Abstract
BACKGROUND
Neurulation requires precise, spatio-temporal expression of numerous genes and coordinated interaction of signal transduction and gene regulatory networks, disruption of which may contribute to the etiology of neural tube (NT) defects. MicroRNAs are key modulators of cell and tissue differentiation. In order to define potential roles of miRNAs in development of the murine NT, miRNA microarray analysis was conducted to establish expression profiles, and identify miRNA target genes and functional gene networks.
METHODS
miRNA expression profiles in murine embryonic NTs derived from gestational days 8.5, 9.0 and 9.5 were defined and compared utilizing miRXplore™ microarrays from Miltenyi Biotech GmbH. Gene expression changes were verified by TaqMan™ quantitative Real-Time PCR. clValid R package and the UPGMA (hierarchical) clustering method were utilized for cluster analysis of the microarray data. Functional associations among selected miRNAs were examined via Ingenuity Pathway Analysis.
RESULTS
miRXplore™ chips enabled examination of 609 murine miRNAs. Expression of approximately 12% of these was detected in murine embryonic NTs. Clustering analysis revealed several developmentally regulated expression clusters among these expressed genes. Target analysis of differentially expressed miRNAs enabled identification of numerous target genes associated with cellular processes essential for normal NT development. Utilization of Ingenuity Pathway Analysis revealed interactive biological networks which connected differentially expressed miRNAs with their target genes, and highlighted functional relationships.
CONCLUSIONS
The present study defined unique gene expression signatures of a range of miRNAs in the developing NT during the critical period of NT morphogenesis. Analysis of miRNA target genes and gene interaction pathways revealed that specific miRNAs may direct expression of numerous genes encoding proteins which have been shown to be indispensable for normal neurulation. This study is the first to identify miRNA expression profiles and their potential regulatory networks in the developing mammalian NT.
Keywords: neural tube, embryo, mouse, microRNA, microarray, neurulation
INTRODUCTION
Morphogenesis of the mammalian neural tube involves elevation of neural folds, from a flat neural plate, followed by neural fold apposition and fusion. The neural folds come into apposition at four discrete closure sites from which fusion of the folds extends in rostral and caudal directions (Copp et al., 2003). Failure of the neural tube to close properly during the first twenty-eight days of human pregnancy results in a spectrum of neural tube defects (NTDs) affecting the brain and spinal cord. These include anencephaly, craniorachischisis, spina bifida, and encephalocele (Botto et al., 1999). NTDs are amongst the most common human malformations, occurring in approximately 1/1000 births in the United States (Copp et al., 2003). The etiology of NTDs is thought to be multifactorial in nature, resulting from a combination of genetic predisposition (susceptibility genes) and interacting environmental factors (Campbell et al., 1986; George and Speer, 2000). Nevertheless, despite the identification of numerous genes which, when functionally deleted in mice, result in NTDs, the cellular and molecular basis of neural tube development, remains ill defined.
MicroRNAs (miRNAs) which represent the largest family of small noncoding RNAs (18–25 bases in length), have been found to execute key functions in silencing expression of specific target genes in both plant and animal systems (for review see, (Wang et al., 2007; Zhang et al., 2007)). MicroRNAs regulate expression of genes post-transcriptionally through binding to target mRNAs. While recent findings suggest that miRNAs can increase translation (Vasudevan et al., 2007), such binding generally results in gene silencing via either inhibition of translation or mRNA destabilization (for review see, (Bartel, 2009; Pan and Chegini, 2008)). Complete complementarity of binding, while rare in animals (Meister, 2007), can lead to degradation of the target, while incomplete complementarity leads to the translational suppression (Kim and Nam, 2006). It is currently estimated that miRNAs account for approximately 1% of predicted genes in higher eukaryotic genomes, and that up to 30% of protein coding genes might be regulated by miRNAs (Ross et al., 2007). It is now well established that miRNAs are critical regulators of proliferation, differentiation, and apoptosis during embryogenesis (for review see, (O’Rourke et al., 2006; Yang and Wu, 2006)), however their role in neural tube development has only recently begun to be examined (Chakrabarty et al., 2007; Song and Tuan, 2006; Wienholds et al., 2005). Indeed, in a recent study, several miRNAs were documented to be expressed in the developing murine neural tube and to target genes crucial for neural tube ontogenesis (Hoesel et al., 2010). Moreover, examination of p53−/− embryos, exhibiting exencephaly, revealed altered expression of a subset of discrete miRNAs implying their involvement in this NTD (Hosako et al., 2009). A profile of miRNA expression during neurulation would identify families of these regulatory molecules and their downstream targets – including unique candidates and known effectors of neural tube development essential for proper development of the neural tube. The present study was thus conducted in order to identify key miRNAs and attendant regulatory signal transduction networks that are associated with the mammalian neural tube during the critical stages of neurulation.
METHODS
Animals
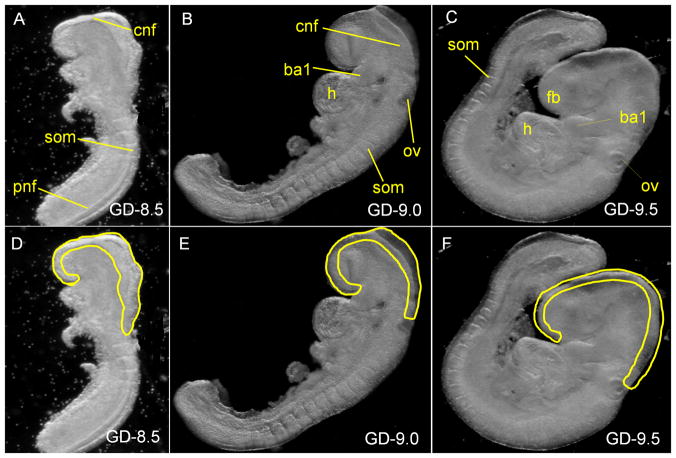
Mature male and female ICR mice (Harlan, Indianapolis, IN), were maintained in an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) approved facility on a 12-hour light/dark cycle, provided ad libitum food and water, and mated overnight. The presence of a vaginal plug the following morning was considered as evidence of mating, and the time designated as gestational day 0 (GD-0). Developmental staging was conducted following the method of Theiler (Theiler, 1989). On GD-8.5, GD-9.0 and GD-9.5, which represent the critical period of neural tube development in the mouse, female mice were euthanized by asphyxiation and embryos were dissected from decidual tissue and placed in ice-cold, sterile calcium/magnesium-free PBS. Embryos (used for microdissection of the neural tube) corresponding to each of the three gestation days were selected based on somite numbers. For example, GD-8.5, GD-9.0 and GD-9.5 embryos were selected based on 8–10 somites, 14–18 somites and 22–27 somites, respectively. Embryonic neural tubes, from the most rostral aspect of the forebrain to the caudal aspect of the hindbrain (above the otic vesicle), were excised as shown in Figure 1. For GD-8.5 and -9.0 embryos, just the edge of the elevated neural plates were dissected (Figure 1) and the microdissected tissues were checked at 60X magnification and further trimmed (if needed) to eliminate any mesoderm, or non-neural tissues as precisely as possible. For GD-9.5 embryos where the neural crest has already migrated out of the neural folds, the dorsal part of the brain containing only the fused neural folds/tube was microdissected (Figure 1). Excised tissue was minced and stored at minus 80°C in PrepProtect™ Stabilization Buffer (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). For each day of gestation, neural tube tissue was collected from 3 independent pools of 15 to 20 staged embryos and extracted to generate 3 distinct pools of RNA from neural tubes of each gestational stage that were independently processed and applied to individual miRXplore™ microRNA Microarray chips (Miltenyi Biotec GmbH).
Figure 1. Photomicrographs of GD-8.5, GD-9.0 and GD-9.5 embryos under darkfield optics.
panels A & D: a GD-8.5 embryo; panels B & E: a GD-9.0 embryo; panels C & F: a GD-9.5 embryo; The neural tubes demarcated by the yellow lines in panels D, E, and F, were excised from GD-8.5, GD-9.0 and GD-9.5 embryos respectively, for total RNA extraction. (cnf) cranial neural folds; (som) somite; (fb) forebrain; (ba1) first branchial arch; (h) heart; (ov) otic vesicle; (pnf) posterior neural folds.
RNA Extraction and Microarray Hybridization
Total RNA (containing miRNAs) from GD-8.5, GD-9.0, or GD-9.5 neural tube tissue was isolated using the miRVANA microRNA isolation kit (Applied Biosystems-Ambion, Foster City, CA). The quality and quantity of total RNA samples were determined using the Agilent 2100 Bioanalyzer (Agilent Technologies, Foster City, CA). The RNA Integrity Numbers (RIN) of all the RNA samples were between 8.6 and 10.0. RNA with a RIN number greater than 6 is of sufficient quality for miRNA microarray experiments (Fleige and Pfaffl, 2006). RNA samples (1 μg) isolated from mouse embryonic neural tube tissues (GD-8.5 – GD-9.5) as well as the miRXplore Universal Reference (control) were fluorescently labeled with Hy5 (red) or Hy3 (green), respectively, and hybridized to miRXplore™ Microarrays (Miltenyi Biotec GmbH) using the a-Hyb™ Hybridization Station (Miltenyi Biotec GmbH). Probes for a total of 1392 mature miRNAs (from human, mouse, rat and virus), including positive control and calibration spots, were spotted in quadruplicate on each microarray. Each array included probes for 609 murine miRNAs. The miRXplore Universal Reference (UR) controls, provided by Miltenyi, represent a defined pool of synthetic miRNAs for comparison of multiple samples. Fluorescence signals of the hybridized miRXplore™ Microarrays were detected using a laser scanner from Agilent Technologies.
Microarray Preprocessing
Mean and median signal and local background intensities for the Hy3 and Hy5 channels were obtained for each spot on each of the nine microarray images using the ImaGene software (BioDiscovery, Inc., El Segundo, CA). Low quality spots were identified and given relative weights which were subsequently used in data analysis and modeling of miRNA expression values. A total of 5,620 spots remained after discarding empty spots, dummy spots, extreme outliers, and those with missing background intensities. The pre-processing procedure consisted of three steps. First, all spots for which the mean foreground intensity was smaller than 1.1 times the mean background intensity in three or more arrays were discarded, unless all three samples came from the same day. Several within-array normalization procedures on the remaining 360 spots were evaluated. Based on visual inspection of the mean log2 expression values versus the log2 expression ratio for each spot (“MA” plots), it was decided that “no within-array” normalization gave the best results and the 72 control spots were subsequently removed. In the second step, a simple background correction was performed by subtracting background from the foreground intensities. Log2 ratios of Hy5 (red, experimental) versus Hy3 (green, control) expression were calculated for each spot. None of the spots (in all nine arrays) exhibited negative intensities after the background procedure. Lastly, the between-array normalization using the quantile method was performed to align the distributions of the log-ratios on each chip (Bolstad et al., 2003; Rao et al., 2008). Visual inspection of heatmaps and hierarchical clustering of the samples revealed that the first replicates from gestational days (GD) 8.5 and 9.0 did not cluster with the other replicates on the same day. Since the other two replicates for these days were highly correlated (r > 0.975), only the two replicates from these days were utilized for subsequent statistical analysis. Since each spot is replicated on each chip 4 times, statistical power was maximized by using a hierarchical linear model which incorporated the replicate information on each chip. The R package limma (Smyth, 2005) - from the Bioconductor project (http://www.bioconductor.org) (Lee et al., 2004) was used for all preprocessing steps, as well as all statistical analyses. The experimental data are available from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc5GSE20879; platform accession no. GPL10178).
Statistical Analysis of miRNA Data
In order to identify miRNAs that are expressed by developing murine neural tube tissue on GD-8.5, GD-9.0, and GD-9.5, data from the four replicates on each chip and from the three arrays corresponding to each of the three gestation days (n=12) were averaged. Since all expression values were represented as log2 ratios of experimental versus universal reference, two criteria were used to determine miRNA expression. The first criterion was that the log-ratio was larger than zero, and the second was that the signal intensity in the experimental sample (red label) was in the 50th percentile or greater of its distribution. Differential expression of miRNAs between different days of gestation was determined by fitting a hierarchical linear model using the limma package (Smyth, 2004) and testing the corresponding contrasts of interest (e.g., GD-9.0 vs. GD-8.5, GD-9.5 vs. GD-9.0, and GD-9.5 vs. GD-8.5) for each miRNA. Each spot was weighted in a fashion inversely proportional to its quality score, and duplicate spots on each chip were taken into account using the duplicate correlation option (Smyth et al., 2005). Fold change, adjusted t-statistic, unadjusted and false discovery rate (FDR) adjusted p-values were calculated for each miRNA for each comparison. All miRNAs with adjusted p-values equal to or below 0.05 were considered as differentially expressed.
After identifying the differentially expressed miRNAs, clustering analysis was performed to group genes with similar temporal patterns of expression. The R package clValid (Ross et al., 2007) was used to evaluate several clustering algorithms (UPGMA (hierarchical clustering), SOM, SOTA, DIANA, and model-based clustering) (Handl et al., 2005; Kaufman, 1990) using a number of validation measures. The clustering algorithm and number of clusters which gave the best results on the validation measures was selected to visually display the data.
MicroRNA Target Analysis
MicroRNA target analysis was performed separately for all genes expressed on either one of the gestation days GD-8.5, GD-9.0, and GD-9.5, and for the miRNAs which were differentially expressed during murine neural tube ontogenesis (GD-8.5 through GD-9.5). Only murine miRNAs (ones with MMU prefix) were selected for analysis. miRNA targets were mined using the miRDB (http://mirdb.org/miRDB/), an online database for miRNA target prediction in animals. The database is populated by the targets predicted by the computational tool MirTarget2 (Wang and El Naqa, 2008). The miRNA target scoring system utilized by the miRDB assigns scores to all 3′UTRs with seed matching sites. If the number of candidate sites per single UTR is larger than one, then all these sites are combined and a single score is computed. The scores were computed using the following the equation:
where Pi represents the p-values for each candidate site as estimated by the SVM, and n represents the number of candidate sites per single UTR. An ordered list of miRNA-Gene Symbol pairs sorted by their target scores was thus obtained. Target analysis of selected differentially expressed miRNAs (whose expression has been verified by TaqMan QRTPCR – described in the Results section) was performed using the miRDB online database (http://mirdb.org/miRDB/). To investigate the probable physiological role(s) of specific differentially expressed miRNAs in neural tube development, gene targets of such miRNAs were also verified with three other miRNA target prediction software programs – PicTar (http://pictar.mdc-berlin.de), Target Scan (http://www.targetscan.org), and miRanda (http://www.microrna.org/microrna/home.do). Predicted targets of differentially expressed miRNAs were assessed for enrichment in GO categories and involvement in biological pathways using DAVID (Dennis et al., 2003; Huang da et al., 2009) and Ingenuity Pathway Analysis (IPA; Ingenuity Systems Inc., Redwood City, CA). In order to predict potential interactions between the differentially expressed genes encoding miRNAs and their downstream targets, a network analysis using Ingenuity Pathway Analysis (IPA; Ingenuity Systems Inc., Redwood City, CA) was performed. IPA utilizes a knowledge base program to create pertinent and interactive biological networks. To further illustrate that the differentially expressed miRNAs may be directly linked to target genes associated with neural tube development, two more gene networks, with miRNAs displaying either enhanced or diminished expression in the neural tube during GD-9.0-9.5 were developed utilizing IPA (Ingenuity Systems Inc.) and the miRDB online database (http://mirdb.org/miRDB/).
