Abstract
Human cytochrome P450c21 (steroid 21-hydroxylase, CYP21A2)1 catalyzes the 21-hydroxylation of progesterone (P4) and its preferred substrate 17α-hydroxyprogestrone (17OHP4). CYP21A2 activities, which are required for cortisol and aldosterone biosynthesis, involve the formation of energetically disfavored primary carbon radicals. Therefore, we hypothesized that the binding of P4 and 17OHP4 to CYP21A2 restricts access of the reactive heme-oxygen complex to the C-21 hydrogen atoms, suppressing oxygenation at kinetically more favorable sites such as C-17 and C-16, which are both hydroxylated by cytochrome P450c17 (CYP17A1). We reasoned that expansion of the CYP21A2 substrate-binding pocket would increase substrate mobility and might yield additional hydroxylation activities. We built a computer model of CYP21A2 based principally on the crystal structure of CYP2C5, which also 21-hydroxylates P4. Molecular dynamics simulations indicate that binding of the steroid nucleus perpendicular to the plane of the CYP21A2 heme ring limits access of the heme oxygen to the C-21 hydrogen atoms. Residues L107, L109, V470, I471, and V359 were found to contribute to the CYP21A2 substate-binding pocket. Mutation of V470 and I471 to alanine or glycine preserved P4 21-hydroxylase activity, and mutations of L107 or L109 were inactive. Mutations V359A and V359G, in contrast, acquired 16α-hydroxylase activity, accounting for 40% and 90% of the P4 metabolites, respectively. We conclude that P4 binds to CYP21A2 in a fundamentally different orientation than to CYP17A1 and that expansion of the CYP21A2 substrate-binding pocket allows additional substrate trajectories and metabolic switching.
The biosynthesis of cortisol requires the concerted action of several enzymes of the adrenal cortex, particularly the steroid dehydrogenases and cytochrome P450 hydroxylases (1). Cytochromes P450c17 (steroid 17-hydroxylase/17,20-lyase, CYP17A1) and P450c21 (steroid 21-hydroxylase, CYP21A2) are microsomal steroid hydroxylases, which participate in cortisol biosynthesis. These enzymes exhibit 29% amino acid identity and share common substrates, and their genes show equivalent intron/exon organization, attesting to the similarity in the two enzymes (2). The biochemistry of CYP17A1 has been studied in detail due to its biological importance (3) and mechanistic intrigue (4), and these studies have been aided by the procedures for its purification after expression in E coli (5). In contrast, the enzymology of CYP21A2 has received little attention, in part because CYP21A2 expression in E coli is relatively poor (6). CYP21A2 and CYP17A1 represent an important pair of similar enzymes for which comparative enzymology studies could yield information of theoretical interest and therapeutic importance.
Despite their similarities, the chemistries of CYP21A2 and CYP17A1 differ in several important respects. CYP17A1 substrates include 3-keto-Δ4-steroids (i.e. progesterone, P4), 3β-hydroxy-Δ5-steroids (i.e. pregnenolone), and 5α-reduced pregnanes (7) as substrates. CYP17A1 catalyzes not only the 17α-hydroxylase reaction but also the 17,20-lyase reaction with 17α-hydroxylated steroids (8), the 16α-hydroxylation of P4 (9), and the formation of 5(16)-androstadien-3β-ol from pregnenolone (4). Computer modeling studies suggest that the geometry of substrate binding, with the 4-ring cyclopenanophenanthrene nucleus parallel to the plane of the heme ring, enables CYP17A1 to execute a diverse repertoire of chemistry using a variety of substrates (10). Energetic considerations suggest that the ~4:1 preference for P4 hydroxylation in the 17α-position versus the 16α-position derives from the lower activation barrier to formation of a tertiary radical at C-17 compared to the less stable secondary radical at C-16, despite comparable proximity of these hydrogen atoms to the heme-oxygen complex (10). Residue A105 enables P4 trajectories favorable for 16α-hydroxylase activity, as substitution of the more bulky leucine in mutation L105A restricts nearly all P4 hydroxylation to the 17α-position (11).
The rich chemistry of CYP17A1 contrasts with the rather restricted chemistry of CYP21A2, which is limited to the 21-hydroxylation of P4 and of its preferred substrate, 17α-hydroxyprogesterone (17OHP4) (12). The exclusive 21-hydroxylation of the complex substrates P4 and 17OHP4 is unusual for a cytochrome P450, because 21-hydroxylation chemistry requires the formation of a relatively unstable and electron-deficient primary carbon radical at C-21 in the presence of more reactive hydrogen atoms within 3 Å of C-21. Computer modeling studies of CYP21A2 have appeared recently (13, 14), but little insight to substrate binding and chemistry was probed with these models. Instead, the models were used to rationalize the activity loss in common mutations causing 21-hydroxylase deficiency, an autosomal recessive disorder that afflicts 1:14,000 newborns (15) and primarily arises by gene conversion from the CYP21A1P pseudogene to the CYP21A2 gene encoding the CYP21A2 enzyme (16). Here we tested the hypotheses that P4 binding to CYP21A2 restricts access of the heme-oxygen complex to the C-21 hydrogen atoms and that expansion of the active site will confer additional substrate trajectories and cause metabolic switching.
EXPERIMENTAL PROCEDURES
Reagents and General Methods
Unless otherwise noted, reagents were obtained from Sigma-Aldrich Inc. (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA). Steroids were obtained from Sigma-Aldrich or Steraloids (Providence, RI). Radiochemicals were obtained from PerkinElmer Life Sciences (Waltham, MA).
