Abstract
Small molecule complexes with DNA that incorporate linking water molecules are rare, and the DB921-DNA complex has provided a unique and well-defined system for analysis of water-mediated binding in the context of a DNA complex. DB921 has a benzimidazole–biphenyl system with terminal amidines that results in a linear conformation that does not possess the appropriate radius of curvature to match the minor groove shape and represents a new paradigm that does not fit the classical model of minor groove interactions. To better understand the role of the bound water molecule observed in the X-ray crystal structure of the DB921 complex, synthetic modifications have been made in the DB921 structure and the interactions of the new compounds with DNA AT sites have been evaluated with an array of methods including DNase I footprinting, biosensor-surface plasmon resonance, isothermal titration microcalorimetry, and circular dichroism. The interaction of a key compound, which has the amidine at the phenyl shifted from the para position in DB921 to the meta position, has also been examined by X-ray crystallography. The detailed structural, thermodynamic and kinetic results provide valuable new information for incorporation of water molecules in the design of new lead scaffolds for targeting DNA in chemical biology and therapeutic applications.
INTRODUCTION
Linked-heterocyclic systems that target the DNA minor groove are promising agents against parasites that cause a number of major infectious diseases in the third world. Compounds that have a concave curvature that closely matches that of the groove with H-bond donating groups can bind strongly to the numerous AT rich sites in parasite mitochondrial kinetoplast DNA. Such binding can induce significant topological changes in AT sequences with destruction of the kinetoplast and cell death. If the compounds are either too curved or not sufficiently curved to match the surface of the groove for interactions with base pair edges at the floor of the groove, they typically have dramatically reduced binding affinity.1–2 The classical model for minor groove binding emphasizes the complementary curvature of DNA and the bound compound, and has been the basis for the design of the polyamide class of DNA sequence-selective agents.3–6 DB921 (Figure 1), however, is a linear heterocyclic diamidine that binds surprisingly strongly to the DNA minor groove.7 Closely related curved compounds, such as DB911 (Figure 1), actually bind more weakly to DNA than DB921.
Fig. 1.
Structure of the compounds and the DNA sequences used in this study. The DNA hairpins were used in SPR with biotin (as shown) and in ITC without biotin.
An X-ray structure (PDB ID: 2B0K) of the DB921 complex with the DNA duplex sequence d(CGCGAATTCGCG)2, which has been used in structural studies with a large number of minor groove binders,1 shows DB921 bound at the AATT site with a water-mediated interaction between the phenylamidine of DB921 and DNA base atoms (Figure 1). This ternary compound-water-DNA interaction serves to complete the curvature of the bound system and results in a very energetically stable complex. Binding of traditional crescent-shaped compounds in the minor groove is generally stabilized by associated water of hydration both in and out of the groove.1 The specific water molecule that links the phenylamidine of DB921 to the bases at the floor of the groove, however, is different from the stabilizing water molecules observed in other minor groove complexes. These waters are generally localized at the ends of the complex or at the outer edge of the minor groove while the linking water for DB921 and DNA is positioned between the compound and N3 of an adenine base. The water molecule in the DB921 binding site can flexibly orient to provide favorable interactions between the compound and DNA.8 The H-bonding ability and adaptability of the bound water appears to account for much of the high binding affinity of DB921. This ability of dynamic water molecules to mediate noncovalent interactions by acting as adaptable hydrogen bond donors or acceptors to stabilize ligand-macromolecule complexes has been observed in both protein and-nucleic acid small molecule complexes.8–9 The classically-curved analog of DB921, DB911, does not require a bound water to optimize DNA interactions but binds to the AATT DNA minor groove site significantly more weakly than DB921.7
Small molecule complexes with DNA that incorporate a linking water molecule are rare and the DB921-DNA complex provides a unique and well-defined system for analysis of water-mediated binding in the context of a DNA complex. The system also provides valuable new information for incorporation of water in the design of new lead scaffolds. To better understand the role of the bound water observed in the X-ray crystal structure of the DB921 complex, and to use such water in compound design, synthetic modifications have been made in the DB921 structure, as described below, and the interactions of the new compounds with DNA AT sites have been evaluated.
Compound Design and DNA Binding Sites
All of the five individual molecular groups of DB921, benzimidazole, two phenyls and two amidines, have been modified to provide additional information about their importance for the exceptionally strong, water-mediated binding of DB921 with DNA (Figure 1). In addition, to help understand the importance of the DNA binding site sequence on the DB921 interaction affinity, the binding of all compounds was evaluated with an ATAT site, which has relatively straight topology, as well as the bent AATT site that has been used previously with DB921 and in a large number of structural and biophysical studies of other minor groove complexes.10 Quantitative analysis of the compound interactions with DNA was done with biosensor-surface plasmon resonance (SPR) methods through streptavidin immobilization of 5′-biotin-labeled DNA to create the biosensor surface. Since this method responds to mass on the surface based on the compound molecular weight, each molecule was evaluated in an identical manner with each DNA on the same sensor chip.11–12 The SPR signal at saturation gives a direct indication of interaction stoichiometry and fitting of the sensorgrams as a function of compound concentration in the flow solution provides the binding constant K and kinetics constants. Additional detailed information was obtained for selected complexes by isothermal titration calorimetry and DNase I footprinting.
The compounds were divided into six groups (Figure 1) based on the modifications made to each molecular unit of DB921:
-
Group I
All of these compounds have an amidine-benzimidazole or an amidine-indole that can donate two H-bonds to DNA as well as a phenyl-amidine that can potentially form the water-mediated interaction with DNA. All of the modifications should change the stacking interactions with the minor groove relative to DB921, either through electronic/dipole moment changes (indole and two pyridines), torsional angle (phenyl to phenol) or shape and stacking (phenyl to allyl).
-
Group II
Both of these compounds have their diamidine groups modified to cyclic structures that maintain the high pK values of the amidines.
-
Group III
These compounds have one of their amidines modified to an uncharged group, either an amide, which should still be able to form the water-mediated interaction with DNA, or to -H which loses the H-bond ability as well as the amidine or amide stacking interactions with the minor groove walls.
-
Group IV
Both of these compounds have the para linkages of DB921 changed to meta (either at the central or terminal phenyl). DB911 is a more classically shaped compound for minor groove binding. DB1055 has the amidine group in the position occupied by the linking water in the DB921 structure. This compound should be able to bind to DNA with similar affinity to DB921 but without the requirement of a water linker. To help understand the DB1055 complex, an X-ray structure that it binds in an AATT site was determined.
-
Group V
All of these compounds have lost the benzimidazole H-bond donating -NH (either by conversion of the benzimidazole to an N-CH3 benzimidazole, to an indole with the -NH pointed in the opposite direction from the amidine, to a naphthyl, or to an imidazopyridine (DB877)). These derivatives should only be able to form one direct H-bond as well as the water mediated interaction with DNA.
