Abstract
The body axis of vertebrates is composed of a serial repetition of similar anatomical modules, termed segments or metameres. This particular mode of organization is especially conspicuous at the level of the periodic arrangement of vertebrae in the spine. The segmental pattern is established during embryogenesis when the somites, the embryonic segments of vertebrates, are rhythmically produced from the paraxial mesoderm. This process involves the segmentation clock, a traveling oscillator that interacts with a maturation wave called the wavefront to produce the periodic series of somites. Recent studies have shed light on several aspects of the segmentation mechanism and provide a conceptual framework to explain human spine malformations, such as congenital scoliosis.
One of the most striking features of the human vertebral column is its periodic organization. This so-called “segmental” or “metameric” arrangement of the vertebrae along the antero-posterior (AP) body axis is established during embryonic development. Structures called somites, which contain the precursors of the vertebrae, form in a rhythmic fashion at the posterior end of the embryo during the process of somitogenesis. Somites are sequentially added to the growing axis, thus, establishing the characteristic periodic pattern of the future vertebral column. In most vertebrate species, somite addition is tightly coordinated bilaterally, occuring simultaneously on the left and the right side. The total number of somites formed during somitogenesis is tightly fixed within a given species but varies significantly among vertebrates. The primary segmentation of the vertebrate embryo displayed by somitic organization also underlies much of the segmental organization of the body, including muscles, nerves and blood vessels. In vertebrates, somites are the major component of the paraxial mesoderm that forms bilaterally along the nerve cord as a result of blastopore/primitive streak and tail bud regression during body axis formation. The dorsal portion of the somite remains epithelial and forms the dermomyotome, which differentiates into muscle and dermis while its ventral moiety undergoes an epithelio-mesenchymal transition, leading to the formation of the sclerotome (Chal and Pourquie, 2009). The sclerotome gives rise to the skeletal elements of the vertebral column: the vertebrae, ribs, intervertebral disks and tendons.
The segmentation clock: a molecular oscillator involved in establishing the periodicity of vertebrae
In all vertebrate species, pairs of somites bud off periodically at the anterior tip of the presomitic mesoderm (PSM) at a defined pace (for example, every 90 minutes in chicken, 120 minutes in mice and 30 minutes in zebrafish) (Aulehla et al., 2008; Palmeirim et al., 1997; Romanoff, 1960; Schroter et al., 2008; Tam, 1981). The rhythmic production of somites from the PSM has inspired theoretical models such as the “clock and wavefront” (Cooke and Zeeman, 1976). This model proposed that the periodicity of somites results from the action of a molecular oscillator (called the clock) traveling along the embryonic axis. The periodic segment formation is triggered during a defined (permissive) phase of the oscillation, while the oscillator is constantly displaced posteriorly by a wave of maturation (the wavefront), hence, ensuring the spacing of the response to the oscillator.
The first evidence of the existence of an oscillator coupled to somitogenesis was provided by the periodic expression of the transcription factor hairy1 mRNA in the chicken embryo PSM (Figure 1A–J) (Palmeirim et al., 1997). During each somitogenesis cycle, hairy1 is first activated in the posterior PSM and then progressively in more anteriorly located cells, thus appearing as a traveling wave of gene expression. Subsequently, similar oscillations of members of the Hes/Her/Hairy family of the basic helix-loop-helix (bHLH) transcriptional repressors have been reported in several species, indicating that the oscillator is conserved in vertebrates (Bessho et al., 2003; Dunwoodie et al., 2002; Henry et al., 2002; Holley et al., 2000; Jouve et al., 2000; Oates and Ho, 2002; Palmeirim et al., 1997). In amniotes, other classes of cyclic genes have been identified based on their rhythmic expression pattern in the PSM. These cyclic genes belong, in their vast majority, to the Notch, Wnt and fibroblast growth factor (FGF) signaling pathways (Figure 1H) (Dequeant and Pourquie, 2008). Recent data also support the role of such an oscillator in invertebrate segmentation (Pueyo et al., 2008).
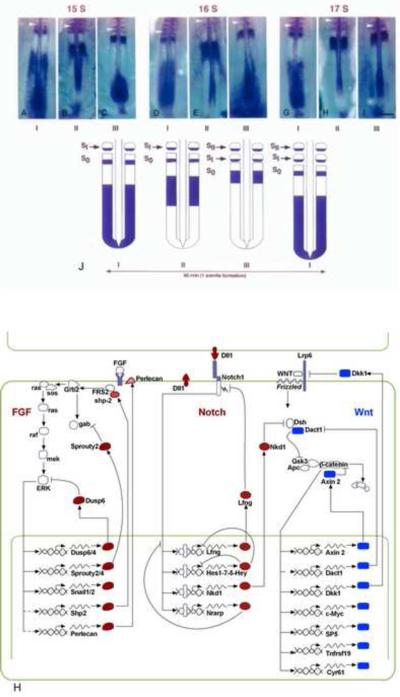
Figure1. The segmentation clock.
(A–I) In situ hybridization with c-hairy1 probe showing the different categories of expression patterns in chicken embryos aged of 15 (A, B, and C), 16 (D, E, and F), and 17 (G, H, and I) somites. Anterior to the top. (J) Schematic representation of the correlation between c-hairy1 expression in the PSM with the progression of somite formation. This highly dynamic sequence of c-hairy1 expression in the PSM was observed at all stages of somitogenesis examined, suggesting a cyclic expression of the c-hairy1 mRNA correlated with somite formation. Arrowheads: last formed somite (somite I: SI). From (Palmeirim et al., 1997)
(H) Cyclic genes belonging to the Notch and FGF pathways (whose products are indicated in red) oscillate in opposite phase to cyclic genes of the Wnt pathway (blue). A large number of the cyclic genes are involved in negative feedback loops. The basic circuitry of the three signaling pathways is represented. Dashed lines correspond to modes of regulation inferred from work in other systems or based on microarray data.
In the mouse PSM, periodic activation of Notch1 can be evidenced by visualizing the rhythmic production of the Notch intracellular domain (NICD) detected using a specific antibody that recognizes the cleaved form of the Notch1 receptor (Huppert et al., 2005; Morimoto et al., 2005). After nuclear translocation, NICD activates transcription of target genes, such as the Hairy/Hes/Her genes, Lunatic fringe (Lfng) or Notch-regulated ankyrin repeat protein (Nrarp) (Figure 1H) (Cole et al., 2002; Dale et al., 2003; Dequeant et al., 2006; Holley et al., 2002; Morales et al., 2002; Niwa et al., 2007; Oates and Ho, 2002; Sewell et al., 2009; Wright et al., 2009). Periodic activation of transcription downstream of Notch is indicated by the rhythmic waves of expression of genes such as Lfng in the PSM (Aulehla and Johnson, 1999; Forsberg et al., 1998; McGrew et al., 1998). Their dynamic expression sequence has now been imaged in real time in live mouse embryos using luciferase or green fluorescent protein-based methods for the Hes1 and Lfng genes (Aulehla et al., 2008; Masamizu et al., 2006). Cyclic expression at the protein level has been demonstrated only for a subset of cyclic genes that includes Hes1 and LFng (Bessho et al., 2003; Dale et al., 2003).
A second pathway rhythmically activated in the PSM is the Wnt signaling pathway. Axin2 is a classical target of the Wnt canonical pathway expressed periodically in the mouse embryo PSM (Aulehla et al., 2003). Other cyclic Wnt targets have been identified in a microarray screen aimed at identifying all cyclic genes in the mouse PSM transcriptome (Figure 1H) (Dequeant et al., 2006). These genes include several negative feedback inhibitors of the pathway such as : Dickkopf homolog 1 (Dkk1), Dapper homolog 1 (Dact1), and Naked cuticle 1 (Nkd1) (Dequeant et al., 2008; Dequeant et al., 2006; Ishikawa et al., 2004; Suriben et al., 2006). Inactivation of several of these inhibitors, such as Dkk1, results in segmentation defects (MacDonald et al., 2004).
The FGF pathway constitutes a third signaling pathway activated periodically in the mouse and chicken PSM. Cyclic expression of FGF negative feedback inhibitors such as Sprouty homolog 2 and 4 (Spry2 and Spry4), and Dual specificity phosphatase 4 and 6 (Dusp4 and 6) indicates that the FGF signaling pathway is activated periodically in the posterior PSM (Figure 1H) (Dequeant et al., 2006; Hayashi et al., 2009; Niwa et al., 2007). This periodic regulation of FGF signaling is further supported by the dynamic phosphorylation of ERK in the mouse PSM (Niwa et al., 2007).
Does the cyclic gene network act as the clock pacemaker?
One of the key challenges in the field has been to identify the molecular circuitry responsible for the generation of the oscillations, ie the segmentation clock pacemaker. In amniotes, the complex epistatic relationships and multiple crosstalks between the Notch, FGF and Wnt signaling pathways in the PSM make the analysis of their respective contributions to the pacemaker particularly challenging. Global analysis of mouse cyclic gene expression reveals that Notch- and FGF-related cyclic genes oscillate mostly in antiphase to Wnt-cyclic genes, suggesting a crosstalk between these signaling pathways (Dequeant et al., 2006; Goldbeter and Pourquie, 2008). This notion is further supported by the oscillations of genes such as Nkd1, a Wnt inhibitor, under Notch control (Ishikawa et al., 2004). Data from genetic and non genetic experiments have been used to create a relational database, which led to the definition of a reference network by combining the data with the KEGG pathway database. This led to a computational representation of the mouse segmentation network containing 36 nodes and 57 interactions (Gonzalez and Kageyama, 2010). Given the ever-increasing complexity of the signaling network associated to the clock, this approach might be useful to provide a comprehensive view of the molecular circuitry associated to the segmentation clock oscillator.