Quantitative Real-Time PCR (TaqMan)
Samples from the same RNA that was used for microarray experiments were used for QRTPCR validation. cDNA was synthesized with miRNA-specific looped reverse transcription (RT) primer using the TaqMan MicroRNA RT kit (Applied Biosystems). Real-time PCR (TaqMan) analysis was performed on a TaqMan ABI Prism 7000 Sequence Detection System (Applied Biosystems). Matching primers and fluorescence probe for each miRNA were purchased from Applied Biosystems. For each miRNA analyzed, 900 nM of forward and reverse primers were used. The final concentration of the fluorescent probe was 200 nM. The PCR reaction was performed in a total volume of 25 μl containing 0.2 mM dATP, dCTP, and dGTP, 0.4 mM dUTP, 0.625 unit of Amplitaq Gold, and 2 μl of cDNA template. Cycling parameters were: 95°C for 10 min for denaturation of DNA strands, followed by 40 cycles of denaturation at 95°C for 15 sec each, and primer extension at 60°C for 1 min. Data were acquired and processed with Sequence Detection System 1.0 software (Applied Biosystems). Each cDNA sample was tested in triplicate and mean Ct values are reported. For purposes of quantification, each miRNA was normalized to the amount of mmu-miR-20b PCR product present in each sample. Primers and fluorescent probe for mmu-miR-20b which was used as a housekeeping standard, were obtained from Applied Biosystems. The QRTPCR validation experiments were performed on the same RNA samples that were used for the microarray experiments. Analysis was performed on 2–3 independent sets of cDNA and each cDNA sample was tested in duplicate. Statistical significance was determined by one-way ANOVA followed by Bonferroni’s Multiple Comparison Test, using GraphPad Prism, v. 4.02 (GraphPad Software, Inc., San Diego, CA). P-values of <0.05 were considered significant.
RESULTS
MicroRNAs derived from three independent sets of GD-8.5, GD-9.0, and GD-9.5 mouse neural tubes (nine total samples) and miRXplore Universal Reference were used to probe separate miRXplore™ microarrays containing oligonucleotide probes representing a total of 1392 mature miRNAs (from human, mouse, rat and virus), of which 609 were of murine origin. After preprocessing of the data, the total number of miRNAs left was 72. Of these 72 miRNAs, 22, 20 and 25 were found to be expressed in the developing neural tube on GD-8.5, GD-9.0 and GD-9.5, respectively (Table 1). Among the expressed miRNAs, 16 were common to all three days of gestation, while 0, 1 (miR-638), and 6 (miR-335, miR-130A, miR-199A-3P, miR-18B, miR-18A, and miR-106A) miRNAs were uniquely expressed on GD-8.5, GD-9.0 and GD-9.5, respectively. Numerous miRNAs exhibited differential temporal expression patterns. Differentially expressed miRNAs with adjusted p-values below 0.05 and with fold changes (in gene expression) are shown in Table 2 for GD-9.0 compared to GD-8.5, and in Table 3 for GD-9.5 compared to GD-9.0. Five miRNAs demonstrated enhanced, and three miRNAs demonstrated reduced, expression in GD-9.0 neural tube tissue when compared to GD-8.5. Eighteen miRNAs demonstrated higher and twelve miRNAs demonstrated lower expression in GD-9.5 neural tubes when compared to GD-9.0. The complete list of fold changes, t-statistics, and p-values associated with each miRNA passing our statistical filter (72 miRNAs in total) for each comparison of gestational days is presented in Supplementary Tables 1–2.
Table 1. List of MicroRNAs Expressed in GD-8.5, GD-9.0 and GD-9.5 Neural Tubes.
Genes encoding miRNAs expressed in the developing neural tube. The average expression signal value (Red) corresponding to each gene was obtained by filtering the miRNA gene expression data sets from neural tubes obtained from each of GD-8.5, GD-9.0 and GD-9.5. Criteria for considering a miRNA as expressed included a log2-ratio of red (experimental sample) vs. green (universal reference) intensity larger than zero, and a signal intensity in the experimental sample greater than or equal to the 50th percentile of the distribution. GD = gestational day.
| GD | miRNA Gene Family: Gene IDs | Expression (Red) | Log Ratio (Red vs. Green) |
|---|---|---|---|
| 8.5 | MIR-1195: MMU-MIR-1195 | 8131.1 | 8.93 |
| 8.5 | MIR-720: MMU-MIR-720; HSA-MIR-720 | 10840.1 | 5.00 |
| 8.5 | MIR-15A*: HSA-MIR-15A* | 2803.5 | 4.27 |
| 8.5 | MIR-709: MMU-MIR-709 | 2755.8 | 3.53 |
| 8.5 | MIR-139-3P: HSA-MIR-139-3P; MMU-MIR-139-3P; RNO-MIR-139-3P | 7665.9 | 3.41 |
| 8.5 | MIR-690: MMU-MIR-690 | 8739.4 | 2.97 |
| 8.5 | MIR-345-3P: MMU-MIR-345-3P; RNO-MIR-345-3P | 5157.7 | 2.67 |
| 8.5 | MIR-20A: HSA-MIR-20A; MMU-MIR-20A; RNO-MIR-20A | 2473.6 | 2.54 |
| 8.5 | MIR-302A: HSA-MIR-302A; MMU-MIR-302A | 1420.2 | 2.48 |
| 8.5 | MIR-302D: HSA-MIR-302D; MMU-MIR-302D | 2423.5 | 2.37 |
| 8.5 | MIR-92A: HSA-MIR-92A; MMU-MIR-92A; RNO-MIR-92A | 797.0 | 2.25 |
| 8.5 | MIR-663: HSA-MIR-663 | 461.6 | 1.71 |
| 8.5 | MIR-347: RNO-MIR-347 | 391.6 | 1.24 |
| 8.5 | MIR-16-1*: HSA-MIR-16-1* | 6463.4 | 1.00 |
| 8.5 | MIR-19B: HSA-MIR-19B; MMU-MIR-19B; RNO-MIR-19B | 8115.1 | 0.98 |
| 8.5 | MIR-26A: HSA-MIR-26A; MMU-MIR-26A; RNO-MIR-26A | 367.7 | 0.90 |
| 8.5 | MIR-16: HSA-MIR-16; MMU-MIR-16; RNO-MIR-16 | 1372.4 | 0.54 |
| 8.5 | MIR-93: HSA-MIR-93; MMU-MIR-93; RNO-MIR-93 | 530.8 | 0.46 |
| 8.5 | MIR-20B: HSA-MIR-20B; MMU-MIR-20B; RNO-MIR-20B-5P | 1593.7 | 0.43 |
| 8.5 | MIR-17: HSA-MIR-17; MMU-MIR-17; RNO-MIR-17 (RNO-MIR-17-5P) | 2333.4 | 0.34 |
| 8.5 | MIR-25: HSA-MIR-25; MMU-MIR-25; RNO-MIR-25 | 562.6 | 0.33 |
| 8.5 | MIR-19A: HSA-MIR-19A; MMU-MIR-19A; RNO-MIR-19A | 2105.9 | 0.12 |
| 9.0 | MIR-1195: MMU-MIR-1195 | 5995.0 | 8.67 |
| 9.0 | MIR-720: MMU-MIR-720; HSA-MIR-720 | 9272.9 | 5.39 |
| 9.0 | MIR-709: MMU-MIR-709 | 1305.7 | 4.32 |
| 9.0 | MIR-15A*: HSA-MIR-15A* | 3362.2 | 3.90 |
| 9.0 | MIR-690: MMU-MIR-690 | 7191.3 | 3.44 |
| 9.0 | MIR-139-3P: HSA-MIR-139-3P; MMU-MIR- 139-3P; RNO-MIR-139-3P | 7415.7 | 3.30 |
| 9.0 | MIR-345-3P: MMU-MIR-345-3P; RNO-MIR- 345-3P | 4610.3 | 2.90 |
| 9.0 | MIR-663: HSA-MIR-663 | 825.7 | 2.74 |
| 9.0 | MIR-92A: HSA-MIR-92A; MMU-MIR-92A; RNO-MIR-92A | 731.0 | 2.19 |
| 9.0 | MIR-347: RNO-MIR-347 | 662.6 | 1.97 |
| 9.0 | MIR-20A: HSA-MIR-20A; MMU-MIR-20A; RNO-MIR-20A | 2681.6 | 1.78 |
| 9.0 | MIR-302A: HSA-MIR-302A; MMU-MIR-302A | 673.4 | 1.75 |
| 9.0 | MIR-638: HSA-MIR-638 | 421.2 | 1.54 |
| 9.0 | MIR-16-1*: HSA-MIR-16-1* | 5353.1 | 1.50 |
| 9.0 | MIR-302D: HSA-MIR-302D; MMU-MIR-302D | 1328.1 | 1.49 |
| 9.0 | MIR-19B: HSA-MIR-19B; MMU-MIR-19B; RNO-MIR-19B | 7415.8 | 0.94 |
| 9.0 | MIR-20B: HSA-MIR-20B; MMU-MIR-20B; RNO-MIR-20B-5P | 1791.0 | 0.57 |
| 9.0 | MIR-17: HSA-MIR-17; MMU-MIR-17; RNO- MIR-17 (RNO-MIR-17-5P) | 2382.3 | 0.51 |
| 9.0 | MIR-19A: HSA-MIR-19A; MMU-MIR-19A; RNO-MIR-19A | 2606.2 | 0.15 |
| 9.0 | MIR-25: HSA-MIR-25; MMU-MIR-25; RNO- MIR-25 | 436.8 | 0.01 |
| 9.5 | MIR-1195: MMU-MIR-1195 | 9923.7 | 8.17 |
| 9.5 | MIR-720: MMU-MIR-720; HSA-MIR-720 | 20709.0 | 5.12 |
| 9.5 | MIR-15A*: HSA-MIR-15A* | 2115.9 | 3.69 |
| 9.5 | MIR-709: MMU-MIR-709 | 5313.7 | 3.38 |
| 9.5 | MIR-20A: HSA-MIR-20A; MMU-MIR-20A; RNO-MIR-20A | 4625.1 | 3.22 |
| 9.5 | MIR-139-3P: HSA-MIR-139-3P; MMU-MIR-139-3P; RNO-MIR-139-3P | 5196.6 | 3.04 |
| 9.5 | MIR-690: MMU-MIR-690 | 13938.3 | 2.55 |
| 9.5 | MIR-345-3P: MMU-MIR-345-3P; RNO-MIR-345-3P | 5134.2 | 2.38 |
| 9.5 | MIR-92A: HSA-MIR-92A; MMU-MIR-92A; RNO-MIR-92A | 1384.2 | 2.36 |
| 9.5 | MIR-19B: HSA-MIR-19B; MMU-MIR-19B; RNO-MIR-19B | 11696.3 | 1.47 |
| 9.5 | MIR-16: HSA-MIR-16; MMU-MIR-16; RNO-MIR-16 | 3572.0 | 1.30 |
| 9.5 | MIR-19A: HSA-MIR-19A; MMU-MIR-19A; RNO-MIR-19A | 3201.2 | 1.29 |
| 9.5 | MIR-26A: HSA-MIR-26A; MMU-MIR-26A; RNO-MIR-26A | 1057.1 | 1.07 |
| 9.5 | MIR-335: HSA-MIR-335; MMU-MIR-335-5P; RNO-MIR-335 | 2600.1 | 1.05 |
| 9.5 | MIR-17: HSA-MIR-17; MMU-MIR-17; RNO-MIR-17 (RNO-MIR-17-5P) | 4292.0 | 1.02 |
| 9.5 | MIR-16-1*: HSA-MIR-16-1* | 6811.7 | 1.00 |
| 9.5 | MIR-20B: HSA-MIR-20B; MMU-MIR-20B; RNO-MIR-20B-5P | 2653.9 | 0.62 |
| 9.5 | MIR-347: RNO-MIR-347 | 906.1 | 0.59 |
| 9.5 | MIR-130A: HSA-MIR-130A; MMU-MIR-130A; RNO-MIR-130A | 4775.0 | 0.57 |
| 9.5 | MIR-199A-3P-199B-3P: HSA-MIR-199A-3P; HSA-MIR-199B-3P; MMU-MIR-199A-3P; MMU-MIR-199B; RNO-MIR-199A-3P | 802.7 | 0.51 |
| 9.5 | MIR-25: HSA-MIR-25; MMU-MIR-25; RNO-MIR-25 | 1185.2 | 0.48 |
| 9.5 | MIR-93: HSA-MIR-93; MMU-MIR-93; RNO-MIR-93 | 1132.3 | 0.38 |
| 9.5 | MIR-18B: HSA-MIR-18B | 1814.3 | 0.23 |
| 9.5 | MIR-18A: HSA-MIR-18A; MMU-MIR-18A; RNO-MIR-18A | 3298.4 | 0.15 |
| 9.5 | MIR-106A: MMU-MIR-106A | 3313.3 | 0.03 |
Table 2. MicroRNAs Differentially Expressed in GD-9.0 vs. GD-8.5 Neural Tubes.