-
Group VI
This is a set of reference compounds whose complexes with the DNA minor groove have been studied in detail and for which crystal structures of the complexes are available. All of these have the classical shape for minor groove interactions.
MATERIALS AND METHODS
Materials
Syntheses of compounds DB75, DB818, DB877, DB911, DB921, DB922, DB1055, DB1798, DB1869, DB1883 and DB1944 have been published.13–19 Syntheses of compounds DB985, DB1177, DB1302, DB1963, DB1964, DB1771, DB1780, DB1781, DB1803 and DB1804 will be published elsewhere. The purity of all synthetic compounds was verified by NMR and elemental analysis.
In SPR experiments, 5′-biotin labeled hairpin DNA oligomers AATT [5′-Biotin-CGAATTCGTCTCCGAATTCG] and ATAT [5′-Biotin-CGATATCGTCTCCGATATCG], with the hairpin loop sequences underlined were used. In ITC and CD experiments either the same DNA hairpin sequences as in SPR (without biotin) or duplex oligomers AATT [5′-GCGAATTCGC-3′] and ATAT [5′-GCGATATCGC-3′] were used. All oligomers were obtained from Integrated DNA Technologies, Inc. with reverse phased HPLC purification and mass spectrometry characterization. The MES20 buffers used in ITC and CD experiments contained 0.01 M [2-(N-morpholino) ethanesulfonic acid] (MES), 0.001 M EDTA, 0.2 M NaCl, pH 6.25. The SPR experiments were performed in filtered, degassed MES20 buffer with 5×10−3% v/v Surfactant P20.
Biosensor-Surface Plasmon Resonance (SPR) studies
SPR measurements were performed with a four–channel Biacore T100 optical biosensor system (GE Healthcare Inc.). 5′-biotin labeled DNA sequences [AATT and ATAT hairpins] were immobilized onto streptavidin-coated sensor chips (Biacore SA) as previously described.7,11–12 Steady state binding analysis was performed with multiple injections of different compound concentrations over the immobilized DNA surface at a flow rate of 25 μl/min. In Biacore instruments the response to binding is reported in RU values which are directly related to the amount of compound bound to the immobilized DNA. RU values from the steady–state region of SPR sensorgrams were converted to r (r = RU/RUmax, where RUmax is the RU value at saturating the DNA by compound) and are plotted against Cfree to obtain the equilibrium binding constant K. Binding results from the SPR experiments were well fitted with a single site interaction model:
| (1) |
where r represents the moles of bound compound per mole of DNA hairpin duplex and Cfree is the free compound concentration in equilibrium with the complex.
Kinetic analysis was also performed with multiple injections of different compound concentrations over the immobilized DNA surface at a flow rate of 50 μl/min. Global fitting of the sensorgram binding data with a mass transport kinetic 1:1 binding model was performed.11,20–21 In the mass transport limitation model, the following Eqs. (2–5) are used for global fitting:
| (2) |
| (3) |
| (4) |
| (5) |
where [A] and [Abulk] are the concentration of the compound at the sensor surface and in the bulk solution respectively; [B] is the concentration of the immobilized DNA without bound compound, [AB] is the concentration of the complex; ka is the association rate constant; kd is the dissociation rate constant, and kt is the mass transport coefficient. 22
Isothermal Titration Calorimetry (ITC) studies
The ITC experiments were performed with a MicroCal VP–ITC (MicroCal Inc., Northampton, MA, USA) interfaced with a computer for instrument control and data collection with Origin 7.0 software. Binding enthalpies for the DNA hairpins and duplexes [AATT and ATAT] with different compounds were determined at different temperatures using a protocol with a high DNA concentration in the calorimeter to ensure that all added compound is effectively bound after each addition in the low ratio region. Specifically, a 10 μm DNA solution was loaded into the sample cell and a 0.025 mM compound solution in MES20 buffer was added from the titration syringe. Usually 20 injections of 7 μl were done with 300 s between injections to ensure equilibration. For the additions where all compound is bound the heat of reaction (Q) is obtained by integration of the peaks after each injection. The dilution heats, determined by injecting drug solution into the same sample cell loaded with buffer alone, were subtracted from the Q value determined for addition into DNA solution to render a corrected value for the binding–induced enthalpy change. An average over all low ratio values gives the ΔH of binding.
DNase I footprinting, electrophoresis and quantitation
DNase I footprinting experiments were performed as described23 with the following modifications. 5′-phosphorylated oligonucleotides containing AATT and ATAT sites (underlined) 5′-CGGTACCAGATCTGGTCTAGAGCCATGGCTGCAATTCTGG CTCGATGAGCAGGATATGCGGCCGCTG -3′ and 5′-GATCCAGCGGCCGCATATCCTGC TCATCGAGCCAGAATTGCAGCCATGGCTCTAGACCAGATCTGGTACCGAGCT-3′ were synthesized by Eurogentec (Belgium). Equimolar concentration of each oligonucleotides were hybridized by heating the samples at 95 °C for 5 min, followed by a slow temperature decrease, until room temperature prior to ligation at SacI and BamHI sites of the pUC19-TTAA vector as described.24 The AATT-ATAT-107 bp DNA fragment was obtained from BglII and PstI double digestion of this new pUC19-AATT-ATAT vector. For footprinting experiments, the generated DNA fragment was 3′-end labeled using α-[32P]-dGTP (3000 Ci/mmol each, GE Healthcare, Buckinghamshire, UK) and 10 units of Klenow enzyme (BioLabs, France) for 30 min at 37 °C. The 265bp DNA fragment was obtained and labeled as previously described.2 The resulting radio-labeled DNA fragments were separated and isolated from a 6% native polyacrylamide gel as previously described.23 Increasing concentrations (as indicated in the legend of Figure 6) of the various tested compounds were incubated for 15 min at 37 °C with the radio-labeled DNA fragments prior to DNase I digestion (0.001 unit/mL, Sigma, France) for 3 min in digestion buffer (20 mM NaCl, 2 mM MgCl2, 2 mM MnCl2, pH 7.3). The reaction was ended by freeze-drying and lyophilization of the digestion solution. The cleaved DNA fragments were then dissolved in 4 μL of formamide-containing denaturing loading buffer, heated for 4 min at 90 °C and rapidly chilled on ice prior to electrophoresis on a denaturing 8% polyacrylamide gel. The gels were then soaked in 10% acetic acid, transferred to Whatman 3 MM paper and dried under vacuum at 80 °C. Dried gels were exposed overnight on storage screens and scanned using a Molecular Dynamics STORM 860. Each resolved band was assigned to a particular bond within the DNA fragments by comparison of its position relative to the guanine ladder obtained from Maxam and Gilbert sequencing standard method. Quantifications of the footprints were performed using Image Quant 4.1 software.
Fig. 6.
