Abstract
Genetic variation in CLU encoding clusterin has been associated with Alzheimer's disease (AD) through replicated genome-wide studies, but the underlying mechanisms remain unknown. Following earlier reports that tightly regulated CLU alternative transcripts have different functions, we tested CLU single-nucleotide polymorphisms (SNPs), including those associated with AD for quantitative effects on individual alternative transcripts. In 190 temporal lobe samples without pathology, we found that the risk allele of the AD-associated SNP rs9331888 increases the relative abundance of transcript NM_203339 (P=4.3 × 10−12). Using an independent set of 115 AD and control samples, we replicated this result (P=0.0014) and further observed that multiple CLU transcripts are at higher levels in AD compared with controls. The AD SNP rs9331888 is located in the first exon of NM_203339 and therefore, it is a functional candidate for the observed effects. We tested this hypothesis by in vitro dual luciferase assays using SK-N-SH cells and mouse primary cortical neurons and found allelic effects on enhancer function, consistent with our results on post-mortem human brain. These results suggest a biological mechanism for the genetic association of CLU with AD risk and indicate that rs9331888 is one of the functional DNA variants underlying this association.
Keywords: Alzheimer's dementia, clusterin, CLU, transcription, splicing, gene regulation
Introduction
Two recent genome-wide association studies independently identified CLU as a risk gene for Alzheimer's disease (AD).1, 2 Follow-up studies and meta analyses have replicated these results, although the strongest associated variant sometimes differed.3, 4, 5, 6, 7 Efforts to identify functional variations through exon sequencing and examining effects of single-nucleotide polymorphisms (SNPs) on CLU expression have not yet provided a functional link between the associated polymorphisms and AD, but they have excluded the involvement of common coding variation.8 The same study examined the effect of SNPs on the gene's expression with negative results; however, the microarray platform used did not examine individual splice variants.
Clusterin, also known as apolipoprotein J, is a glycoprotein first identified in 19889 and discussed as a candidate gene for AD for more than 15 years.10, 11 Its multiple functions include roles in apoptosis, complement regulation, lipid transport, sperm maturation, endocrine secretion, membrane protection, promotion of cell interactions and as a chaperone.12, 13, 14, 15, 16 Secreted soluble and nuclear forms of clusterin have been described and their production is likely regulated by use of alternative transcription start sites17 or alternative splicing.13 This is achieved through use of discrete translation initiation sites, alternatively introducing an endoplasmic reticulum-targeting signal upstream of a nuclear localization signal. The nuclear form of clusterin is specifically induced in epithelial cells by tumor growth factor-β,17 whereas in prostate cells, different CLU isoforms have been shown to have different responses to androgens and opposing functions with regard to apoptosis.18, 19
The importance of CLU alternative splicing on its function led us to the hypothesis that the reported association with AD, although it is shown not to have a significant impact on the overall transcript levels as measured by microarrays,8 might reflect a disruption of the balance between transcripts. We tested our hypothesis on a set of 190 temporal lobe samples without brain pathology (controls) and followed up in another set of 115 temporal lobe samples from AD cases and controls.
Materials and methods
Samples
Tissue samples were acquired from the Harvard Brain Tissue Resource Center (HBTRC) and the Johns Hopkins Brain Resource Center, dissected from the superior temporal lobe (Brodmann area 22) of flash-frozen brain slices from donors, without macroscopically visible brain pathology or with definite AD (replication set), and stored at −80 °C. Detailed information on all individual samples including age at death, sex and post-mortem tissue collection interval (PMI) are provided in Supplementary Table 1. Genomic DNA was extracted from 10 mg of tissue using the Gentra Puregene Tissue Kit (Qiagen, Valencia, CA, USA) following manufacturer's protocol. RNA was extracted from 30 mg of tissue using the RNeasy Lipid Tissue Mini Kit (Qiagen). Reverse transcription reactions on total RNA were performed using GeneAmp RNA PCR Kit (Applied Biosystems, Carlsbad, CA, USA) and random hexamer primers following standard protocols. All real-time PCR experiments on each set of samples (discovery or replication) were done on the same set of reverse-transcribed RNAs to assure template consistency across transcripts minimizing experimental noise.
