaluate the statistical significance of the data. By comparing the ratio of the survival of the wild-type vs. the Δrsr strains, the differences in survival rates were significant at 490 J/m2 and higher doses (P < 0.007, using the Bernoulli correction for multiple comparisons). Furthermore, the differences between the survival rates of the wild-type and Δrsr/pMD66(rsr+) cells were not significant at any of the depicted doses, although the survival rate of Δrsr/pMD66(rsr+) was always less than wild-type cells at 912 J/m2 and higher doses. (D) Wild-type cells (lanes 1–3), Δrsr cells (lanes 4–6), and Δrsr/pMD66(rsr+) cells were irradiated with 490 J/m2 of UV light. Following irradiation, cells were allowed to recover in TGY broth for either 30 min (lanes 2,5,8) or 2 hr (lanes 3,6,9). Cell extracts were prepared and subjected to immunoblotting using anti-Rsr antibodies. Extracts from unirradiated cells were loaded in lanes 1, 4, and 7. Because antibodies to other deinococcal proteins have not been described, the blot was standardized by loading equal amounts of protein as judged by Coomassie blue staining.
To determine whether Rsr contributes to the resistance of D. radiodurans to irradiation, we subjected the wild-type and Δrsr strains to UV irradiation. For the wild-type strain, the survival rate as a function of UV dose (Fig. 2C, squares) was similar to published data for D. radiodurans (e.g., Battista 1997). At levels of irradiation >490 J/m2, strains lacking Rsr were significantly less resistant to UV than wild-type strains. This decreased resistance ranged from 3.5-fold at 490 J/m2 to ∼100-fold at 912 J/m2 and above (Fig. 2C). However, these cells were still more resistant to UV irradiation than E. coli, which have a survival rate of <0.1% upon exposure to 200 J/m2 of irradiation (Battista 1997).
To verify that the decreased resistance in Δrsr strains was due to loss of Rsr, we examined the UV sensitivity of a Δrsr strain carrying a plasmid in which rsr was expressed under the control of a heterologous deinococcal promoter. Expression of rsr increased the resistance of Δrsr cells to UV irradiation [Δrsr/pMD66(rsr+) cells, Fig. 2C], confirming that the heightened sensitivity of Δrsr cells was at least partially due to loss of Rsr. At lower levels of irradiation (770 J/m2 or less), Δrsr/pMD66(rsr+) cells behaved similarly to wild-type cells. However, at higher doses, the survival of these cells was intermediate between Δrsr and wild-type cells.
Because expression of rsr on the plasmid did not fully restore the radiation resistance of Δrsr cells, we examined the levels of Rsr in the various strains following exposure to UV light. The levels of Rsr in wild-type cells increased fourfold within 30 min of exposure to 490 J/m2 of UV light (Fig. 2D, cf. lanes 1 and 2). However, although the levels of Rsr were slightly higher in the Δrsr/pMD66(rsr+) cells in the absence of irradiation, the protein did not increase following exposure to UV light (lanes 7–9). Thus, the fact that rsr on the plasmid does not fully restore the radiation resistance of Δrsr cells may be due to a failure of the heterologous promoter to be up-regulated following UV irradiation.
Several small RNAs accumulate in rsr+ cells following UV irradiation
To determine the consequences of UV irradiation on RNA metabolism, we subjected wild-type and Δrsr strains to irradiation with 490 J/m2 and examined the RNA profiles during recovery. Several small RNAs accumulated following UV irradiation (Fig. 3A). Immediately after irradiation, a band appeared in both wild-type and Δrsr strains that was slightly smaller than mature 5S rRNA (designated 5S* in Fig. 3A). Using direct RNA sequencing and Northern hybridization, we identified this band as a form of the D. radiodurans 5S rRNA. As this faster-migrating form of 5S rRNA appeared immediately upon exposure to UV irradiation, it may represent 5S rRNA containing an RNA:RNA cross-link similar to the 5S rRNA cross-link that occurs in eukaryotic cells following UV irradiation (Branch et al. 1985).