Computer Modeling
The model of CYP21A2 was built by the procedure used for CYP17A1 (10) using an Octane workstation (Silicon Graphics, Mountain View, CA). The amino acid sidechains of CYP21A2 (residues 24-494) were substituted onto the backbone structure of rabbit CYP2C5 (17) (residues 12-473, pdbid 1DT6) using MidasPlus version 2.1, and energy minimization and molecular dynamics were performed with Amber 7 (both University of California, San Francisco). The alignments (18) of human CYP21A2 and rabbit CYP2C5, as well as human CYP17A1 for comparison, is shown in Figure 1. Direct side-chain substitution only was used for CYP21A2 residues 24-97 and 119-183. For residues 184-253, 254-272, and 280-460, and 461-494, length discrepancies were accommodated by manually adding or deleting residues and turns, aided by the backbone fragments of the corresponding regions in the P450BMP (CYP101, a fatty acid hydroxylase) structure and human CYP17A1 model (pdbid 2HPD and 2C17, respectively). The P450BMP structure was used as template for residues 98-118 due to the long B-C region predicted to contain a B’ helix. The C-terminus was extended by adding residues 489-494 in β-sheet conformation, and all modified segments were subjected to energy minimization. The model was assembled by manually displaying and bonding the fragments, followed by energy minimization and molecular dynamics using the sander module of Amber 7 to obtain the final structure. Docking of P4 and 17OHP4, followed by molecular dynamics, was performed using the sander and carnal modules as described for CYP17A1 to obtain distances from the modeled ferryl oxene to steroid hydrogen atoms (10). Structure analysis with PROCHECK showed that 94% of non-glycine mainchain residues scored in either the most favored or additionally allowed regions of the Ramachandran plot.
Figure 1.
Alignment of human CYP21A2 and CYP17A1 with modified CYP2C5 (17). Residues chosen for mutagenesis in this study are underlined.
Mutagenesis and Enzyme Assays
Site-directed mutagenesis was performed using overlapping PCR reactions followed by sequencing of the amplicons and subcloning all or part of the cDNAs into eukaryotic and yeast expression vectors pcDNA3 and V60 (19), respectively. CYP21A2 mutations were screened for activity by expression in transiently transfected HEK-293 cells (human embryonic kidney cells, ATCC #1573), and active mutations were studied with human P450-oxidoreductase in microsomes prepared from yeast using strain YiV(B) as described (19). Enzyme incubations, extraction, thin-layer chromatography (TLC), and quantitation of steroid metabolites were performed as previously described (20). In a typical experiment, steroid (0.1-10 μM, 100,000-400,000 cpm when [3H]-labeled) was incubated with 10-40 μg of microsomal protein (~1 pmol P450) in 0.2 mL 50 mM potassium phosphate (pH 7.4) at 37C for 30 min, and the products were extracted with 0.4 mL 1:1 ethyl acetate:isooctane. High-performance liquid chromatography (HPLC) was performed on a Breeze 1525 system with a C18 Symmetry column (150 × 4.6 mm, 5 μm, Waters, Woburn, MA) and methanol/water gradients (7, 21). Data were exported as ACII files and graphed with Origin version 7.5 (OriginLab, Northampton, MA). Protein determinations used the Coomassie Plus Reagent (Pierce, Rockford, IL). P450 content was determined by substrate-induced difference spectroscopy, the difference in absorbance at 386 nm and 420 nm using ε = 110 mM−1•cm−1, with versus without 10 μM substrate for wild-type CYP17A1 and CYP21A2 (21). Some CYP21A2 mutations could not be quantified accurately by difference spectroscopy and tended to be unstable with freeze-thawing of microsomes; consequently, rates for all enzymes are expressed per mg protein. Apparent Km and Vmax values were obtained by fitting means of 2-3 experiments to the Michaelis-Menton equation using Origin 7.5 (all r values exceeded 0.89). DNA sequencing was performed at the McDermott Center for Molecular Genetics core facility at UT Southwestern.
Preparation of 4-Pregnene-16α,17α-diol-3,20-dione and 4-Pregnene-16β-ol-3,20-dione (16β-hydroxyprogesterone)
The 4-pregnene-16α,17α-diol-3,20-dione was prepared by refluxing the acetonide of this compound with p-toluenesulfonic acid in refluxing methanol. The 16β-hydroxyprogesterone was prepared by oxidation of protected 16α-hydroxyprogesterone and stereoselective reduction. Full details of chemical synthesis and characterization are given in the Supporting Information.