The molecular oscillations of Notch signaling could be triggered by the periodic expression of the Notch ligands, since they exhibit cyclic expression in the mouse (Dll1) and zebrafish (deltaC) PSM (Jiang et al., 2000; Maruhashi et al., 2005). Alternatively, periodic Notch activation could also be triggered by intracellular negative feedback loops. Hes7 codes for an unstable protein of the bHLH family of transcriptional repressor, acting downstream of Notch and FGF, and which can repress its own transcription, as well as that of Lfng (Bessho et al., 2003; Chen et al., 2005; Morimoto et al., 2005; Niwa et al., 2007). The Hes7 negative autoregulatory loop was proposed to play a critical role in the control of cyclic gene oscillations in the PSM (Hirata et al., 2004). However, whereas oscillations of Notch targets such as Lfng are disrupted in the Hes7 null mutant, Axin2 cyclic expression is still observed thus arguing against a role for Hes7 as a key element of the clock pacemaker (Ferjentsik et al., 2009; Hirata et al., 2004). In zebrafish, homologs of the Hes7 gene – Her1 and Her7 – display a cyclic expression in the PSM, and their overexpression or their knockdown leads to segmentation defects {Holley, 2000 #2144;Holley, 2002 #2397; Oates, 2002 #2396;Henry, 2002 #2395;Ozbudak, 2008 #4783;Giudicelli, 2007 #3862}. A simple oscillator model that essentially relies on the Her1/Her7 transcriptional repressors acting as a central pacemaker has been proposed (Figure 2 A–C) (Holley et al., 2002; Lewis, 2003; Oates and Ho, 2002). In this model, oscillations are generated by a negative feedback loop in which her genes are directly repressed by their own protein products. To generate oscillations, the model takes into account a defined time delay in the auto-inhibitory circuit that occurs from the beginning of her RNA transcription until the Her protein binds to the her gene promoter (Lewis, 2003). Using plausible numerical values for the model parameters, oscillations exhibiting a period consistent with that observed in zebrafish have been obtained. This Her1/Her7 intracellular oscillator was proposed to be linked to an intercellular oscillator involving the Notch-signaling pathway (Figure 2C) (Lewis, 2003). Her1/Her7 negatively regulate deltaC, thus, triggering oscillations of this Notch ligand, which should, in turn, result in periodic Notch activation in neighboring cells. Such a coupling could provide a basis for maintaining synchrony among the oscillations of neighboring cells (see below) (Jiang et al., 2000; Riedel-Kruse et al., 2007). By combining immunostaining and fluorescent in situ hybridization, the measured translational delay of DeltaC has been shown to be consistent with the constraint set by the delay model for DeltaC to entrain the oscillations of neighboring cells (Giudicelli et al., 2007).
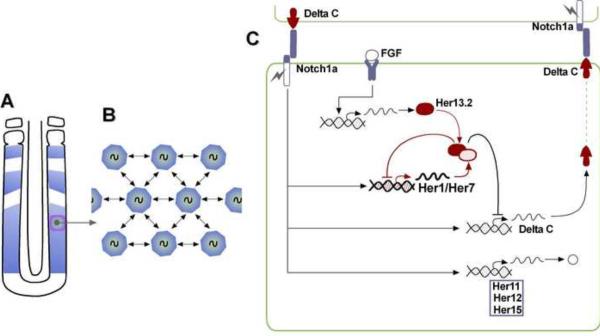
Figure 2. Synchronization of the presomitic mesoderm cellular oscillators.
(A) Smooth transcriptional waves of cyclic gene expression sweeping through the zebrafish PSM (shown in blue).
(B) Schematic representation of the PSM cells as coupled phase oscillators.
(C) Model of the zebrafish oscillator. The Notch pathway has been proposed to synchronize oscillations by coupling by connecting the zebrafish Her1/Her7 intracellular oscillator to the Notch/DeltaC intercellular loop. The transcription factors Her1/Her7 establish a negative feedback loop controling the periodic repression of DeltaC, allowing the synchronous activation of Notch signaling in neighboring cells. In addition to receiving inputs from Notch signaling, the Her1/Her7 oscillator requires the Her13.2 partner, which is downstream of FGF signaling. The coupling between cellular oscillators provided by the Notch/Delta intercellular loop is thought to confer robustness to the clock synchronized oscillations against developmental noise such as cell proliferation, cell movement or stochastic gene expression.
A second negative feedback loop relying on Lfng and regulating Notch activation has been described in amniotes (Figure 1H) (Dale et al., 2003; Morimoto et al., 2005). Lfng is a glycosyltransferase that modifies the Notch and Delta extracellular domain during its trafficking to the cell surface (Haines and Irvine, 2003). Lfng cyclic expression in the PSM, is observed only in amniotes and is controlled at the transcriptional level (Aulehla and Johnson, 1999; Cole et al., 2002; Forsberg et al., 1998; McGrew et al., 1998; Morales et al., 2002; Prince et al., 2001). In the PSM, Lfng periodically inhibits Notch signaling and participates in a negative feedback loop involved in the control of the Notch pathway oscillation and somite boundary positioning (Dale et al., 2003; Morimoto et al., 2005; Serth et al., 2003; Shifley et al., 2008; Stauber et al., 2009). Driving Lfng from the Hes7 promoter which only recapitulates the posterior cyclic expression pattern is able to fully rescue the segmentation defects of the Lfng null mutant (Oginuma et al., 2010). Interestingly, selective deletion of the regulatory sequence driving the oscillatory expression of Lfng in the posterior PSM results in disruption of the segmental organization of the vertebral column all the way to the lumbar region, but leaves the sacral and caudal vertebrae intact (Shifley et al., 2008; Stauber et al., 2009). Therefore, this indicates that the segmentation clock might exhibit different regional requirements along the AP axis. Nonetheless, dynamic NICD production and rhythmic expression of Hes7 are still observed in the Lfng null mutants suggesting that this loop is not essential for clock oscillations (Ferjentsik et al., 2009; Niwa et al., 2007).
Whether Notch oscillations act as a pacemaker triggering the oscillations of the three pathways in mouse is controversial. On one hand, blocking Notch signaling with DAPT or by mutating both components of the gamma-secretase complex PS1 and PS2, leads to a complete loss of cyclic genes oscillations (Donoviel et al., 1999; Ferjentsik et al., 2009; Gibb et al., 2009; Huppert et al., 2005), suggesting that Notch is a key element of the clock pacemaker. However, the γ-secretase complex activity is not restricted to Notch signaling and it could be that in these experiments, the cleavage of key proteins involved in the function of the pacemaker is prevented. On the other hand, several experiments argue against a role for Notch in the clock pacemaker. For instance, oscillations of Axin2 and Spry2 are maintained in mouse mutants for the essential Notch co-activator RBP-jk (although it has been argued that residual Notch activity is found in these mutants) (Aulehla et al., 2003; Dequeant et al., 2006; Ferjentsik et al., 2009; Niwa et al., 2007). Also, in transgenic mouse embryos constitutively activating Notch in the PSM, Axin2 but not Spry2 were still detected as cyclic (Feller et al., 2008). Lastly, Lfng and Axin2 oscillations are disrupted in the mouse Wnt3a hypomorphic vestigial tail (vt) mutant, indicating that in mouse, Wnt signaling acts upstream of Notch oscillations (Aulehla et al., 2003).
In zebrafish embryos, the role of Notch in the clock pacemaker has been ruled out based on the observations that i. anterior somites do form in Notch mutants, ii. when using the γ-secretase inhibitor DAPT to block Notch signaling, somite boundary defects are only observed after a long delay, iii. the initial Her genes oscillations during gastrulation remain synchronized even if Notch signaling is blocked by DAPT treatment (Holley et al., 2000; Holley et al., 2002; Jiang et al., 2000; Mara et al., 2007; Ozbudak and Lewis, 2008; Riedel-Kruse et al., 2007). Therefore, the role of Notch in the control of the clock oscillations still needs to be clarified.
The FGF pathway has also been implicated in the control of the clock oscillations. In zebrafish, the Her1/Her7 oscillations require the hairy and enhancer of Split6 (Hes6)-related gene, Her13.2 (Figure 2C) (Kawamura et al., 2005b). This transcription factor is regulated by FGF signaling and can form a heterodimer with Her1, enhancing the ability of Her1 to negatively regulate its own promoter (Kawamura et al., 2005b). In mouse, conditional deletion of Fgfr1 in the PSM abolishes oscillations of Hes7, Lfng, Axin2 and Spry2 (Niwa et al., 2007; Wahl et al., 2007), suggesting that FGF acts upstream of the three pathways. In vitro treatment of mouse embryos with the FGF inhibitor SU5402 leads to a rapid arrest of Axin2 and Spry2 oscillations (Niwa et al., 2007; Wahl et al., 2007). FGF signaling was proposed to control the initiation of Hes7 oscillations in the tail bud, while Notch signaling would maintain these oscillations in the PSM (Niwa et al., 2007). In vitro, FGF treatment induces periodic expression of the cyclic gene Hes1 in 10T1/2 cells with a 2 h cycle and trigger oscillation of RAS activity mediated by periodic ERK phosphorylation regulating the function of SOS (Nakayama et al., 2008). However, introducing a constitutively stable version of β-catenin in a mutant mouse embryo in which Fgfr1 is conditionally deleted in the PSM, restores the formation of the Lfng stripes of expression (Aulehla et al., 2008). This experiment challenges the role for an FGF-based negative feedback loop acting as the pacemaker of the segmentation clock.
Wnt ligands mRNAs are distributed in a graded fashion along the PSM. Thus, oscillations of Wnt targets are observed despite the presence of a constant (non-periodic) signaling input in the PSM. In theory, these oscillations could be triggered by the periodic activation of unstable negative feedback inhibitors (Goldbeter and Pourquie, 2008). Since Wnt acts genetically upstream of Notch and FGF in the PSM (Aulehla et al., 2003), this raises the possibility that the periodic destabilization of β-catenin expected to be triggered by these negative feedback loops could act as the clock pacemaker that controls oscillations of the Notch and FGF pathways. However, Lfng has been shown to oscillate with the same periodicity in β-catenin gain-of-function mutants and in wild-type mice (Aulehla et al., 2008). Therefore, periodic β-catenin production is not required to control the rhythmicity of Notch activation in the PSM. In the chick embryo, a role for Wnt signaling in the control of the period of Notch-dependent oscillations has been proposed based on the slowing down of the Lfng wave when treating half embryos with the casein kinase inhibitor CK-7 (Gibb et al., 2009). Such a role has also been recently proposed for the Sonic Hedgehog pathway (SHH) (Resende et al., 2010).
Taken together, these data suggest that none of the three signaling pathways periodically activated in the PSM individually acts as a global clock pacemaker. Hence, this raises doubts about models in which the periodic gene expression associated with the segmentation clock results from the dynamic properties of the cyclic gene network (Dequeant and Pourquie, 2008). In amniotes, it is possible that each subnetwork has the capacity to generate its own oscillations – independent of the oscillations of the other subnetworks – while coupling among the subnetworks entrain them to each other (Goldbeter and Pourquie, 2008; Ozbudak and Lewis, 2008). Alternatively, it is possible that the network of cyclic genes that underlie the segmentation clock is entrained by an outside pacemaker that remains to be identified.