1 Gene expression in neural tubes from each of GD-8.5 and GD-9.0 embryos was filtered and the average fold change for each gene was calculated. Only those genes which demonstrated a statistically significant (adjusted p < 0.05) increase or decrease in expression for the GD-9.0 vs. GD-8.5 expression comparison, were included in this table. Note that GD-9.0 vs. GD-8.5 means that expression on gestation day 8.5 was utilized as the baseline. Therefore, ratios below 1.0 indicate a decrease in expression, whereas ratios above 1.0 indicate an increase in expression. GD = gestational day.
| GD 9.0 vs. GD 8.5 -- Overexpressed | Fold Change1 | t-stat | P-Value | adjusted P-Value2 |
|---|---|---|---|---|
| miRNA Gene Family: Gene IDs | ||||
| MIR-638: HSA-MIR-638 | 4.11 | 3.81 | 0.001 | 0.017 |
| MIR-124: HSA-MIR-124; MMU-MIR-124; RNO-MIR-124 | 3.70 | 5.90 | 0.000 | 0.000 |
| MIR-762: MMU-MIR-762 | 2.88 | 3.12 | 0.004 | 0.044 |
| MIR-17*: MMU-MIR-17* | 2.35 | 3.13 | 0.004 | 0.044 |
| MIR-663: HSA-MIR-663 | 2.05 | 3.02 | 0.005 | 0.049 |
| GD 9.0 vs. GD 8.5 -- Underexpressed | Fold Change1 | t-stat | P-Value | adjusted P-Value2 |
| miRNA Gene Family: Gene IDs | ||||
| MIR-20A*: MMU-MIR-20A* | 0.15 | −5.52 | 0.000 | 0.000 |
| MIR-182_1: HSA-MIR-182; MMU-MIR-182; RNO- MIR-182 | 0.52 | −3.61 | 0.001 | 0.022 |
| MIR-20A: HSA-MIR-20A; MMU-MIR-20A; RNO-MIR-20A | 0.59 | −3.23 | 0.003 | 0.044 |
empirical Bayes t-statistic from the hierarchical linear model fitted by limma. See Methods for description
P-value corrected for the false-discovery rate (FDR)
Table 3. MicroRNAs Differentially Expressed in GD-9.5 vs. GD-9.0 Neural Tubes.
1 Gene expressions from neural tubes from each of GD-9.0 and GD-9.5 embryos were filtered and the average fold change for each gene was calculated. Only those genes which demonstrated a statistically significant (adjusted p < 0.05) increase or decrease in expression for the GD-9.5 vs. GD-9.0 expression comparison, were included in this table. Note that GD-9.5 vs. GD-9.0 means that expression on gestation day 9.0 was utilized as the baseline. Therefore, ratios below 1.0 indicate a decrease in expression, whereas ratios above 1.0 indicate an increase in expression. GD = gestational day.
| GD 9.5 vs GD 9.0 -- Overexpressed | Fold Change1 | t-stat | P- Value | adjusted P-Value2 |
|---|---|---|---|---|
| miRNA Gene Family: Gene IDs | ||||
| MIR-199A-5P: HSA-MIR-199A-5P; MMU-MIR-199A-5P; RNO-MIR-199A-5P | 10.05 | 2.64 | 0.014 | 0.037 |
| MIR-24: HSA-MIR-24; MMU-MIR-24; RNO-MIR-24 | 8.65 | 2.57 | 0.016 | 0.040 |
| MIR-199A-3P-199B-3P: HSA-MIR-199A-3P; HSA-MIR- 199B-3P; MMU-MIR-199A-3P; MMU-MIR-199B; RNO- MIR-199A-3P | 4.02 | 2.45 | 0.021 | 0.050 |
| MIR-20A*: MMU-MIR-20A* | 3.74 | 4.22 | 0.000 | 0.001 |
| MIR-335: HSA-MIR-335; MMU-MIR-335-5P; RNO-MIR-335 | 3.60 | 4.42 | 0.000 | 0.001 |
| MIR-30A: HSA-MIR-30A; MMU-MIR-30A; RNO-MIR-30A | 3.52 | 4.94 | 0.000 | 0.000 |
| MIR-16: HSA-MIR-16; MMU-MIR-16; RNO-MIR-16 | 2.81 | 5.56 | 0.000 | 0.000 |
| MIR-20A: HSA-MIR-20A; MMU-MIR-20A; RNO-MIR-20A | 2.72 | 6.67 | 0.000 | 0.000 |
| MIR-301A: HSA-MIR-301A; MMU-MIR-301A; RNO-MIR-301A | 2.70 | 7.52 | 0.000 | 0.000 |
| MIR-130A: HSA-MIR-130A; MMU-MIR-130A; RNO-MIR-130A | 2.45 | 5.51 | 0.000 | 0.000 |
| MIR-19A: HSA-MIR-19A; MMU-MIR-19A; RNO-MIR-19A | 2.20 | 3.88 | 0.001 | 0.003 |
| MIR-126-3P: HSA-MIR-126; MMU-MIR-126-3P; RNO-MIR-126 | 2.07 | 4.26 | 0.000 | 0.001 |
| MIR-103: HSA-MIR-103; MMU-MIR-103; RNO-MIR-103 | 1.72 | 3.67 | 0.001 | 0.005 |
| MIR-107: HSA-MIR-107; MMU-MIR-107; RNO-MIR-107 | 1.71 | 4.26 | 0.000 | 0.001 |
| MIR-106A: HSA-MIR-106A | 1.70 | 4.15 | 0.000 | 0.002 |
| MIR-106A: MMU-MIR-106A | 1.63 | 3.30 | 0.003 | 0.010 |
| MIR-15B: HSA-MIR-15B; MMU-MIR-15B; RNO-MIR-15B | 1.57 | 3.54 | 0.001 | 0.006 |
| MIR-17: HSA-MIR-17; MMU-MIR-17; RNO-MIR-17 (RNO-MIR-17-5P) | 1.50 | 2.69 | 0.012 | 0.034 |
| GD 9.5 vs GD 9.0 -- Underexpressed | Fold Change1 | t-stat | P- Value | adjusted P-Value2 |
| miRNA Gene Family: Gene IDs | ||||
| MIR-302A: HSA-MIR-302A; MMU-MIR-302A | 0.15 | −2.99 | 0.006 | 0.019 |
| MIR-638: HSA-MIR-638 | 0.16 | −5.36 | 0.000 | 0.000 |
| MIR-712*: MMU-MIR-712* | 0.17 | −2.59 | 0.015 | 0.040 |
| MIR-762: MMU-MIR-762 | 0.22 | −4.80 | 0.000 | 0.000 |
| MIR-663: HSA-MIR-663 | 0.26 | −6.12 | 0.000 | 0.000 |
| MIR-494: HSA-MIR-494; MMU-MIR-494; RNO-MIR-494 | 0.31 | −4.12 | 0.000 | 0.002 |
| MIR-347: RNO-MIR-347 | 0.38 | −2.58 | 0.015 | 0.040 |
| MIR-17*: MMU-MIR-17* | 0.41 | −3.57 | 0.001 | 0.005 |
| MIR-487B: HSA-MIR-487B; MMU-MIR-487B; RNO-MIR-487B | 0.41 | −2.69 | 0.012 | 0.034 |
| MIR-298: MMU-MIR-298; RNO-MIR-298 | 0.60 | −2.77 | 0.010 | 0.031 |
| MIR-16-1*: HSA-MIR-16-1* | 0.69 | −3.04 | 0.005 | 0.018 |
| MIR-345-3P: MMU-MIR-345-3P; RNO-MIR-345-3P | 0.69 | −3.59 | 0.001 | 0.005 |
empirical Bayes t-statistic from the hierarchical linear model fitted by limma. See Methods for description
P-value corrected for the false-discovery rate (FDR)
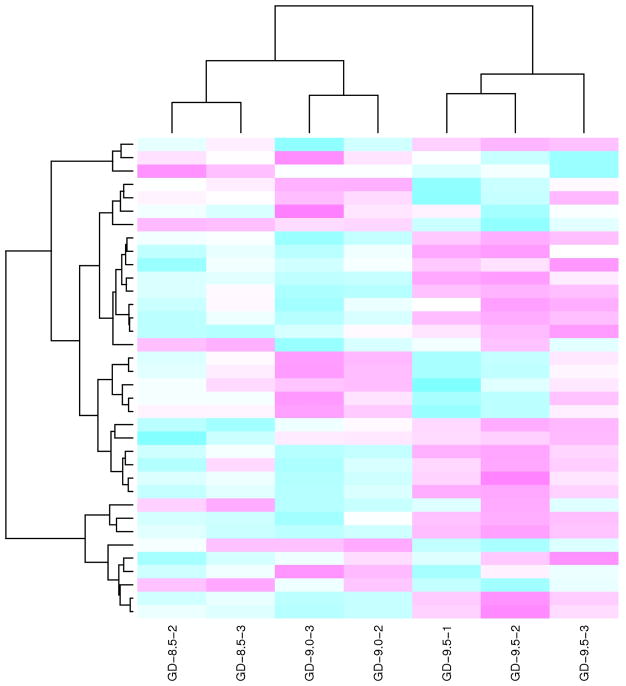
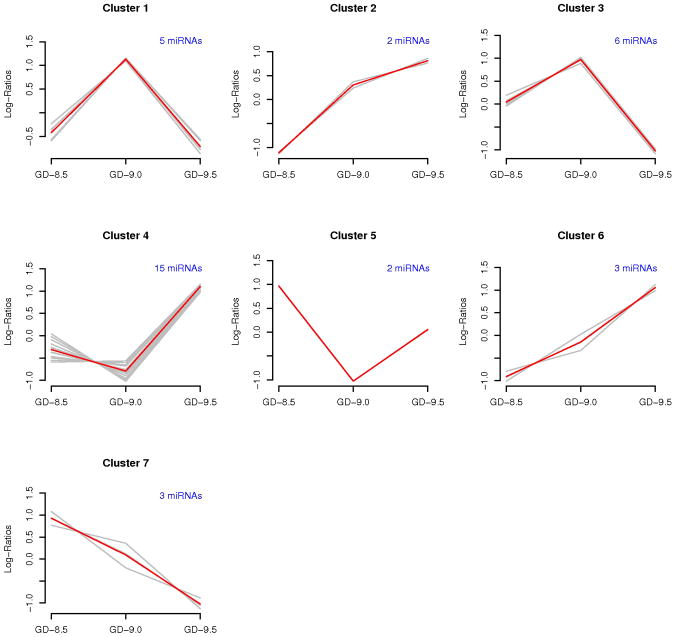
Clustering analysis revealed distinct patterns of microRNA expression during murine neural tube morphogenesis. Several recognizable temporal clusters of differentially expressed miRNAs (36 in total) were identified (Fig. 2). Seven separate cluster profiles representing distinguishable patterns of miRNA expression were defined (Fig. 3). MicroRNAs comprising each cluster have been tabulated on Supplementary Table 4. Expressed miRNAs include members of the miR-15, -16, -18, -19, miR-20, -25, -26, miR-92-93, -106, -130, -139, -199, -302, -335, -345, -347, -638, -663, -690, -709, -720 and -1195 miRNA families (Table 1). The expression profile of selected differentially expressed miRNAs was verified by TaqMan QRTPCR (Table 4). Prediction of downstream mRNA targets of these miRNAs was performed using the miRDB online database (http://mirdb.org/miRDB/). This analysis revealed a panoply of genes encoding transcriptional regulators, signaling mediators, cytoskeletal and extracellular matrix proteins, as well as growth and differentiation factors (Supplementary Data Files 1 and 2). Notably, many of these target genes have been documented to be either linked to, or indispensable for, development of the mammalian neural tube (Table 5). In addition, utilization of enriched gene ontology biological processes (GO BP), to identify putative miRNA target genes predicted by the DAVID software (http://david.abcc.ncifcrf.gov/), revealed many that were either documented as being linked, or likely to be linked, with cellular processes key to morphogenesis of the neural tube (Table 6).
Figure 2. Heat maps (hierarchical clusters) of differentially expressed miRNAs in the developing murine neural tube.
Heat maps (hierarchical clusters) of all 36 miRNAs found to be significantly differentially expressed (adjusted p-value < 0.05) for at least one of the comparisons between gestational days (GD-9.0 vs. GD-8.5 and GD-9.5 vs. GD-9.0, see Tables 2 and 3 respectively) in developing murine neural tube. Each row of the heat map represents a gene, and each column represents a time point in development (as labeled at the bottom; “GD-8.5-1” stands for “Gestation Day 8.5 - sample replicate 1” and so on). Pink indicates an increase in miRNA gene expression (relative to the other expression measurements in the same row), whereas blue indicates a decrease.
Figure 3. Hierarchical cluster analysis illustrating seven patterns of expression of microRNAs that exhibit significant alteration in expression during murine neural tube development.
Gene expression data sets from neural tubes from each of the critical days of neural tube development (GD-8.5, GD-9.0, and GD-9.5) were filtered and differentially expressed miRNAs were clustered as detailed in the Methods section. The solid red line on each graph represents the average miRNA expression pattern for the miRNA cluster, while the lighter grey lines on each graph represent the individual expression patterns for each miRNA. The expression pattern for each gene was normalized to mean zero and standard deviation one to better reflect similarities between patterns based on the correlation measure (the measure used for clustering). The number above each graph indicates the number of miRNAs in the respective cluster. The lists of genes (36 genes) comprising each cluster can be found in tabular form as supplementary material (Supplementary Table 4).
Table 4. TaqMan™ Verification of Differentially Expressed Genes Encoding miRNAs in the Developing Mouse Neural Tube.
Differential expression of genes encoding miRNAs in the murine neural tube (GD-8.5, GD-9.0 and GD-9.5). Gene expression was compared using Miltenyi miRexplore™ miRNA arrays and TaqMan® quantitative real-time PCR as detailed in Materials and Methods.
| Gene Name1 | Microarray | TaqMan QRTPCR | Concordance2 | ||
|---|---|---|---|---|---|
| FC 9.0 vs. 8.5 | FC 9.5 vs. 9.0 | FC 9.0 vs. 8.5 | FC 9.5 vs. 9.0 | ||
| mmu-miR-124 | + 3.70 | + 1.41 | + 1.10 | + 3.65 | +/+ |
| mmu-miR-199A-3p | + 4.37 | + 4.02 | + 1.21 | + 8.22 | +/+ |
| mmu-miR-199A-5p | + 1.00 | + 10.05 | + 1.61 | + 16.60 | +/+ |
| mmu-miR-16 | − 1.67 | + 2.81 | − 2.50 | + 1.80 | +/+ |
| mmu-miR-17 | − 1.13 | + 1.50 | −1.56 | + 4.14 | +/+ |
| mmu-miR-762 | + 2.88 | − 4.55 | + 1.33 | − 2.64 | +/+ |
| mmu-miR-302a | − 1.64 | − 6.70 | − 1.15 | −1.71 | +/+ |
| hsa-miR-638 | + 4.11 | − 6.25 | + 3.00 | − 2.83 | +/+ |
| mmu-miR-210 | − 1.47 | − 2.10 | − 1.64 | − 1.71 | +/+ |
| mmu-miR-20a* | − 6.70 | + 3.74 | + 2.30 | + 1.01 | −/+ |
Target genes encoding miRNAs were selected based on results from Miltenyi miRexplore™ miRNA arrays.