RESULTS
Computer modeling of CYP21A2 and P4 docking
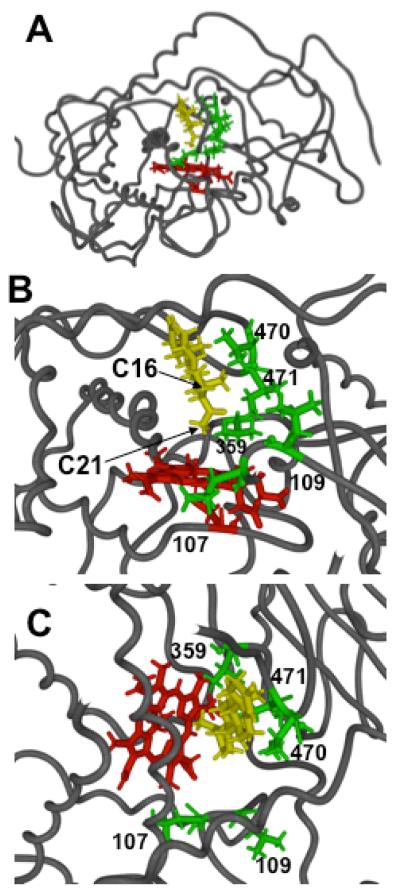
A computer model of CYP21A2 was generated using the x-ray structure of rabbit CYP2C5, which is also a P4 21-hydroxylase (22), as the primary template. We manually docked P4 with the planes of the heme ring and the steroid nucleus parallel, perpendicular, or obliquely oriented. We then performed molecular dynamics calculations, while monitoring the distances between hydrogen atoms of P4 and the modeled ferryl oxene during the last 40 ps of a 100 ps simulation. Only docking with the steroid nucleus perpendicular to the heme ring (Figure 2A) placed the C-21 hydrogen atoms closest to the ferryl oxene and excluded the more reactive hydrogen atoms at C-17 and C-16 (Table 1).
Figure 2.
Computer model of CYP21A2 with heme and ferryl oxene in red, docked P4 in yellow, and sidechains of the labeled residues studied here in green. Panel A, entire structure showing backbone atoms of protein, looking down axis of I-helix, plus all atoms of heme, P4, and residues L107, L109, V359, V470, and I471. Panels B and C, expanded views of active site with P4 hydrogen atoms at C-21 and C-16 labeled as viewed from side (Panel B) or top (Panel C). Distances in Table 1 are derived from the last 40 ps of a 100 ps molecular dynamics simulation with P4 docked in the orientation shown.
Table 1.
Distances (Å) of P4 hydrogen atoms to ferryl oxene oxygen of CYP21A2 model in molecular dynamics simulations
| Wild-type | V359A | V359G | ||||
|---|---|---|---|---|---|---|
| Atom | Average | Minimum | Average | Minimum | Average | Minimum |
| H-21 | 3.4 | 2.3 | 3.9 | 2.9 | 7.2 | 6.1 |
| H-17 | 5.8 | 4.6 | 6.2 | 4.9 | 6.3 | 5.3 |
| H-16α | 7.8 | 6.8 | 6.6 | 5.9 | 4.6 | 3.1 |
| H-16β | 7.6 | 6.9 | 5.8 | 4.5 | 3.6 | 2.1 |
Values for H-21 are the mean of the three hydrogen atoms, which are within 0.05 Å
The model suggested that V470 and I471 at the turn of β-sheet 4, L107 and L109 in the B’-helix, and V359 in a loop following the K-helix all participate in P4 binding (Figure 2B,C). These amino acids reside within the substrate-recognition sequences (SRSs) SRS-6, SRS-1, and SRS-5, respectively (23). We endeavored to increase substrate mobility by reducing the size of these large, hydrophobic residues using site-directed mutagenesis.
Mutagenesis of L107, L109, V470 and I471
When expressed in HEK-293 cells or in yeast strain YiV(B) with human P450-oxidoreductase (19), wild-type CYP21A2 21-hydroxylates 17OHP4 at a maximal rate 28 times faster than for P4, yielding 11-deoxycortisol and 11-deoxycorticosterone, respectively (21). Mutations L107A and L109A were inactive when expressed in HEK-293 cells. Mutation V470A retained 21-hydroxylase activity, and the double mutation V470A+I471A displayed a reversal of substrate preference from wild-type CYP21A2. Mutation V470A+I471A 21-hydroxylates P4 2.4 times faster than 17OHP4, both due to increased turnover of P4 and to much reduced activity with 17OHP4 (Table 2). Mutations V470G and I471G also retained 21-hydroxylase activity with P4 and 17OHP4 substrates (not shown). Thus, we were unable to engineer additional P4 hydroxylase activities onto CYP21A2 by mutating residues L107, L109, V470, or I471.
Table 2.