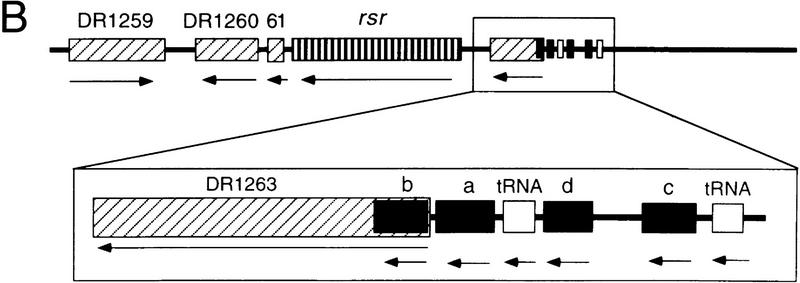
Figure 3.
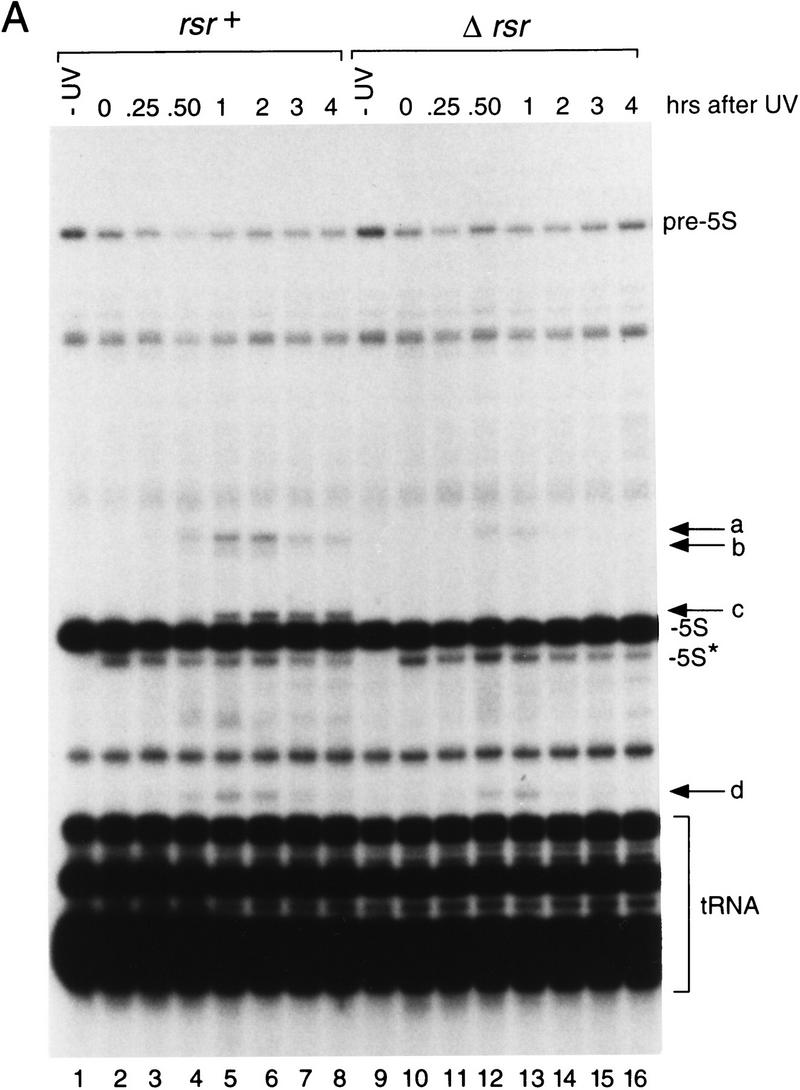
Small RNAs encoded upstream of rsr accumulate during recovery from irradiation with UV light. (A) Wild-type (lanes 1–8) and Δrsr (lanes 9–16) cells were either not irradiated (lanes 1,9) or irradiated with 490 J/m2 of UV light (lanes 2–8,10–16). At the times indicated, cell aliquots were removed and total RNA extracted and labeled at the 3′ end. (pre-5S) 5S rRNA that is mature at the 3′ end and contains spacer sequences at the 5′ end; (5S*) a form of mature 5S rRNA. Consistent with the presence of a UV cross-link, several attempts to synthesize cDNA complementary to the 5S* RNA were unsuccessful. (B) Map of the rsr locus, showing the positions of small RNAs a–d. The tRNA gene between a and d encodes tRNATyr; tRNAAsp is encoded 5′ of RNA c. (GenBank accession nos. are AF233654 (a), AF233655 (b), AF233656 (c), and AF233657 (d).