Genotyping
Genotyping was performed at the Johns Hopkins SNP center on a custom SNP panel, using the Illumina GoldenGate platform (Illumina Inc., San Diego, CA, USA). We attempted 76 SNPs, and the SNP center released 70 SNPs after considering adequate clustering definitions, SNP call rate and intensity. Two released SNPs were flagged for atypical clustering and we removed them from analysis. Among the SNPs not released was rs11136000, which we wanted to analyze, as it is the most consistently associated SNP with AD. We used the Beadstudio software (lllumina Inc.) and found that the separation of alleles was clear (Supplementary Figure 1). Nevertheless, we re-genotyped this SNP using an ApoI restriction enzyme digestion assay and after confirming the genotypes, we included them in our analysis (see primers in Supplementary Table 2). Despite good quality data, we also decided to re-genotype and confirm rs9331888 by nucleotide sequencing, as it is important to our conclusions. In the replication sample, rs9331888 was genotyped by BslI restriction digestion, using the primers shown in Supplementary Table 2. All restriction enzyme digestion assays included control restriction sites that confirmed complete digestion. All SNPs are shown in Supplementary Table 3 with their location, genotype frequencies, Hardy–Weinberg equilibrium and allele identities.
Real-time PCR
Real-time PCR reactions were performed in triplicate using the SYBR Green qPCR Detection System (Invitrogen, Carlsbad, CA, USA). SYBR Green was preferred to the TaqMan chemistry (Applied Biosystems, Carlsbad, CA, USA), because it provides more flexibility in primer design, which was necessary for splice variant-specific assays. All PCR products were designed with one primer's 3′ end, overlapping by a few nucleotides the isoform-specific splice junction. All primer sequences are reported in Supplementary Table 2. Amplicon sequences were verified by Sanger sequencing. Gel electrophoresis and melting curves further confirmed that a single product was amplified and measured by the assays. Samples in triplicate in the discovery set were run on two separate 384-well plates, where they were randomly distributed (as shown in Supplementary Table 1). Plate identity was accounted for in the analyses as described below. Real-time measurements were made on an Applied Biosystems 7900HT sequence detection system and measurements were converted to relative quantities using six serial twofold dilutions in triplicate. Triplicates for all samples and standards were examined for outlier measurements that were removed if present. On the basis of the literature on optimal normalization controls, we chose three genes, ACTB, POLR2F and MRIP, to control for variations in RNA input and reverse transcription efficiency.20, 21 We performed real-time experiments for all three, compared their inter-sample variability and their pair-wise correlations, and used for normalization the mean of the least variable and most correlated pair, ACTB and MRIP. The normalized values were log transformed (base 2) to achieve a normal distribution before proceeding to statistical analysis.
Statistical analyses
SNP genotypes were coded in a quantitative manner (0, 1 or 2 alleles B—see Supplementary Table 3 for allele identities). Principal component (PC) analyses of the ancestry informative SNP marker SNPs were performed in R, using the ‘principal()' function in the ‘psych' package (Revelle, W. 2011, Northwestern University (Chicago, IL, USA), R package version 1.0–95), replacing missing genotypes with the population's mean for the corresponding SNP. Statistical analyses of normalized log-transformed expression data were also performed in R, using the ‘glm' function for fitting generalized linear models with formulas as described in the text, an identity link function and a Gaussian distribution. Log-transformed transcript measurements were tested for normality of the distributions, using the Kolmogorov–Smirnov test. All were normally distributed (P>0.1) in the discovery data set. In the replication set, small deviations from normality were seen for transcripts NM_001171138 and CR617497 (P=0.03 and P=0.01, respectively), not significant after correcting for six tests. Statistical comparisons of intensities between constructs in the dual luciferase reporter assays were performed by Student's t-test, comparing the two alleles of each construct across the four observations from quadruplicate experiments. All results are reported without corrections for multiple comparisons, deemed unnecessary, given the robustness of all P-values for the reported main effects and the consistency of results, both in the original analyses and in replications.