Kinetic constant for CYP21A2 and mutations
| Progesterone | 17OHProgesterone | ||||
|---|---|---|---|---|---|
| Enzyme | Km | Vmax | Km | Vmax | Vmax Ratio (P4/17OHP4) |
| Wild-type | 1.1 | 0.12 | 1.6 | 3.3 | 0.036 |
| V470A | 1.0 | 0.03 | 2.5 | 0.48 | 0.063 |
| V470A+I471A | 1.5 | 0.58 | 1.6 | 0.24 | 2.4 |
| V359A | 0.48 | 0.09 | 0.82 | 1.0 | 0.09 |
| V359G | 3.2 | 0.08 | 0.61 | 1.0 | 0.08 |
Apparent Km in μM, Vmax in pmol•min−1•mg protein−1
Mutations V359A and V359G
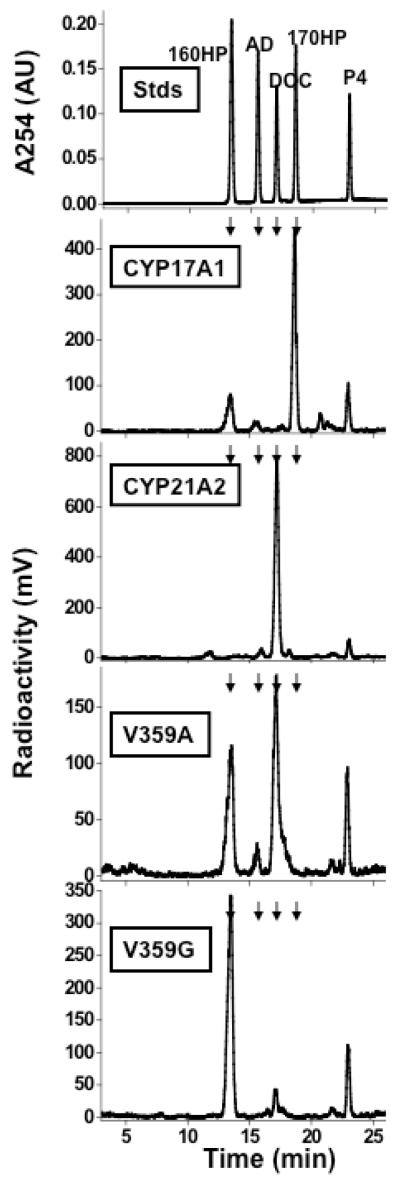
Mutations V359A and V359G both produced an additional metabolite of P4, which was identified as 16α-hydroxyprogesterone (16OHP4) by co-chromatography with standard both on TLC and reverse-phase HPLC (Figure 3) and was also distinguished from 16β-hydroxyprogesterone (Figure S1 of the Supporting Information). Mutation V359A yielded a 60:40 mix of 21- and 16α-hydroxylated products, whereas mutation V359G gave 90% 16OHP4 (Figure 3). When mutation V359G was expressed in transfected HEK-293 cells, 16OHP4 was again the dominant product (not shown). The identity of the new product was confirmed as 16OHP4 by liquid chromatography-mass spectrometry (LC-MS), with molecular ion of m/z = 331 (Figure S2 of the Supporting Information). These data demonstrate that expansion of the CYP21A2 substrate-binding pocket by substituting the less bulky A and G for V359 progressively shifts hydroxylation from C-21 to the more reactive hydrogen atoms at C-16, as predicted from computer modeling studies and theoretical considerations. Control experiments indicated that this product was not formed by control microsomes (prepared from YiV(B) yeast strain without CYP21A2 expression), that these products were not formed in the presence of the P450 inhibitor ketoconazole, and that neither catalase nor superoxide dismutase inhibited their formation (Table S1 and Figure S3 of the Supporting Information). These control experiments indicate that the products were formed by the CYP21A2 mutations and not an artifact of endogenous yeast enzymes or nonenzymatic products derived from reactive oxygen species released into the incubations.
Figure 3.
HPLC chromatograms of products formed upon incubation of yeast microsomes containing CYP17A1 or CYP21A2 and mutations with P4. The top panel shows absorbance at 254 nm from standards (Stds) of P4, 17OHP4, 11-deoxycorticosterone (DOC), androstenedione (AD), and 16OHP4. Lower four panels show radioactivity derived from P4 metabolism by CYP17A1, wild-type CYP21A2, CYP21A2 mutation V359A, and CYP21A2 mutation V359G. Small arrows atop each panel track elution positions of standards.
Metabolism of 17OHP4 by CYP21A2 Mutations
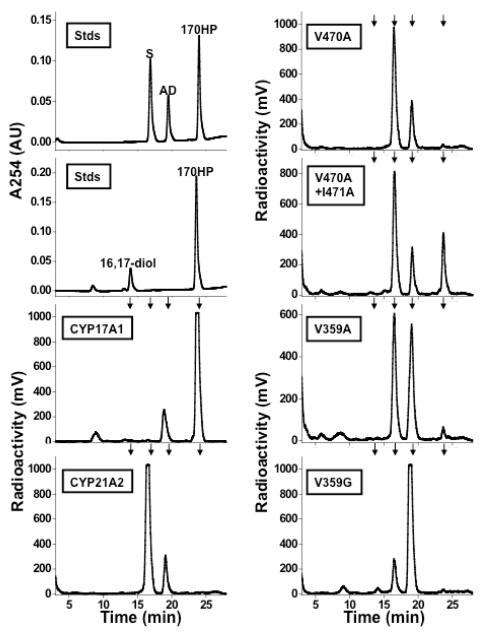
Because 17OHP4 is the preferred substrate for CYP21A2, we also studied 17OHP4 metabolism by the CYP21A2 mutations. The 17OHP4 metabolites formed by mutations of V470 and I471 included a second, unidentified product. Both mutations V359A and V359G also metabolized 17OHP4 to 11-deoxycortisol and this second product, and for mutation V359G, the unidentified compound was the major product (Figure 4). The second compound was not androstenedione and was neither the 16α- nor 6β-hydroxylation products (4-pregnene-16α,17α-diol-3,20-dione and 4-pregnene-6β,17α-diol-3,20-dione, respectively), as the mobilities of the unknown compound on TLC and HPLC are different than those of standards. This unknown compound appears to be a hydroxylated metabolite of 17OHP4, based on the molecular ion of m/z = 347 (Figure S2 of Supporting Information). These data show that expansion of the CYP21A2 substrate-binding pocket also affords an additional metabolism pathway for 17OHP4, but the identity of this product is unknown, and the physiochemical basis of this result is not readily rationalized as are results with P4 metabolism.
Figure 4.