+/+ Indicates full concordance whereas −/+ indicates partial concordance between the pattern/level of gene expression from Miltenyi miRexplore™ miRNA arrays and TaqMan® quantitative real-time PCR.
Table 5.
Partial list of Target Genes (of selected differentially expressed miRNAs) Known to be Associated with, or Indispensable for Development of the Mammalian Neural Tube.
| miRNA Type1 | Target Gene2 | Gene Name | Score3 |
|---|---|---|---|
| miR-199A-3p | Map3k44 | Mitogen activated protein kinase kinase kinase 4 (Mekk4) | 95 |
| miR-712 | Zeb25 | Zinc finger E-box binding homeobox 2 (Zfhx1b or Sip1) | 92 |
| miR-199A-3p | Lrp26 | Low density lipoprotein receptor-related protein 2 | 92 |
| miR-494 | Zic2 | Zinc finger protein of the cerebellum 2 | 89 |
| miR-494 | Gli3 | GLI-Kruppel family member | 83 |
| miR-124 | Meox2 | Mesenchyme homeobox 2 | 80 |
| miR-15a | Nf17 | Neurofibromatosis 1 | 81 |
| miR-199A-5p | TGFβ2 | Transforming Growth Factor-β2 | 79 |
| miR-16 | Nf17 | Neurofibromatosis 1 | 79 |
| miR-124 | Gli3 | GLI-Kruppel family member | 78 |
| miR-199A-5p | Grlf1 | Glucocorticoid receptor DNA binding factor 1 | 78 |
| miR-712 | Zic3 | Zinc finger protein of the cerebellum 3 | 76 |
| miR-199A-5p | Celsr18 | Cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) | 75 |
| miR-130a | Tsc1 | Tuberous sclerosis 1 | 75 |
| miR-124 | Dhfr | Dihydrofolate reductase | 75 |
| miR-124 | Snai2 | Snail homolog 2 (Drosophila) | 75 |
| miR-709 | Smad5 | MAD homolog 5 (Drosophila) | 74 |
| miR-301a | Tsc19 | Tuberous sclerosis 1 | 73 |
| miR-301b | Tsc19 | Tuberous sclerosis 1 | 73 |
| miR-103/107 | Nf17 | Neurofibromatosis 1 | 72 |
| miR-182 | Gli2 | GLI-Kruppel family member GLI2 | 72 |
| miR-17 | Rgma | RGM domain family, member A | 71 |
| miR-199A-5p | Ets1 | E26 avian leukemia oncogene 1, 5′ domain | 69 |
| miR-15a | Nup50 | Nucleoporin 50 | 69 |
| miR-16 | Nup50 | Nucleoporin 50 | 67 |
| miR-376b | Vasp | Vasodilator-stimulated phosphoprotein | 63 |
| miR-199A-3p | Cited2 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | 62 |
These miRNAs exhibited differential expression during neural tube development (GD-8.5 GD-9.5) and the ones shown in bold were also verified by TaqMan QRTPCR (see Table 5).
Target analysis of these miRNAs was performed using the miRDB online database (http://mirdb.org/miRDB/), as described in the Methods section.
miRNA target scoring was done as described in the Methods section. Only those gene targets with scores above 60 are listed (see Supplementary Data Files 1 and 2). Higher scores represent higher sequence similarity between the mature miRNA transcript and the 3′UTR of the target gene.
Some representative references:
Chi H, Sarkisian MR, Rakic P, Flavell RA. 2005. Loss of mitogen-activated protein kinase kinase kinase 4 (MEKK4) results in enhanced apoptosis and defective neural tube development. Proc Natl Acad Sci USA 102(10):3846–3851.
Nitta KR, Takahashi S, Haramoto Y, Fukuda M, Tanegashima K, et al. 2007. The N-terminus zinc finger domain of Xenopus SIP1 is important for neural induction, but not for suppression of Xbra expression. Int J Dev Biol 51(4):321–325.
Spoelgen R, Hammes A, Anzenberger U, Zechner D, Andersen OM, et al. 2005. LRP2/megalin is required for patterning of the ventral telencephalon. Development. 132(2):405–414.
Lakkis MM, Golden JA, O’Shea KS, Epstein JA. 1999. Neurofibromin deficiency in mice causes exencephaly and is a modifier for Splotch neural tube defects. Dev Biol 212(1):80–92.
Curtin JA, Quint E, Tsipouri V, Arkell RM, Cattanach B, et al. 2003. Mutation of Celsr1 disrupts planar polarity of inner ear hair cells and causes severe neural tube defects in the mouse. Curr Biol 13(13):1129–1133.
Kobayashi T, Minowa O, Sugitani Y, Takai S, Mitani H, et al. 2001. A germ-line Tsc1 mutation causes tumor development and embryonic lethality that are similar, but not identical to, those caused by Tsc2 mutation in mice. Proc Natl Acad Sci USA 98(15):8762–8767.
Table 6.
Partial list of Target Genes (of selected differentially expressed miRNAs) Known or likely to be Associated with Biological Processes Indispensable for Mammalian Neural Tube Development.
| Target Genes1,2,3 | Cellular Process4,5 |
|---|---|
| Bcam, Cadm3, Pcdhac2, Pvrl1, Col12a1, Celsr1, Col4a2, Shc1, Pcdha10, Pcdha5, Grlf1, Pcdha2, Camk2n1, Pvrl4, Pcdhb18, Pcdh7, Pik3r1, Itga7, Dsg2, Pcdha1, Reln, Flrt3, B4galt1, Cdh2 (Ncad), Olr1, Nf1, Pcdh8, Bcl6, Pdpn, Pvrl2, Pcdhb4, Jmy, Astn1, Colec10, Neo1, Lama3, Col19a1, Pcdha8, Nid2, Myh9, Pcdh1, Eda, Pcdha3, Col24a1, Pcdha7, Cxadr, Thbs4, Itgb8, Cdh9, Pten, Itga3, Cd164, Rs1, Rgma, Pcdhb17, Cldn12, Celsr2, Pcdhb16, Pcdha4, Itgb1, Flot2, Pcdha12, Pcdha9, Rgmb, Lamc1, Traf6, Pcdha6, Col4a1, Ror2, Fn1, Pcdhac1, Col4a4, Col5a1 | Adhesion6 |
| Kif5c, Sema5a, Epha4, Atoh1, Neurog2, Myh10, Lrp6, Pafah1b1, Camk2n1, Ndel1, Gab1, Epha7, Pik3r1, Pten, Reln, Mapk8ip3, Itga3, B4galt1, Cdh2, Btg1, Dpysl5, Nf1, Podxl, Pdpn, Tgfb2, Dclk1, Bdnf, Lamc1, Ednrb, Astn1, Prkca, Hif1a, Lama3, Sema3a, Etv1, Gna13, Myh9, Wasf2 | Migration7 |
| Timp2, Gng2, Rb1, Gnai2, Shc1, Myh10, Apc, E2f1, E2f2, E2f7, Mfn2, Ndel1, Cops2, B4galt1, Cd274, Mycn, Foxp2, Nf1, Trim35, Bcl6, Pdpn, Tgfb2, Bdnf, Fshb, Pdcd1lg2, Topors, Igsf8, Cd3e, Lrp2, Mab21l1, Ppap2a, Lhx9, Acvr1c, Pafah1b1, Gsk3b, Ncapg2, Pten, Btg1, Gli3, Tsg101, Acvr2a, Tgfbr2, St8sia1, Ereg, Itgb1, Derl2, Lamc1, Zfp91, Cebpa, Nras, Cdkn1a, Lgr4, Krtap16-7, Pgf, A730098P11Rik, | Proliferation8 |
| Ap3b1, Atoh1 (Math1), Neurog2, Rb1, Myh10, Ube2b, Dmrta1, Plxnb2, Ndel1, Pik3r1, Mapk8ip3, Thoc1, Dpysl5, Myo5a, Calcr, Dlx5, Nf1, Bcl6, Raf1, Tgfb2, Slitrk4, E2f2, Il25, Snai2, Traf3, Myt1l, Ednrb, Sox8, Ntng1, Aff4, Col19a1, Sema3a, Cd3e, Ube4b, Pak7, Myh9, Serpine2, Eda, Egln3, Mmp9, Sema5a, Nfatc1, Btg2, Mbnl1, Ank2, Acvr1c, Sgpp1, Gsk3b, Eif2b2, Stk4, Stat3, Ywhah, Son, Epha7, Sema4b, Wwp1, Pten, Rock1, Cep57, Ptprj, Mcl1, Ppl, E2f1, Rnf6, Gli3, Rb1cc1, Neurod6, Pldn, 2610018G03Rik, Dclk1, Ereg, Slitrk6, Itgb1, Camk1d, Ercc6, Cebpa, Atp7a, Birc2, Efna1, Cdkn1a, Sema7a, Pappa2, Timp2, Kif5c, Wnt3a, Cbfa2t3, Ubn1, Erap1, Epas1, Reln, Socs7, Atg7, Trib3, B4galt1, Trim35, Cln8, Pappa, Dzip1l, Ets1, Bdnf, Cdx2, Zfp488, Twist2, Jmy, Sgms1, Frap1, Tns4, Ppp3r1, Sema6d, Tnfsf10, Topors, Qk, Neo1, Hif1a, Lilrb3, Mapk14, Eya1, Lhx8, Tnfrsf22, Txndc1, Epha4, Klrb1b, Siah1a, Rfng, Gmcl1, Pafah1b1, Strbp, Atp2b2, Creb1, Zfp36l1, Phf17, Ulk1, Tshr, Itga3, Zfpm2, Btg1, Mef2c, Rnf144b, Tsg101, Dapk1, Tgfbr2, Nuak2, Dclre1c, Sall1, Pdcd6, Tnfrsf21, Zfp91, Traf6, Rad9, Txnip, Olig2, Suv39h1, Eif4g2, Bag2, Ror2, Mink1, Prkca, Mixl1, Nras, Limk1, Wrn, Etv1, Gna13, Pgf | Differentiation9 |
| Tnfrsf21, Tnfrsf22, Tnfrsf23, Traf3, Map3k4 (Mekk4), Traf6, Tnfsf10, Tnfaip1, Bag2, Bcl6, Bcl7a, Pdcd6, Pdcd1lg2, Dapk1, Tgfbr2, Tgfbr3, Bmpr2, Bambi, Dmtf1, Pak7, Stk4, Rgma, Rgmb, E2f1, E2f2, Smad7, Rb1, Tgfb2, Apc, Cdkn1a | Apoptosis10 |
| Rab10, Neurog2, Dmrta1, Meis2, E2f5, Med9, Zeb1, Phb2, Npas3, Rela, Pbx3, Znfx1, Ankib1, Tshz1, Dlx5, Apol6, Bcl6, Whsc1l1, Cited2, Tox3, Zkscan1, Aff4, Zbtb33, Cbx4, Taf1, Asb3, Jazf1, Trak1, Ythdc1, Stat3, Polr3f, Klf12, Brms1l, Mllt6, Tcfeb, Rnf6, Rab8b, Hivep1, Neurod6, Chaf1a, Rb1cc1, A630089N07Rik, Zfp300, Atrx, Mll1, Ash1l, Gtf2h2, Cphx, Zfhx4, Nfatc3, Atf6, Zfp9, Mdm2, Ncoa3, EG210853, A730098P11Rik, Tle4, Ascc2, Csdc2, Tcfap2d, Zfp426, Zfp92, Grlf1, Epas1, Rreb1, Lhx6, Cask, Mycn, Foxp2, Ankrd57, Asf1a, Ccnl1, Zfp810, Ets1, Zfp93, Twist2, Homez, Zfp697, Gcdh, Tbl1xr1, Arid4b, Elk4, Zfp800, Hmga1, Prrx1, Mapk14, Lin28b, Pcdh1, Lhx8, Eya1, Mier1, Nr3c1, Nfix, Pou2af1, Nfya, Gata6, Hif1an, Zfp238, Mtf1, Crebl2, Tanc2, Taf9b, Zfpm2, Meox2, Npas2, Pcdhb16, Onecut2, Sirt4, Rbbp7, Txnip, Fosb, Mixl1, Zhx2, Etv1, Atoh1 (Math1), Rb1, Preb, Zfp367, 9230110K08Rik, Suhw4, Irf9, Zbtb5, Mga, Irf2bp2, E2f2, Smad7, Rfc1, Snai2, Myt1l, Dnajb6, Pou6f1, Ccnt2, Chd9, Sox8, Lama3, Mybl2, Csde1, Elk3, Foxq1, Nfatc1, Lhx9, Tox, Btg2, Hdx, Fem1b, Ezh1, Zik1, E2f1, Nfat5, Gli3, Nkx6-3, Hoxa10, Nfia, Abt1, Cebpa, Zfp148, Pappa2, Zkscan3, Klf16, Prdm4, Sertad2, Cbfa2t3, Fubp1, Rab11a, E2f7, Ncoa7, Trib3, Rps6ka5, Usf2, Irf2, Zbtb44, Clock, Cnot7, Cdx2, Jmy, Hif1a, Tbx22, Alx1, Tshz3, Rbl1, Mysm1, Ankrd52, Rfx5, Creb1, Tgif2, A930001N09Rik, Cbx6, Phf17, Htr5a, Mxi1, Zfp239, Zfp592, Mef2c, Tsg101, Sall1, Zfp91, Smarce1, Olig2, Zfp617, Suv39h1, Phf19, Hivep2, Foxb1, Zbtb41, Tcfap2e, Trpv6 | Regulation of transcription, DNA-dependent11 |
Target genes of miRNA that are differentially expressed during neural tube development (GD-8.5 – GD-9.5). Only those gene targets with scores above 60 are listed (see Supplementary Data Files 1 and 2).