Luciferase reporter assays
Constructs: The upstream (U), short (S) and downstream (D) inserts (shown in Figure 1c were PCR amplified from genomic DNA from individuals homozygous for the reference or risk alleles for rs9331888 (primers in Supplementary Table 2). Sequencing revealed no other variation in the amplified sequence. Fragments were amplified using PfuTurbo Polymerase (Stratagene, Santa Clara, CA, USA) high-fidelity taq, and A' overhangs were added by incubating the products for 5 min with Taq DNA polymerase (Invitrogen) and excess deoxyadenosine triphosphate. The amplicons were TA cloned into pCR 8/GW/TOPO (lnvitrogen) entry vector containing attL1 and attL2 recombination sites. Inserts were subcloned via recombination, using the Gateway LR Clonase Enzyme Mix (Invitrogen) to a pDSma_promoter vector.22 This plasmid is a pGL3 firefly luciferase reporter vector containing an SV40 promoter (Promega, Madison, WI, USA), modified to contain the Gateway cassette containing the attA and attB recombination sites as described in Grice et al.22 Inserts were verified by Sanger sequencing using the commercial primer RVprimer3 (Promega) and pGLR 5′ (Supplementary Table 2). Neuroblastoma (SK-N-SH, ATCC no. HTB-11) cells were grown in an ATCC-suggested medium (http://www.atcc.org), without antibiotics. Approximately 0.8 × 105 cells were plated 24 h before transfection for cells to reach 90–95% confluency. Primary cortical neuron cultures were prepared from day 16–18 embryonic C57BL/6 mice, following established protocols.23 Animal protocols were approved by Johns Hopkins University Animal Care and Use Committee. Primary cortical neurons were plated in 24-well plates at a density of 2 × 105 in 1 ml of growth medium per well. The neurons were switched to 500 μl antibiotic-free medium on the third day in vitro and transfected on day 4. SK-N-SH and primary cortical neurons were co-transfected (Lipofectamine 2000, Invitrogen) using 0.8 μg of plasmid DNA and 0.08 μg phRL-SV40 control renilla plasmid, using standard 24-well protocol. Medium was replaced 5 h post transfection. Dual luciferase assays (Promega) were performed in quadruplicate 24 h post transfection, following the manufacturers standard protocol (Tecan Genios Microplate Reader, Männedorf, Switzerland). Relative luciferase units were calculated by dividing the firefly luciferase values by the renilla control values for each transfection reaction.
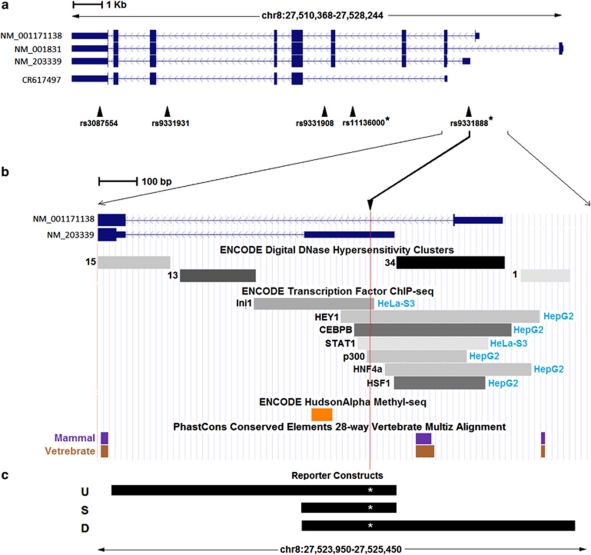
Figure 1.
(a) CLU alternative transcripts from reference sequence (RefSeq) and UCSC examined in this study. Single-nucleotide polymorphisms (SNPs) genotyped within the gene are shown and SNPs reported in recent genome-wide association studies (GWAS) are marked with an asterisk. (b) Enlargement of the region around rs9331888. The SNP location is marked by a vertical red line. Selected tracks from the UCSC genome browser ENCODE data, as well as phylogenetic conservation data are shown. For clarifications, see the UCSC genome database website. (c) The sequences inserted into reporter constructs are shown named U (upstream) S (short) and D (downstream). Each sequence was made to carry either a reference or a risk allele at rs9331888, shown here by a white asterisk and found to influence Alzheimer's disease (AD) risk in the Lambert et al.2 GWAS.
Results
Three different CLU splice forms with different transcription start sites (Figure 1a) are reported in RefSeq24 (NM_001831, NM_203339 and NM_001171138) and can be differentiated using sequence-specific primers for quantitative real-time PCR. All RefSeq transcripts include coding sequence for both the endoplasmic reticulum-targeting signal and the nuclear localization signal, but whether they all use the upstream translation start site and include both signals in the protein is unknown. In our experiments using brain RNA, we did not observe the transcript lacking exon 2 described in a breast cancer cell line,13 but we found a transcript reported in the UCSC genome browser (GenBank acc# CR617497) lacking exons 1, 3 and 4, thus missing both the endoplasmic reticulum-targeting signal and nuclear localization signal. We designed specific primers (Supplementary Table 2) and successfully amplified NM_203339, NM_001171138 and CR617497. We confirmed that single and specific amplicons were amplified from each transcript by Sanger sequencing, agarose gel electrophoresis and DNA melting curves. Using real-time PCR, we quantified the transcripts in tissue from the superior temporal lobe of 190 individuals without gross brain pathogy (‘controls') obtained from the HBTRC (see Supplementary Table 2 for sample details). We extracted brain DNA from the same individuals and successfully genotyped 42 HapMap SNPs in and around CLU, chosen to be inter-correlated at r2⩽0.8 and including the associated SNPs reported in the two first genome-wide association studies reporting CLU.1, 2 Further, we genotyped 27 ancestry informative SNP markers,25 as the HBTRC did not retain ancestry information (Supplementary Table 3 describes all study SNPs).