During recovery from irradiation, at least four small RNAs accumulated in wild-type cells (Fig. 3A, a–d). These RNAs ranged in size from 146 (RNA a) to 96 nucleotides (RNA d). Interestingly, two of these RNAs appeared less prominent in Δrsr cells (Fig. 3A, a,d), whereas the remaining two RNAs were undetectable (b,c). The identities of the RNAs were determined by synthesizing and sequencing cDNAs complementary to the RNAs. This revealed that all four RNAs were encoded upstream of rsr (Fig. 3B). One of these RNAs overlaps the coding sequence of an ORF encoded upstream of rsr. Although this ORF, DR1263, is homologous to a predicted ORF in the eubacterium Streptomyces coelicolor, its function is unknown.
Rsr binds a small RNA that resembles a Y RNA
To examine whether Rsr bound any deinococcal RNAs, we performed immunoprecipitations with anti-Rsr antibodies. In unirradiated cells, no RNAs were detected (Fig. 4A, lanes 1–4). However, when wild-type cells were irradiated and then allowed to recover for 2 hr, several RNAs were present in the immunoprecipitate (Fig. 4A, lane 6). The most intense bands corresponded to two small RNAs that accumulate in rsr+ cells following irradiation (b and c). In addition, RNAs a and d were present, although in far lower amounts.
Figure 4.
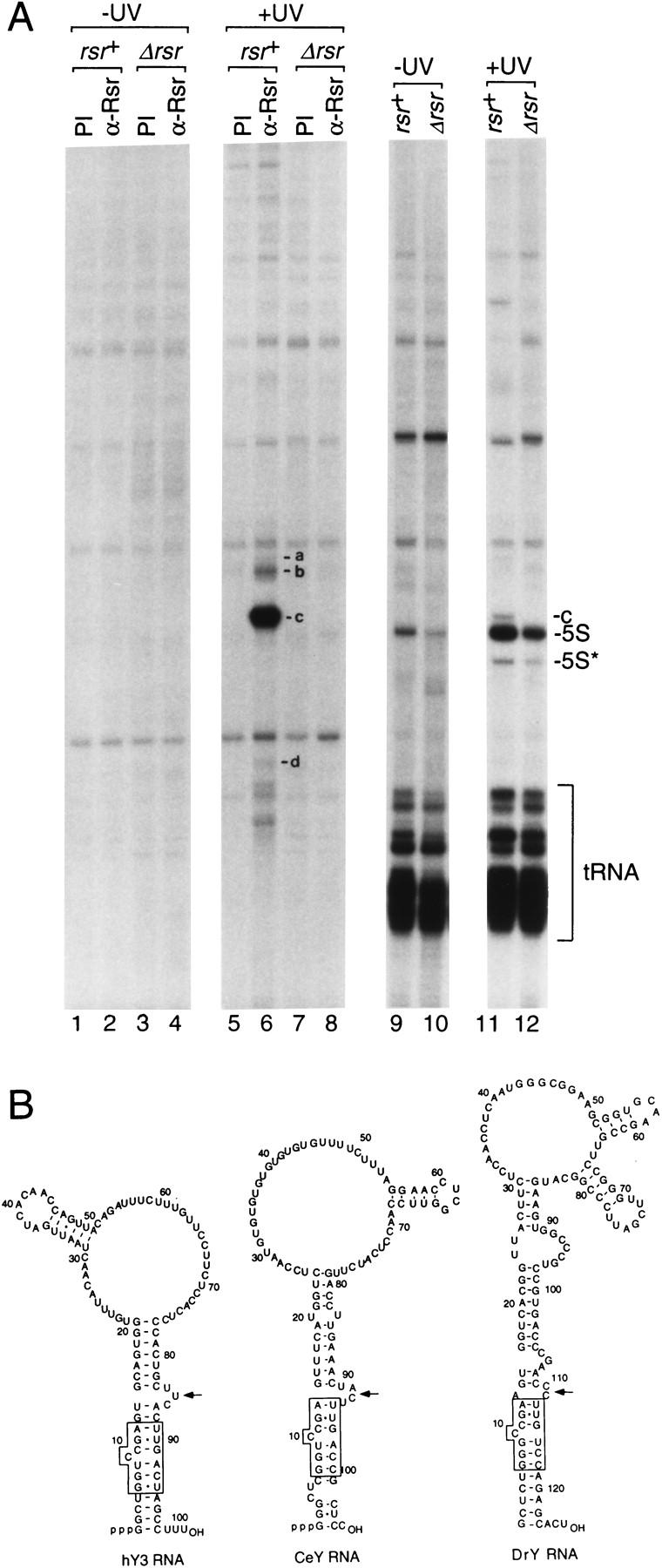
The major RNA bound by Rsr resembles a Y RNA. (A) Wild-type (lanes 1,2,5,6,9,11) and Δrsr (lanes 3,4,7,8,10,12) cells were either not irradiated (lanes 1–4,9–10) or irradiated with 490 J/m2 of UV light (lanes 5–8,11–12). Following recovery, extracts were prepared and subjected to immunoprecipitation with the indicated antibodies. RNAs within immunoprecipitates (lanes 1–8) and total RNA (lanes 9–12) were extracted and labeled at the 3′ end with [32P]pCp. RNAs a–d in lane 6 were identified by sequencing cDNAs made from the RNAs. The two bands migrating below d were subjected to cDNA synthesis and found to be degradation products of c. (B) Potential secondary structures of human, C. elegans, and D. radiodurans Y RNAs. The human Y3 RNA structure was proposed by O'Brien et al. (1993) and the CeY RNA structure was proposed by Van Horn et al. (1995). The D. radiodurans structure was derived using MFOLD (Mathews et al. 1999).
Remarkably, the major RNA in the immunoprecipitate, c, can be folded to resemble a Y RNA, the RNAs bound by Ro proteins in higher eukaryotes (Fig. 4B). Specifically, this RNA can be drawn as a structure containing a large internal loop and a long stem formed by base-pairing the 5′ and 3′ ends. A defining feature of Y RNAs, a conserved bulged helix that is the binding site of the X. laevis Ro protein on the Xenopus Y3 RNA (Green et al. 1998), is largely present in the D. radiodurans RNA (Fig. 4B, box). The only difference is that one of the conserved basepairs in the helix, U-A, has been changed to G · U. Furthermore, all Y RNAs contain either a 3- to 4-nucleotide bulge or a small internal loop adjacent to the helix. In Xenopus Y3 RNA, this bulge is required for binding by the Ro protein, most likely because it widens the major groove of the helix, making it accessible to protein side chains (Green et al. 1998). Similarly, a small internal loop is present adjacent to the conserved helix in the D. radiodurans RNA (Fig. 4B, arrow). Finally, as was observed for the C. elegans Y RNA (Labbe et al. 1999), this RNA fails to accumulate in Δrsr cells (Fig. 3A), consistent with the idea that binding by the Ro protein stabilizes Y RNAs. We designate RNA c as a D. radiodurans Y RNA.