Target analysis of the miRNAs was performed using the miRDB [(http://mirdb.org/miRDB/)], as described in the Methods section.
Target genes known to be associated with neural tube development are underlined and in bold.
Enriched gene ontology biological processes (GO BP) for the putative target genes, as predicted by the DAVID software, as described in the Methods section.
For the full list of biological processes associated with the target genes, including p-value and fold enrichment, see Supplementary Data Files 1 and 2.
Some representative references:
Radice GL, Rayburn H, Matsunami H, Knudsen KA, Takeichi M, et al. 1997. Developmental defects in mouse embryos lacking N-cadherin. Dev Biol. 181(1):64–78.
Tsuda S, Kitagawa T, Takashima S, Asakawa S, Shimizu N, et al. 2010. FAK-mediated extracellular signals are essential for interkinetic nuclear migration and planar divisions in the neuroepithelium. J Cell Sci 123(Pt 3):484–496.
Kim TH, Goodman J, Anderson KV, Niswander L. 2007. Phactr4 regulates neural tube and optic fissure closure by controlling PP1-, Rb-, and E2F1-regulated cell-cycle progression. Dev Cell 13(1):87–102.
Canzoniere D, Farioli-Vecchioli S, Conti F, Ciotti MT, Tata AM, et al. 2004. Dual control of neurogenesis by PC3 through cell cycle inhibition and induction of Math1. J Neurosci 24(13):3355–3369.
Chi H, Sarkisian MR, Rakic P, Flavell RA. 2005. Loss of mitogen-activated protein kinase kinase kinase 4 (MEKK4) results in enhanced apoptosis and defective neural tube development. Proc Natl Acad Sci USA 102(10):3846–3851.
Bamforth SD, Bragança J, Eloranta JJ, Murdoch JN, Marques FI, et al. 2001. Cardiac malformations, adrenal agenesis, neural crest defects and exencephaly in mice lacking Cited2, a new Tfap2 co-activator. Nat Genet 29(4):469–474.
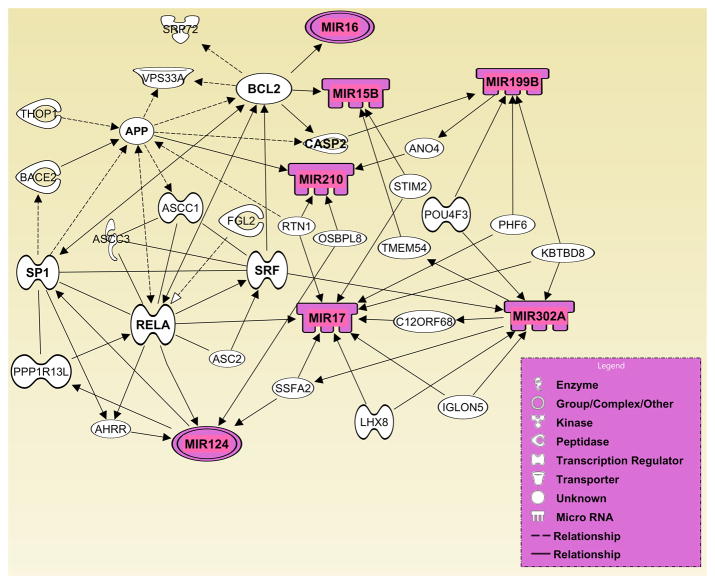
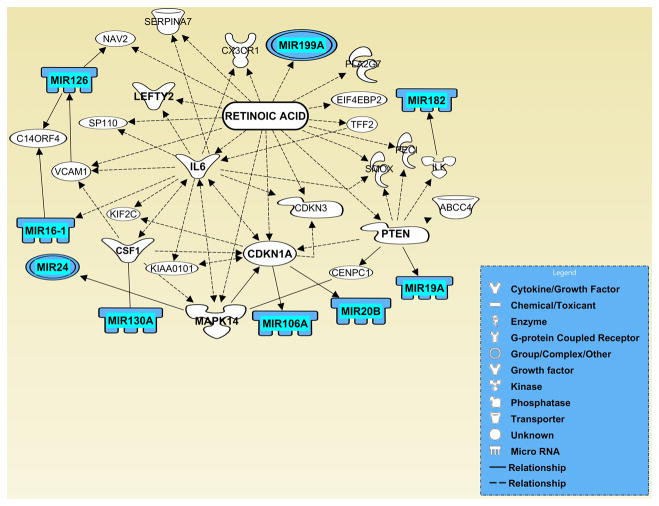
Network analysis using IPA (Ingenuity Systems Inc.) for predicting potential interactions between the differentially expressed genes encoding miRNAs and their downstream gene targets, generated two networks with the highest IPA analysis scores (12 and 15), and with the most QRTPCR-confirmed miRNAs. One representative network (IPA analysis score: 12) with seven differentially expressed miRNAs – miR-15b, miR-16, miR-17, miR-124, miR-199B, miR-210, and miR-302a, and another (IPA analysis score: 15) connecting nine differentially expressed miRNAs – miR-16, miR-19a, miR-20b, miR-24, miR-106a, miR-126, miR-130, miR-182, and miR-199a, are presented in Figures 4 and 5, respectively. Overlay of these networks with relevant biological functions highlighted numerous cellular processes crucial for neurulation (Supplementary Figs. 1–3). Similarly, overlay of these networks with canonical pathways detected a range of key signal transduction pathways (e.g. BMP-, Wnt-, FGF-, EGF-, PDGF-, VEGF-, Retinoic acid receptor-, JAK/Stat-, ERK/Mapk-, p38 Mapk-, PI3K/AKT-, p53-, and tight junction signaling cascades) operative within the gene networks (Supplementary Figs. 4 and 5). These findings emphasize the notion that diverse signal transduction pathways coordinately operate to regulate neural tube morphogenesis. Finally, the network presented in (Supplementary Fig. 6) highlights various target genes associated with a variety of developmental disorders as well as cellular events essential for embryogenesis, including neural tube development.
Figure 4. Computational gene interaction predictions: selected microRNA gene network (#1) in the developing neural tube.
A network of selected genes that encode differentially regulated microRNAs was constructed with the Ingenuity Systems Pathway Analysis (IPA) software. Solid lines specify direct relationships whereas dotted lines indicate indirect interactions.
Figure 5. Computational gene interaction predictions: selected microRNA gene network (#2) in the developing neural tube.
A network of selected genes that encode differentially regulated microRNAs was constructed with the Ingenuity Systems Pathway Analysis (IPA) software. Solid lines specify direct relationships whereas dotted lines indicate indirect interactions.
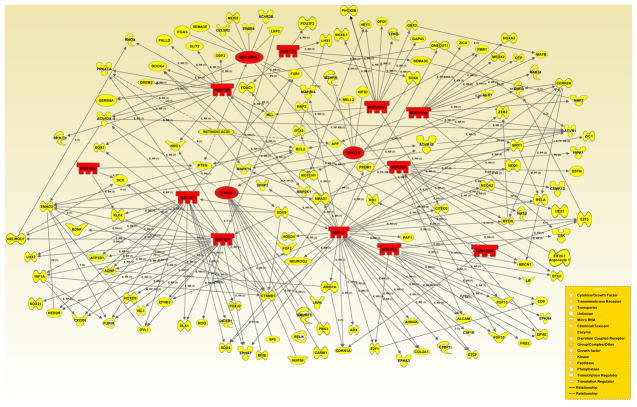
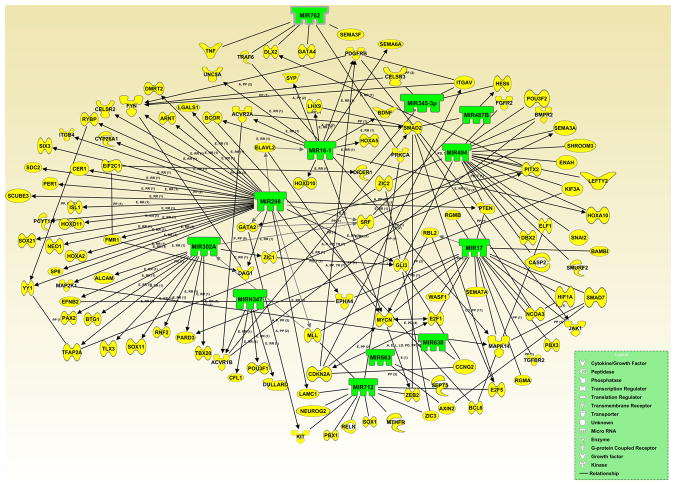
In addition, two other networks, one with miRNAs exhibiting enhanced expression (Fig. 6), and the other with miRNAs displaying diminished expression (Fig. 7) in the neural tube during GD-9.0-9.5, developed utilizing IPA (Ingenuity Systems Inc.) and the miRDB online database (http://mirdb.org/miRDB/), further highlighted direct linkage between the differentially expressed miRNAs and their gene targets associated with neural tube development. Note that most target genes of the differentially expressed miRNAs have been documented as playing a key role in various aspects of neurulation. Moreover, several of these target genes have been associated with syndromes involving dysmorphologies of the neural tube.
Figure 6. Computational gene interaction predictions: gene network with microRNAs demonstrating enhanced expression in developing neural tube (GD-9.5 vs. GD-9.0) and their target genes.
A network with selected genes encoding microRNAs (orange) with increased expression on GD-9.5 vs. GD-9.0 in the developing neural tube and their known or predicted target genes (yellow) critical for neural tube ontogenesis was constructed with Ingenuity Systems Pathway Analysis (IPA) software and the miRDB (http://mirdb.org/miRDB/) database. Solid lines specify direct relationships whereas dotted lines indicate indirect interactions.
Figure 7. Computational gene interaction predictions: gene network with microRNAs demonstrating diminished expression in developing neural tube tissue (GD-9.5 vs. GD-9.0) and their target genes.
A network with selected genes encoding microRNAs (green) with decreased expression on GD-9.5 vs. GD-9.0 in the developing neural tube and their known or predicted target genes (yellow) critical for neural tube ontogenesis was constructed with Ingenuity Systems Pathway Analysis (IPA) software and the miRDB (http://mirdb.org/miRDB/) database. Solid lines specify direct relationships whereas dotted lines indicate indirect interactions.
DISCUSSION
Discovered nearly two decades ago (Lee et al., 1993; Wightman et al., 1993), microRNAs (miRNAs) represent the largest family of noncoding RNAs involved in gene silencing. MicroRNAs are found in polycistronic clusters or residing in the introns of protein coding genes (for reviews see, (Bartel, 2009; Chuang and Jones, 2007)). They are transcribed as long primary miRNAs (pri-miRNA), processed in the nucleus by a protein complex containing a RNase III (Drosha) into double-stranded stem-loop precursor miRNAs (pre-miRNA) (Lee et al., 2004), exported into the cytoplasm (Lund et al., 2004), and further processed by another RNase III (Dicer) into mature, functional miRNAs which are incorporated into an RNA-induced silencing complex (RISC) (Lee et al., 2002). Each RISC-miRNA complex can then silence multiple, often hundreds (Lim et al., 2005) of genes, by binding to specific sequences in the 3′ UTRs, or even the coding regions (Tay et al., 2008), of target mRNAs. A number of studies have established that miRNAs are critical regulators of cell and tissue differentiation during embryogenesis (for reviews see, (Conrad et al., 2006; Lee et al., 2006; Mineno et al., 2006)).
Maintenance and regulation of endogenous miRNA levels is necessary for proper mammalian neurogenesis and neurodevelopment (Giraldez et al., 2005; Hosako et al., 2009; Maller Schulman et al., 2008; Singh, 2007; Zhao et al., 2008). Development of the mammalian neural tube into the CNS requires proper orchestration of cellular proliferation, adhesion, migration, differentiation, and apoptosis directed by numerous genes encoding a range of transcriptional regulators, growth factors/morphogens, and extracellular matrix proteins (Copp et al., 2003; Richman and Tickle, 1989). Moreover, recent experimental findings convincingly documented that a panoply of miRNAs potentially govern these cellular processes by modulating the expression of discrete target genes. During organogenesis expression of avian (Darnell et al., 2006; Xu et al., 2006), zebrafish (Giraldez et al., 2005) and murine (Takada et al., 2006) miRNAs are tightly regulated in a tissue- and developmental stage-specific manner (Saetrom et al., 2007; Wienholds et al., 2005). Since such definition of miRNA expression patterns offers revealing insights into biological function, we examined the expression profiles of 1392 mature miRNAs in the developing neural tube. Expression profiles of 67 miRNAs, of which 48 were distributed in specific temporal patterns, are reported in the present study. Gene ontology (GO) enrichment analysis [(exploiting the GO database (http://www.geneontology.org/)] and the DAVID software (http://david.abcc.ncifcrf.gov/) was utilized in our study to verify that genes, targeted by differentially expressed miRNAs in the developing neural tube, are associated with cellular processes central to neural tube morphogenesis such as proliferation, adhesion, migration, differentiation and apoptosis (Table 6).
MiR-124
MiR-124 is highly expressed in differentiating and mature neurons during neurogenesis (Deo et al., 2006), where it is thought to inhibit the expression of progenitor genes in neuronal differentiation during neural tube development (Cao et al., 2007). MicroRNA target prediction software programs – miRDB, PicTar, Target Scan, and miRanda – predicted several integrins (such as Itgβ1, Itgα7, Itgα3 and Itgα11), known to play a critical role in morphogenesis of the neural tube (Fournier-Thibault et al., 2009; Nagy et al., 2009), as potential targets of miR-124, implicating this miRNA in the regulation of neural tube development via modulation of integrin signaling. In fact, direct targeting of Itgb1 by miR-124 and resulting reduction of adherence and motility of oral squamous cell carcinoma cells have been documented (Hunt et al.). The brain-specific miRNAs miR-124a (miR-124) and miR-9 also target the signal transducer and activator of transcription (STAT) 3 which, in turn, mediates the morphogenetic effects of the hepatocyte growth factor (HGF)/c-met system, known to play a defining role in neurogenesis (Trovato et al., 2007). Krichevsky et al. (Krichevsky et al., 2006) reported modulation of neurogenesis of embryonic stem cells by miR-124 targeting Stat3. Moreover, all components of the p53/HGF/c-met/STAT3 cascade exhibited reduced expression in the neural tubes of fetuses presenting with NTDs (Trovato et al., 2007). Finally, numerous additional miR-124 targets such as Gli3, Meox2, Dlx2, Dlx5, Sox8, Sox9, Snail2 (Slug), and Dihydrofolate reductase (Dhfr) are known to execute key roles in normal and abnormal neural tube development. Consistent with these observations is our demonstration that miR-124 exhibited significantly elevated expression within the developing neural tube (GD-8.5-GD-9.5) suggesting a vital role for miR-124a in gene regulation during neurulation (Tables 2–5).