HPLC chromatograms of products formed upon incubation of yeast microsomes containing CYP17A1 or CYP21A2 and mutations with 17OHP4 as labeled. Chromatographic mobilities of standards (Stds) are indicated by arrows, including 17OHP4, 11-deoxycortisol (S), androstenedione (AD), and 4-pregnene-16α,17α-diol-3,20-dione (P4diol). The unknown metabolite at 19 min does not co-chromatograph with any of the known standards, and the mobility of 4-pregnene-6β,17α-diol-3,20-dione is similar to that of P4diol (not shown).
Other Substrates for CYP21A2
Pregnenolone, 17α-hydroxypregnenolone, and 5α-pregnane-3α-ol-20-one (allopregnanolone) are all are excellent substrates for human CYP17A1 (7). In contrast, wild-type CYP21A2 in yeast microsomes did not appreciably metabolize these substrates, and none of these compounds significantly inhibited 21-hydroxylation of P4. However, we found that CYP21A2 metabolizes 5α-pregnane-3,20-dione (5α-dihydroprogesterone) with an apparent Km of 2.6 μM (not shown). These results suggest that substrate binding to CYP21A2 requires a 3-keto group, at least for common pregnanes.
DISCUSSION
The engineering of novel activities to hepatic cytochromes P450 has received considerable attention after the cDNAs for these enzymes were isolated. These studies are useful for confirming which residues comprise SRS regions and for understanding the metabolism of xenobiotics (24). The capacity of xenobiotic-metabolizing P450s to utilize a variety of substrates and to tolerate both naturally-occurring and artificial amino acid substitutions while retaining activity is consistent with the broad scope of catalysis embodied by these versatile enzymes. In contrast, the activities of the P450s essential for the biosynthesis of steroids, sterols, and bile acids are considered highly restricted and vulnerable to mutations (25, 26), as evidenced by human genetic diseases such as 21- and 17-hydroxylase deficiencies (3, 27). The major new finding of this study is that human CYP21A2 can be re-engineered to a P4 16α-hydroxylase with a single amino acid substitution. Our data are consistent with a model in which, as the steric bulk provided by V359 is progressively reduced in mutations V359A and V359G, the mobility of bound substrate increases, which thus changes the trajectories of P4 and enables 16α-hydroxylation. Consistent with this model and the observed shift from 21- to 16α-hydroxylation in this series, the average distances from the ferryl oxene to hydrogen atoms of P4 at C-21 increases (3.4 to 3.9 to 7.2 Å), while the average distances to H16α-falls (7.8 to 6.6 to 4.6 Å) based on molecular dynamics calculations (Table 1). To our knowledge, our data provide the first example of metabolic switching to change the product distribution for the natural substrate (P4) of a steroidogenic P450 using rational mutagenesis.
We used CYP2C5 as principal template for the CYP21A2 model because CYP2C5 is a P4 21-hydroxylase (22), although 16OHP4 accounts for 3% of its P4 metabolites. In addition, the major activities of other rabbit CYP2C isoforms using P4 substrate are 16α- and 6β-hydroxylation (28). P4 metabolism by human CYP2C9 and CYP2C19 is dominated by 21-hydroxylation, and CYP2C19 has significant 16α- and 17α-hydroxylase activities with P4 while CYP2C9 does not (29). Thus, it is not surprising that the first additional activity that we could engineer onto CYP21A2 is 16α-hydroxylation. Apparently, elimination of some steric bulk in SRS-5 enables P4 to tip sufficiently in the CYP21A2 substrate-binding pocket (Figure 2) to allow access of the 16α-hydrogen atom to the ferryl oxene. Our data also suggest that P4 binds to the active site of CYP2C5 and probably other CYP2C family enzymes that 21-hydroxylate P4 in a similar orientation.
Despite their similarities, the chemistries of human CYP17A1 and CYP21A2 reflect fundamentally different catalytic strategies. The major 17α-hydroxylase and minor 16α-hydroxylase activities of CYP17A1 are consistent with preferential formation of the tertiary carbon radical at C-17, which is more energetically favorable than the secondary carbon radical at C-16. We proposed that these activities are a consequence of steroid docking into the substrate binding pocket parallel to the plane of the heme ring, and molecular dynamics calculations confirmed that both the 17α-hydrogen and 16α-hydrogen atoms are accessible to a ferryl oxene in this docking orientation (10). In fact, these studies demonstrated that the hydrogen atoms at C-21 of P4 are also within 4 Å of the reactive oxygen center. We rationalized that CYP17A1 lacks 21-hydroxylase activity because formation of a primary carbon radical at C-21 is far less energetically favorable compared to abstraction of the more reactive hydrogen atoms at C-17 and C-16, which are simultaneously accessible.
We reasoned that CYP21A2 must bind P4 in a fundamentally different orientation than CYP17A1 and force 21-hydroxylation by precluding access of other hydrogen atoms. Our modeling studies of CYP21A2 identified one orientation, with the four-ring steroid nucleus of P4 perpendicular to the plane of the heme ring, which favors 21-hydroxylation (Figure 2), and molecular dynamics calculations support the preferential access of the 21-hydrogen atoms to the ferryl oxene (Table 1). This binding orientation, which restrains the majority of the steroid away from the heme and dangles only one methyl group adjacent to the ferryl oxene, might also explain why the chemistries of CYP21A2 are so limited. Of other common pregnanes that we studied, the 3β-hydroxy-Δ5-steroids pregnenolone and 17α-hydroxypregnenolone, as well as the 3α-hydroxysteroid allopregnanolone were neither substrates nor high-affinity inhibitors of CYP21A2, consistent with earlier studies of bovine CYP21A2 (12), and only 5α-pregnane-3,20-dione was found to be a fair substrate. In addition, previous studies have shown that bovine CYP21A2 has 20β-oxdiase activity with pregn-4-en-20β-ol-3-one and pregn-4-ene-17α,20β-diol-3-one substrates (30). Although we did not study 20β-hydroxypreganes directly, our modeling studies suggest that hydrogen atoms on C-20 might also be accessible to the ferryl oxene of human CYP21A2, enabling 20β-oxidase activity at this more reactive carbon center for these substrates versus the 21-methyl group.