Functional implications
We do not yet know the precise roles of Rsr and its associated RNAs in assisting in the recovery of D. radiodurans from UV irradiation. However, it is intriguing that a protein that binds misfolded, defective 5S rRNA precursors has been acquired by an organism capable of surviving massive radiation damage. Interestingly, although Δrsr cells display decreased resistance to UV irradiation, they are identical to wild-type cells in their sensitivity to X-ray damage (J.R. Battista, pers. comm.). Thus, the role of Rsr may be specialized for recovery from UV-induced damage. UV irradiation has well-documented effects on both RNA molecules and RNA–protein complexes, such as the introduction of both RNA: RNA and RNA–protein cross-links (Doudney 1968). Thus, Rsr could have a role in a pathway by which defective cross-linked RNAs are recognized and degraded. Alternatively, Rsr and its associated RNAs may function to repair DNA damage, such as pyrimidine dimers, or other cellular damage that is unique to UV irradiation. Whatever the mechanism, our results reveal that in addition to previously described DNA-repair proteins such as DNA polymerase I and RecA (Gutman et al. 1993, 1994), RNA-binding proteins and their associated RNAs contribute to the radiation resistance of this extraordinary organism. Moreover, although the Rsr-associated RNAs accumulate following UV irradiation, we can detect RNA c in unirradiated cells by Northern blotting (X. Chen and S. Wolin, unpubl.), suggesting that both Rsr and Y RNA may function in normal cellular metabolism.
The finding that Ro RNPs contribute to the resistance of D. radiodurans to UV irradiation suggests that these RNPs could function similarly in the response to UV light in higher eukaryotes. In preliminary experiments, we did not detect an increase in the levels of the Ro 60-kD protein following irradiation of cultured mouse cells with UV light (X. Chen and S. Wolin, unpubl.). However, following UV irradiation-induced apoptosis of keratinocytes, anti-Ro antibodies stain small blebs on the cell surface that also contain ribosomes and ER markers (Casciola-Rosen et al. 1994). Thus, the cellular distribution of the Ro 60-kD protein may change as a consequence of UV damage. In this regard, it is intriguing that the presence of anti-Ro antibodies in patients suffering from rheumatic disease is highly correlated with photosensitive dermatitis (von Muhlen and Tan 1995). Although it remains controversial whether autoantibodies enter cells in vivo (see McCauliffe 1996), it is a tantalizing possibility that the observed photosensitivity may be due to loss of Ro RNP function. Further experiments, in which higher eukaryotic cells and organisms that lack the 60-kD Ro protein are assayed for their response to UV irradiation, will be required to address this question.
Materials and methods
Strains, plasmids, and media
D. radiodurans RI was purchased from the American Type Culture Collection and grown at 30°C in TGY (0.8% tryptone, 0.1% glucose, 0.4% yeast extract). Transformation was performed in liquid culture (Udupa et al. 1994). Plasmids pI3 (Masters and Minton 1992) and pMD66 (Daly et al. 1994) were gifts of K. Minton and M. Daly.
Antisera
To prepare antibodies, the oligonucleotides 5′-AATTGGATCCATGAAGAACTTGCTCCGTGCCATCAAC-3′ and 5′-AATTCTGCAGATCTCGCCCCGCGCAAAAGCCGTCATCAC-3′ were used to amplify DNA encoding amino acids 1–531 of Rsr. After digestion with BamHI and PstI, the DNA was inserted into pTrcHisA (Invitrogen Corp., Carlsbad, CA). The recombinant protein was solubilized in 6 m guanidine-HCl and purified from E. coli lysates under denaturing conditions using a ProBond column (Invitrogen) as described by the manufacturer. Because the protein precipitated during dialysis, it was subjected to a denaturation and renaturation procedure described previously (Van Horn et al. 1995), except that heparin was added to 100 μg/ml in the renaturation and dialysis buffers. Following dialysis against renaturation buffer containing 5 μg/ml heparin, the protein was concentrated by dialysis against dry sucrose and used to immunize rabbits.