MiR-638, -663, and -762
Genes encoding miR-638, -663, and -762 demonstrated interesting pattern of expression in the developing neural tube in the present study. All three genes displayed transiently enhanced expression in the earlier stage (GD-9.0 vs. -8.5) followed by significantly lower expression in the later phase of neurulation (GD-9.5 vs. -9.0) (Tables 2 and 3). Although nothing is presently known about the role of these miRNAs in embryogenesis, a recent study highlighted miR-638 and -663 as key regulators of cell proliferation and apoptosis - two cellular events that are crucial for neural tube development (Maes et al., 2009). In addition, gene targets of miR-638 and -663 are yet to be predicted or identified whereas, miR-762 is predicted to target genes such as, Epha1, Mdm2, TGFβRI, Msx1 and Ncam, among others, which are known to be involved in neural tube development (miRDB).
MiR-199a-5p and miR-199a-3p
The SWI/SNF complex, a chromatin remodeling factor executing key roles in epigenetic gene regulation and comprised of about 10 protein subunits, also contains a single molecule of either Brm or BRG1 as the catalytic subunit responsible for DNA-dependent ATPase activity (Sakurai et al.). SWI/SNF can interact with a range of transcriptional activators and repressors, which can in turn, recruit the complex to different gene promoters leading to transcriptional activation or repression (Sakurai et al.). Sakurai et al. (2010) demonstrated that Brm mRNA, shown to be expressed in the developing neural tube (Schofield et al., 1999), is a target of two miRNAs, miR-199a-5p and miR-199a-3p, both of which are processed from pre-miR-199a, and the expression patterns of mature miR-199a-5p, -199a-3p, and Brm protein, are mutually exclusive in many human cancer cells. Interestingly, Twist1, which is involved in cranial neural tube closure (Chen et al., 2007), has been recently documented to regulate miR-199a-5p and miR-199a-3p - expressed within developing neural tissues (Lee et al., 2009). Robust increase in expression of these two miRNAs (>16-fold and >8-fold) in the developing murine tube further reinforce their potential role(s) in epigenetic regulation of neural tube morphogenesis (Table 4).
MiR-17~92, miR-106b~25 and miR-106a~363 clusters
miR-17~92, miR-106b~25 and miR-106a~363 represent a family of highly conserved miRNA clusters that, in the current study, were shown to contain miRNAs that were differentially expressed in the developing neural tube (Tables 2–3). Interestingly, a recent study reported expression of several members of these miRNA families (e.g. miR-106b, miR-25, miR-93; miR-92, miR-19b and miR-17) in cells derived from the GD-8.5 murine neural tube (Hosako et al., 2009).
Alterations in cell proliferation, cell survival, patterning, actin cytoskeletal regulation of the developing neuroepithelium can cause neural tube defects (NTDs) (for a review see, (Copp et al., 2003)). Dysregulation of normal apoptosis is a particularly well established mechanism underlying perturbation of neural tube morphogenesis. For instance, NTDs in caspase-9-deficient mice are associated with decreased apoptosis (Kuida et al., 1998), while NTDs in Pax3-deficient mice occur as a consequence of enhanced cell death in the neuroepithelium (Pani et al., 2002). The mitogen-activated protein kinase (MAPK) cascade is a well conserved module of signaling pathways involved in regulating an array of cellular functions, including apoptosis. Stress-activated MAP kinases, including p38 and c-Jun N-terminal kinase (JNK), exert pro- or anti-apoptotic effects, in accordance with cellular context (for a review see, (Kyriakis and Avruch, 2001)). Jnk1/Jnk2 double mutant mice, exhibit both NTDs and alterations in apoptosis within their neuroepithelium, supporting the notion of JNK-regulated region-specific apoptosis (Sabapathy et al., 1999). Since various MAP kinase kinases (MAP2Ks) and MAP kinase kinase kinases (MAP3Ks) function upstream of JNK, it is not surprising that mice deficient in MEKK4, a MAP3K in the JNK/p38 pathway, develop NTDs associated with enhanced apoptosis in the developing neuroepithelium (Giraldez et al., 2005). Moreover, several miRNAs belonging to the three paralogous clusters: miR-17~92, miR-106a~363 and miR-106b~25, are predicted to target a range of MAPKs such as MAPK9 or JNK2 and MAP3K-1, -2, -5, -8, -9, -13 (miRDB).
The Janus kinase/signal transducers and activators of transcription (JAK/STAT) pathway is one of the foremost signaling mechanism for cytokines and growth factors during mammalian development and represents a pleiotropic cascade transducing a panoply of signals throughout morphogenesis. Two members of this pathway, Jak1 and Stat3, are associated with neural tube development (Trovato et al., 2007; Zhao et al., 2008), and are potential targets of miR-17, -20a, -20b, -106a, -106b and -93 (miRDB) – all of which are differentially expressed during neural tube development. Experimental confirmation of such target predictions is reinforced by direct targeting of Stat3 and Mapk14 (p38) by miR-17, -20a, and -106b in embryonic cells (Carraro et al., 2009).
The inhibitory Smads, Smad6 and Smad7, antagonize TGFβ superfamily signaling. Furthermore, when misexpressed in the dorsal neural tube, Smad6 or Smad7 can cause spina bifida (Nakayama et al., 2001). Interestingly, Smad7 is a common target of miR-20a, -20b, -17, -93, -106a, and -106b (miRDB), and thus, maintenance of proper level of expression of Smad7 during normal neural tube development could potentially be governed by some of these miRNAs. Recent studies also demonstrated miR-106b-25 and miR-17-92 clusters as key modulators of TGFβ signaling and targeting crucial members such as TGFβRII, among others, affecting proliferation and apoptosis – cellular events indispensible for neural tube morphogenesis (Petrocca et al., 2008; Volinia et al., 2006).
Both the aforementioned evidence, and our data indicating that miR-20a, -20b, -17, -93, -106a, and -106b are either expressed and/or differentially expressed in the developing neural tube, provide support for the premise that these miRNAs represent key modulators of discrete cellular processes essential for normal neural tube morphogenesis.
MiR-126, miR-103 and miR-107
Genes encoding these three miRNAs displayed developmentally regulated expression in the murine neural tube in the current study (Tables 2 and 3). While functions of these miRNAs have been reported to include involvement in pathogenesis of Alzheimer’s disease (miR-107) (Wang et al., 2008), regulation of erythroid differentiation (miR-126 and miR-103) (Yang and Wu, 2006), angiogenesis (miR-126) (Fish and Srivastava, 2009), and self-renewal and lineage differentiation of stem cells (miR-103 and miR-107) (Wang et al., 2007), the precise role of these miRNAs in morphogenesis of the neural tube remains unclear. miR-126 has a binding site in 3′-untranslated region of the VEGFA mRNA, and transfection of miR-126 can decrease the expression of VEGFA resulting in inhibition of cell proliferation (Wang et al., 2007). Furthermore, VEGFA signaling is a necessary component of vascular patterning in the neural tube which represents the first midline signaling center for embryonic vascular pattern formation in higher vertebrates (Hogan et al., 2004; James et al., 2009). Thus, miR-126 regulated, neural-derived VEGFA is likely to play a key role in neurovascular development and patterning in the neural tube.
The secreted factor, epidermal growth factor-like domain 7 (EGFL7), documented to be involved in cell migration and blood vessel formation, is another potential miR-126 target effecting inhibition of cellular proliferation (Sun et al.). EGFL7 secreted by neurons in the brain (Schmidt et al., 2009), acts as an antagonist of Notch signaling—indispensable for neural tube development (le Roux et al., 2003; Wang et al., 2007; Williams et al., 1995) reducing proliferation and self-renewal of NSCs. These observations support the possibility that miR-126 can regulate morphogenesis of the neural tube via modulating EGFL7-regulated Notch signaling.
The Sonic hedgehog (Shh) and insulin-like growth factor (IGF) signaling pathways are also known to play an important role in neurulation (Anlar et al., 1999; Ruiz i Altaba et al., 2003). Insulin receptor substrate 1 (Irs1), a component of the IGF pathway and effector of SHH signaling (Parathath et al., 2008), is targeted by miR-126 (Zhang et al., 2008) and can function as a component of the neurotrophin signaling pathway. This latter pathway is, in turn, essential for early embryogenesis including ontogenesis of the neural tube (Bernd, 2008; McDougall et al., 2001; Yamada et al., 1997). Interestingly, Hoxa9, another experimentally-verified target gene of miR-126, has been reported to induce the expression of insulin-like growth factor-1 receptor (IGF-1R) (Shen et al., 2008; Whelan et al., 2008). Thus, miR-126 could potentially regulate neural tube development by interacting with several important signaling pathways (Shh-, IGF-and Neurotrophin).
The aforementioned data all strengthens the case that miR-126 plays a potentially key role in normal neural tube ontogenesis by regulating a variety of developmentally important signal transduction pathways (Tables 2–4).
MiR-103 and miR-107 are expressed in a number of tissues and organs with highest expression in brain tissue (Babak et al., 2004). Moreover, miR-103 and miR-107 are differentially expressed during embryogenesis (Wienholds et al., 2005). By targeting genes such as transforming growth factor, beta receptor III (TGFBR3), Wnt3a, Dishevelled homolog 1 (Dvl1) and Bdnf, miR-103 and miR-107 can be thought of as potential regulators of neural tube development by modulating TGFβ-BMP, Wnt, and Neurotrophin signaling pathways, all recognized as contributing to neurulation (Dono et al., 1998; Etheridge et al., 2008; Jungbluth et al., 1997; Niederkofler et al., 2004; Samad et al., 2005).
Several studies document that incorrect DNA methylation patterns can result in neural tube defects (Kakutani et al., 1996; Prater et al., 2006; Rogner et al., 2002; van der Put et al., 1995). The importance of the DNA methyltransferases in morphogenesis of the neural tube is illustrated by the fact that Dnmt3b−/− embryos display multiple developmental defects including growth impairment and rostral neural tube defects (Okano et al., 1999). A potential target of miR-103/-107, ZHX1, a member of the zinc-finger and homeobox protein families, has been found to interact with, and silence, DNMT3B in vivo and in vitro (Kim and Nam, 2006). Thus, these miRNAs can directly effect DNA methylation and hence, can act as epigenetic regulators of neural tube ontogenesis.
The glypicans represent a family of glycosylphosphatidylinositol-anchored heparan sulfate proteoglycans. The prime function of membrane-attached glypicans is to regulate Wnt, Hedgehog, FGF and BMP signaling – all of which are central to neural tube development (for a review see, (Filmus et al., 2008)). Since glypican-6, represents a potential target of miR-103/106 (miRDB), one could speculate that these signaling pathways, and by extension, neurulation, may be regulated, in part, by miR-103/107 (Tables 2–4). Finally, a recent study highlighted potential roles of several miRNAs including miR-103 and -107, (as well as miR-130a and -130b, also detected in the present study) in orchestrating murine neural tube development, via targeting a key gene, the Notch ligand, Delta-like 1 (Hoesel et al., 2010) (Tables 2–4).
MiR-19a and miR-19b
The miR-19 family of miRNAs, miR-19a, miR-19b-1 and miR-19b-2 are encoded by two separate yet paralogous miRNA clusters. Based on target analysis (miRDB), LRP2 (Megalin), a member of the low-density lipoprotein (LDL) receptor-related protein (LRP) family, represents a downstream mRNA target for both miR-19a and miR-19b. Initially considered as endocytic receptors involved in lipoprotein metabolism (Nykjaer and Willnow, 2002), recent studies have demonstrated that the LRPs interact with a variety of adaptor and scaffold proteins implicated in signaling of several signal transduction pathways including the Wnt pathway (Herz and Bock, 2002; Mao et al., 2001; Nykjaer and Willnow, 2002). LRP2, or megalin, expressed in the neuroepithelium of embryos at mid-gestation, is essential for neurodevelopment inasmuch as targeted disruption of the murine Lrp2 gene results in perinatal lethality and holoprosencephaly (Willnow et al., 1996).
Functionality of miR-19a and -19b in neural tube development is further suggested by the fact that these miRNAs target RhoB, which has a role in the delamination of neural crest cells from the dorsal neural tube (Wang et al., 2007), qkl, which when ablated results in an open neural tube (Li et al., 2003), TGFB-induced factor homeobox 1 (TGIF1), a transcriptional repressor exhibiting mutations in patients with holoprosencephaly (Gripp et al., 2000) and exencephaly in mice with either heterozygous or homozygous deletion of Tgif (Kuang et al., 2006), and Id2, which when overexpressed in cranial neural folds and migrating cranial neural crest cells results in overgrowth and premature neurogenesis of the neural tube (Martinsen and Bronner-Fraser, 1998).
In addition, an array of genes encoding signaling mediators, morphogens and transcription factors, known to be directly or indirectly involved in neural tube development are also predicted targets of miR-19a and -19b. These include, but are not limited to genes encoding members of the IGF (IGF1, IGF2R, IGFBP3), TGFβ (BMPR2, TGFβRII, TGFβRIII, BAMBI, Snip1 and SnoN), Wnt (Wnt3a and Wnt7b) and MAP kinase (Taok1, Mapk6, Map3k2, Map3k7ip3) signal transduction pathways (miRDB).
We have shown that the gene encoding miR-19a exhibited statistically significant and consistently elevated expression whereas, the gene encoding miR-19b demonstrated statistically significant, constitutive expression during neural tube development (Tables 2 and 3). Collectively, these data support the notion that miR-19a and miR-19b, contribute a key role in regulating the differential expression of numerous target genes, known to play a defining role in neural tube morphogenesis.