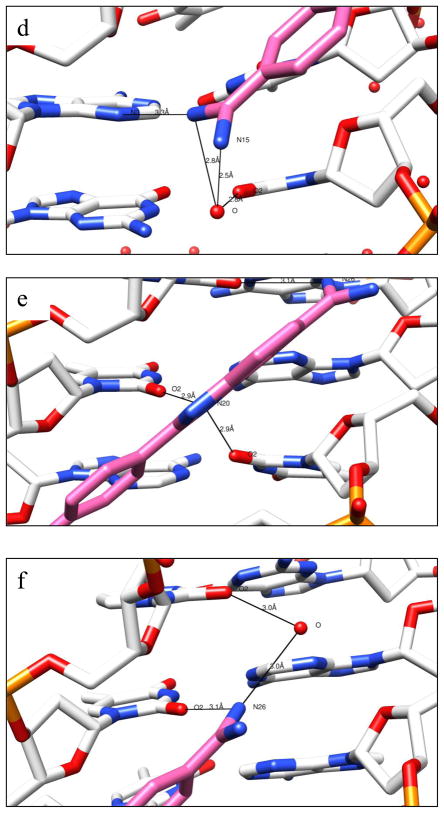
X-ray crystallography: a. The crystal structure of the DB1055-d(CGCGAATTCGCG)2 complex, with the DNA in cartoon ribbon representation and the ligand shown bound in the minor groove as a solvent-accessible surface colored according to B factor values; b. Overlay of the crystal structure positions of DB1055 (blue sticks) and DB921 (green sticks), together with the associated water molecule, colored in the same way; c. Detailed view of the meta and para amidinium ends of DB1055 and DB921, using the same color representations as in Figure 6b, showing hydrogen-bond interactions with the DNA and water molecules; d-f. Views of the hydrogen bonding between the three regions of DB1055 and the minor groove substituents: d. shows the meta amidinium group, e. shows the central imidazole group and f. shows the benzimidazole amidinium group.
CD titration studies
CD spectra were recorded using a Jasco J–810 instrument with a 1cm cell and a scan speed of 50 nm/min with a response time of 1 s. The spectra from 500 to 220 nm were averaged over five scans. A buffer baseline scan was collected in the same cuvette and subtracted from the average scan for each sample. The titration experiments were performed at 25 ºC. The desired ratios of compound to DNA were obtained by adding compound to the cell containing a constant amount of DNA. Data processing and plotting were performed with Kaleidagraph software.
X-ray crystallography
The HPLC-purified oligonucleotide d(CGCGAATTCGCG) (Eurogentec) at 6 mM single-stranded DNA solution in 30 mM sodium cacodylate buffer at pH 7.0 was annealed into a duplex before use by incubation in a heating block at 85 °C for 15 min and then left to cool gradually at room temperature for about 1 hour. After one month, the complex crystallized as elongated rods by the sitting-drop vapor diffusion method from a 24 μl drop containing 31.25 mM MgCl2, 1.0417% v/v (±)-2-methyl-2,4-pentanediol (MPD), 0.125 mM double-stranded DNA, 0.167 mM DB1055, and 5.0 mM sodium cacodylate buffer at pH 7.0 at 20 °C. The DNA: DB1055 ratio was 3:4. The drop was equilibrated against a reservoir of 0.5 ml of 50% v/v MPD solution in water.
X-ray diffraction data were collected at −168 °C on a Rigaku R-AXIS IV image plate detector with CuKα radiation emitted from a Rigaku RU200 rotating anode generator and an Osmic focusing mirror system (Rigaku/MSC). The crystal-to-detector distance was set at 90 mm. Each frame was exposed for 30 min with the crystal oscillating at a range of 2°. Data was collected to a maximum resolution of 1.65 Å. Indexing and data processing were carried out using XdisplayF, DENZO and SCALEPACK of the HKL package v.1.97.2 (HKL Research, Inc.).25
The structure was solved by molecular replacement using the isomorphous DNA structure from the berenil-d(CGCGAATTCGCG)2 complex (PDB id 2DBE).26 Unambiguous electron density for the DB1055 molecule was seen in 2Fo - Fc and Fo - Fc maps. The structure was refined to final R and Rfree values of 20.1% and 28.7% respectively at a resolution of 1.65 Ǻ. Atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank with entry code 215A.
RESULTS
Biosensor-SPR Binding Analysis: Determination of K and ΔG
As described above, to quantitatively compare the interactions of a number of different small molecules, which have quite different structures and physical properties, with different DNA sequences, biosensor-SPR methods are very attractive since each interaction is detected in the same way.11, 12 Essentially, the same moles of each DNA oligomer were immobilized on the sensor chips so that the sensorgram saturation levels (RUmax) can be compared directly for stoichiometry differences.11–12 Representative SPR sensorgrams for the injection of DB921, DB911, DB1055, DB1963 and DB985 over the surfaces with AATT and ATAT sequences are compared in Figure 2A.
Fig. 2.
SPR binding affinity: (A) Representative SPR sensorgrams for the interaction of selected compounds with AATT (upper panel) and ATAT (lower panel) DNA sequences. In all cases the compound concentrations from bottom to top are 0 to 1μM. (B) Comparison of the SPR binding affinity for AATT and ATAT DNA sequences with different compounds. RU values from the steady–state region of SPR sensorgrams, such as those in (A), were converted to r (r = RU/RUmax) and are plotted against the unbound compound concentration (flow solution) for DB921 (circles), DB911 (squares), DB1055 (up triangle), DB1963 (down triangle) and DB985 (diamond) binding with AATT and ATAT DNA sequences. The lines are the best fit values to a single site interaction model as described in the text.
Under these conditions, the sensorgrams generally reach a steady state plateau where the on and off rates are equal, except at the lowest concentrations. These steady-state values do not depend on binding kinetics or mass transfer effects12 and can be used to determine binding constants without a kinetic model. Mass transfer does not affect the steady-state RU and possible mass transfer corrections and more complex fitting models20–22 are unnecessary for steady-state analysis. The steady-state RU values, at each compound injection concentration were converted to r as described in the Method Section.
For these compounds the results can be fitted to a single-site binding model well within experimental error and the representative steady-state fitting for the selected compounds binding with AATT and ATAT DNA sequences are shown in Figure 2B. All K values for compounds binding with AATT and ATAT sequences are collected in Table 1. A disadvantage of steady-state analysis is that it cannot be used for cases where the reaction does not reach a steady state within the instrument operating association reaction time limit and a kinetic fit is the only possibility. In many cases, however, this limitation can be overcome. Since the association reaction must at least have a stoichiometry of 1, for a 1:1 complex, the association must be at least bimolecular and the rate of association will increase with injected concentration and at some point, except for the very slowest reactions, will reach a steady state. All sensorgrams above this point can be used in a steady state fit. It is also possible to use some lower concentration points by extrapolating the association region of the sensorgrams to the steady state time limit. Although this must be done with caution, it can be quite accurate if RUmax is known and the curves are close to the steady-state limit12. The biosensor method is used to determine binding information on a surface in the presence of a small amount of neutral detergent to help prevent extensive surface absorption of the organic compounds. It is important to question how valid these results are in comparison to more classical solution experiments. Fortunately, the agreement for experiments that are conducted properly is excellent and a brief summary of the extensive, available comparisons, with a focus on small molecule-DNA interactions, is presented in the Supplementary Materials. Given that the DNA is tethered to the surface through an alkyl tail and biotin that effectively put the DNA in a solution environment, it is perhaps not surprising that the SPR results are in agreement with those from solution.