We first examined the ancestry informative SNP markers in a PC analysis and identified two outlier individuals for PC-1, likely of African ancestry, who were removed from further analysis. No additional PCs showed outliers. Using log-transformed transcript quantity as an outcome, we applied a generalized linear model including age, sex, PMI, PCR plate ID (identity) (to account for plate effects as we used two 384-well real-time plates) and the first PC of the ancestry informative SNP markers to account for possible residual admixture. We found a highly significant increase for all transcripts with increasing age (about 1% increase per year, all P-values <0.0002, see Table 1), consistent with our recent report of significant overlap between genes changing expression in AD and with normal aging.26 Transcript CR617497 was significantly lower in males (∼17%, P=0.001) and NM_203339 approached significance for an approximate 13% decrease in males (P=0.058). A significant plate effect was identified only for transcript NM_001171138 and no effects of PMI were identified. Plate ID and PMI were both included in all analyses to account for possible small effects, regardless of significance. Transcript expression levels showed strong positive inter-correlations persisting after correcting for all the effects in our model, possibly suggesting significant common regulation (all P<10−4). We then included each SNP genotype sequentially as a predictor in the model. The results for the three transcripts for SNPs showing at least nominally significant effects on transcription are shown on the left side of Table 2. The SNP variants had little or no effect on NM_001171138 and CR617497. In contrast, NM_203339 showed multiple, nominally significant correlations with genotypes, the strongest with rs9331888. This SNP, located in exon 1 of NM_203339 (Figure 1b) ranked third in the recent genome-wide association studies on Caucasians by Lambert et al.2 and was the only one of the three SNPs that replicated in an independent study of a Chinese population,6 although contradicting results have been reported,27 providing strongest support for a different SNP, rs11136000. The risk alleles of rs11136000 and rs9331888 are in strong linkage disequilibrium (D'=0.96, r2=0.224 based on our data). We then examined the effect of SNP genotypes on each transcript, adjusting for the amount of the other two transcripts, which were added as predictors in the model. This analysis reflects regulation of alternative splicing by removing variability common to all transcripts and highlighting their differences. The results of this analysis (Table 2, right side) showed a highly significant effect of rs9331888 on the relative levels of NM_203339 (P=4.3 × 10−12). To test whether other less significant effects on NM_203339 observed for SNPs rs11136000, rs9331908, rs9331931, rs3087554 and rs17466684 were due to linkage disequilibrium, we analyzed each in a model that also included rs9331888. In all cases, only rs9331888 remained significant, suggesting that other SNP effects on splicing likely reflect their linkage disequilibrium with rs9331888.
Table 1. Effects of age, sex and AD (replication sample only) on each examined CLU transcript and on their relative levels. Sex (M) signifies that the reported effect is for males and Dx (Ctrl) that the reported effect is for controls, negative effects meaning lower transcription in males and controls.
|
Discovery sample |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Overall transcript quantity |
Controlled for other CLU transcripts |
|||||||||||
|
CR617497 |
NM_1171138 |
NM_203339 |
CR617497 |
NM_1171138 |
NM_203339 |
|||||||
| Estimate | P-value | Estimate | P-value | Estimate | P-value | Estimate | P-value | Estimate | P-value | Estimate | P-value | |
| Age | 0.014 | 5.62 × 10−7 | 0.010 | 9.94 × 10−7 | 0.014 | 0.0002 | 0.003 | 0.04 | 0.002 | 0.07 | −0.002 | 0.45 |
| Sex (M) | −0.26 | 0.0011 | −0.07 | 0.22 | −0.20 | 0.06 | −0.15 | 0.0012 | 0.06 | 0.11 | 0.03 | 0.67 |
| |
Replication sample |
|||||||||||
| Age | 0.010 | 0.021 | −0.002 | 0.75 | 0.004 | 0.40 | 0.009 | 0.007 | −0.006 | 0.10 | −0.001 | 0.77 |
| Sex (M) | −0.17 | 0.08 | −0.06 | 0.58 | 0.04 | 0.71 | −0.17 | 0.011 | −0.03 | 0.73 | 0.16 | 0.04 |
| Dx (Ctrl) | −0.36 | 0.0007 | −0.40 | 0.0011 | −0.53 | 4.87 x10−5 | −0.04 | 0.5904 | −0.07 | 0.48 | −0.17 | 0.06 |
Abbreviation: AD, Alzheimer's disease.
Significant P-values are shown in bold.