We mutated residues adjacent to the docked P4 in our CYP21A2 model: L107 and L109, V470 and I471, and V359, which distribute to the SRS-1, SRS-6, and SRS-5, respectively. SRS-1 appears critical for CYP21A2 function, based on the absence of detectable activity in the relatively conservative mutations L107A and L109A. Consistent with this finding, residues in SRS-1 have been shown to account for much of the variability in P4 21-hydroxylase activity for CYP2C1, 2, and 5 (31). The residues corresponding to V359 are L in CYP2C5 (Figure 1) and N in CYP2C9 and CYP2C19; consequently, the V359G substitution is not one that would have been predicted to change activity from homology considerations alone. Alignment of CYP21A2 sequences from several species reveals complete conservation of not only V359 but also the adjacent residues, supporting the importance of SRS-5 in restricting activities with P4 substrate to 21-hydroxylation (Figure S4 of Supporting Information).
In contrast, results with 17OHP4 metabolism by our CYP21A2 mutations are not as readily explained as the results of P4 metabolism. CYP21A2 mutations with A or G in place of V470, I471, or V359 all 21-hydroxylated 17OHP4 and yielded an additional metabolite, which is neither the 16α- nor 6β-hydroxylation product. Furthermore, mutation V470A+I471A has significantly increased activity with P4 substrate such that the Vmax for P4 exceeded that of 17OHP4 (Table 2). Mutagenesis and chimerism (32) of the corresponding residues in SRS-6 of rabbit CYP2C2 is sufficient to confer P4 21-hydroxylase activity to this enzyme, without altering SRS-1. In particular, CYP2C2 mutation S473V has high P4 21-hydroxylase activity (33); however, exhaustive mutagenesis experiments, including mutations S473A and S473G, were not reported. In addition, residues in SRS-6 appear to constrain the motion of the F-G loop, which regulates substrate entry and exit in CYP2C5 and CYP2C9 (34, 35). Results with CYP21A2 mutation V470A+I471A reflect a combination of impaired 17OHP4 oxygenation and augmented P4 oxygenation, which derives from several of these factors. Although other mechanisms might explain these results with 17OHP4, our data and interpretations regarding P4 metabolism are internally consistent.
Congenital adrenal hyperplasia due to 21-hydroxylase deficiency is one of the most common autosomal recessive human diseases, afflicting 1:14,000 newborns throughout most of the world (15), with higher prevalence in certain isolated populations (36). Despite years of study, treatments remain inadequate, mechanisms of therapeutic benefits are poorly understood, and optimal management of children who survive to adulthood has received little attention (37). Most cases of 21-hydroxylase deficiency derive from gene conversion events (16), although several de novo mutations have been described (14, 38). None of the human mutations in CYP21A2 so far identified have been shown to possess a second activity besides 21-hydroxylation, although this mechanism is unlikely to be significant for the common alleles derived from gene conversions. Nevertheless, a better understanding of the structural basis for the 21-hydroxylase activity of CYP21A2 and the comparative biochemistry of related enzymes might enable strategies to better treat 21-hydroxylase deficiency by exploiting and optimizing the 21-hydroxylase activities of other human cytochromes P450.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mr. Richard Hall and Ms. Mahboubeh Papari-Zareei for assistance with yeast transformation and microsome preparation, Mr. Francis Yoshimoto for assistance with chemical syntheses, Dr. David J. Mangelsdorf for use of the Agilent 1100 LC/MSD SL mass spectrometer, and Drs. Julian Peterson and Ronald Estabrook for helpful discussions.
Footnotes
SUPPORTING INFORMATION AVAILABLE Chemical syntheses and characterizations are described in detail. Table S1 gives results of incubations in the presence of ketoconazole, catalase, and superoxide dismutase. Figure S1 shows chromatographic properties of 16OHP4 versus 16β-hydroxyprogesterone, the 16-epimer. Figure S2 shows LC-MS data confirming the hydroxylation state of the products, and Figure S3 shows results of prolonged incubations of P4 and 17OHP4 with microsomal enzymes, with or without further incubation with microsomes from mock-transformed yeast. Figure S4 shows alignment of CYP21A2 sequences from 5 species in the region containing V359. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by grants #I-1493 from the Robert A. Welch Foundation and R01GM086596 from the National Institutes of Health (both to R.J.A.), and C.R.D. was a UT Southwestern SURF research fellow.