Notch signaling synchronizes oscillations among presomitic mesoderm cells
An alternative role for the Notch pathway was suggested by Lewis and colleagues, who proposed that Notch signaling synchronizes the cell-autonomous oscillations of neighboring cells in the PSM (Figure 2A–B) (Jiang et al., 2000). Thus, the delayed disruption in somite boundary formation evident in Notch mutants would result from the gradual desynchronization of oscillations in neighboring cells in the absence of cell-to-cell coupling (Jiang et al., 2000). In Notch mutants, oscillations of neighboring cells initially would begin as synchronized; however, in the absence of coupling, synchrony is progressively lost, resulting in a salt-and-pepper expression pattern of the cyclic genes in the PSM and in an arrest of somite formation (Jiang et al., 2000) . To test the role of Notch signaling in the coupling of the oscillations, zebrafish PSM cells injected with morpholinos against her1 and her7 were transplanted into a wild type host (Horikawa et al., 2006). This led to the acceleration of the oscillation phase of neighboring wild-type cells in the host zebrafish embryo, resulting in an anterior shift of the somite boundaries. The transplanted cells show an elevated deltaC level due to the release of the Her-mediated inhibition. The phenotype is rescued by co-injection of deltaC or deltaD morpholinos, together with her1/7 morpholinos, showing that the effect is due to interference with Notch signaling. Also, when groups of PSM cells from donor embryos were transplanted into the PSM of host embryos, the transplanted cells synchronized their oscillations with the host cells within three clock cycles. The authors also reported variation in the oscillation phases of neighboring cells in wild-type embryos due to stochastic gene expression and cell division. Together, these data indicate that Notch provides a robust coupling system which ensures the synchronization of segmentation clock oscillations in the PSM despite this intrinsic noise.
Theoretical models based on the mean-field approximation have been proposed to explain the synchronization of cells in the PSM despite the presence of noise (Riedel-Kruse et al., 2007). In these models, the PSM is considered as composed of a collection of coupled phase oscillators which oscillate with a noisy autonomous frequency. The slowing down of the oscillations along the PSM together with the coupling between the oscillators results in the production of periodic smooth oscillatory waves traveling along the tissue (Figure 2A–B) (Jiang et al., 2000; Morelli et al., 2009; Riedel-Kruse et al., 2007). Several non-intuitive predictions derive from such models, namely that the apparent frequency of the oscillatory wave is different from the autonomous frequency of the oscillators (Herrgen et al., 2010). Notch is the major candidate for the intercellular coupling between oscillators and its role in this process has been tested experimentally in the PSM (Herrgen et al., 2010; Horikawa et al., 2006; Ozbudak and Lewis, 2008; Riedel-Kruse et al., 2007). The fact that the first 5–6 somites are spared in Notch mutants is interpreted as reflecting the time required to lose the synchrony among the oscillators (decay time). This idea was further tested by treating embryos with DAPT at different somitic stages to disrupt Notch signaling. A lag of 5–6 somites was always observed between the number of somites at time of treatment and the first morphological boundary disruption (Ozbudak and Lewis, 2008; Riedel-Kruse et al., 2007). These results suggest that, in vivo, the oscillators are initially set simultaneously by a Notch independent process during gastrulation and that synchronization is actively maintained by a Notch-dependent coupling mechanism during somitogenesis. Interestingly, this coupling system also endows the system with self-organizing properties hence contributing to its robustness. Coupling-dependent self organization of the oscillatory pattern was demonstrated by pulse chase DAPT treatments, showing that normal segment formation recovers after around 10 oscillations after the wash (Riedel-Kruse et al., 2007). Based on these findings, Riedel-Kruse et al. developed a model in which the balance between the strength of Notch signaling (coupling agent) and the noise in gene expression (desynchronizing agent) dictates whether the neighboring cells will or will not oscillate in synchrony (Riedel-Kruse et al., 2007). The simultaneous initiation of the oscillations appears to involve FGF signaling which was shown to be able to trigger ectopic waves of Her1 expression in the early zebrafish embryo prior to somite formation (Ishimatsu et al., 2010). Disruption of the coupling as in the Notch pathway mutants in zebrafish or using DAPT treatment increases the period of somitogenesis leading to the formation of larger segments (Herrgen et al., 2010). Such experiments were used to extract quantitative parameters allowing to predict the autonomous period of the oscillator as well as the coupling strength and the coupling delay (Herrgen et al., 2010). One potential caveat to these coupling models is the high motility of the cells in the posterior PSM (Benazeraf et al., 2010; Mara et al., 2007). However, numerical simulations show that synchronized oscillations can be maintained despite the random cell movements, providing the system with robustness to perturbations (Uriu et al., 2010).
A direct demonstration, using real-time imaging, of the role of Notch signaling in the synchronization of the oscillations among the PSM cells, however, is still lacking. Desynchronization was, nevertheless, observed by in situ hybridization in dissociated PSM cells in the chick embryo (Maroto et al., 2005). In mouse, single-cell oscillations in dissociated PSM cells were demonstrated using a real-time Hes1-luciferase transgenic reporter (Masamizu et al., 2006). Unlike in the intact PSM, the period and the amplitude of single-cell oscillations in cell culture were quite noisy (Masamizu et al., 2006). These data provide strong evidence supporting the existence of cell-autonomous oscillations, at least in mouse. Thus far, no evidence has been provided to support the role of Notch signaling in the synchronization of the oscillations in any amniote species.
The wavefront: translating the periodic signal into repeated segments
A key step in segment formation is the establishment of the first segmental prepattern in the PSM. This prepattern appears as a stripe of expression of genes such as Mesp2, of approximately one somite in length (Figure 3D–E). This stripe would form in the anterior PSM in response to the periodic clock signal at a level defined as the determination front and it defines the future somitic boundaries. The determination front is positioned by antagonistic gradients of FGF, Wnt and retinoic acid (RA) signaling and regresses posteriorly as the embryo elongates along the AP axis (Figure 3A–E) (Aulehla et al., 2003; Aulehla et al., 2008; Diez del Corral et al., 2003; Dubrulle et al., 2001; Moreno and Kintner, 2004). The parallel posterior-to-anterior gradients of FGF and Wnt signaling (evidenced by graded phosphorylated ERK and nuclear β-catenin, respectively), are established in response to the graded expression of secreted ligands, such as Fgf8 and Wnt3a, along the PSM (Figure 3A–B) (Aulehla et al., 2003; Aulehla et al., 2008; Delfini et al., 2005; Dubrulle et al., 2001; Dubrulle and Pourquie, 2004; Sawada et al., 2001). In the posterior PSM, cells are exposed to a high level of FGF and Wnt activity, and are maintained in an immature, undifferentiated state (Aulehla et al., 2008; Dubrulle et al., 2001; Dunty et al., 2008; Sawada et al., 2001). The transition from the immature to the competent state of PSM cells can be visualized by the downregulation of the Mesogenin1 (Msgn1) gene that precedes activation of Mesp2 (Yoon et al., 2000). The level of the determination front defines a major transitional region in gene regulation (Ozbudak et al., 2010) and cellular properties along the PSM (Dale et al., 2006).
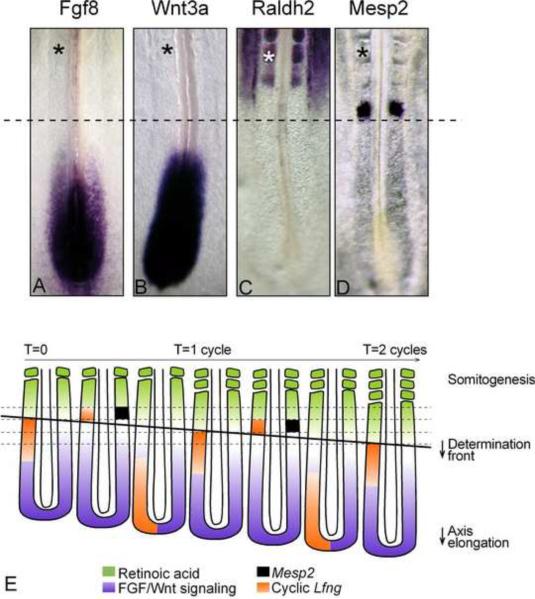
Figure 3. A model for vertebrate somitogenesis.
(A–D) Expression pattern of key components of the segmentation system during somitogenesis in 2-day chicken embryos. (A) Wnt3a, (B) Fgf8, (C) Raldh2, (D) Mesp2 expression by in situ hybridization. (*) last formed somite; The dotted line marks the approximate position of the determination front
(I) A segmentation model. Antagonistic gradients of FGF/Wnt signaling (purple) and retinoic acid signaling (green) position the determination front (thick, black line). The periodic signal of the segmentation clock is shown in orange (represented on the left side only). As the embryo extends posteriorly, the determination front moves caudally. Cells that reach the determination front are exposed to the periodic clock signal, initiating the segmentation program and activating simultaneously expression of genes such as Mesp2 (black stripes, represented on the right side only), in a stripe domain that prefigures the future segment. This establishes the segmental pattern of the presumptive somites. Dorsal views, anterior to the top.
The role of the FGF signaling gradient in positioning the determination front was first demonstrated by experiments challenging the slope of the signaling gradient in chicken and fish embryos by grafting FGF8-soaked beads next to the PSM or by overexpressing an Fgf8-expressing construct in the PSM by electroporation. This resulted in an anterior extension of posterior PSM markers (such as Brachyury) and in downregulation of segmentation and differentiation markers (such as Paraxis, Mesp2 or myogenic differentiation 1 [MyoD]) and in the formation of smaller somites (Delfini et al., 2005; Dubrulle et al., 2001; Sawada et al., 2001). Conversely, inhibition of FGF signaling was achieved by treating chicken or fish embryos with pharmacological inhibitors (Dubrulle et al., 2001; Sawada et al., 2001). This resulted in a posterior shift of the anterior boundary of genes associated with a posterior PSM identity (such as Fgf8), and in the formation of larger somites. Conditional deletion of Fgfr1 in the mouse PSM also results in transient formation of larger somites (Wahl et al., 2007). These data led to the idea that the progressive decrease in FGF signaling activity along the PSM defines a specific threshold below which the cells become competent to respond to the signaling pulse delivered by the segmentation clock. Changing the position of the threshold by acting on FGF levels leads to a change in the position of the somite boundary position resulting in smaller or larger somites. The position of this threshold was proposed to correspond to the determination front (Dubrulle et al., 2001).
The posterior gradients and, thus, the determination front constantly move posteriorly in the wake of the body axis elongation (Figure 3E). The posterior displacement of the FGF gradient relies on an original mechanism involving Fgf8 mRNA decay (Dubrulle and Pourquie, 2004). Transcription of the Fgf8 mRNA is restricted to the PSM precursors in the tail bud, and it ceases when their descendents enter the posterior PSM. Thus, as the axis elongates, the relative position of the cells in the PSM becomes gradually more anterior, and their Fgf8 mRNA content progressively decays. This promotes the posterior movement of the anterior boundary of the Fgf8 mRNA gradient and hence of the determination front (Goldbeter et al., 2007). This mechanism generates a dynamic gradient of FGF activity along the PSM which accompanies the elongation movements (Delfini et al., 2005; Dubrulle and Pourquie, 2004; Sawada et al., 2001). A similar mechanism is assumed to be responsible for establishing the Wnt gradient (Aulehla et al., 2003). In zebrafish, chicken, mouse and snake, the regression speed of the determination front (marked by the anterior boundary of the Msgn1 expression domain) during somitogenesis is similar to the speed of somite formation but different from the speed of axis elongation (Gomez et al., 2008). The slowing down of the axis elongation speed results in a progressive shrinking of the PSM, ultimately leading to the arrest of segment formation (Gomez et al., 2008). Thus the coupling of segmentation with axis elongation downstream of FGF signaling plays a key role in the control of segment numbers in the embryo (Gomez et al., 2008; Schroter and Oates, 2010).