Table 1.
Summary of binding affinity for the interaction of all compounds with AATT and ATAT DNA sequences using steady-state analysis.
| Group | Comp. Structure | AATT KA (×107) | ATAT KA (×107) | Comp. Structure | AATT KA (×107) | ATAT KA (×107) | ||
|---|---|---|---|---|---|---|---|---|
| I | DB921 |
|
14.2 | 2.5 | DB922 |
|
6.8 | 1.1 |
| DB1883 |
|
24.6 | 5.5 | DB1771 |
|
4.2 | 1.9 | |
| DB1869 |
|
0.17 | 0.079 | DB985 |
|
4.5 | 1.0 | |
| II | DB1804 |
|
48.2 | 7.9 | DB1803 |
|
13.3 | 2.4 |
| III | DB1944 |
|
<0.01 | <0.01 | DB1964 |
|
2.0 | 0.54 |
| DB1963 |
|
3.9 | 0.56 | |||||
| IV | DB1055 |

|
5.8 | 1.74 | DB911 |

|
1.2 | 0.89 |
| V | DB1302 |

|
0.35 | 0.15 | DB1798 |
|
1.9 | 1.0 |
| DB1781 |
|
1.0 | 0.46 | DB1780 |
|
2.1 | 0.36 | |
| DB1177 |

|
2.1 | 0.66 | DB877 |
|
2.0 | 0.51 | |
| VI | DB75 |
|
0.32 | 1.0 | Netropsin |

|
17.5 | 8.0 |
| DB818 |

|
11 | 5.6 | |||||
The single-site fitting errors in these experiments are less than 10%. The reproducibility errors are larger and are between 15 – 20%.
SPR results show that the linear compounds DB921 and DB1883, which have two amidine and a benzimidazole or indole group that can H-bond with DNA, bind very strongly. Reversing the indole in DB1798 (Group II) so that a -CH rather than an -NH faces the floor of the DNA minor groove results in approximately a factor of 10 decrease in K. A similar effect is seen with other modified compounds in Group V that lose one potential H-bonding group. Cyclization of the amidine system (Group II) has little effect on K values relative to the amidine parent. Surprisingly, replacing the central phenyl with an acetylene group (DB1869 in Group I) results in a large 100 fold decrease in K. Replacement of the amidine with an amide in Group III (dication to monocation) causes a relatively small, 4–5 fold decrease in K. The amide maintains the ability to H-bond with A and T bases at the floor of the DNA minor groove and the primary difference in the amide compounds is the loss of a single charge. With DB1944 the amidine is replaced with an -H and the binding constant drops to a value too low to determine accurately under these conditions. In this case, the H-bond and the charge, as well as the amidine stacking interactions in the minor groove, are lost.
Interestingly, the curved compound DB911, which has a more classical shape to match the minor groove, has a 10 fold decrease in K for AATT relative to linear compounds such as DB921, in agreement with previous results,7 but the decrease for ATAT is less than for the linear compounds. This appears to be a characteristic of curved compounds in general since the standard compounds in Group V all bind more similarly to ATAT and AATT than the strong binding linear DB921 analogs. Netropsin, which forms a strong H-bond network with the bases at the floor of the minor groove, and DB818, which has a shape that is close to an ideal match to optimize H-bond and van der Waals contacts with the minor groove, bind in the same range as DB921, K ~ 108 M−1 with AATT. They bind only about a factor of 2 more weakly to ATAT in agreement with their classical concave shape.
Full kinetic analyses of the interaction of the AATT DNA sequence with DB921, DB911 and DB1055 are compared in Figure 3. Association (ka) and dissociation (kd) rate constants were determined by global fitting of the entire sensorgrams binding data. The kinetic binding constant (KA) was derived from the ratio of the association and dissociation rate constants (Table 2). As can be seen from Table 1 and 2, comparisons of the binding affinities derived from the kinetic model are in good agreement with the equilibrium K from steady state fitting. The kinetic fitting results for these three compounds binding with AATT meet the criteria previously outlined (ka×RUmax/kt ≤ 5)21 to minimize transport effects under the experimental conditions of Figure 3. As can be seen in Table 2, DB921 and DB1055 have slower “on” (ka) and much slower “off” (kd) rate kinetics than DB911. As a result of the slow dissociation, their binding affinities are almost 10 times greater than that for DB911.
Fig. 3.
SPR sensorgrams to evaluate the kinetics of the interactions of DB921, DB911 and DB1055 with the AATT DNA sequence in MES20 buffer at a 50 μl/min flow rate are shown. The black lines are best fit curves determined by global kinetic fitting. The concentrations from bottom to top are 0.001, 0.003, 0.006, 0.012, 0.03 and 0.065 μM for DB921, and 0.004, 0.015, 0.03, 0.08, 0.15, 0.9 μM for DB911, and 0.003, 0.009, 0.015, 0.045, 0.1, 0.15, 0.75 μM for DB1055, respectively.
Table 2.
Comparison of SPR kinetics results of AATT DNA sequence binding with DB921, DB911 and DB1055.
| Comp. | Flow rate (μl/min) | RUmax (RU) | ka (M−1s−1) | kd (s−1) | KA= ka/kd (M−1) | kt [RU/(Ms)] | ka× RUmax/kt | χ2 (RU)2 |
|---|---|---|---|---|---|---|---|---|
| DB921 | 50 | 18.8 | 2.0 × 106 | 0.014 | 1.5 × 108 | 0.84 × 107 | 4.6 | 0.059 |
| DB911 | 50 | 20.9 | 3.5 × 106 | 0.24 | 1.5 × 107 | 1.9 × 107 | 3.8 | 0.047 |
| DB1055 | 50 | 18.5 | 1.1 × 106 | 0.016 | 0.70 × 108 | 1.2 × 107 | 1.8 | 0.27 |
The single-site fitting errors in these experiments are similar to those in the steady-state analysis (less than 10%). The reproducibility errors are somewhat larger, 20–25%), probably due to the larger number of variables in the fitting equations (Methods Section).