Abbreviations: 16OHP4, 16α-hydroxyprogesterone; 17OHP4, 17α-hydroxyprogesterone; CYP, cytochrome P450; HPLC, high-performance liquid chromatography; LC-MS, liquid chromatography-mass spectrometry; P4, progesterone; SRS, substrate recognition sequence; TLC, thin-layer chromatography
REFERENCES
- 1.Miller WL, Auchus RJ. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr Rev. 2011;32:81–151. doi: 10.1210/er.2010-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Picado-Leonard J, Miller WL. Cloning and sequence of the human gene encoding P450c17 (steroid 17α-hydroxylase/17,20 lyase): Similarity to the gene for P450c21. DNA. 1987;6:439–448. doi: 10.1089/dna.1987.6.439. [DOI] [PubMed] [Google Scholar]
- 3.Auchus RJ. The genetics, pathophysiology, and management of human deficiencies of P450c17. Endocrinol Metab Clin North Am. 2001;30:101–119. doi: 10.1016/s0889-8529(08)70021-5. [DOI] [PubMed] [Google Scholar]
- 4.Lee-Robichaud P, Shyadehi AZ, Wright JN, Akhtar ME, Akhtar M. Mechanistic kinship between hydroxylation and desaturation reactions: Acyl carbon bond cleavage promoted by pig and human CYP17 (P-45017α; 17α-hydroxylase-17,20-lyase) Biochemistry. 1995;34:14104–14113. doi: 10.1021/bi00043a015. [DOI] [PubMed] [Google Scholar]
- 5.Imai T, Globerman H, Gertner JM, Kagawa N, Waterman MR. Expression and purification of functional human 17α-hydroxylase/17,20-lyase (P450c17) in Escherichia coli. Use of this system for study of a novel form of combined 17α-hydroxylase/17,20-lyase deficiency. J Biol Chem. 1993;268:19681–19689. [PubMed] [Google Scholar]
- 6.Harnastai IN, Gilep AA, Usanov SA. The development of an efficient system for heterologous expression of cytochrome P450s in Escherichia coli using hemA gene co-expression. Protein Expr Purif. 2006;46:47–55. doi: 10.1016/j.pep.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Gupta MK, Guryev OL, Auchus RJ. 5α-reduced C21 steroids are substrates for human cytochrome P450c17. Arch Biochem Biophys. 2003;418:151–160. doi: 10.1016/j.abb.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Zuber MX, Simpson ER, Waterman MR. Expression of bovine 17α-hydroxylase cytochrome P450 cDNA in non-steroidogenic (COS-1) cells. Science. 1986;234:1258–1261. doi: 10.1126/science.3535074. [DOI] [PubMed] [Google Scholar]
- 9.Swart P, Swart AC, Waterman MR, Estabrook RW, Mason JI. Progesterone 16α-hydroxylase activity is catalyzed by human cytochrome P450 17α-hydroxylase. J Clin Endocrinol Metab. 1993;77:98–102. doi: 10.1210/jcem.77.1.8325965. [DOI] [PubMed] [Google Scholar]
- 10.Auchus RJ, Miller WL. Molecular modeling of human P450c17 (17α-hydroxylase/17,20-lyase): Insights into reaction mechanisms and effects of mutations. Mol Endocrinol. 1999;13:1169–1182. doi: 10.1210/mend.13.7.0326. [DOI] [PubMed] [Google Scholar]
- 11.Swart AC, Storbeck KH, Swart P. A single amino acid residue, Ala 105, confers 16α-hydroxylase activity to human cytochrome P450 17α-hydroxylase/17,20 lyase. J Steroid Biochem Mol Biol. 2010;119:112–120. doi: 10.1016/j.jsbmb.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 12.Lorence MC, Trant JM, Mason JI, Bhasker CR, Fujii-Kuriyama Y, Estabrook RW, Waterman MR. Expression of a full-length cDNA encoding bovine adrenal cytochrome P450C21. Arch Biochem Biophys. 1989;273:79–88. doi: 10.1016/0003-9861(89)90164-1. [DOI] [PubMed] [Google Scholar]
- 13.Krone N, Riepe FG, Grotzinger J, Partsch CJ, Sippell WG. Functional characterization of two novel point mutations in the CYP21 gene causing simple virilizing forms of congenital adrenal hyperplasia due to 21-hydroxylase deficiency. J Clin Endocrinol Metab. 2005;90:445–454. doi: 10.1210/jc.2004-0813. [DOI] [PubMed] [Google Scholar]
- 14.Robins T, Carlsson J, Sunnerhagen M, Wedell A, Persson B. Molecular model of human CYP21 based on mammalian CYP2C5: structural features correlate with clinical severity of mutations causing congenital adrenal hyperplasia. Mol Endocrinol. 2006;20:2946–2964. doi: 10.1210/me.2006-0172. [DOI] [PubMed] [Google Scholar]
- 15.Therrell BLJ, Berenbaum SA, Manter-Kapanke V, Simmank J, Korman K, Prentice L, Gonzalez J, Gunn S. Results of screening 1.9 million Texas newborns for 21-hydroxylase-deficient congenital adrenal hyperplasia. Pediatrics. 1998;101:583–590. doi: 10.1542/peds.101.4.583. [DOI] [PubMed] [Google Scholar]
- 16.Morel Y, Miller WL. Clinical and molecular genetics of congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Adv Hum Genet. 1991;20:1–68. doi: 10.1007/978-1-4684-5958-6_1. [DOI] [PubMed] [Google Scholar]