Body axis elongation was recently shown to be controlled by the same FGF gradient that controls segmentation (Benazeraf et al., 2010; Delfini et al., 2005). Tissue ablation experiments in the chicken embryo demonstrated that the caudal region of PSM contains highly motile cells which play a key role in axis elongation. A clear posterior-to-anterior gradient of cell motility and directionality in the PSM was evidenced using time-lapse microscopy. However, when subtracting the extracellular matrix movement from the global motion of cells, cell motility remained graded but lacked directionality, indicating that the posterior cell movements associated with axis elongation reflect tissue deformation. The gradient of cell motion along the PSM parallels the fibroblast growth factor (FGF)/mitogen-activated protein kinase (MAPK) gradient, which has been implicated in the control of cell motility in this tissue (Delfini et al., 2005). Both FGF signaling gain- and loss-of-function experiments lead to disruption of the motility gradient and a slowing down of axis elongation. Furthermore, embryos treated with cell movement inhibitors (Blebbistatin or RhoK inhibitor), but not cell cycle inhibitors, show a slower axis elongation rate. This led to propose that the gradient of random cell motility, ie “cell diffusion”, downstream of FGF signaling in the PSM controls posterior elongation in the amniote embryo. Tissue elongation is an emergent property that arises from the collective regulation of graded, random cell motion, rather than by the regulation of directionality of individual cellular movements.
Disruption of the Wnt/β-catenin gradient in mouse embryos demonstrates that the level of nuclear β-catenin also controls segmentation (Aulehla et al., 2008; Dunty et al., 2008). Gain of function of Wnt/ β-catenin signaling leads to an anterior expansion of the expression domain of posterior genes, such as Brachyury, Mesogenin1 or Tbx6. In these experiments, both segmentation and the onset of the differentiation program in the PSM are blocked (Aulehla et al., 2008; Dunty et al., 2008). The most striking effect in the β-catenin gain-of-function mouse mutant is an AP extension of the oscillatory domain that results in a multi-stripe, oscillatory expression pattern in the enlarged PSM (Aulehla et al., 2008; Dunty et al., 2008). These results imply that Wnt/β-catenin signaling maintains the immature oscillating state of posterior cells and, thus, controls the onset of segmentation and differentiation in the PSM (Aulehla et al., 2008).
The Wnt and FGF pathways have been shown to simultaneously control the expression of a number of transcription factors such as Brachyury/T, or Tbx6 which in turn, control different aspects of PSM maturation (Wahl et al., 2007). In mice, Fgf8 expression is absent from the Wnt3a hypomorph vt mutants, suggesting that Wnt signaling acts upstream of FGF signaling in the tail bud (Aulehla et al., 2003; Aulehla et al., 2008). However, gain of function of β-catenin in the PSM of mouse embryos results only in a partial gain of function of FGF signaling, suggesting that Wnt signaling is necessary but insufficient for FGF signaling in the PSM (Aulehla et al., 2008). How FGF and Wnt signalings elicit a response, which is graded for some genes and cyclic for others, is not understood.
In amniotes, RA signaling forms a decreasing rostrocaudal gradient that is opposite to the FGF and Wnt gradients in the PSM (Diez del Corral et al., 2003; Moreno and Kintner, 2004; Sirbu and Duester, 2006). Raldh2 (Aldh1a2), which catalyzes the last step of RA biosynthesis, is expressed in the anterior PSM and somites (Figure 3C) and is the major source of RA during early embryogenesis (Niederreither et al., 1999). Using a RARE-LacZ reporter mouse, RA signaling was found to be restricted to the anterior PSM and segmented region, and was absent from the posterior PSM and tail bud where Cyp26A1, a cytochrome P450 family protein involved in RA catabolism, is expressed downstream of FGF (Vermot et al., 2005; Wahl et al., 2007). Analysis of mice deficient for RA production (null for Raldh2) or of Vitamin A-deficient (VAD) quail embryos reveals that perturbation of RA signaling leads to an anterior expansion of the Fgf8 domain along the PSM and in the formation of smaller somites, consistant with an FGF signaling gain of function (Diez del Corral et al., 2003; Vermot et al., 2005; Vermot and Pourquie, 2005). In the chicken embryo, treatment of posterior PSM explants with RA agonists can downregulate Fgf8 expression, while a graft of a FGF8-soaked bead in the PSM represses Raldh2 expression (Diez del Corral et al., 2003). Experiments in chicken and Xenopus embryos indicate that RA can activate transcription of key segmentation genes, such as the Mesp2 homologs, either directly or by counteracting FGF signaling that represses their expression (Delfini et al., 2005; Moreno and Kintner, 2004). In the Fgfr1 conditional knockout, no significant posterior shift of the RARE-lacZ domain is observed, suggesting that FGF is not the only antagonist to the RA gradient (Wahl et al., 2007). Interestingly, mice mutant for Cyp26a1 exhibit downregulation of Wnt signaling targets such as T (Abu-Abed et al., 2001; Sakai et al., 2001). In zebrafish, Brachyury was shown to be involved in an autoregulatory loop controling Wnt expression in the tail bud and also acting upstream of Cyp26 to create a niche protecting mesodermal precursors from RA differentiating action (Martin and Kimelman, 2010).Together, these data led to the proposal that the mutual inhibition of the FGF, Wnt and RA gradients is involved in the control of the spatial response to the segmentation clock.
A new model for vertebrate segmentation
A new segmentation model can be proposed based on these results (Figure 3E). During one segmentation clock oscillation, the determination front moves posteriorly along the AP axis by a distance corresponding to approximately one somite (Figure 3E, black line). When PSM cells become located anterior to the determination front, they become competent to respond to a periodic signal delivered by the segmentation clock such as a pulse of Notch activation (Dubrulle et al., 2001; Oginuma et al., 2010). In response to this signal, the cohort of cells located between the determination front level and the anterior boundary of the Tbx6 expression domain, simultaneously activate Mesp2, resulting in the formation of a stripe of Mesp2 expression that prefigures the future segment (Oginuma et al., 2008). During the next oscillation cycle, the newly specified segmental domain becomes located more anteriorly in the PSM by one somite. Cells in this territory begin to activate a complex genetic program downstream of Mesp2. These cells activate transcriptional repressors of the Ripply family that establish a negative feedback loop shutting down Tbx6 protein expression by a post-transcriptional mechanism, leading to the downregulation of Mesp2 expression in the future posterior somite compartment (Kawamura et al., 2005a; Morimoto et al., 2007; Oginuma et al., 2008; Takahashi et al., 2010). The establishment of the stripe of Mesp2 expression was explored by performing computer simulations, which revealed that oscillating Notch signaling induces not only the periodic activation of Mesp2 but also a rostral-caudal gradient of Mesp2 in the absence of striped Notch activity in the anterior PSM (Oginuma et al., 2010). This suggests that the oscillating wave of Notch activity is translated into the rostral-caudal polarity of a somite by regulating Mesp2 expression in the anterior PSM. This complex genetic cascade ultimately results in the specification of the anterior and posterior somite compartments and of the somite boundaries.
A striking feature of segmentation is the highly coordinated gene activation occurring in the stripes of cells that define the future segments. This coordination was proposed to reflect a molecular switch that simultaneously triggers Mesp2 gene activation in the cohort of competent cells that passed the determination front (Goldbeter et al., 2007). Mathematical modeling shows that the mutual inhibition of FGF and RA signaling can define a bistability domain along the PSM in which such a switch behavior could be observed(Goldbeter et al., 2007). In response to an appropriate signal, cells located in the bistability domain (in fact, in the area between the determination front and the last specified segment) could abruptly switch from the FGF-dominated steady state to the other RA-dominated steady state (Goldbeter et al., 2007). The signaling pulse generated by the segmentation clock is a good candidate to trigger the switch-like transition. This transition would result in a simultaneous exposure of cells of the future segmental domain to RA signaling, hence explaining the collective gene activation in the stripe of cells. This hypothesis is consistent with the observation that Mesp2/thylacine expression is repressed by FGF and is activated by Notch and RA signaling (Delfini et al., 2005; Moreno and Kintner, 2004; Takahashi et al., 2000). This model needs however to be reconciled with the fact that somites do form in the mouse Raldh2 mutant in which RA signaling is not detected (Niederreither et al., 2002). Other dynamic properties of the gradients system such as positive feedback loops could also account for such a bistable behavior. Remarkably, such a bistable behavior working together with an autonomous clock is systematically observed with in silico simulations of segmentation controlled by a moving gradient as in the clock and wavefront model (Francois et al., 2007).
Molecular control of bilateral symmetry of somitogenesis
Whereas the spine is a symmetrical structure, obvious left-right asymmetries are observed in the positioning and structure of vertebrate internal organs, such as the heart and liver (Raya and Izpisua Belmonte, 2008; Shiratori and Hamada, 2006). Establishment of these asymmetries is downstream of a left-right (LR) signaling pathway that is active during gastrulation (Shiratori and Hamada, 2006). In all vertebrate species examined so far, a secreted factor of the Nodal family acts on the left side of the embryo to activate the expression of specific genes, such as Paired-like homeodomain transcription factor 2 (Pitx2), in the left lateral plate (Shiratori and Hamada, 2006). The way the nodal factors are activated in the left lateral plate significantly differs among species, involving ciliae in the node or Kupffer vesicle in mouse and fish respectively and asymmetrical cell movements in chicken (Cui et al., 2009; Gros et al., 2009). These initial asymmetries define the situs of the animal and the specific left-right development of the internal organs.