Table 2. SNPs with at least one P<0.05 are shown.
|
Discovery sample |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Overall transcript quantity |
Controlled for other CLU transcripts |
||||||||||||
|
CR617497 |
NM_1171138 |
NM_203339 |
CR617497 |
NM_1171138 |
NM_203339 |
||||||||
| SNP | Position | Estimate | P-value | Estimate | P-value | Estimate | P-value | Estimate | P-value | Estimate | P-value | Estimate | P-value |
| rs1873933 | 27,473,339 | 0.01 | 0.81 | −0.01 | 0.71 | −0.07 | 0.24 | 0.04 | 0.09 | 0.00 | 0.87 | −0.07 | 0.048 |
| rs2741352 | 27,473,876 | −0.04 | 0.41 | 0.01 | 0.88 | 0.03 | 0.60 | −0.05 | 0.044 | 0.01 | 0.53 | 0.06 | 0.12 |
| rs1316801 | 27,485,145 | 0.06 | 0.18 | 0.03 | 0.42 | 0.15 | 0.016 | −0.01 | 0.82 | −0.02 | 0.29 | 0.09 | 0.0187 |
| rs7012217 | 27,504,805 | −0.04 | 0.39 | −0.03 | 0.47 | −0.13 | 0.044 | 0.02 | 0.48 | 0.01 | 0.55 | −0.09 | 0.028 |
| rs17466684 | 27,508,764 | 0.01 | 0.88 | −0.02 | 0.74 | −0.18 | 0.02327 | 0.09 | 0.013 | 0.02 | 0.58 | −0.18 | 0.00017 |
| rs3087554 | 27,511,359 | −0.10 | 0.11 | −0.07 | 0.15 | 0.12 | 0.16 | −0.12 | 0.0019 | −0.06 | 0.06 | 0.24 | 4.05 × 10−6 |
| rs9331931 | 27,514,021 | −0.04 | 0.48 | −0.03 | 0.45 | −0.15 | 0.031 | 0.03 | 0.26 | 0.01 | 0.61 | −0.10 | 0.010 |
| rs9331908 | 27,519,535 | 0.06 | 0.27 | 0.04 | 0.30 | −0.18 | 0.0076 | 0.11 | 0.00020 | 0.06 | 0.015 | −0.24 | 5.56 × 10−10 |
| rs11136000a | 27,520,436 | −0.10 | 0.050 | −0.07 | 0.07 | 0.02 | 0.77 | −0.07 | 0.015 | −0.04 | 0.11 | 0.13 | 0.0012 |
| rs9331888a | 27,524,779 | 0.05 | 0.33 | 0.05 | 0.18 | −0.21 | 0.0027 | 0.12 | 0.00031 | 0.09 | 7.28 × 10−4 | −0.28 | 4.26 × 10−12 |
| rs484458 | 27,540,871 | −0.11 | 0.048 | −0.10 | 0.023 | −0.09 | 0.21 | −0.03 | 0.42 | −0.04 | 0.16 | 0.04 | 0.39 |
| rs17467992 | 27,552,600 | 0.00 | 0.96 | 0.05 | 0.19 | −0.03 | 0.66 | −0.02 | 0.51 | 0.05 | 0.017 | −0.05 | 0.17 |
| rs3824098 | 27,578,549 | −0.14 | 0.049 | −0.06 | 0.30 | −0.24 | 0.0083 | −0.02 | 0.61 | 0.04 | 0.19 | −0.12 | 0.038 |
| rs559251 | 27,595,289 | 0.05 | 0.28 | 0.03 | 0.42 | 0.14 | 0.026 | −0.02 | 0.56 | −0.02 | 0.47 | 0.09 | 0.021 |
| rs7001584 | 27,604,084 | 0.06 | 0.25 | 0.02 | 0.58 | −0.04 | 0.55 | 0.06 | 0.031 | 0.01 | 0.77 | −0.09 | 0.020 |
| rs11778402 | 27,607,553 | −0.06 | 0.16 | −0.03 | 0.42 | 0.01 | 0.82 | −0.05 | 0.039 | −0.01 | 0.75 | 0.07 | 0.041 |
|
Replication sample |
|||||||||||||
| rs9331888 | 27,524,779 | 0.04 | 0.61 | 0.01 | 0.86 | -0.16 | 0.08 | 0.11 | 0.032 | 0.08 | 0.24 | −0.19 | 0.0014 |
Abbreviations: AD. Alzheimer's disease; GWAS, genome-wide association studies; SNP, single-nucleotide polymorphism.
SNPs that were reported among the best results in recent GWAS for AD.
Significant P-values are shown in bold.