Disruption of rsr+
Targeted disruption was performed as described (Gutman et al. 1993). DNA containing 193 bp of 5′ coding sequence of rsr and 1.8 kb of 5′-flanking sequence was amplified using 5′-AATTTCTAGACGCTTGGTAATCTCTGTGTGCC-3′, and 5′-AATTTCGCGACCTGCACGGTGTGTTTCTGC-3′ and digested with XbaI and NruI. DNA containing 226 bp of 3′ coding sequence of rsr and 2.7 kb of 3′-flanking region was amplified with 5′-AATTTCGCGATCGTCGTCTACACCGACACAAC-3′ and 5′-AATTGGATCCACTTCGAGAAGATTCCGAAC-3′ and digested with BamHI and NruI. These fragments were cloned into the XbaI–BamHI sites of pBluescriptSK (Stratagene) to generate pDRoR1. A 3.1-kb StuI–AviII fragment containing a promoterless cat gene fused to a deinococcal-promoting region was excised from pI3 (Masters and Minton 1992) and inserted into pDRoR1 at the NruI site, generating pDRoR15. This plasmid was introduced into D. radiodurans cells and recombinants selected by plating on TGY containing 3 μg/ml chloramphenicol. Because pDRoR15 cannot replicate in D. radiodurans, transformation conferred Cmr by recombination. After further selection on chloramphenicol to encourage replacement of the multiple copies of the wild-type chromosome by Δrsr chromosomes, strains in which the Cmr marker had correctly inserted into the rsr locus were identified by Southern analysis.
To express rsr in the Δrsr strain, the rsr coding sequence was amplified using 5′-ATGAAGAACTTGCTCCGTGCCATCAAC-3′ and 5′-AATTGGATCCTCAAACCTCGCCCCGCGCAAAAGCCGTC-3′. The DNA was digested with BamHI and ligated adjacent to the deinococcal-promoting sequences in the kanamycin-resistant plasmid pMD66 (Daly et al. 1994). For this construct, pMD66 was digested with EcoRI, modified with Klenow to produce blunt ends, and digested further with BamHI. Ligation of rsr resulted in plasmid pMDRRo1, in which Rsr was expressed under control of the deinococcal promoter. Plasmid pMDRRo1 was introduced into the Δrsr strain and transformants selected on TGY containing 8 μg/ml kanamycin.
Survival measurements
Survival measurements were performed as described (Gutman et al. 1993). Cells were grown to OD600 = 0.8, pelleted, and resuspended in PBS (137 mm NaCl, 2.7 mm KCl, 5.3 mm Na2HPO4, 1.8 mm KH2PO4 at pH 7.4) at an OD600 of 0.06. Cells were placed in plastic petri dishes (with the lids off) and irradiated at room temperature with constant agitation using a germicidal UV lamp (254 nm) at a dose rate of 1.0 J/m2/sec. At intervals, aliquots were removed, diluted, plated on TGY agar, and incubated at 30°C for 3 days.
RNA extraction and analysis
Total RNA was extracted from D. radiodurans using the hot phenol method described for Saccharomyces cerevisiae by Ausubel et al. (1998). Briefly, cells were pelleted, washed with water, and resuspended in 10 mm Tris-HCl (pH 7.5), 10 mm EDTA, and 0.5% SDS, and an equal volume of acid phenol was added. After vortexing, the mixture was incubated for 30 min at 65°C with occasional vortexing. Following a second phenol extraction and a chloroform extraction, RNA was precipitated with ethanol. RNAs were labeled with [32P]pCp and T4 RNA ligase (England et al. 1980) and fractionated in 5% polyacrylamide/8 m urea gels. Direct RNA sequencing using base-specific ribonucleases was performed as described (Donis-Keller et al. 1977). To prepare cDNA, a deoxyoligonucleotide was ligated to the 3′ end of the desired RNA and subjected to reverse transcription and PCR amplification (O'Brien and Wolin 1994).
Immunoprecipitation and immunoblotting
For immunoprecipitations, D. radiodurans cells were grown to OD600 = 0.8, washed with water and resuspended in 40 mm Tris-HCl (pH 7.4), 150 mm NaCl, 0.1% NP-40, 0.5 mm PMSF, 1 mm EGTA, 1 mm MgCl2, 1 mm DTT, 200 U/ml RNasin, 10 mm vanadyl ribonucleoside complexes, and 125 ng/ml each leupeptin, chymostatin, pepstatin, and antipain. Cells were lysed using a Bead Beater (Biospec Products, Bartlesville, OK) in the presence of glass beads (0.1 mm, Biospec Products). Following sedimentation at 10,000g for 10 min to remove debris, lysates were cleared by centrifugation at 100,000g in a Beckman TL100.3 rotor for 30 min at 4°C. The supernatants were incubated with antisera and processed as described (Wolin and Steitz 1984). RNAs in immunoprecipitates were labeled using [32P]pCp and T4 RNA ligase (England et al. 1980) and fractionated in 5% polyacrylamide/8 m urea gels. For immunoblotting, cells were resuspended in SDS-PAGE buffer (83 mm Tris base, 30% glycerol, 6.8% SDS, 120 mm DTT), disrupted by vortexing with glass beads, and heated to 95°C for 5 min. After fractionating the extracts in SDS–polyacrylamide gels and transferring to nitrocellulose, the membranes were blocked and incubated with antisera as described (Yoo and Wolin 1994).