ITC: Determination of ΔH° values for Binding
To determine full thermodynamic profiles for representative compounds, DB921, DB911, DB1055, DB1963 and DB985, ITC experiments were conducted with DB75 and netropsin as controls (Figure S1). Although it is also possible to determine ΔG values for binding by ITC, experimental limitations for large binding constants and relatively low heats, prevented accurate ΔG determination by this method for DB921.27 To obtain the most accurate binding enthalpies, an excess of DNA was used in the calorimeter cell and ΔH was determined at low ratios of the compounds to DNA. For these compounds under ITC experimental conditions, essentially all added compound is bound to DNA. In this type of experiment the binding ΔH can be directly determined for AATT and ATAT sites without any model assumptions by simply averaging the blank subtracted heat per mole for each of the injections at low ratio.28–29 The average binding enthalpy values for these compounds with the AATT and ATAT DNA sequences are collected in Table S2. The biosensor experiments were conducted with DNA hairpin duplex sequences to prevent dissociation in the continuous flow. ITC experiments with DB921 were conducted with both duplex structures to test the assumption that the AATT and ATAT binding sites, in the same flanking sequence context, are reporting on essentially equivalent bound and free binding sites in these two cases (Figure S2). As can be seen, the values obtained under these conditions are in excellent agreement. These results indicate that the binding sites in hairpins and duplexes are reporting the same thermodynamic information under these conditions and the hairpin loop does not make a significant contribution to the strong binding of DB921. The text in Supplementary Materials provides additional literature validation for use of duplex and hairpin comparisons.
From the SPR binding free energy and ITC enthalpy values determined above, the entropy (ΔS) of complex formation could be calculated from equation:
| (6) |
The thermodynamics parameters are summarized in Table S1. To visualize the different thermodynamic quantities more easily, the overall thermodynamic profiles for the interactions of AATT and ATAT DNA sequences with these compounds are compared in Figure 4. The thermodynamics driving these compounds to bind to AATT and ATAT at 25 °C have favorable enthalpy and entropy components that vary significantly among the compounds. The ΔH values for binding of the compounds range from about −2 to −7 kcal/mole while the TΔS values vary from 4 to 8 kcal/mole. The ΔG variations are much smaller, approximately 2 kcal/mol, indicating that the various compounds present quite similar free energies resulting from different enthalpy via entropy components.
Fig. 4.
Comparison of binding thermodynamics values for selected compound with AATT and ATAT DNA sequences at 25 °C.
DNase I Footprinting: Results with High Molecular Weight DNA
To compare the biosensor-SPR results with immobilized oligomer DNA to those with long DNA sequences in solution, DNase I footprinting was first conducted with DNA complexes of five representative compounds with several DNA fragments. DNase I footprinting is an excellent method to sample a large conformational space with a reasonable sized fragment. There are 256 possible 4 bp sequences of the size recognized by the minor groove compounds in Figure 1. In footprinting these can be considered as overlapping 4bp sites (for example, two 4bp sites in a 5bp sequence), suggesting that a 257 bp (256bp +1bp for lagging) DNA fragment could be enough to have all 256 possible tetrabases. We use more than one DNA sequence because the sequences are not designed to contain all possible 4bp sites and the full length sequence cannot be evaluated on a single gel. The 265 bp and 107 bp sequences (Figure 5) could only be analyzed up to 130 and 100 bp respectively but this still covers a large variety of 4 bp fragments. The binding selectivity of those compounds was also addressed using other DNA sequences of 198bp, 119bp, 121bp and 178bp that are not show here because they do not provide additional binding information. This corresponds to a total of 635 bp that were quantified to cover possible 4 bp sequences.
Fig. 5.
DNase I footprinting of compounds at different concentrations with (A) the 265 bp or (B) the AATT-ATAT-107 bp DNA fragments. The concentrations (μM) of the compound tested are shown at the top of the gel lanes. Tracks labelled “G” represent dimethyl sulfate-piperidine markers specific for guanine. Numbers in the bottom figure of cleavage plots refer to the sequence and nucleotide position of the different fragments. Negative values in the densitometry analyses correspond to a ligand-protected site and positive values represent enhanced cleavage.
In agreement with the biosensor results DB921 and DB1883 have very pronounced footprints in the 265 bp sequence at AATT sites (base pair positions 79-76 and 125-122) while the reverse indole (DB1798) and especially DB1869 (Group I) have much weaker footprints at the same concentration range. The result with the pyridine (DB1771) is more similar to that for DB921. An ATTA sequence (position 95-92) also shows strong footprints while an AAAA site (near position 55) binds more weakly in this sequence context. Footprinting with DB75 and DB911 (densitometric analyses presented in Table S3) require higher concentrations to produce a significant footprint, also in agreement with SPR results. The AATT-ATAT-107 bp DNA fragment (Figure 5B) was then designed and used to compare the sequence selectivity of this series of compounds between an AATT site (position 25-22), a TTAA site (71-68) or the alternate ATAT site (46-43). All those compounds clearly evidenced a greater binding affinity for the AATT site and no clear footprint at the TTAA position whereas binding at the ATAT site requires higher concentration of compounds to obtain a full protection from DNase I cleavage. DB921 and DB1883 are more potent than the reverse compounds DB1798 and DB1869. Comparison of the DNase I protection in the presence of a larger series of compounds is presented in Figure S3. Particularly, compounds DB921, DB922 and DB1055 present the strongest footprints at the AATT site and weaker binding at the ATAT sequence, contrasting with DB75 which binds to both ATAT and AATT sites with nearly similar efficiencies. These comparisons also indicate that the biosensor-SPR method is in excellent agreement with solution results as has been found in numerous quantitative comparisons.30
Circular Dichroism: Probing the binding mode and binding ratio
CD titration experiments as a function of compound concentration were evaluated to monitor the binding mode and saturation limit for compound binding with AATT and ATAT DNA (Figure S4, Supporting information). CD spectra monitor the asymmetric environment of the compounds when bound to DNA and therefore can be used to obtain information on the binding mode.31–32 The free compounds do not exhibit CD signals, however, upon addition of the compounds to DNA, substantial positive induced CD signals arise between 300 and 400 nm. These positive induced CD signals for the complexes are characteristic patterns for a minor grove binding mode in AT sequences. As can be seen in Figure S4, at the maximum adsorption wavelengths of the compounds, 325 nm (DB921), 315 nm (DB911), 325 nm (DB1055), 330 nm (DB1963) and 315 nm (DB985), where the DNA signals do not interfere, positive induced CD signals were observed. The induced CD signal of DB921, DB1055 and DB1963 is not as large as that of DB911, although these compounds have much higher binding affinity than DB911 to AATT sequences. Smaller induced CD changes on binding to DNA may be characteristic of linear minor-groove-binding compounds.33–34 Interestingly, DB985 also shows a much larger induced CD signal compared with DB921. The reason for this may be that the biphenyl torsional angle of DB985 is changed with the introduction of a phenyl hydroxide group to a more twisted structure than with DB921. For all compounds, the saturated maximum compound to DNA ratios are 1 compound/hairpin for AATT and ATAT respectively, and these values are consistent with results from SPR experiments.