To exclude artifacts, we performed multiple quality-control steps. We excluded nucleotide variation under a PCR primer by sequencing the regions under the NM_203339-specific primers in all individuals. Additionally, we designed a new set of transcript-specific primers for NM_203339 and CR617497, and performed new quantitative PCR experiments starting from RNA, obtaining similar highly significant results. We only used CR617497 in this experiment, because the original data showed that correcting NM_203339 for just one other transcript revealed the effect almost equally well as correcting for both. We further tested for genotyping errors and confirmed by nucleotide sequencing all rs9331888 genotypes.
We proceeded to replicate our result in an independent set of samples, which included AD cases allowing us to test for possible disease-related transcript variation. This replication set included 24 controls from the HBTRC and 29 controls from the Johns Hopkins Brain Resource Center, as well as 22 and 40 AD cases, respectively. We quantified NM_203339, NM_001171138 and CR617497 as above, genotyped rs9331888 by sequencing and by BslI restriction enzyme digestion and applied a similar generalized linear model, with NM_203339 as a dependent variable, age, sex, PMI, sample source, diagnosis and rs9331888 genotype as predictors. The quantitative PCR plate variable no longer applied, as a single plate was used in this experiment. We used existing ancestry information on the Johns Hopkins Brain Resource Center samples and included only Caucasian individuals. Ancestry information was not available for HBTRC samples, but as we identified only two genetic outliers in the previous larger HBTRC sample and observed no significant effects of ethnicity, we included all HBTRC samples. The effect of rs9331888 on relative NM_203339 levels was strongly replicated (P=0.0014, Table 2). As with the initial sample, the significance of the effect on total mRNA levels was more modest, this time only suggestive (P=0.076). Cases had higher levels of all transcripts (Table 1) and suggestively higher levels of relative levels of NM_203339. After correcting for the effect of rs9331888, the difference in relative levels of NM_203339 between AD cases and controls was diminished. Many of the previously observed effects of age and sex on the various transcripts were replicated in this sample as shown in Table 1.
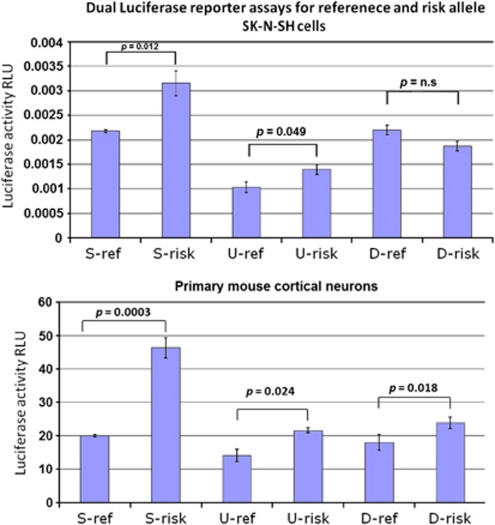
The location of rs9331888 in exon 1 of NM_203339, together with ChIP-seq and DNase hypersensitivity data from ENCODE (encyclopedia of DNA elements; see Figure 1b), suggest that this variant might be directly responsible for regulation of this alternative transcription start. We used the Promega Dual-Luciferase Reporter Assay System to test the six expression constructs shown in Figure 1c (S-ref, S-risk, U-ref, U-risk, D-ref and D-risk; ref and risk indicate the rs9331888 alleles) for enhancer activity in SK-N-SH cells and primary mouse cortical neurons. We observed significantly higher activity of the risk allele for all constructs in primary neurons and the S- and U- constructs in SK-N-SH cells (Figure 2), consistent with our post-mortem brain expression results.
Figure 2.
Dual luciferase reporter assays comparing constructs carrying reference (ref) or risk alleles in the three different constructs shown in Figure 1c in front of an SV40 promoter. Firefly, relative to renilla luciferase levels, is shown with s.e. bars based on four replicates. All constructs show significant differences between the two rs9331888 alleles in primary neuron culture and two of three also show differences in SK-N-SH cells. As expected, the risk allele shows higher activity. RLU, relative luciferase units.
As shown in Figure 1, rs9331888 lies in a region with significant evidence of regulatory potential. We used the bioinformatics program rVista 2.0 (http://rvista.dcode.org/)28 and found that the risk variant of rs9331888 eliminates binding sites for nuclear factor kappa B and early B-cell factor, whereas it generates a new binding site for heat shock factor protein-1. Pending experimental verifications, this interesting result could provide a guide for further dissection of the relationship between this SNP, this transcript and AD.