Acknowledgments
We are extremely grateful to J. Battista for performing the X-ray survival analysis on the wild-type and Δrsr strains. We also thank D. Noe for assistance with statistical analyses, M. Daly and K. Minton for providing plasmids and advice, and K.B. Low for use of his UV light meter and helpful advice. We thank K.B. Low, M. Solomon, and E. Ullu for comments on the manuscript. S.L.W. is an Associate Investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sandra.wolin@yale.edu; FAX (203) 737-1761.
References
- Agostini HJ, Carroll JD, Minton KW. Identification and characterization of uvrA, a DNA repair gene of Deinococcus radiodurans. J Bacteriol. 1996;178:6759–6765. doi: 10.1128/jb.178.23.6759-6765.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: John Wiley & Sons; 1998. [Google Scholar]
- Battista JR. Against all odds: The survival strategies of Deinococcus radiodurans. Annu Rev Microbiol. 1997;51:203–224. doi: 10.1146/annurev.micro.51.1.203. [DOI] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch AD, Benenfeld BJ, Robertson HD. Ultraviolet light-induced crosslinking reveals a unique region of local tertiary structure in potato spindle tuber viroid and HeLa 5S RNA. Proc Natl Acad Sci. 1985;82:6590–6594. doi: 10.1073/pnas.82.19.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casciola-Rosen LA, Anhalt G, Rosen A. Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J Exp Med. 1994;179:1317–1330. doi: 10.1084/jem.179.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly MJ, Ouyang L, Fuchs P, Minton KW. In vivo damage and recA-dependent repair of plasmid and chromosomal DNA in the radiation-resistant bacterium Deinococcus radiodurans. J Bacteriol. 1994;176:3508–3517. doi: 10.1128/jb.176.12.3508-3517.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher SL, Harley JB, Keene JD. Molecular analysis of the 60-kDa human Ro ribonucleoprotein. Proc Natl Acad Sci. 1988;85:9479–9483. doi: 10.1073/pnas.85.24.9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H, Maxam AM, Gilbert W. Mapping adenines, guanines and pyrimidines in RNA. Nucleic Acids Res. 1977;4:2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudney CO. Ultraviolet light effects on the bacterial cell. Curr Top Microbiol Immunol. 1968;46:116–175. doi: 10.1007/978-3-642-46121-7_4. [DOI] [PubMed] [Google Scholar]
- England TE, Bruce AG, Uhlenbeck OC. Specific labeling of 3′ termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65:65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. Life with 6000 genes. Science. 1996;27:546. doi: 10.1126/science.274.5287.546. , 563–567. [DOI] [PubMed] [Google Scholar]
- Green CD, Long KS, Shi H, Wolin SL. Binding of the 60-kDa Ro autoantigen to Y RNAs: Evidence for recognition in the major groove of a conserved helix. RNA. 1998;4:750–765. doi: 10.1017/s1355838298971667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman PD, Fuchs P, Ouyang L, Minton KW. Identification, sequencing, and targeted mutagenesis of a DNA polymerase gene required for the extreme radioresistance of Deinococcus radiodurans. J Bacteriol. 1993;175:3581–3590. doi: 10.1128/jb.175.11.3581-3590.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman PD, Carroll JD, Masters CI, Minton KW. Sequencing, targeted mutagenesis and expression of a recA gene required for extreme radioresistance of Deinococcus radiodurans. Gene. 1994;141:31–37. doi: 10.1016/0378-1119(94)90124-4. [DOI] [PubMed] [Google Scholar]