In mouse and zebrafish, asymmetric signals produced by the node cross the forming paraxial mesoderm without affecting the perfect symmetry of paraxial mesoderm patterning. In contrast, in chicken embryos, initiating the left-right asymmetry pathway leads to the transient desynchronization of the segmentation clock as evidenced by asymmetric Lfng stripes (Raya et al., 2004). In these three species, no somitic asymmetries are observed in embryos, because the desynchronizing action of the LR machinery is buffered by RA (Kawakami et al., 2005; Sirbu and Duester, 2006; Vermot and Pourquie, 2005). Mice mutant for Raldh2 exhibit a shorter segmental region on the right side in the cervical region (Vermot et al., 2005), while chicken embryos treated with Disulfiram (which blocks RA synthesis) exhibit similar defects but on the left side (Vermot and Pourquie, 2005) (Figure 4). A phenotype similar to mouse is observed in zebrafish embryos lacking Raldh2 activity (Kawakami et al., 2005). In the three species, this asymmetry of somite formation is downstream of the left-right machinery, as it can be reversed by changing the situs of the embryos (Kawakami et al., 2005; Vermot and Pourquie, 2005). Thus, RA signaling acts in the paraxial mesoderm by buffering the asymmetric signal generated by the left-right machinery. Asymmetry defects in the positioning of somite boundaries can also be observed when inhibiting Notch signaling with RBPjk morpholinos in fish embryos (Echeverri and Oates, 2007), or in several mouse mutants in the Notch pathway (Conlon et al., 1995; Hrabe de Angelis et al., 1997). Unlike the defects seen in RA deficient embryos, these defects are not lateralized, suggesting that they rather reflect a global dysfunctioning of the segmentation clock mechanism.
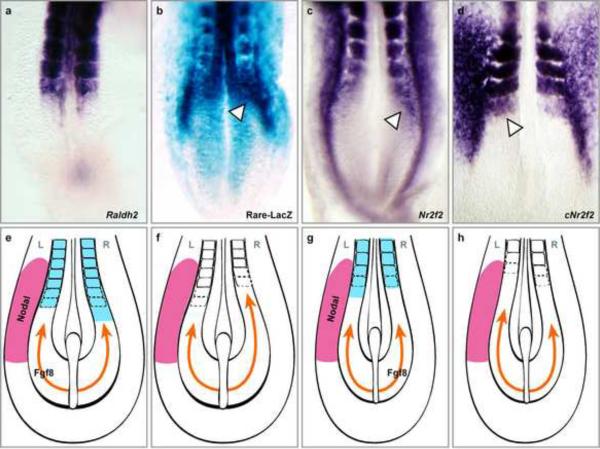
Figure 4. Role of Retinoic acid in the control of left-right symmetry of somitogenesis.
Retinoic acid signaling activity and NR2F2 expression are asymmetric across the left-right axis in the presomitic mesoderm of early-somite stage embryos. In situ hybridization for mouse Raldh2 (a), mouse NR2F2 (c) and chicken NR2F2 (d). Asymmetric expression is indicated by the white arrowheads. (b) RARE-LacZ mouse embryo indicating the asymmetric RA signaling activity (white arrowheads) (dorsal views). (e-h) Schematic interpretation of the role of RA signaling and Fgf8 in the control of somite bilateral symmetry in early mouse and chicken embryos. Asymmetrical RA signaling in the anterior PSM and somites is shown in blue and antagonizes FGF signaling (orange). Nodal signaling is indicated on the left side (pink). The embryonic side for which Fgf8 acts as a left-right determinant is indicated with the label Fgf8. (e) Wild-type mouse embryo. (f) Rere or Raldh2 mouse mutant. (g) Wild-type chicken embryo. (h) RA-deprived chicken embryo. L (left) and R (right). From (Vilhais-Neto et al., 2010)
The chromatin remodelling protein Rere (Atrophin2) was recently shown to act as a co-factor for RA signaling involved in the control of somite bilateral symmetry(Vilhais-Neto et al., 2010). Mutation of the Rere protein in the Openmind mouse mutant leads to the asymmetrical formation of somites. Rere forms a complex with NR2F2 (CoupTF2), p300 and the retinoic acid receptor (RAR), which is recruited to the retinoic acid regulatory element (RARE) of RA targets, such as the RAR-beta promoter. Knockdown of NR2F2 and/or Rere decreases RA signaling, suggesting that this complex is required to promote transcriptional activation of RA targets. NR2F2 is asymmetrically expressed in the right PSM in a domain which overlaps with the PSM domain showing asymmetrical RA signaling detected in the RARE-LacZ reporter mouse (Figure 4). Remarkably, the asymmetry of NR2F2 expression in chicken is reversed, being more strongly expressed in the left PSM (Figure 4D). This inversion provides an explanation for the opposite sides of the somite symmetry defects observed in mouse and chicken (Vermot and Pourquie, 2005). Because in mouse and chicken, Nodal is always detected in the left side (Raya and Izpisua Belmonte, 2008), these inversions suggest that Nodal is not the left-right determinant buffered by RA (Figure 4E–H). Fgf8 has been shown to act as a right determinant in chicken and as a left one in mouse (Boettger et al., 1999; Meyers and Martin, 1999). Crossing the mouse Rere mutant with the Fgf8 mutant increases the frequency of somite asymmetry defects in the compound mutants thus supporting the idea that RA shields the PSM from the desynchronizing action of Fgf8 rather than from that of Nodal (Vilhais-Neto et al., 2010). Genetic interactions between Rere and the Fgf pathway have also been reported in zebrafish (Plaster et al., 2007). This RA-dependent buffering mechanism could be involved in symmetry defects of the human spine, such as those observed in scoliosis. In humans, a majority of patients with idiopathic scoliosis exhibit a spine curvature toward the right, suggesting an underlying defect in the left-right symmetry (Ahn et al., 2002). While the molecular mechanisms underlying these diseases have not been identified, pathways controlling the left-right symmetry of the spine during embryogenesis, such as the RA pathway, are indeed attractive candidates.
4. Defects in the segmentation clock lead to congenital scoliosis in humans
In humans, congenital abnormalities in vertebral segmentation occur in a wide variety of rare but well-characterized disorders, encompassing many diverse and poorly understood phenotypic patterns (Turnpenny et al., 2007). Congenital forms of scoliosis involve structural malformations of the spine that are visible on radiographs and include segmental abnormalities such as hemivertebrae, wedge-shaped vertebrae, vertebral fusions and bars. In contrast, vertebrae are essentially normal in patients with idiopathic scoliosis, who are characterized by an abnormal torsion of the vertebral column. Observations in patients with congenital scoliosis are suggestive of various abnormalities that may occur during early developmental patterning particularly during somitogenesis. A new nomenclature, developed by the International Consortium for Vertebral Anomalies and Scoliosis (ICVAS), has been created to better describe congenital scoliosis patients (Offiah et al., 2010; Turnpenny et al., 2007).
Manifestations leading to a diagnosis of congenital scoliosis include: (1) generalized vertebral segmentation abnormalities as observed in patients with spondylocostal dysostosis (SCD) or spondylothoracic dysostosis (STD), (2) regionalized conditions, such as Klippel-Feil syndrome which affect only the cervical region and (3) conditions that involve only one or two vertebrae. Congenital scoliosis can also be associated with anomalies in other organ systems, most frequently involving the renal, cardiac or neural systems. In some cases, patients with congenital scoliosis with abnormalities of the chest wall present a major surgical challenge. A better understanding and increased knowledge of the disease mechanism(s) will aid in improving the prediction of the clinical course of the disease, particularly in children.
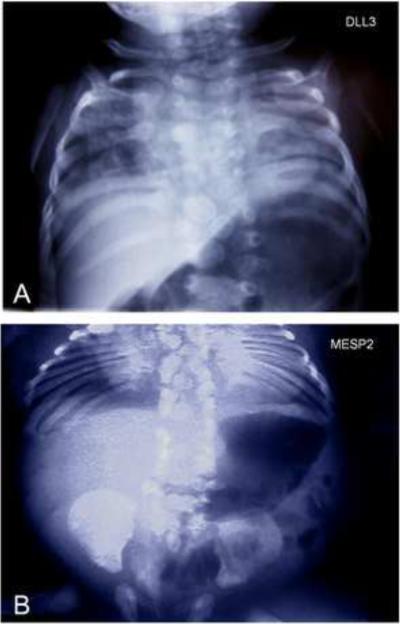
Although most cases of congenital scoliosis were previously thought to be sporadic, recent evidence suggests that a considerable genetic component may be involved. Strikingly, thus far, the four mutations associated with congenital scoliosis in humans have been identified in genes associated with the segmentation clock mechanism. These mutations result in monogenic autosomal recessive forms of SCD and STD. Homozygosity mapping and linkage analysis in consanguineous Arab-Israeli and Pakistani pedigrees with a particular form of SCD (SCDO1 [MIM 277300]) led to the discovery of multiple mutations in the Notch ligand DLL3 (Bulman et al., 2000). These DLL3 mutations result in abnormal vertebral segmentation throughout the entire spine with all vertebrae losing their normal form and regular three-dimensional shape (Figure 5A). A second, milder form of SCD (SCDO2 [MIM 608681]) has been associated with a 4-base-pair duplication in the MESP2 gene that is essential for normal segmentation (Figure 3) (Whittock et al., 2004). A mutation in LFNG was identified in members of one family with SCD (SCDO3 [MIM 609813]) (Sparrow et al., 2006). Several mutations in the HES7 gene were also found to be associated with SCD (SCDO4) (Sparrow et al., 2008; Sparrow et al., 2010). The identified mutations prevent HES7 protein to heterodimerize with the E47 cofactor or block DNA binding. In addition, a recessive null mutation in the MESP2 gene was also identified in patients with STD (Jarcho-Levin syndrome [MIM 277300]) by sequencing candidate genes associated to the segmentation clock (Figure 5B) (Cornier et al., 2008). Also, various mutations of the Notch ligand JAGGED1 have been associated with the autosomal dominant Alagille syndrome in which misshaped vertebrae (butterfly vertebrae) are observed (AGS [MIM 118450]) (Li et al., 1997; Oda et al., 1997). Polymorphisms in the Tbx6 gene involved in somitogenesis have also been associated to congenital scoliosis (Fei et al., 2010). Together, these data strongly suggest that human somitogenesis also relies on molecular mechanisms similar to those identified in mouse, chicken and fish embryos. Currently, the identified mutations explain only a minor fraction of the congenital scoliosis cases and no mutations have been identified in other cyclic pathways than Notch. Given the segmentation phenotypes of several of mouse Wnt and FGF cyclic gene mutants, mutations of orthologous genes in humans are likely to result in segmentation defects of the vertebral column.
Figure 5. Mutation of human orthologues of the segmentation clock genes leads to congenital scoliosis.
(A) Radiograph of a patient with Spondylocostal dysostosis (SCDO1) showing the severe axial skeletal malformations associated with a mutation in the DLL3 gene. Courtesy of Peter Turnpenny
(B) Radiograph of patient with spondylothoracic dysostosis (STD) illustrating severe vertebral and rib malformations associated with a mutation in the MESP2 gene. Courtesy of Alberto Cornier.