Discussion
We have shown that the minor allele of rs9331888, previously associated with increased risk of AD, is associated with increased-relative levels of NM_203339 and is likely the functional variant responsible for this effect. Given the prior genetic association results for this SNP and the distinct roles of CLU transcripts,18 we hypothesize that alternative splicing is the etiological link between rs9331888 and AD. However, the functional properties of the protein products and whether or how their production varies across the three transcripts are unclear. At least two transcripts, NM_001171138 and NM_203339, potentially contain both the endoplasmic reticulum-targeting signal and nuclear localization signal, but it is unknown whether there is preferential utilization of a specific translation start site, which could dramatically change the functional outcome.18, 19 Finally, the function of CR617497 which we were able to reliably detect in the temporal lobe transcriptome is unknown. An alternative hypothesis is that the increased risk is not the result of the different transcript functions, but rather of their specific responses to different signals, responses that might be aberrantly lost or gained for carriers of the rs9331888 risk allele, which abolishes two and introduces one new transcription factor binding site. Regardless of what the true underlying biology will turn out to be, in view of our results and the reported genome-wide association studies, clarifying the properties of these transcripts, the corresponding proteins and their regulation is of great importance for AD research.
As we mentioned in the introduction, although rs9331888 has been independently reported in two populations, it is another CLU SNP, rs11136000 that has shown overall the most consistent associations. As the two risk alleles are in near complete linkage disequilibrium with each other (D'=0.96), it is likely that the functional effect of rs9331888 is responsible for only part of the observed association of the gene with AD. Other rare or common regulatory variants might underlie the remaining risk attributed to CLU and the inconsistencies in association patterns described in the literature. Further clarification of these effects remains a significant task, which will be facilitated by this and future work, testing the new hypotheses and moving translational research forward. Together with the recent many advances in AD through new gene discovery, our understanding of the disease biology will quickly improve, hopefully leading to significant benefits for the patients and those at risk.
Acknowledgments
We thank Dr Francine Benes of the Harvard Brain Tissue resource center and Dr Juan Troncoso of the Johns Hopkins Brain Resource Center for providing brain tissue. We thank Dr Andrew McCallion for conversations, advice and expertise in multiple aspects of the project and Dr David Valle and Mariela Zeledon for providing mouse primary cortical neurons. This work was supported by NIA grants to DA and SSB (RO1AG022099 and RO1AG021804) and an award from the Neurosciences Education and Research Foundation to DA.
Author contributions.Megan Szymanski performed the large majority of experimental work and helped with data analysis and manuscript preparation. Ruihua Wang performed post-mortem tissue preparations and provided support in the daily functions of the laboratory. Susan Bassett provided clinical expertise on Alzheimer's disease and led the post-mortem collection of patient brains by the JHBRC. Dimitrios Avramopoulos supervised the experimental work, performed data analyses and prepared the manuscript assisted by the co-authors.
The authors declare no conflicts of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corneveaux JJ, Myers AJ, Allen AN, Pruzin JJ, Ramirez M, Engel A, et al. Association of CR1, CLU and PICALM with Alzheimer's disease in a cohort of clinically characterized and neuropathologically verified individuals. Hum Mol Genet. 2010;19:3295–3301. doi: 10.1093/hmg/ddq221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F, et al. Replication of CLU, CR1, and PICALM associations with alzheimer disease. Arch Neurol. 2010;67:961–964. doi: 10.1001/archneurol.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JT, Li L, Zhu QX, Zhang Q, Zhang W, Wu ZC, et al. Implication of CLU gene polymorphisms in Chinese patients with Alzheimer's disease. Clin Chim Acta. 2010;411:1516–1519. doi: 10.1016/j.cca.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Barral S, Reitz C, Medrano M, Lantigua R, et al. Identification of Novel Loci for Alzheimer Disease and Replication of CLU, PICALM, and BIN1 in Caribbean Hispanic Individuals. Arch Neurol. 2011;68:320–328. doi: 10.1001/archneurol.2010.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro RJ, Beck J, Gibbs JR, Santana I, Rossor MN, Schott JM, et al. Genetic variability in CLU and its association with Alzheimer's disease. PLoS One. 2010;5:e9510. doi: 10.1371/journal.pone.0009510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy BF, Kirszbaum L, Walker ID, d'Apice AJ. SP-40,40, a newly identified normal human serum protein found in the SC5b-9 complex of complement and in the immune deposits in glomerulonephritis. J Clin Invest. 1988;81:1858–1864. doi: 10.1172/JCI113531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlokovic BV. Cerebrovascular transport of Alzheimer's amyloid beta and apolipoproteins J and E: possible anti-amyloidogenic role of the blood-brain barrier. Life Sci. 1996;59:1483–1497. doi: 10.1016/0024-3205(96)00310-4. [DOI] [PubMed] [Google Scholar]