MiR-16 and miR-15b
MicroRNAs miR-16-1 (miR-16) and miR-15b have been reported to function as tumor suppressors and negative regulators of cell growth (Aqeilan et al., 2010). A number of studies support the existence of a complex interplay between autophagy, cell growth and apoptosis required for mammalian neural tube development. Bcl2 and the Bcl2-like anti-apoptotic proteins BCL2-associated athanogene 5 (Bag5) and BCL2-like 2 (Blc2l2) are targets of miR-16 and miR-15a (miRDB). Indeed, Cimmino et al. (Cimmino et al., 2005) experimentally validated Bcl2 as a common target of miR-15a and miR-16, documenting these two miRNAs as pro-apoptotic. Moreover, miR-16 and miR-15b target a host of genes, including cyclin D, -D3, -E1, -M2, -T3, cdcA4, cdc37l1, cdc42se2, cdc25a, cdc14a, cdc14b, cdc23 and cdc27, that regulate cell cycle progression. Since precise balance between cell proliferation, death and apoptosis in the neural tube is necessary for neurulation, one can speculate that these miRNAs (miR-16 and miR-15b), displaying developmentally regulated expression in the neural tube in the present study, are potential candidates for orchestrating the balance of these cellular processes during morphogenesis of the neural tube (Table 2 and 3).
The neuroectodermal tissue close to the midbrain-hindbrain boundary called the isthmic organizer (IsO), is a critical secondary organizer in the developing neural tube. This region secretes signaling molecules, such as members of the fibroblast growth factor and Wnt families, which regulate cellular survival, patterning and proliferation in the midbrain-hindbrain region of the developing neural tube (Partanen, 2007). Indeed, genes encoding ligands, receptors and inhibitors of the FGF, Wnt and TGFβ/BMP/Nodal signal transduction pathways are predicted targets of miR-16 and miR-15b (miRDB). Functional verification for their developmental role comes from the observation that these miRNAs modulate Nodal/activin signaling by targeting the Nodal type II receptor Acvr2a, to restrict the size of Spemann’s organizer in early Xenopus embryos (Martello et al., 2007).
Mouse gene knockout studies have revealed that gene expression in a particular tissue does not necessarily imply that the knockout phenotype will be reflected in that tissue. By the same token, expression of particular miRNAs in specific tissues, does not necessarily imply that reduction of miRNA expression in that tissue will have a direct consequence for the phenotype of that tissue. Since miRNAs generally target multiple mRNAs, the downstream effects of silencing these mRNAs, and the proteins they encode, could easily be in other tissues with far-reaching developmental consequences.
Pathway analysis
To facilitate further investigation of cause-consequence relationships between developmentally regulated miRNAs and their predicted target genes, network/pathway reconstruction from our miRNA microarray data was performed utilizing Ingenuity Pathway Analysis (IPA; Ingenuity Systems) to map these miRNA genes and their miRNA gene targets to specific cellular pathways. This pathway analysis program identifies and constructs biological relationships among genes/proteins of interest. Two such gene networks, one involving seven (miR-15b, miR-16, miR-17, miR-124, miR-199b (miR-199a-3p), miR-210, miR-302a, and miR-140) and another connecting nine (miR-16, miR-19a, miR-20b, miR-24, miR-106a, miR-126, miR-130, miR-182, and miR-199a) differentially expressed miRNAs (whose expression was examined by microarray and subsequently verified by TaqMan QRTPCR), are presented in Figures 4 and 5. These networks are noteworthy for revealing biological processes known to be essential for neural tube development/morphogenesis (Supplementary Figs. 1 – 3 and 6). In addition, a range of signal transduction pathways operative during normal neural tube development, can be linked directly or indirectly to various miRNAs and genes present in the two networks (Supplementary Figs. 4 and 5). Such pathway analyses emphasize the potential significance of these developmentally expressed miRNAs in regulating specific cellular signal transduction systems known to orchestrate normal growth and morphogenesis of the neural tube.
Lastly, two gene networks are presented in Figures 6 and 7 that illustrate networks comprising miRNAs demonstrating enhanced (Fig. 6) or diminished (Fig. 7) expression in the GD-9.5 vs. GD-9.0 embryonic neural tube, and an array of their potential target genes – almost all of which have been documented to be indispensable for neural tube development. Moreover, altered expression of several of these target genes has been associated with syndromes involving dysmorphologies of the neural tube. These two gene regulatory networks convincingly highlight the potential significance of the differentially expressed miRNAs in orchestrating the complexity of gene expression which occurs during neural tube ontogenesis.
CONCLUSIONS
The present study is the first to conduct a comprehensive analysis of miRNA expression and identify candidate target genes in the developing mammalian neural tube. Differential expression patterns of miRNAs during three key stages of neurulation have been identified as have the most biologically relevant miRNA target gene candidates. Elucidation of embryonic neural tube miRNA signatures, and their changing patterns, during cranial neurulation, coupled with a bioinformatics-based modeling of associated target genes and signaling pathways, facilitates the identification of known and unique regulatory networks governing normal neural tube morphogenesis and broadens our understanding concerning the etiology of neural tube defects.
Supplementary Material
Supplementary Data (Excel) File #1: Target Analysis_Selected miRNAs_List 01
Supplementary Data (Excel) File #2: Target Analysis_Selected miRNAs_List 02
Supplementary Table 1: Testing for MicroRNAs Differential Expression in GD-9.0 vs. GD-8.5 Neural Tube Tissue.
Supplementary Table 2: Testing for MicroRNAs Differential Expression in GD-9.5 vs. GD-8.5 Neural Tube Tissue.
Supplementary Table 3: Testing for MicroRNAs Differential Expression in GD-9.5 vs. GD-9.0 Neural Tube Tissue.
Supplementary Table 4: List of MicroRNAs Belonging to Each Cluster in the Hierarchical Clusters (Figure 3).
Acknowledgments
This research was supported in part by NIH grants DE018215, HD053509, and P20 RR017702 from the COBRE program of the National Center for Research Resources.
Footnotes
AUTHOR CONTRIBUTIONS
PM was involved in project development, procurement of embryonic tissue, experimental conduct of miRNA gene expression profiling, performance of pathway analysis and drafted the manuscript. GB and SA were responsible for bioinformatic analysis of microarray data, miRNA target analysis and assisted in drafting the manuscript. MMP and RMG were responsible for overall project development, review of the data, financial administration of the project, and review/editing of the manuscript. RMG is the corresponding author. All contributing authors reviewed and approved the final copy of this manuscript.
References
- Anlar B, Sullivan KA, Feldman EL. Insulin-like growth factor-I and central nervous system development. Horm Metab Res. 1999;31(2–3):120–125. doi: 10.1055/s-2007-978708. [DOI] [PubMed] [Google Scholar]
- Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17(2):215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10(11):1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernd P. The role of neurotrophins during early development. Gene Expr. 2008;14(4):241–250. doi: 10.3727/105221608786883799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Botto LD, Moore CA, Khoury MJ, Erickson JD. Neural-tube defects. N Engl J Med. 1999;341(20):1509–1519. doi: 10.1056/NEJM199911113412006. [DOI] [PubMed] [Google Scholar]
- Campbell LR, Dayton DH, Sohal GS. Neural tube defects: a review of human and animal studies on the etiology of neural tube defects. Teratology. 1986;34(2):171–187. doi: 10.1002/tera.1420340206. [DOI] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21(5):531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro G, El-Hashash A, Guidolin D, Tiozzo C, Turcatel G, Young BM, De Langhe SP, Bellusci S, Shi W, Parnigotto PP, Warburton D. miR-17 family of microRNAs controls FGF10-mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E-Cadherin distribution. Dev Biol. 2009;333(2):238–250. doi: 10.1016/j.ydbio.2009.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A, Tranguch S, Daikoku T, Jensen K, Furneaux H, Dey SK. MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc Natl Acad Sci U S A. 2007;104(38):15144–15149. doi: 10.1073/pnas.0705917104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YT, Akinwunmi PO, Deng JM, Tam OH, Behringer RR. Generation of a Twist1 conditional null allele in the mouse. Genesis. 2007;45(9):588–592. doi: 10.1002/dvg.20332. [DOI] [PubMed] [Google Scholar]
- Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res. 2007;61(5 Pt 2):24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R, Barrier M, Ford LP. Role of miRNA and miRNA processing factors in development and disease. Birth Defects Res C Embryo Today. 2006;78(2):107–117. doi: 10.1002/bdrc.20068. [DOI] [PubMed] [Google Scholar]
- Copp AJ, Greene ND, Murdoch JN. The genetic basis of mammalian neurulation. Nat Rev Genet. 2003;4(10):784–793. doi: 10.1038/nrg1181. [DOI] [PubMed] [Google Scholar]
- Darnell DK, Kaur S, Stanislaw S, Konieczka JH, Yatskievych TA, Antin PB. MicroRNA expression during chick embryo development. Dev Dyn. 2006;235(11):3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- Deo M, Yu JY, Chung KH, Tippens M, Turner DL. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev Dyn. 2006;235(9):2538–2548. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- Dono R, Texido G, Dussel R, Ehmke H, Zeller R. Impaired cerebral cortex development and blood pressure regulation in FGF-2-deficient mice. EMBO J. 1998;17(15):4213–4225. doi: 10.1093/emboj/17.15.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge SL, Ray S, Li S, Hamblet NS, Lijam N, Tsang M, Greer J, Kardos N, Wang J, Sussman DJ, Chen P, Wynshaw-Boris A. Murine dishevelled 3 functions in redundant pathways with dishevelled 1 and 2 in normal cardiac outflow tract, cochlea, and neural tube development. PLoS Genet. 2008;4(11):e1000259. doi: 10.1371/journal.pgen.1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filmus J, Capurro M, Rast J. Glypicans. Genome Biol. 2008;9(5):224. doi: 10.1186/gb-2008-9-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JE, Srivastava D. MicroRNAs: opening a new vein in angiogenesis research. Sci Signal. 2009;2(52):pe1. doi: 10.1126/scisignal.252pe1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27(2–3):126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Fournier-Thibault C, Blavet C, Jarov A, Bajanca F, Thorsteinsdottir S, Duband JL. Sonic hedgehog regulates integrin activity, cadherin contacts, and cell polarity to orchestrate neural tube morphogenesis. J Neurosci. 2009;29(40):12506–12520. doi: 10.1523/JNEUROSCI.2003-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George TM, Speer MC. Genetic and embryological approaches to studies of neural tube defects: a critical review. NTD Collaborative Group. Neurol Res. 2000;22(1):117–122. doi: 10.1080/01616412.2000.11741046. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308(5723):833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Gripp KW, Wotton D, Edwards MC, Roessler E, Ades L, Meinecke P, Richieri-Costa A, Zackai EH, Massague J, Muenke M, Elledge SJ. Mutations in TGIF cause holoprosencephaly and link NODAL signalling to human neural axis determination. Nat Genet. 2000;25(2):205–208. doi: 10.1038/76074. [DOI] [PubMed] [Google Scholar]
- Handl J, Knowles J, Kell DB. Computational cluster validation in post-genomic data analysis. Bioinformatics. 2005;21(15):3201–3212. doi: 10.1093/bioinformatics/bti517. [DOI] [PubMed] [Google Scholar]
- Herz J, Bock HH. Lipoprotein receptors in the nervous system. Annu Rev Biochem. 2002;71:405–434. doi: 10.1146/annurev.biochem.71.110601.135342. [DOI] [PubMed] [Google Scholar]
- Hoesel B, Bhujabal Z, Przemeck GK, Kurz-Drexler A, Weisenhorn DM, Angelis MH, Beckers J. Combination of in silico and in situ hybridisation approaches to identify potential Dll1 associated miRNAs during mouse embryogenesis. Gene Expr Patterns. 2010;10(6):265–273. doi: 10.1016/j.gep.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Hogan KA, Ambler CA, Chapman DL, Bautch VL. The neural tube patterns vessels developmentally using the VEGF signaling pathway. Development. 2004;131(7):1503–1513. doi: 10.1242/dev.01039. [DOI] [PubMed] [Google Scholar]
- Hosako H, Martin GS, Barrier M, Chen YA, Ivanov IV, Mirkes PE. Gene and microRNA expression in p53-deficient day 8.5 mouse embryos. Birth Defects Res A Clin Mol Teratol. 2009;85(6):546–555. doi: 10.1002/bdra.20565. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hunt S, Jones AV, Hinsley EE, Whawell SA, Lambert DW. MicroRNA-124 suppresses oral squamous cell carcinoma motility by targeting ITGB1. FEBS Lett. 585(1):187–192. doi: 10.1016/j.febslet.2010.11.038. [DOI] [PubMed] [Google Scholar]
- James JM, Gewolb C, Bautch VL. Neurovascular development uses VEGF-A signaling to regulate blood vessel ingression into the neural tube. Development. 2009;136(5):833–841. doi: 10.1242/dev.028845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungbluth S, Koentges G, Lumsden A. Coordination of early neural tube development by BDNF/trkB. Development. 1997;124(10):1877–1885. doi: 10.1242/dev.124.10.1877. [DOI] [PubMed] [Google Scholar]
- Kakutani T, Jeddeloh JA, Flowers SK, Munakata K, Richards EJ. Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc Natl Acad Sci U S A. 1996;93(22):12406–12411. doi: 10.1073/pnas.93.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman L, Rousseeuw PJ. Finding Groups in Data: An Introduction to Cluster Analysis. New York: Wiley; 1990. [Google Scholar]
- Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22(3):165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24(4):857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang C, Xiao Y, Yang L, Chen Q, Wang Z, Conway SJ, Chen Y. Intragenic deletion of Tgif causes defects in brain development. Hum Mol Genet. 2006;15(24):3508–3519. doi: 10.1093/hmg/ddl427. [DOI] [PubMed] [Google Scholar]
- Kuida K, Haydar TF, Kuan CY, Gu Y, Taya C, Karasuyama H, Su MS, Rakic P, Flavell RA. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell. 1998;94(3):325–337. doi: 10.1016/s0092-8674(00)81476-2. [DOI] [PubMed] [Google Scholar]
- Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev. 2001;81(2):807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- le Roux I, Lewis J, Ish-Horowicz D. Notch activity is required to maintain floorplate identity and to control neurogenesis in the chick hindbrain and spinal cord. Int J Dev Biol. 2003;47(4):263–272. [PubMed] [Google Scholar]
- Lee CT, Risom T, Strauss WM. MicroRNAs in mammalian development. Birth Defects Res C Embryo Today. 2006;78(2):129–139. doi: 10.1002/bdrc.20072. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. Embo J. 2002;21(17):4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YB, Bantounas I, Lee DY, Phylactou L, Caldwell MA, Uney JB. Twist-1 regulates the miR-199a/214 cluster during development. Nucleic Acids Res. 2009;37(1):123–128. doi: 10.1093/nar/gkn920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Takakura N, Oike Y, Imanaka T, Araki K, Suda T, Kaname T, Kondo T, Abe K, Yamamura K. Defective smooth muscle development in qkI-deficient mice. Dev Growth Differ. 2003;45(5–6):449–462. doi: 10.1111/j.1440-169x.2003.00712.x. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Maes OC, Sarojini H, Wang E. Stepwise up-regulation of microRNA expression levels from replicating to reversible and irreversible growth arrest states in WI-38 human fibroblasts. J Cell Physiol. 2009;221(1):109–119. doi: 10.1002/jcp.21834. [DOI] [PubMed] [Google Scholar]
- Maller Schulman BR, Liang X, Stahlhut C, DelConte C, Stefani G, Slack FJ. The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle. 2008;7(24):3935–3942. doi: 10.4161/cc.7.24.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Wang J, Liu B, Pan W, Farr GH, 3rd, Flynn C, Yuan H, Takada S, Kimelman D, Li L, Wu D. Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol Cell. 2001;7(4):801–809. doi: 10.1016/s1097-2765(01)00224-6. [DOI] [PubMed] [Google Scholar]
- Martello G, Zacchigna L, Inui M, Montagner M, Adorno M, Mamidi A, Morsut L, Soligo S, Tran U, Dupont S, Cordenonsi M, Wessely O, Piccolo S. MicroRNA control of Nodal signalling. Nature. 2007;449(7159):183–188. doi: 10.1038/nature06100. [DOI] [PubMed] [Google Scholar]
- Martinsen BJ, Bronner-Fraser M. Neural crest specification regulated by the helix-loop-helix repressor Id2. Science. 1998;281(5379):988–991. doi: 10.1126/science.281.5379.988. [DOI] [PubMed] [Google Scholar]
- McDougall K, Kubu C, Verdi JM, Meakin SO. Developmental expression patterns of the signaling adapters FRS-2 and FRS-3 during early embryogenesis. Mech Dev. 2001;103(1–2):145–148. doi: 10.1016/s0925-4773(01)00337-9. [DOI] [PubMed] [Google Scholar]
- Meister G. miRNAs get an early start on translational silencing. Cell. 2007;131(1):25–28. doi: 10.1016/j.cell.2007.09.021. [DOI] [PubMed] [Google Scholar]
- Mineno J, Okamoto S, Ando T, Sato M, Chono H, Izu H, Takayama M, Asada K, Mirochnitchenko O, Inouye M, Kato I. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34(6):1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy N, Mwizerwa O, Yaniv K, Carmel L, Pieretti-Vanmarcke R, Weinstein BM, Goldstein AM. Endothelial cells promote migration and proliferation of enteric neural crest cells via beta1 integrin signaling. Dev Biol. 2009;330(2):263–272. doi: 10.1016/j.ydbio.2009.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Berg LK, Christian JL. Dissection of inhibitory Smad proteins: both N- and C-terminal domains are necessary for full activities of Xenopus Smad6 and Smad7. Mech Dev. 2001;100(2):251–262. doi: 10.1016/s0925-4773(00)00533-5. [DOI] [PubMed] [Google Scholar]
- Niederkofler V, Salie R, Sigrist M, Arber S. Repulsive guidance molecule (RGM) gene function is required for neural tube closure but not retinal topography in the mouse visual system. J Neurosci. 2004;24(4):808–818. doi: 10.1523/JNEUROSCI.4610-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykjaer A, Willnow TE. The low-density lipoprotein receptor gene family: a cellular Swiss army knife? Trends Cell Biol. 2002;12(6):273–280. doi: 10.1016/s0962-8924(02)02282-1. [DOI] [PubMed] [Google Scholar]
- O’Rourke JR, Swanson MS, Harfe BD. MicroRNAs in mammalian development and tumorigenesis. Birth Defects Res C Embryo Today. 2006;78(2):172–179. doi: 10.1002/bdrc.20071. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Pan Q, Chegini N. MicroRNA signature and regulatory functions in the endometrium during normal and disease states. Semin Reprod Med. 2008;26(6):479–493. doi: 10.1055/s-0028-1096128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pani L, Horal M, Loeken MR. Rescue of neural tube defects in Pax-3-deficient embryos by p53 loss of function: implications for Pax-3- dependent development and tumorigenesis. Genes Dev. 2002;16(6):676–680. doi: 10.1101/gad.969302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parathath SR, Mainwaring LA, Fernandez LA, Campbell DO, Kenney AM. Insulin receptor substrate 1 is an effector of sonic hedgehog mitogenic signaling in cerebellar neural precursors. Development. 2008;135(19):3291–3300. doi: 10.1242/dev.022871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partanen J. FGF signalling pathways in development of the midbrain and anterior hindbrain. J Neurochem. 2007;101(5):1185–1193. doi: 10.1111/j.1471-4159.2007.04463.x. [DOI] [PubMed] [Google Scholar]
- Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68(20):8191–8194. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- Prater MR, Johnson VJ, Germolec DR, Luster MI, Holladay SD. Maternal treatment with a high dose of CpG ODN during gestation alters fetal craniofacial and distal limb development in C57BL/6 mice. Vaccine. 2006;24(3):263–271. doi: 10.1016/j.vaccine.2005.07.105. [DOI] [PubMed] [Google Scholar]
- Rao Y, Lee Y, Jarjoura D, Ruppert AS, Liu CG, Hsu JC, Hagan JP. A comparison of normalization techniques for microRNA microarray data. Stat Appl Genet Mol Biol. 2008;7(1):Article22. doi: 10.2202/1544-6115.1287. [DOI] [PubMed] [Google Scholar]
- Richman JM, Tickle C. Epithelia are interchangeable between facial primordia of chick embryos and morphogenesis is controlled by the mesenchyme. Dev Biol. 1989;136(1):201–210. doi: 10.1016/0012-1606(89)90142-5. [DOI] [PubMed] [Google Scholar]
- Rogner UC, Danoy P, Matsuda F, Moore GE, Stanier P, Avner P. SNPs in the CpG island of NAP1L2: a possible link between DNA methylation and neural tube defects? Am J Med Genet. 2002;110(3):208–214. doi: 10.1002/ajmg.10453. [DOI] [PubMed] [Google Scholar]
- Ross JS, Carlson JA, Brock G. miRNA: the new gene silencer. Am J Clin Pathol. 2007;128(5):830–836. doi: 10.1309/2JK279BU2G743MWJ. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Nguyen V, Palma V. The emergent design of the neural tube: prepattern, SHH morphogen and GLI code. Curr Opin Genet Dev. 2003;13(5):513–521. doi: 10.1016/j.gde.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Sabapathy K, Jochum W, Hochedlinger K, Chang L, Karin M, Wagner EF. Defective neural tube morphogenesis and altered apoptosis in the absence of both JNK1 and JNK2. Mech Dev. 1999;89(1–2):115–124. doi: 10.1016/s0925-4773(99)00213-0. [DOI] [PubMed] [Google Scholar]
- Saetrom P, Snove O, Jr, Rossi JJ. Epigenetics and microRNAs. Pediatr Res. 2007;61(5 Pt 2):17R–23R. doi: 10.1203/pdr.0b013e318045760e. [DOI] [PubMed] [Google Scholar]
- Sakurai K, Furukawa C, Haraguchi T, Inada KI, Shiogama K, Tagawa T, Fujita S, Ueno Y, Ogata A, Ito M, Tsutsumi Y, Iba H. microRNAs miR-199a-5p and -3p target the Brm subunit of SWI/SNF to generate a double-negative feedback loop in a variety of human cancers. Cancer Res. doi: 10.1158/0008-5472.CAN-10-2345. [DOI] [PubMed] [Google Scholar]
- Samad TA, Rebbapragada A, Bell E, Zhang Y, Sidis Y, Jeong SJ, Campagna JA, Perusini S, Fabrizio DA, Schneyer AL, Lin HY, Brivanlou AH, Attisano L, Woolf CJ. DRAGON, a bone morphogenetic protein co-receptor. J Biol Chem. 2005;280(14):14122–14129. doi: 10.1074/jbc.M410034200. [DOI] [PubMed] [Google Scholar]
- Schmidt MH, Bicker F, Nikolic I, Meister J, Babuke T, Picuric S, Muller-Esterl W, Plate KH, Dikic I. Epidermal growth factor-like domain 7 (EGFL7) modulates Notch signalling and affects neural stem cell renewal. Nat Cell Biol. 2009;11(7):873–880. doi: 10.1038/ncb1896. [DOI] [PubMed] [Google Scholar]
- Schofield J, Isaac A, Golovleva I, Crawley A, Goodwin G, Tickle C, Brickell P. Expression of Drosophila trithorax-group homologues in chick embryos. Mech Dev. 1999;80(1):115–118. doi: 10.1016/s0925-4773(98)00207-x. [DOI] [PubMed] [Google Scholar]
- Shen WF, Hu YL, Uttarwar L, Passegue E, Largman C. MicroRNA-126 regulates HOXA9 by binding to the homeobox. Mol Cell Biol. 2008;28(14):4609–4619. doi: 10.1128/MCB.01652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK. miRNAs: from neurogeneration to neurodegeneration. Pharmacogenomics. 2007;8(8):971–978. doi: 10.2217/14622416.8.8.971. [DOI] [PubMed] [Google Scholar]
- Smyth G. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21(9):2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- Song L, Tuan RS. MicroRNAs and cell differentiation in mammalian development. Birth Defects Res C Embryo Today. 2006;78(2):140–149. doi: 10.1002/bdrc.20070. [DOI] [PubMed] [Google Scholar]
- Sun Y, Bai Y, Zhang F, Wang Y, Guo Y, Guo L. miR-126 inhibits non-small cell lung cancer cells proliferation by targeting EGFL7. Biochem Biophys Res Commun. 391(3):1483–1489. doi: 10.1016/j.bbrc.2009.12.098. [DOI] [PubMed] [Google Scholar]
- Takada S, Berezikov E, Yamashita Y, Lagos-Quintana M, Kloosterman WP, Enomoto M, Hatanaka H, Fujiwara S, Watanabe H, Soda M, Choi YL, Plasterk RH, Cuppen E, Mano H. Mouse microRNA profiles determined with a new and sensitive cloning method. Nucleic Acids Res. 2006;34(17):e115. doi: 10.1093/nar/gkl653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Theiler K. The House Mouse. Springer-Verlag; 1989. [Google Scholar]
- Trovato M, D’Armiento M, Lavra L, Ulivieri A, Dominici R, Vitarelli E, Grosso M, Vecchione R, Barresi G, Sciacchitano S. Expression of p53/HGF/c-met/STAT3 signal in fetuses with neural tube defects. Virchows Arch. 2007;450(2):203–210. doi: 10.1007/s00428-006-0356-5. [DOI] [PubMed] [Google Scholar]
- van der Put NM, Steegers-Theunissen RP, Frosst P, Trijbels FJ, Eskes TK, van den Heuvel LP, Mariman EC, den Heyer M, Rozen R, Blom HJ. Mutated methylenetetrahydrofolate reductase as a risk factor for spina bifida. Lancet. 1995;346(8982):1070–1071. doi: 10.1016/s0140-6736(95)91743-8. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28(5):1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24(3):325–332. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- Wang Y, Stricker HM, Gou D, Liu L. MicroRNA: past and present. Front Biosci. 2007;12:2316–2329. doi: 10.2741/2234. [DOI] [PubMed] [Google Scholar]
- Whelan JT, Ludwig DL, Bertrand FE. HoxA9 induces insulin-like growth factor-1 receptor expression in B-lineage acute lymphoblastic leukemia. Leukemia. 2008;22(6):1161–1169. doi: 10.1038/leu.2008.57. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309(5732):310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Williams R, Lendahl U, Lardelli M. Complementary and combinatorial patterns of Notch gene family expression during early mouse development. Mech Dev. 1995;53(3):357–368. doi: 10.1016/0925-4773(95)00451-3. [DOI] [PubMed] [Google Scholar]
- Willnow TE, Hilpert J, Armstrong SA, Rohlmann A, Hammer RE, Burns DK, Herz J. Defective forebrain development in mice lacking gp330/megalin. Proc Natl Acad Sci U S A. 1996;93(16):8460–8464. doi: 10.1073/pnas.93.16.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Wang X, Du Z, Li N. Identification of microRNAs from different tissues of chicken embryo and adult chicken. FEBS Lett. 2006;580(15):3610–3616. doi: 10.1016/j.febslet.2006.05.044. [DOI] [PubMed] [Google Scholar]
- Yamada M, Ohnishi H, Sano S, Nakatani A, Ikeuchi T, Hatanaka H. Insulin receptor substrate (IRS)-1 and IRS-2 are tyrosine-phosphorylated and associated with phosphatidylinositol 3-kinase in response to brain-derived neurotrophic factor in cultured cerebral cortical neurons. J Biol Chem. 1997;272(48):30334–30339. doi: 10.1074/jbc.272.48.30334. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wu J. Small RNAs and development. Med Sci Monit. 2006;12(7):RA125–129. [PubMed] [Google Scholar]
- Zhang B, Wang Q, Pan X. MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol. 2007;210(2):279–289. doi: 10.1002/jcp.20869. [DOI] [PubMed] [Google Scholar]
- Zhang J, Du YY, Lin YF, Chen YT, Yang L, Wang HJ, Ma D. The cell growth suppressor, mir-126, targets IRS-1. Biochem Biophys Res Commun. 2008;377(1):136–140. doi: 10.1016/j.bbrc.2008.09.089. [DOI] [PubMed] [Google Scholar]
- Zhao JJ, Sun DG, Wang J, Liu SR, Zhang CY, Zhu MX, Ma X. Retinoic acid downregulates microRNAs to induce abnormal development of spinal cord in spina bifida rat model. Childs Nerv Syst. 2008;24(4):485–492. doi: 10.1007/s00381-007-0520-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data (Excel) File #1: Target Analysis_Selected miRNAs_List 01
Supplementary Data (Excel) File #2: Target Analysis_Selected miRNAs_List 02
Supplementary Table 1: Testing for MicroRNAs Differential Expression in GD-9.0 vs. GD-8.5 Neural Tube Tissue.
Supplementary Table 2: Testing for MicroRNAs Differential Expression in GD-9.5 vs. GD-8.5 Neural Tube Tissue.
Supplementary Table 3: Testing for MicroRNAs Differential Expression in GD-9.5 vs. GD-9.0 Neural Tube Tissue.
Supplementary Table 4: List of MicroRNAs Belonging to Each Cluster in the Hierarchical Clusters (Figure 3).