Acknowledgements
I am grateful to A. Aulehla and members of the Pourquié lab for helpful comments. Research in my laboratory is supported by the Association Française contre les Myopathies, the Chaire d'excellence ANR, an ERC advanced grant and an NIH grant R02 HD043158. I apologize to authors whose work was not cited due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu-Abed S, Dolle P, Metzger D, Beckett B, Chambon P, Petkovich M. The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev. 2001;15:226–240. doi: 10.1101/gad.855001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn UM, Ahn NU, Nallamshetty L, Buchowski JM, Rose PS, Miller NH, Kostuik JP, Sponseller PD. The etiology of adolescent idiopathic scoliosis. American journal of orthopedics (Belle Mead, NJ. 2002;31:387–395. [PubMed] [Google Scholar]
- Aulehla A, Johnson RL. Dynamic expression of lunatic fringe suggests a link between notch signaling and an autonomous cellular oscillator driving somite segmentation. Dev Biol. 1999;207:49–61. doi: 10.1006/dbio.1998.9164. [DOI] [PubMed] [Google Scholar]
- Aulehla A, Wehrle C, Brand-Saberi B, Kemler R, Gossler A, Kanzler B, Herrmann BG. Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev Cell. 2003;4:395–406. doi: 10.1016/s1534-5807(03)00055-8. [DOI] [PubMed] [Google Scholar]
- Aulehla A, Wiegraebe W, Baubet V, Wahl MB, Deng C, Taketo M, Lewandoski M, Pourquie O. A beta-catenin gradient links the clock and wavefront systems in mouse embryo segmentation. Nat Cell Biol. 2008;10:186–193. doi: 10.1038/ncb1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benazeraf B, Francois P, Baker RE, Denans N, Little CD, Pourquie O. A random cell motility gradient downstream of FGF controls elongation of an amniote embryo. Nature. 2010;466:248–252. doi: 10.1038/nature09151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho Y, Hirata H, Masamizu Y, Kageyama R. Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes Dev. 2003;17:1451–1456. doi: 10.1101/gad.1092303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettger T, Wittler L, Kessel M. FGF8 functions in the specification of the right body side of the chick. Curr Biol. 1999;9:277–280. doi: 10.1016/s0960-9822(99)80119-5. [DOI] [PubMed] [Google Scholar]
- Bulman MP, Kusumi K, Frayling TM, McKeown C, Garrett C, Lander ES, Krumlauf R, Hattersley AT, Ellard S, Turnpenny PD. Mutations in the human delta homologue, DLL3, cause axial skeletal defects in spondylocostal dysostosis. Nat Genet. 2000;24:438–441. doi: 10.1038/74307. [DOI] [PubMed] [Google Scholar]
- Chal J, Pourquie O. Patterning and differentiation of the vertebrate spine. In: Pourquie O, editor. The Skeletal System. Cold Spring Harbor Laboratory Press; 2009. pp. 41–116. [Google Scholar]
- Chen J, Kang L, Zhang N. Negative feedback loop formed by Lunatic fringe and Hes7 controls their oscillatory expression during somitogenesis. Genesis. 2005;43:196–204. doi: 10.1002/gene.20171. [DOI] [PubMed] [Google Scholar]
- Cole SE, Levorse JM, Tilghman SM, Vogt TF. Clock Regulatory Elements Control Cyclic Expression of Lunatic fringe during Somitogenesis. Dev Cell. 2002;3:75–84. doi: 10.1016/s1534-5807(02)00212-5. [DOI] [PubMed] [Google Scholar]
- Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- Cooke J, Zeeman EC. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J Theor Biol. 1976;58:455–476. doi: 10.1016/s0022-5193(76)80131-2. [DOI] [PubMed] [Google Scholar]
- Cornier AS, Staehling-Hampton K, Delventhal KM, Saga Y, Caubet JF, Sasaki N, Ellard S, Young E, Ramirez N, Carlo SE, et al. Mutations in the MESP2 Gene Cause Spondylothoracic Dysostosis/Jarcho-Levin Syndrome. Am J Hum Genet. 2008;82:1334–1341. doi: 10.1016/j.ajhg.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C, Little CD, Rongish BJ. Rotation of organizer tissue contributes to left-right asymmetry. Anat Rec (Hoboken) 2009;292:557–561. doi: 10.1002/ar.20872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale JK, Malapert P, Chal J, Vilhais-Neto G, Maroto M, Johnson T, Jayasinghe S, Trainor P, Herrmann B, Pourquie O. Oscillations of the snail genes in the presomitic mesoderm coordinate segmental patterning and morphogenesis in vertebrate somitogenesis. Dev Cell. 2006;10:355–366. doi: 10.1016/j.devcel.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Dale JK, Maroto M, Dequeant ML, Malapert P, McGrew M, Pourquie O. Periodic Notch inhibition by Lunatic Fringe underlies the chick segmentation clock. Nature. 2003;421:275–278. doi: 10.1038/nature01244. [DOI] [PubMed] [Google Scholar]
- Delfini MC, Dubrulle J, Malapert P, Chal J, Pourquie O. Control of the segmentation process by graded MAPK/ERK activation in the chick embryo. Proc Natl Acad Sci USA. 2005;102:11343–11348. doi: 10.1073/pnas.0502933102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequeant ML, Ahnert S, Edelsbrunner H, Fink TM, Glynn EF, Hattem G, Kudlicki A, Mileyko Y, Morton J, Mushegian AR, et al. Comparison of pattern detection methods in microarray time series of the segmentation clock. PLoS ONE. 2008;3:e2856. doi: 10.1371/journal.pone.0002856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequeant ML, Glynn E, Gaudenz K, Wahl M, Chen J, Mushegian A, Pourquie O. A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science. 2006;314:1595–1598. doi: 10.1126/science.1133141. [DOI] [PubMed] [Google Scholar]
- Dequeant ML, Pourquie O. Segmental patterning of the vertebrate embryonic axis. Nat Rev Genet. 2008;9:370–382. doi: 10.1038/nrg2320. [DOI] [PubMed] [Google Scholar]
- Diez del Corral R, Olivera-Martinez I, Goriely A, Gale E, Maden M, Storey K. Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron. 2003;40:65–79. doi: 10.1016/s0896-6273(03)00565-8. [DOI] [PubMed] [Google Scholar]
- Donoviel DB, Hadjantonakis AK, Ikeda M, Zheng H, Hyslop PS, Bernstein A. Mice lacking both presenilin genes exhibit early embryonic patterning defects. Genes Dev. 1999;13:2801–2810. doi: 10.1101/gad.13.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrulle J, McGrew MJ, Pourquie O. FGF signaling controls somite boundary position and regulates segmentation clock control of spatiotemporal Hox gene activation. Cell. 2001;106:219–232. doi: 10.1016/s0092-8674(01)00437-8. [DOI] [PubMed] [Google Scholar]
- Dubrulle J, Pourquie O. fgf8 mRNA decay establishes a gradient that couples axial elongation to patterning in the vertebrate embryo. Nature. 2004;427:419–422. doi: 10.1038/nature02216. [DOI] [PubMed] [Google Scholar]
- Dunty WC, Jr., Biris KK, Chalamalasetty RB, Taketo MM, Lewandoski M, Yamaguchi TP. Wnt3a/beta-catenin signaling controls posterior body development by coordinating mesoderm formation and segmentation. Development. 2008;135:85–94. doi: 10.1242/dev.009266. [DOI] [PubMed] [Google Scholar]
- Dunwoodie SL, Clements M, Sparrow DB, Sa X, Conlon RA, Beddington RS. Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene Dll3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development. 2002;129:1795–1806. doi: 10.1242/dev.129.7.1795. [DOI] [PubMed] [Google Scholar]
- Echeverri K, Oates AC. Coordination of symmetric cyclic gene expression during somitogenesis by Suppressor of Hairless involves regulation of retinoic acid catabolism. Dev Biol. 2007;301:388–403. doi: 10.1016/j.ydbio.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Fei Q, Wu Z, Wang H, Zhou X, Wang N, Ding Y, Wang Y, Qiu G. The association analysis of TBX6 polymorphism with susceptibility to congenital scoliosis in a Chinese Han population. Spine (Phila Pa 1976) 2010;35:983–988. doi: 10.1097/BRS.0b013e3181bc963c. [DOI] [PubMed] [Google Scholar]
- Feller J, Schneider A, Schuster-Gossler K, Gossler A. Noncyclic Notch activity in the presomitic mesoderm demonstrates uncoupling of somite compartmentalization and boundary formation. Genes Dev. 2008;22:2166–2171. doi: 10.1101/gad.480408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferjentsik Z, Hayashi S, Dale JK, Bessho Y, Herreman A, De Strooper B, del Monte G, de la Pompa JL, Maroto M. Notch is a critical component of the mouse somitogenesis oscillator and is essential for the formation of the somites. PLoS genetics. 2009;5:e1000662. doi: 10.1371/journal.pgen.1000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg H, Crozet F, Brown NA. Waves of mouse Lunatic fringe expression, in four-hour cycles at two-hour intervals, precede somite boundary formation. CurrBiol. 1998;8:1027–1030. doi: 10.1016/s0960-9822(07)00424-1. [DOI] [PubMed] [Google Scholar]
- Francois P, Hakim V, Siggia ED. Deriving structure from evolution: metazoan segmentation. Mol Syst Biol. 2007;3:154. doi: 10.1038/msb4100192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb S, Zagorska A, Melton K, Tenin G, Vacca I, Trainor P, Maroto M, Dale JK. Interfering with Wnt signalling alters the periodicity of the segmentation clock. Dev Biol. 2009;330:21–31. doi: 10.1016/j.ydbio.2009.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicelli F, Ozbudak EM, Wright GJ, Lewis J. Setting the Tempo in Development: An Investigation of the Zebrafish Somite Clock Mechanism. PLoS Biol. 2007;5:e150. doi: 10.1371/journal.pbio.0050150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbeter A, Gonze D, Pourquie O. Sharp developmental thresholds defined through bistability by antagonistic gradients of retinoic acid and FGF signaling. Dev Dyn. 2007;236:1495–1508. doi: 10.1002/dvdy.21193. [DOI] [PubMed] [Google Scholar]