X-ray structure of the DB1055 complex: Probing the water binding region
The crystal structure of DB1055 as a complex with the dodecamer duplex d(CGCGAATTCGCG)2 has been solved and compared with DB921 bound to the same sequence (PDB 2B0K). The structure (Figure 6a) confirms that DB1055 binds in the AT region of the DNA minor groove in the same manner as many other ligands with the core benzimidazole-phenyl-phenyl aromatic planar moieties in close non-bonded contact with the walls of the groove. Figure 6b shows a superposition of DB1055 and DB921 in their observed positions in the two crystal structures. The left-hand parts of the two molecules are in identical positions, whereas there is a small but significant difference (0.09 nm) at the right-hand extremities of the two phenyl rings (Figure 6c, d). This is most likely due to the direct hydrogen-bond interaction (0.33 nm) that occurs between the amidinium atom substituent (N15) of DB1055 and N3 of the adjacent adenine. This amidine group is at the meta position on the terminal phenyl ring, bringing it closer to the position that is occupied by the bridging water in the DB921 complex (Figure 6c); this water molecule is required in order for the DB921 para amidinium substituent to interact with N3.
At the other end of the ligands the benzimidazole-amidine group, which is a strong DNA binding motif, locks the benzimidazole end of both compounds into the AATT site of the minor groove. Both ligands have a pair of strong hydrogen bonds between the inner imidazole nitrogen atom to O2 atoms of the central two thymines, one on each strand (Figure 6e). There is also a hydrogen-bond contact from the benzimidazole-amidine group of DB1055 (0.31 nm) to the O2 atom of thymine8 (Figure 6f) with an identical contact involving DB921 (0.30 nm). This amidinium group also contacts a water molecule in the groove (Figure 6f), which in turn interacts with a cytosine O2 substituent. No such secondary contact was observed in the DB921 complex. Crystallographic data for DB1055 are summarized in Table S2 (Supporting information). An electron density map calculated with the final refined coordinates is drawn in the plane of the DB1055 molecule, showing the asymmetric fit of the drug in the groove and the excellent fit of DB1055 to the density in the groove at AATT (Figure 7).
Fig. 7.
An electron-density contour at 1.3σ of a 2Fo - Fc map with the DB1055 structure overlaid in the DNA minor groove.
DISCUSSION
DB921, its isomers, DB911, DB1055, and other analogs (Figure 1) form an excellent model system to investigate (i) the influence of a single interfacial water molecule on DNA interactions, (ii) the detailed requirements for incorporation of a mimetic group to replace water molecules in the design of drugs to selectively target the minor groove, and (iii) evaluate the differential effects of charge, specific H-bonds, and group stacking contributions to minor groove binding energetics. The quite high binding constant and favorable binding entropy for DB921, for example, illustrates that a rigid view of a bound water molecule as entropically unfavorable in complex formation is not correct when viewed in the context of the total entropy of binding (Figure 4 and Table S1). The ΔS for DB921 binding to the AATT sequence is only slightly less than that for the meta-substituted compounds, DB911 and DB1055, which do not require a linking water for complex function. The ΔS for DB921 is actually slightly greater for binding to the ATAT site. The structural and thermodynamic results (Figure 4 and Table S1) thus illustrate that inclusion of an interfacial water molecule in an optimal site can provide a favorable contribution to the Gibbs energy of complex formation relative to very similar systems that do not incorporate a water. Analogous observations have been made by Cooper and coworkers for protein-ligand interactions.9 This energetic enhancement effect is possible since an interfacial water molecule can have a number of favorable features in a complex: (i) bound water molecules can still maintain considerable motion with the possibility of several low energy binding orientations; (ii) the multiple H-bonding orientations and positions of water make it a highly adaptable H-bonding group in a complex while covalently-fixed groups in a similar complex are much more limited in structural-energetic adaptability and Gibbs energy minimization and (iii) concerted motions of the bound water, compound and DNA provide many possibilities for optimization of complex energetics with a favorable entropy.
The Group I compounds are all amidines that are closely related in structure to DB921. DB985 has a hydroxy group attached to the biphenyl system of DB921 and this causes increased twist of the biphenyl group. DB985 has slightly lower binding constants than DB921 for both AATT and ATAT and this is due to less favorable binding entropies. The -OH group on DB985 decreases the bound configurations relative to DB921, the bulky -OH must point away from the minor groove, and the increased twist may make the DB985 complex more rigid than with DB921, thus reducing the binding ΔS. This result suggests that the –OH group can form an H-bond with DNA, perhaps at the edge of the minor groove to a phosphate oxygen. Future structural studies should be able to help resolve the difference between DB921 and DB985.
DB1055 has the terminal amidine group of DB921 moved from the para to the meta position and the structural results clearly show that this removes the need for the interfacial water in the complex. The compound binds slightly more weakly than DB921 (loss of 0.5 kcal/mol in ΔG) with a significant decrease in the binding entropy relative to DB921, but with a higher ΔH of binding. The x-ray results in Figure 6 show that the terminal phenyl-meta-amidine of DB1055 forms a more rigid H-bonded system with the floor of the minor groove and perhaps better stacking with the groove walls than the water-linked complex of DB921. This leads to an improved binding enthalpy for DB1055 but a lower entropy with a slight loss in the Gibbs energy. The type of change from DB921 to DB1055 has frequently been considered to be a way to improve binding energetics through replacement of a linking water with a fixed compound group. The results show that DB1055 is indeed able to replace the interfacial water of DB921 but with slightly reduced energetics. Clearly incorporation of water, at least in DNA minor groove complexes can be favorable and is a feature that deserves more attention.
Replacement of the central phenyl of DB921 with an acetylene group (DB1869) causes a large drop in affinity, approximately a 100 fold decrease in K. The increased rotational flexibility of the acetylene relative to the phenyl is clearly not sufficient to overcome the loss in stacking energetics of the phenyl relative to the acetylene group. In addition, the distance from the phenyl and benzimidazole linking atoms is about 25% less with the acetylene group than with the phenyl (Figure S5, Supporting information). This moves the phenyl amidine away from the optimum H-bonding position and may disrupt the compound-water-DNA stabilizing interaction. Conversion of the benzimidazole to an indole gives a 2-fold increase in K while conversion of a phenyl to a pyridine results in a 3-fold reduction in K relative to DB921 binding. In both of these cases conversion of a CH to an N group results in a 2–3 fold decrease in K. This may be due to a less favorable water interaction with the N when it is in the minor groove than when it is in water.