- 17.Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol Cell. 2000;5:121–131. doi: 10.1016/s1097-2765(00)80408-6. [DOI] [PubMed] [Google Scholar]
- 18.Graham SE, Peterson JA. Sequence alignments, variabilities, and vagaries. Methods Enzymol. 2002;357:15–28. doi: 10.1016/s0076-6879(02)57661-8. [DOI] [PubMed] [Google Scholar]
- 19.Sherbet DP, Tiosano D, Kwist KM, Hochberg Z, Auchus RJ. CYP17 mutation E305G causes isolated 17,20-lyase deficiency by selectively altering substrate binding. J Biol Chem. 2003;278:48563–48569. doi: 10.1074/jbc.M307586200. [DOI] [PubMed] [Google Scholar]
- 20.Auchus RJ, Lee TC, Miller WL. Cytochrome b5 augments the 17,20 lyase activity of human P450c17 without direct electron transfer. J Biol Chem. 1998;273:3158–3165. doi: 10.1074/jbc.273.6.3158. [DOI] [PubMed] [Google Scholar]
- 21.Auchus RJ, Kumar AS, Boswell CA, Gupta MK, Bruce K, Rath NP, Covey DF. The enantiomer of progesterone (ent-progesterone) is a competitive inhibitor of human cytochromes P450c17 and P450c21. Arch Biochem Biophys. 2003;409:134–144. doi: 10.1016/s0003-9861(02)00491-5. [DOI] [PubMed] [Google Scholar]
- 22.Dieter HH, Muller-Eberhard U, Johnson EF. Identification of rabbit microsomal cytochrome P-450 isozyme, form 1, as a hepatic progesterone 21-hydroxylase. Biochem Biophys Res Commun. 1982;105:515–520. doi: 10.1016/0006-291x(82)91465-6. [DOI] [PubMed] [Google Scholar]
- 23.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 24.Domanski TL, Halpert JR. Analysis of mammalian cytochrome P450 structure and function by site-directed mutagenesis. Curr Drug Metab. 2001;2:117–137. doi: 10.2174/1389200013338612. [DOI] [PubMed] [Google Scholar]
- 25.Hall PF. Role of cytochromes P-450 in the biosynthesis of steroid hormones. Vitam Horm. 1985;42:315–368. doi: 10.1016/s0083-6729(08)60065-5. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez FJ. Human cytochromes P450: problems and prospects. Trends Pharmacol Sci. 1992;13:346–352. doi: 10.1016/0165-6147(92)90107-h. [DOI] [PubMed] [Google Scholar]
- 27.White PC, Speiser PW. Congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Endocr Rev. 2000;21:245–291. doi: 10.1210/edrv.21.3.0398. [DOI] [PubMed] [Google Scholar]
- 28.Dieter HH, Johnson EF. Functional and structural polymorphism of rabbit microsomal cytochrome P-450 form 3b. J Biol Chem. 1982;257:9315–9323. [PubMed] [Google Scholar]
- 29.Yamazaki H, Shimada T. Progesterone and testosterone hydroxylation by cytochromes P450 2C19, 2C9, and 3A4 in human liver microsomes. Arch Biochem Biophys. 1997;346:161–169. doi: 10.1006/abbi.1997.0302. [DOI] [PubMed] [Google Scholar]
- 30.Tsubaki M, Matsumoto N, Tomita S, Ichikawa Y, Hori H. 20β-hydroxy-C21-steroid 20β-oxidase activity of cytochrome P450c21 purified from bovine adrenocortical microsomes. Biochim Biophys Acta. 1998;1390:197–206. doi: 10.1016/s0005-2760(97)00175-6. [DOI] [PubMed] [Google Scholar]
- 31.Kronbach T, Kemper B, Johnson EF. A hypervariable region of P450IIC5 confers progesterone 21-hydroxylase activity to P450IIC1. Biochemistry. 1991;30:6097–6102. doi: 10.1021/bi00239a003. [DOI] [PubMed] [Google Scholar]
- 32.Straub P, Lloyd M, Johnson EF, Kemper B. Differential effects of mutations in substrate recognition site 1 of cytochrome P450 2C2 on lauric acid and progesterone hydroxylation. Biochemistry. 1994;33:8029–8034. doi: 10.1021/bi00192a006. [DOI] [PubMed] [Google Scholar]
- 33.Ramarao M, Kemper B. Substitution at residue 473 confers progesterone 21-hydroxylase activity to cytochrome P450 2C2. Mol Pharmacol. 1995;48:417–424. [PubMed] [Google Scholar]
- 34.Cojocaru V, Winn PJ, Wade RC. The ins and outs of cytochrome P450s. Biochim Biophys Acta. 2007;1770:390–401. doi: 10.1016/j.bbagen.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Mo SL, Zhou ZW, Yang LP, Wei MQ, Zhou SF. New insights into the structural features and functional relevance of human cytochrome P450 2C9. Part I. Curr Drug Metab. 2009;10:1075–1126. doi: 10.2174/138920009790820129. [DOI] [PubMed] [Google Scholar]
- 36.Speiser PW, New MI, Tannin GM, Pickering D, Yang SY, White PC. Genotype of Yupik Eskimos with congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Hum Genet. 1992;88:647–648. doi: 10.1007/BF02265290. [DOI] [PubMed] [Google Scholar]
- 37.Auchus RJ. Congenital adrenal hyperplasia in adults. Curr Opin Endocrinol Diabet Obes. 2010;17:210–216. doi: 10.1097/MED.0b013e32833961d7. [DOI] [PubMed] [Google Scholar]
- 38.Speiser PW, White PC. Congenital adrenal hyperplasia. N Engl J Med. 2003;349:776–788. doi: 10.1056/NEJMra021561. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.