- Hendrick JP, Wolin SL, Rinke J, Lerner MR, Steitz JA. Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: Further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol Cell Biol. 1981;1:1138–1149. doi: 10.1128/mcb.1.12.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe JC, Hekimi S, Rokeach LA. The levels of the RoRNP-associated Y RNA are dependent upon the presence of ROP-1, the Caenorhabditis elegans Ro60 protein. Genetics. 1999;151:143–150. doi: 10.1093/genetics/151.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters CI, Minton KW. Promoter probe and shuttle plasmids for Deinococcus radiodurans. Plasmid. 1992;28:258–261. doi: 10.1016/0147-619x(92)90057-h. [DOI] [PubMed] [Google Scholar]
- Matera AG, Frey MR, Margelot K, Wolin SL. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J Cell Biol. 1995;129:1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters provides robust prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- McCauliffe DP. Antibody penetration into the cells of mice and men. J Invest Dermatol. 1996;107:3–4. doi: 10.1111/1523-1747.ep12293642. [DOI] [PubMed] [Google Scholar]
- O'Brien CA, Wolin SL. A possible role for the 60-kD Ro autoantigen in a discard pathway for defective 5S ribosomal RNA precursors. Genes & Dev. 1994;8:2891–2903. doi: 10.1101/gad.8.23.2891. [DOI] [PubMed] [Google Scholar]
- O'Brien CA, Margelot K, Wolin SL. Xenopus Ro ribonucleoproteins: Members of an evolutionarily conserved class of cytoplasmic ribonucleoproteins. Proc Natl Acad Sci. 1993;90:7250–7254. doi: 10.1073/pnas.90.15.7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RC, Doering JL, Brown DD. Characterization of two Xenopus somatic 5S DNAs and one minor oocyte-specific 5S DNA. Cell. 1980;20:131–141. doi: 10.1016/0092-8674(80)90241-x. [DOI] [PubMed] [Google Scholar]
- Shi H, O'Brien CA, Van Horn DJ, Wolin SL. A misfolded form of 5S rRNA is complexed with the Ro and La autoantigens. RNA. 1996;2:769–784. [PMC free article] [PubMed] [Google Scholar]
- Udupa KS, O'Cain PA, Mattimore V, Battista JR. Novel ionizing radiation-sensitive mutants of Deinococcus radiodurans. J Bacteriol. 1994;176:7439–7446. doi: 10.1128/jb.176.24.7439-7446.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelder CWG, Thijssen JPHM, Klaassen ECJ, Sturchler C, Krol A, van Venrooij WJ, Pruijn GJM. Common structural features of the Ro RNP associated hY1 and hY5 RNAs. Nucleic Acids Res. 1994;22:2498–2506. doi: 10.1093/nar/22.13.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn DJ, Eisenberg D, O'Brien CA, Wolin SL. Caenorhabditis elegans embryos contain only one major species of Ro RNP. RNA. 1995;1:293–303. [PMC free article] [PubMed] [Google Scholar]
- von Muhlen CA, Tan EM. Autoantibodies in the diagnosis of systemic rheumatic diseases. Semin Arthritis Rheum. 1995;24:323–358. doi: 10.1016/s0049-0172(95)80004-2. [DOI] [PubMed] [Google Scholar]
- White O, Eisen JA, Heidelberg JF, Hickey EK, Peterson JD, Dodson RJ, Haft DH, Gwinn ML, Nelson WC, Richardson DL, et al. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Steitz JA. The Ro small cytoplasmic ribonucleoproteins: Identification of the antigenic protein and its binding site on the Ro RNAs. Proc Natl Acad Sci. 1984;81:1996–2000. doi: 10.1073/pnas.81.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CJ, Wolin SL. La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: A yeast homolog of the La autoantigen is dispensable for growth. Mol Cell Biol. 1994;14:5412–5424. doi: 10.1128/mcb.14.8.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]