The conversion of the terminal phenyl-amidine of DB921 to an amide in Group III yields a monocation and causes a change in the underlying thermodynamics of complex formation. The amide has a relatively small decrease in binding affinity, relative to DB921 (loss of less than 1 kcal/mol), with a drop in binding ΔH but a compensating increase in binding ΔS. The outer nitrogen of the phenyl-amidine of DB921 is located near the edge of the minor groove and between two phosphate groups that may provide electrostatic and H-bonding contributions to the binding ΔH but reduce the dynamics and ΔS. Conversion of the same amidine to a simple -H, DB1944 in Table 1, causes a huge decrease in binding affinity as would be expected if H-bonding, electrostatic and stacking interactions are lost. The results define the benefits of an amidine or amide group contribution to binding affinity, which can both form an H-bond with the interfacial water. The charge on the amidine clearly is a minor contributor, but the H-bonding and stacking ability of the amidine and amide are very important for affinity with the DNA minor groove. Since the experiments reported here are done at 0.2 M added NaCl, the contributions of both electrostatic effects as well as ion release on compound binding are expected to be relatively small. The relatively small ΔG difference between the +1 amide and +2 amidine support this conclusion. These observations suggest that monoamidines should be investigated in more detail in drug applications where monoamidines may have increased permeability in some cell types. Both the five and six-member cyclic amidine systems in the Group II compounds bind as well as the basic amidine group and these groups may also give advantages in drug design for uptake in some cell types.
The importance of the H-bonding ability of the benzimidazole and indole -NH groups is shown by the significant decrease in DNA affinity of the compounds in Group V that cannot form the benzimidazole or indole H-bond to DNA. An indole group with the -NH pointed in the same direction as the amidine is, however, is very effective in DNA binding (Group I). This effect of the benzimidazole, or inner facing indole, is completely consistent with the structures of both compounds which show excellent H-bonding of both benzimidazole and amidine –NH groups to DNA (Figure 6). In Group VI, the classically curved compound, netropsin, binds significantly better to AATT and ATAT than the similarly curved DB75 and DB911. In all cases the improved affinity is due to a more favorable binding ΔH but is accompanied by a lower binding ΔS. Netropsin can form many more H-bonds in a DNA minor groove complex than DB75 or DB911 and this source of binding ΔH clearly makes a major contribution to its minor groove affinity.
The general loss in affinity for the compounds in Table 1 for binding to ATAT relative to AATT is not surprising. The ATAT sequence is known to have a wider minor groove than AATT and we have found that, for compounds of the type in Table 1, complex formation causes narrowing of the groove and bending of the ATAT sequence into the minor groove.35 The narrow groove in AATT is a more suitable fit to the same compounds and binding causes much smaller topological changes. The conformational changes with ATAT decrease the binding energetics and the intermolecular interactions in the complex are not enough in general to give a binding equilibrium constant in the same range as with AATT.
The differences in compound binding affinities for AATT and ATAT are also very dependent on compound structure. DB921, for example, binds 40–50 times more strongly to AATT than DB75 but less than 3 times more strongly to ATAT. In these groups of compounds the linear compounds generally bind much better to AATT than ATAT while the more classically shaped compounds for minor groove binding, DB75, netropsin, DB911 and DB818, bind more similarly to the two DNAs. Given the generally similar behavior of the compound structural types, the primary source of the binding differences must reside in a slightly different structure of the two DNAs. Analysis of X-ray structures suggests that compounds such as netropsin and DB75 are slightly more curved than the DNA minor groove in AATT sites.26,36 The linear compounds are thus able to better adapt to the AATT site geometry than the more curved analogs. On a comparative scale the linear compounds still bind quite well to the ATAT site. Most of them still bind better to ATAT than DB75, for example, even though DB75 binds to ATAT more strongly than to AATT. A reasonable explanation for this behavior is that the base pair helical twist and net curvature of the minor groove is slightly greater for ATAT than for AATT under our solution conditions.35 This will allow a smaller loss in binding energetics, or an actual gain as with DB75, for the more highly curved compounds. It should also be emphasized that the solution footprinting results in Figure 5, with a much longer DNA sequence relative to the SPR studies, provide strong support for the binding affinity observations from the chip-based SPR method. This is true for all of the compounds and both AATT and ATAT.
An earlier study with a longer AATT sequence d(CGCGAATTCGCG) gave very similar entropy for DB75 binding as observed in this work. This result suggests that end effects and differential electrostatic contributions are not very important for the thermodynamics of the centrally located binding sites in these complexes. This is probably due to the fact that we are primarily comparing dications that bind in the four base pair sequence at a relatively high added NaCl concentration (0.2 M). This high salt and compound similarity makes differential contributions from ion release and bound water changes on complex formation less important to variations in the thermodynamics of binding. It should be noted, however, that this similarity may not be true in all systems37.
The kinetics results in Figure 3 and Table 2 provide some insight into the differences in binding mechanisms and energetics for the isomeric compounds, DB921, DB1055 and DB911. DB911 has a more favorable binding ka, almost twice the DB921 value and three times the DB1055 value. It is thus faster to bind either without water, as required with DB921, or without the more complicated compound geometry of DB1055. When DB921 and DB1055 are bound, however, they interact with the minor groove much more favorably than DB911. The kd values for DB921 and DB1055 are around 15 times lower and more favorable for binding affinity than for DB911. The curvature of the DB911 systems is slightly too large to make optimum contacts with the DNA minor groove and this clearly is much more detrimental than a linear shape that can more readily accommodate itself to the groove shape. It is interesting that both the linear DB921 with a water, and the curved DB1055 with a shape change and direct H-bond, are both able to bind to the minor groove significantly better than DB911. The lower ka for the stronger binding compounds is counterintuitive but is overcome by a larger reduction in kd for the stronger binding compounds. The greater affinity of DB921 and DB1055 is completely due to the kd.
In conclusion, the detailed structural, thermodynamics and kinetics results offer very important information for identification of the key groups responsible for the strong binding of the linear compounds. These studies provide patterns for designing new non-classical sequence-specific groove DNA-binding agents. The results also provide information about compound structure and sequence-dependent DNA binding affinity.
Supplementary Material
Acknowledgments
Research at Georgia State University is supported by NIH grant AI 064200 (WDW and DWB). Work at the School of Pharmacy, University of London was supported by a program grant to SN from CRUK (No. C129/A4489). M.-H. D.-C. thanks the Association Laurette Fugain, the Ligue contre le Cancer (Comité du Pas-de-Calais), the Fonds Européen de Développement Régional (FEDER, European Community) and the Région Nord-Pas de Calais for grants, the CHRU de Lille-Conseil Régional Nord-Pas de Calais for a Ph.D fellowship to R.N, and the Institut de Médecine Prédictive et de Recherche Thérapeutique (IMPRT) – IFR114 for access to the Molecular Dynamics STORM 860 equipment.