- Goldbeter A, Pourquie O. Modeling the segmentation clock as a network of coupled oscillations in the Notch, Wnt and FGF signaling pathways. J Theor Biol. 2008;252:574–585. doi: 10.1016/j.jtbi.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Gomez C, Ozbudak EM, Wunderlich J, Baumann D, Lewis J, Pourquie O. Control of segment number in vertebrate embryos. Nature. 2008;454:335–339. doi: 10.1038/nature07020. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Kageyama R. Automatic reconstruction of the mouse segmentation network from an experimental evidence database. Bio Systems. 2010;102:16–21. doi: 10.1016/j.biosystems.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Gros J, Feistel K, Viebahn C, Blum M, Tabin CJ. Cell movements at Hensen's node establish left/right asymmetric gene expression in the chick. Science. 2009;324:941–944. doi: 10.1126/science.1172478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines N, Irvine KD. Glycosylation regulates Notch signalling. Nat Rev Mol Cell Biol. 2003;4:786–797. doi: 10.1038/nrm1228. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Shimoda T, Nakajima M, Tsukada Y, Sakumura Y, Dale JK, Maroto M, Kohno K, Matsui T, Bessho Y. Sprouty4, an FGF inhibitor, displays cyclic gene expression under the control of the notch segmentation clock in the mouse PSM. PLoS ONE. 2009;4:e5603. doi: 10.1371/journal.pone.0005603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry CA, Urban MK, Dill KK, Merlie JP, Page MF, Kimmel CB, Amacher SL. Two linked hairy/Enhancer of split-related zebrafish genes, her1 and her7, function together to refine alternating somite boundaries. Development. 2002;129:3693–3704. doi: 10.1242/dev.129.15.3693. [DOI] [PubMed] [Google Scholar]
- Herrgen L, Ares S, Morelli LG, Schroter C, Julicher F, Oates AC. Intercellular coupling regulates the period of the segmentation clock. Curr Biol. 2010;20:1244–1253. doi: 10.1016/j.cub.2010.06.034. [DOI] [PubMed] [Google Scholar]
- Hirata H, Bessho Y, Kokubu H, Masamizu Y, Yamada S, Lewis J, Kageyama R. Instability of Hes7 protein is crucial for the somite segmentation clock. Nat Genet. 2004;36:750–754. doi: 10.1038/ng1372. [DOI] [PubMed] [Google Scholar]
- Holley SA, Geisler R, Nusslein-Volhard C. Control of her1 expression during zebrafish somitogenesis by a delta-dependent oscillator and an independent wavefront activity. Genes Dev. 2000;14:1678–1690. [PMC free article] [PubMed] [Google Scholar]
- Holley SA, Julich D, Rauch GJ, Geisler R, Nusslein-Volhard C. her1 and the notch pathway function within the oscillator mechanism that regulates zebrafish somitogenesis. Development. 2002;129:1175–1183. doi: 10.1242/dev.129.5.1175. [DOI] [PubMed] [Google Scholar]
- Horikawa K, Ishimatsu K, Yoshimoto E, Kondo S, Takeda H. Noise-resistant and synchronized oscillation of the segmentation clock. Nature. 2006;441:719–723. doi: 10.1038/nature04861. [DOI] [PubMed] [Google Scholar]
- Hrabe de Angelis M, McIntyre J, Gossler A. Maintenance of somite borders in mice requires the Delta homologue DII1. Nature. 1997;386:717–721. doi: 10.1038/386717a0. [DOI] [PubMed] [Google Scholar]
- Huppert SS, Ilagan MX, De Strooper B, Kopan R. Analysis of Notch function in presomitic mesoderm suggests a gamma-secretase-independent role for presenilins in somite differentiation. Dev Cell. 2005;8:677–688. doi: 10.1016/j.devcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Ishikawa A, Kitajima S, Takahashi Y, Kokubo H, Kanno J, Inoue T, Saga Y. Mouse Nkd1, a Wnt antagonist, exhibits oscillatory gene expression in the PSM under the control of Notch signaling. Mech Dev. 2004;121:1443–1453. doi: 10.1016/j.mod.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Ishimatsu K, Takamatsu A, Takeda H. Emergence of traveling waves in the zebrafish segmentation clock. Development. 2010;137:1595–1599. doi: 10.1242/dev.046888. [DOI] [PubMed] [Google Scholar]
- Jiang YJ, Aerne BL, Smithers L, Haddon C, Ish-Horowicz D, Lewis J. Notch signalling and the synchronization of the somite segmentation clock. Nature. 2000;408:475–479. doi: 10.1038/35044091. [DOI] [PubMed] [Google Scholar]
- Jouve C, Palmeirim I, Henrique D, Beckers J, Gossler A, Ish-Horowicz D, Pourquie O. Notch signalling is required for cyclic expression of the hairy-like gene HES1 in the presomitic mesoderm. Development. 2000;127:1421–1429. doi: 10.1242/dev.127.7.1421. [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Raya A, Raya RM, Rodriguez-Esteban C, Belmonte JC. Retinoic acid signalling links left-right asymmetric patterning and bilaterally symmetric somitogenesis in the zebrafish embryo. Nature. 2005;435:165–171. doi: 10.1038/nature03512. [DOI] [PubMed] [Google Scholar]
- Kawamura A, Koshida S, Hijikata H, Ohbayashi A, Kondoh H, Takada S. Groucho-associated transcriptional repressor ripply1 is required for proper transition from the presomitic mesoderm to somites. Dev Cell. 2005a;9:735–744. doi: 10.1016/j.devcel.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Kawamura A, Koshida S, Hijikata H, Sakaguchi T, Kondoh H, Takada S. Zebrafish hairy/enhancer of split protein links FGF signaling to cyclic gene expression in the periodic segmentation of somites. Genes Dev. 2005b;19:1156–1161. doi: 10.1101/gad.1291205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. Autoinhibition with transcriptional delay: a simple mechanism for the zebrafish somitogenesis oscillator. Curr Biol. 2003;13:1398–1408. doi: 10.1016/s0960-9822(03)00534-7. [DOI] [PubMed] [Google Scholar]
- Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, Trask BJ, Kuo WL, Cochran J, et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Adamska M, Meisler MH. Hypomorphic expression of Dkk1 in the doubleridge mouse: dose dependence and compensatory interactions with Lrp6. Development. 2004;131:2543–2552. doi: 10.1242/dev.01126. [DOI] [PubMed] [Google Scholar]
- Mara A, Schroeder J, Chalouni C, Holley SA. Priming, initiation and synchronization of the segmentation clock by deltaD and deltaC. Nat Cell Biol. 2007;9:523–530. doi: 10.1038/ncb1578. [DOI] [PubMed] [Google Scholar]
- Maroto M, Dale JK, Dequeant ML, Petit AC, Pourquie O. Synchronised cycling gene oscillations in presomitic mesoderm cells require cell-cell contact. Int J Dev Biol. 2005;49:309–315. doi: 10.1387/ijdb.041958mm. [DOI] [PubMed] [Google Scholar]
- Martin BL, Kimelman D. Brachyury establishes the embryonic mesodermal progenitor niche. Genes Dev. 2010;24:2778–2783. doi: 10.1101/gad.1962910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruhashi M, Van De Putte T, Huylebroeck D, Kondoh H, Higashi Y. Involvement of SIP1 in positioning of somite boundaries in the mouse embryo. Dev Dyn. 2005;234:332–338. doi: 10.1002/dvdy.20546. [DOI] [PubMed] [Google Scholar]
- Masamizu Y, Ohtsuka T, Takashima Y, Nagahara H, Takenaka Y, Yoshikawa K, Okamura H, Kageyama R. Real-time imaging of the somite segmentation clock: revelation of unstable oscillators in the individual presomitic mesoderm cells. Proc Natl Acad Sci USA. 2006;103:1313–1318. doi: 10.1073/pnas.0508658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew MJ, Dale JK, Fraboulet S, Pourquie O. The lunatic fringe gene is a target of the molecular clock linked to somite segmentation in avian embryos. Curr Biol. 1998;8:979–982. doi: 10.1016/s0960-9822(98)70401-4. [DOI] [PubMed] [Google Scholar]
- Meyers EN, Martin GR. Differences in left-right axis pathways in mouse and chick: functions of FGF8 and SHH. Science. 1999;285:403–406. doi: 10.1126/science.285.5426.403. [DOI] [PubMed] [Google Scholar]
- Morales AV, Yasuda Y, Ish-Horowicz D. Periodic Lunatic fringe Expression Is Controlled during Segmentation by a Cyclic Transcriptional Enhancer Responsive to Notch Signaling. Dev Cell. 2002;3:63–74. doi: 10.1016/s1534-5807(02)00211-3. [DOI] [PubMed] [Google Scholar]
- Morelli LG, Ares S, Herrgen L, Schroter C, Julicher F, Oates AC. Delayed coupling theory of vertebrate segmentation. HFSP J. 2009;3:55–66. doi: 10.2976/1.3027088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno TA, Kintner C. Regulation of segmental patterning by retinoic acid signaling during Xenopus somitogenesis. Dev Cell. 2004;6:205–218. doi: 10.1016/s1534-5807(04)00026-7. [DOI] [PubMed] [Google Scholar]
- Morimoto M, Sasaki N, Oginuma M, Kiso M, Igarashi K, Aizaki K, Kanno J, Saga Y. The negative regulation of Mesp2 by mouse Ripply2 is required to establish the rostro-caudal patterning within a somite. Development. 2007;134:1561–1569. doi: 10.1242/dev.000836. [DOI] [PubMed] [Google Scholar]
- Morimoto M, Takahashi Y, Endo M, Saga Y. The Mesp2 transcription factor establishes segmental borders by suppressing Notch activity. Nature. 2005;435:354–359. doi: 10.1038/nature03591. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Satoh T, Igari A, Kageyama R, Nishida E. FGF induces oscillations of Hes1 expression and Ras/ERK activation. Curr Biol. 2008;18:R332–334. doi: 10.1016/j.cub.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Fraulob V, Garnier JM, Chambon P, Dolle P. Differential expression of retinoic acid-synthesizing (RALDH) enzymes during fetal development and organ differentiation in the mouse. Mech Dev. 2002;110:165–171. doi: 10.1016/s0925-4773(01)00561-5. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Subbarayan V, Dolle P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet. 1999;21:444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- Niwa Y, Masamizu Y, Liu T, Nakayama R, Deng CX, Kageyama R. The initiation and propagation of Hes7 oscillation are cooperatively regulated by Fgf and notch signaling in the somite segmentation clock. Dev Cell. 2007;13:298–304. doi: 10.1016/j.devcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Oates AC, Ho RK. Hairy/E(spl)-related (Her) genes are central components of the segmentation oscillator and display redundancy with the Delta/Notch signaling pathway in the formation of anterior segmental boundaries in the zebrafish. Development. 2002;129:2929–2946. doi: 10.1242/dev.129.12.2929. [DOI] [PubMed] [Google Scholar]
- Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, Meltzer PS, Spinner NB, Collins FS, et al. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- Offiah A, Alman B, Cornier AS, Giampietro PF, Tassy O, Wade A, Turnpenny PD. Pilot assessment of a radiologic classification system for segmentation defects of the vertebrae. American journal of medical genetics. 2010;152A:1357–1371. doi: 10.1002/ajmg.a.33361. [DOI] [PubMed] [Google Scholar]
- Oginuma M, Niwa Y, Chapman DL, Saga Y. Mesp2 and Tbx6 cooperatively create periodic patterns coupled with the clock machinery during mouse somitogenesis. Development. 2008;135:2555–2562. doi: 10.1242/dev.019877. [DOI] [PubMed] [Google Scholar]
- Oginuma M, Takahashi Y, Kitajima S, Kiso M, Kanno J, Kimura A, Saga Y. The oscillation of Notch activation, but not its boundary, is required for somite border formation and rostral-caudal patterning within a somite. Development. 2010;137:1515–1522. doi: 10.1242/dev.044545. [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Lewis J. Notch signalling synchronizes the zebrafish segmentation clock but is not needed to create somite boundaries. PLoS genetics. 2008;4:e15. doi: 10.1371/journal.pgen.0040015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak EM, Tassy O, Pourquie O. Spatiotemporal compartmentalization of key physiological processes during muscle precursor differentiation. Proc Natl Acad Sci USA. 2010;107:4224–4229. doi: 10.1073/pnas.0909375107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmeirim I, Henrique D, Ish-Horowicz D, Pourquié O. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell. 1997;91:639–648. doi: 10.1016/s0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- Plaster N, Sonntag C, Schilling TF, Hammerschmidt M. REREa/Atrophin-2 interacts with histone deacetylase and Fgf8 signaling to regulate multiple processes of zebrafish development. Dev Dyn. 2007;236:1891–1904. doi: 10.1002/dvdy.21196. [DOI] [PubMed] [Google Scholar]
- Prince VE, Holley SA, Bally-Cuif L, Prabhakaran B, Oates AC, Ho RK, Vogt TF. Zebrafish lunatic fringe demarcates segmental boundaries. Mech Dev. 2001;105:175–180. doi: 10.1016/s0925-4773(01)00398-7. [DOI] [PubMed] [Google Scholar]
- Pueyo JI, Lanfear R, Couso JP. Ancestral Notch-mediated segmentation revealed in the cockroach Periplaneta americana. Proc Natl Acad Sci USA. 2008;105:16614–16619. doi: 10.1073/pnas.0804093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya A, Izpisua Belmonte JC. Insights into the establishment of left-right asymmetries in vertebrates. Birth Defects Res C Embryo Today. 2008;84:81–94. doi: 10.1002/bdrc.20122. [DOI] [PubMed] [Google Scholar]
- Raya A, Kawakami Y, Rodriguez-Esteban C, Ibanes M, Rasskin-Gutman D, Rodriguez-Leon J, Buscher D, Feijo JA, Izpisua Belmonte JC. Notch activity acts as a sensor for extracellular calcium during vertebrate left-right determination. Nature. 2004;427:121–128. doi: 10.1038/nature02190. [DOI] [PubMed] [Google Scholar]
- Resende TP, Ferreira M, Teillet MA, Tavares AT, Andrade RP, Palmeirim I. Sonic hedgehog in temporal control of somite formation. Proc Natl Acad Sci USA. 2010;107:12907–12912. doi: 10.1073/pnas.1000979107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel-Kruse IH, Muller C, Oates AC. Synchrony dynamics during initiation, failure, and rescue of the segmentation clock. Science. 2007;317:1911–1915. doi: 10.1126/science.1142538. [DOI] [PubMed] [Google Scholar]
- Romanoff A. The Avian Embryo: Structural and Functional Development. Vol 1. The Macmillan Company; New York: 1960. [Google Scholar]
- Sakai Y, Meno C, Fujii H, Nishino J, Shiratori H, Saijoh Y, Rossant J, Hamada H. The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio-posterior axis within the mouse embryo. Genes Dev. 2001;15:213–225. doi: 10.1101/gad.851501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada A, Shinya M, Jiang YJ, Kawakami A, Kuroiwa A, Takeda H. Fgf/MAPK signalling is a crucial positional cue in somite boundary formation. Development. 2001;128:4873–4880. doi: 10.1242/dev.128.23.4873. [DOI] [PubMed] [Google Scholar]
- Schroter C, Herrgen L, Cardona A, Brouhard GJ, Feldman B, Oates AC. Dynamics of zebrafish somitogenesis. Dev Dyn. 2008;237:545–553. doi: 10.1002/dvdy.21458. [DOI] [PubMed] [Google Scholar]
- Schroter C, Oates AC. Segment number and axial identity in a segmentation clock period mutant. Curr Biol. 2010;20:1254–1258. doi: 10.1016/j.cub.2010.05.071. [DOI] [PubMed] [Google Scholar]
- Serth K, Schuster-Gossler K, Cordes R, Gossler A. Transcriptional oscillation of Lunatic fringe is essential for somitogenesis. Genes Dev. 2003;17:912–925. doi: 10.1101/gad.250603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewell W, Sparrow DB, Smith AJ, Gonzalez DM, Rappaport EF, Dunwoodie SL, Kusumi K. Cyclical expression of the Notch/Wnt regulator Nrarp requires modulation by Dll3 in somitogenesis. Dev Biol. 2009;329:400–409. doi: 10.1016/j.ydbio.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifley ET, Vanhorn KM, Perez-Balaguer A, Franklin JD, Weinstein M, Cole SE. Oscillatory lunatic fringe activity is crucial for segmentation of the anterior but not posterior skeleton. Development. 2008;135:899–908. doi: 10.1242/dev.006742. [DOI] [PubMed] [Google Scholar]
- Shiratori H, Hamada H. The left-right axis in the mouse: from origin to morphology. Development. 2006;133:2095–2104. doi: 10.1242/dev.02384. [DOI] [PubMed] [Google Scholar]
- Sirbu IO, Duester G. Retinoic-acid signalling in node ectoderm and posterior neural plate directs left-right patterning of somitic mesoderm. Nat Cell Biol. 2006;8:271–277. doi: 10.1038/ncb1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow DB, Chapman G, Wouters MA, Whittock NV, Ellard S, Fatkin D, Turnpenny PD, Kusumi K, Sillence D, Dunwoodie SL. Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. Am J Hum Genet. 2006;78:28–37. doi: 10.1086/498879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow DB, Guillen-Navarro E, Fatkin D, Dunwoodie SL. Mutation of HAIRY-AND-ENHANCER-OF-SPLIT-7 in Humans Causes Spondylocostal Dysostosis. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn272. [DOI] [PubMed] [Google Scholar]
- Sparrow DB, Sillence D, Wouters MA, Turnpenny PD, Dunwoodie SL. Two novel missense mutations in HAIRY-AND-ENHANCER-OF-SPLIT-7 in a family with spondylocostal dysostosis. Eur J Hum Genet. 2010;18:674–679. doi: 10.1038/ejhg.2009.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber M, Sachidanandan C, Morgenstern C, Ish-Horowicz D. Differential axial requirements for lunatic fringe and Hes7 transcription during mouse somitogenesis. PLoS ONE. 2009;4:e7996. doi: 10.1371/journal.pone.0007996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suriben R, Fisher DA, Cheyette BN. Dact1 presomitic mesoderm expression oscillates in phase with Axin2 in the somitogenesis clock of mice. Dev Dyn. 2006;235:3177–3183. doi: 10.1002/dvdy.20968. [DOI] [PubMed] [Google Scholar]
- Takahashi J, Ohbayashi A, Oginuma M, Saito D, Mochizuki A, Saga Y, Takada S. Analysis of Ripply1/2-deficient mouse embryos reveals a mechanism underlying the rostro-caudal patterning within a somite. Dev Biol. 2010;342:134–145. doi: 10.1016/j.ydbio.2010.03.015. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Koizumi K, Takagi A, Kitajima S, Inoue T, Koseki H, Saga Y. Mesp2 initiates somite segmentation through the Notch signalling pathway. Nat Genet. 2000;25:390–396. doi: 10.1038/78062. [DOI] [PubMed] [Google Scholar]
- Tam PP. The control of somitogenesis in mouse embryos. JEmbryolExpMorphol. 1981;65(Suppl):103–128. [PubMed] [Google Scholar]
- Turnpenny PD, Alman B, Cornier AS, Giampietro PF, Offiah A, Tassy O, Pourquie O, Kusumi K, Dunwoodie S. Abnormal vertebral segmentation and the notch signaling pathway in man. Dev Dyn. 2007;236:1456–1474. doi: 10.1002/dvdy.21182. [DOI] [PubMed] [Google Scholar]
- Uriu K, Morishita Y, Iwasa Y. Random cell movement promotes synchronization of the segmentation clock. Proc Natl Acad Sci U S A. 2010;107:4979–4984. doi: 10.1073/pnas.0907122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermot J, Llamas JG, Fraulob V, Niederreither K, Chambon P, Dolle P. Retinoic acid controls the bilateral symmetry of somite formation in the mouse embryo. Science. 2005;308:563–566. doi: 10.1126/science.1108363. [DOI] [PubMed] [Google Scholar]
- Vermot J, Pourquie O. Retinoic acid coordinates somitogenesis and left-right patterning in vertebrate embryos. Nature. 2005;435:215–220. doi: 10.1038/nature03488. [DOI] [PubMed] [Google Scholar]
- Vilhais-Neto GC, Maruhashi M, Smith KT, Vasseur-Cognet M, Peterson AS, Workman JL, Pourquie O. Rere controls retinoic acid signalling and somite bilateral symmetry. Nature. 2010;463:953–957. doi: 10.1038/nature08763. [DOI] [PubMed] [Google Scholar]
- Wahl MB, Deng C, Lewandoski M, Pourquie O. FGF signaling acts upstream of the NOTCH and WNT signaling pathways to control segmentation clock oscillations in mouse somitogenesis. Development. 2007;134:4033–4041. doi: 10.1242/dev.009167. [DOI] [PubMed] [Google Scholar]
- Whittock NV, Sparrow DB, Wouters MA, Sillence D, Ellard S, Dunwoodie SL, Turnpenny PD. Mutated MESP2 causes spondylocostal dysostosis in humans. Am J Hum Genet. 2004;74:1249–1254. doi: 10.1086/421053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D, Ferjentsik Z, Chong SW, Qiu X, Jiang YJ, Malapert P, Pourquie O, Van Hateren N, Wilson SA, Franco C, et al. Cyclic Nrarp mRNA expression is regulated by the somitic oscillator but Nrarp protein levels do not oscillate. Dev Dyn. 2009;238:3043–3055. doi: 10.1002/dvdy.22139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JK, Moon RT, Wold B. The bHLH class protein pMesogenin1 can specify paraxial mesoderm phenotypes. Dev Biol. 2000;222:376–391. doi: 10.1006/dbio.2000.9717. In Process Citation. [DOI] [PubMed] [Google Scholar]