- Bertram L, Tanzi RE. Alzheimer disease: New light on an old CLU. Nat Rev Neurol. 2010;6:11–13. doi: 10.1038/nrneurol.2009.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp J, French LE. Clusterin: modulation of complement function. Clin Exp Immunol. 1994;97 (Suppl 2:11–14. doi: 10.1111/j.1365-2249.1994.tb06256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leskov KS, Klokov DY, Li J, Kinsella TJ, Boothman DA. Synthesis and functional analyses of nuclear clusterin, a cell death protein. J Biol Chem. 2003;278 (13:11590–11600. doi: 10.1074/jbc.M209233200. [DOI] [PubMed] [Google Scholar]
- Buttyan R, Olsson CA, Pintar J, Chang C, Bandyk M, Ng PY, et al. Induction of the TRPM-2 gene in cells undergoing programmed death. Mol Cell Biol. 1989;9:3473–3481. doi: 10.1128/mcb.9.8.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MR, Easterbrook-Smith SB. Clusterin is a secreted mammalian chaperone. Trends Biochem Sci. 2000;25:95–98. doi: 10.1016/s0968-0004(99)01534-0. [DOI] [PubMed] [Google Scholar]
- Rosenberg ME, Silkensen J. Clusterin: physiologic and pathophysiologic considerations. Int J Biochem Cell Biol. 1995;27:633–645. doi: 10.1016/1357-2725(95)00027-m. [DOI] [PubMed] [Google Scholar]
- Reddy KB, Jin G, Karode MC, Harmony JA, Howe PH. Transforming growth factor beta (TGF beta)-induced nuclear localization of apolipoprotein J/clusterin in epithelial cells. Biochemistry. 1996;35:6157–6163. doi: 10.1021/bi952981b. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zhou W, Kundu S, Jang TL, Yang X, Pins M, et al. The leader sequence triggers and enhances several functions of clusterin and is instrumental in the progression of human prostate cancer in vivo and in vitro. BJU Int. 2006;98:452–460. doi: 10.1111/j.1464-410X.2006.06263.x. [DOI] [PubMed] [Google Scholar]
- Cochrane DR, Wang Z, Muramaki M, Gleave ME, Nelson CC. Differential regulation of clusterin and its isoforms by androgens in prostate cells. J Biol Chem. 2007;282:2278–2287. doi: 10.1074/jbc.M608162200. [DOI] [PubMed] [Google Scholar]
- Hoerndli FJ, Toigo M, Schild A, Gotz J, Day PJ. Reference genes identified in SH-SY5Y cells using custom-made gene arrays with validation by quantitative polymerase chain reaction. Anal Biochem. 2004;335:30–41. doi: 10.1016/j.ab.2004.08.028. [DOI] [PubMed] [Google Scholar]
- Johansson S, Fuchs A, Okvist A, Karimi M, Harper C, Garrick T, et al. Validation of endogenous controls for quantitative gene expression analysis: application on brain cortices of human chronic alcoholics. Brain Res. 2007;1132:20–28. doi: 10.1016/j.brainres.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Grice EA, Rochelle ES, Green ED, Chakravarti A, McCallion AS. Evaluation of the RET regulatory landscape reveals the biological relevance of a HSCR-implicated enhancer. Hum Mol Genet. 2005;14:3837–3845. doi: 10.1093/hmg/ddi408. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Kamiya A, Yokota Y, Prikulis I, Kano S, Hayashi-Takagi A, et al. Disrupted-in-Schizophrenia-1 expression is regulated by beta-site amyloid precursor protein cleaving enzyme-1-neuregulin cascade. Proc Natl Acad Sci USA. 2010;107:5622–5627. doi: 10.1073/pnas.0909284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33 (Database issue:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C, Salas A, Sanchez JJ, Fondevila M, Gomez-Tato A, Alvarez-Dios J, et al. Inferring ancestral origin using a single multiplex assay of ancestry-informative marker SNPs. Forensic Sci Int Genet. 2007;1:273–280. doi: 10.1016/j.fsigen.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Avramopoulos D, Szymanski M, Wang R, Bassett S. Gene expression reveals overlap between normal aging and Alzheimer's disease genes. Neurobiol Aging. 2010. [DOI] [PMC free article] [PubMed]
- Jun G, Naj AC, Beecham GW, Wang LS, Buros J, Gallins PJ, et al. Meta-analysis confirms CR1, CLU, and PICALM as alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch Neurol. 2010;67:1473–1484. doi: 10.1001/archneurol.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots GG, Ovcharenko I. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004;32 (Web Server issue:W217–W221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.