Footnotes
Supporting Information. Figures for comparison of ITC results, including duplex-hairpin results; footprinting results; models of several small molecules; CD spectra; detail procedures for crystallization and model building; tables for crystallographic data for DB1055 (PDB id 2I5A); thermodynamic parameters. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Nguyen B, Neidle S, Wilson WD. Acc Chem Res. 2009;42:11. doi: 10.1021/ar800016q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munde M, Ismail MA, Arafa R, Peixoto P, Collar CJ, Liu Y, Hu L, David-Cordonnier MH, Lansiaux A, Bailly C, Boykin DW, Wilson WD. J Am Chem Soc. 2007;129:13732. doi: 10.1021/ja074560a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchmueller KL, Staples AM, Howard CM, Horick SM, Uthe PB, Le NM, Cox KK, Nguyen B, Pacheco KA, Wilson WD, Lee M. J Am Chem Soc. 2005;127:742. doi: 10.1021/ja044359p. [DOI] [PubMed] [Google Scholar]
- 4.Lajiness J, Sielaff A, Mackay H, Brown T, Kluza J, Nguyen B, Wilson WD, Lee M, Hartley JA. Med Chem. 2009;5:216. doi: 10.2174/157340609788185945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chenoweth DM, Dervan PB. Proc Natl Acad Sci USA. 2009;106:13175. doi: 10.1073/pnas.0906532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dervan PB, Burli RW. Curr Opin Chem Biol. 1999;3:688. doi: 10.1016/s1367-5931(99)00027-7. [DOI] [PubMed] [Google Scholar]
- 7.Miao Y, Lee MP, Parkinson GN, Batista-Parra A, Ismail MA, Neidle S, Boykin DW, Wilson WD. Biochemistry. 2005;44:14701. doi: 10.1021/bi051791q. [DOI] [PubMed] [Google Scholar]
- 8.Athri P, Wilson WD. J Am Chem Soc. 2009;131:7618. doi: 10.1021/ja809249h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper A. Biophys Chem. 2005;115:89. doi: 10.1016/j.bpc.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Tevis DS, Kumar A, Stephens CE, Boykin DW, Wilson WD. Nucleic Acids Res. 2009;37:5550. doi: 10.1093/nar/gkp558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Wilson WD. Methods Mol Biol. 2010;613:1. doi: 10.1007/978-1-60327-418-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen B, Tanious FA, Wilson WD. Methods. 2007;42:150. doi: 10.1016/j.ymeth.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Das BP, Boykin DW. J Med Chem. 1977;20:531. doi: 10.1021/jm00214a014. [DOI] [PubMed] [Google Scholar]
- 14.Hall JE, Kerrigan JE, Ramachandran K, Bender BC, Stanko JP, Jones SK, Patrick DA, Tidwell RR. Antimicrob Agents Chemother. 1998;42:666. doi: 10.1128/aac.42.3.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mallena S, Lee MP, Bailly C, Neidle S, Kumar A, Boykin DW, Wilson WD. J Am Chem Soc. 2004;126:13659. doi: 10.1021/ja048175m. [DOI] [PubMed] [Google Scholar]
- 16.Ismail MA, Batista-Parra A, Miao Y, Wilson WD, Wenzler T, Brun R, Boykin DW. Bioorg Med Chem. 2005;13:6718. doi: 10.1016/j.bmc.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 17.Ismail MA, Brun R, Wenzler T, Tanious FA, Wilson WD, Boykin DW. Bioorg Med Chem. 2004;12:5405. doi: 10.1016/j.bmc.2004.07.056. [DOI] [PubMed] [Google Scholar]
- 18.Ismail MA, Brun R, Wenzler T, Tanious FA, Wilson WD, Boykin DW. J Med Chem. 2004;47:3658. doi: 10.1021/jm0400092. [DOI] [PubMed] [Google Scholar]
- 19.Farahat AA, Kumar A, Say M, Barghash Ael D, Goda FE, Eisa HM, Wenzler T, Brun R, Liu Y, Mickelson L, Wilson WD, Boykin DW. Bioorg Med Chem. 2011;18:557. doi: 10.1016/j.bmc.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Myszka DG, He X, Dembo M, Morton TA, Goldstein B. Biophys J. 1998;75:583. doi: 10.1016/S0006-3495(98)77549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlsson R. J Mol Recognit. 1999;12:285. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<285::AID-JMR469>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 22.Myszka DG. Methods Enzymol. 2000;323:325. doi: 10.1016/s0076-6879(00)23372-7. [DOI] [PubMed] [Google Scholar]
- 23.Peixoto P, Liu Y, Depauw S, Hildebrand MP, Boykin DW, Bailly C, Wilson WD, David-Cordonnier MH. Nucleic Acids Res. 2008;36:3341. doi: 10.1093/nar/gkn208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munde M, Kumar A, Nhili R, Depauw S, David-Cordonnier MH, Ismail MA, Stephens CE, Farahat AA, Batista-Parra A, Boykin DW, Wilson WD. J Mol Biol. 2010;402:847. doi: 10.1016/j.jmb.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski ZM, Minor W. Methods Enzymol. 1997;276:307. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Brown DG, Sanderson MR, Skelly JV, Jenkins TC, Brown T, Garman E, Stuart DI, Neidle S. Embo J. 1990;9:1329. doi: 10.1002/j.1460-2075.1990.tb08242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiseman T, Williston S, Brandts JF, Lin LN. Anal Biochem. 1989;179:131. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 28.Ren J, Jenkins TC, Chaires JB. Biochemistry. 2000;39:8439. doi: 10.1021/bi000474a. [DOI] [PubMed] [Google Scholar]
- 29.Barcelo F, Capo D, Portugal J. Nucleic Acids Res. 2002;30:4567. doi: 10.1093/nar/gkf558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Day YS, Baird CL, Rich RL, Myszka DG. Protein Sci. 2002;11:1017. doi: 10.1110/ps.4330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ardhammar MNB, Kurucsev T. Circular Dichroism: Principles and Applications. Wiley-Vch; New York: 2000. [Google Scholar]
- 32.Garbett NC, Ragazzon PA, Chaires JB. Nat Protoc. 2007;2:3166. doi: 10.1038/nprot.2007.475. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen B, Lee MP, Hamelberg D, Joubert A, Bailly C, Brun R, Neidle S, Wilson WD. J Am Chem Soc. 2002;124:13680. doi: 10.1021/ja027953c. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Collar CJ, Kumar A, Stephens CE, Boykin DW, Wilson WD. The J Phys Chem B. 2008;112:11809. doi: 10.1021/jp804048c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hunt RA, Munde M, Kumar A, Ismail MA, Farahat AA, Arafa RK, Say M, Batista-Parra A, Tevis D, Boykin DW, Wilson WD. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkq1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nunn CM, Garman E, Neidle S. Biochemistry. 1997;36:4792. doi: 10.1021/bi9628228. [DOI] [PubMed] [Google Scholar]
- 37.Mazur S, Tanious FA, Ding D, Kumar A, Boykin DW, Simpson IJ, Neidle S, Wilson WD. J Mol Biol. 2000;300:321. doi: 10.1006/jmbi.2000.3869. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.