Abstract
Oncogenic mutations in NOTCH1 are present in over 50% of T-cell lymphoblastic leukemias (T-ALLs). Activation of NOTCH1 requires a double proteolytic processing in the extracellular region of the receptor (S2) and in the transmembrane domain (S3). Currently, anti-NOTCH1 therapies based on inhibition of S3 processing via small molecule γ-secretase inhibitors are in development. Here we report on the characterization of the protease system responsible for S2 processing of NOTCH1 in T-ALL. Analysis of NOTCH1 HD class I, NOTCH1 HD class II and NOTCH1 JME alleles characterized by increased and aberrant S2 processing shows that both ADAM10, a metalloprotease previously implicated in activation of wild type NOTCH1 in mammalian cells, and ADAM17, a closely related protease capable of processing NOTCH1 in vitro, contribute to the activation of oncogenic forms of NOTCH1. However, and despite this apparent functional redundancy, inhibition of either ADAM10 is sufficient to blunt NOTCH1 signaling in T-ALL lymphoblasts. These results provide further insight on the mechanisms that control the activation of oncogenic NOTCH1 mutants and identify ADAM10 as potential therapeutic target for the inhibition of oncogenic NOTCH1 in T-ALL.
Keywords: ADAM10, ADAM17, NOTCH1, T-ALL, leukemia
Introduction
The NOTCH1 signaling pathway plays a critical role in promoting multiple steps of T-cell development and berrant NOTCH1 signaling is a major oncogenic event in the pathogenesis of T-cell acute lymphoblastic leukemia (T-ALL).(1–3)
The NOTCH1 receptor is a type I transmembrane protein which functions as a ligand-activated transcription factor.(4) In resting conditions, the extracellular heterodimerization (HD) and LNR repeat domains of NOTCH1 located in the extracellular portion of the receptor form a molecular lock that precludes spontaneous NOTCH1 activation. Physiologic NOTCH1 signaling is triggered by interaction of the receptor with Delta-like and Jagged ligand proteins expressed on the surface of nearby cells. This ligand-receptor interaction induces a conformational change in the HD-LNR complex and exposes the otherwise cryptic C-terminal part of the HD domain to protease cleavage at the so called S2 site. This initial cut in the extracellular portion of the receptor primes NOTCH1 for further proteolytic processing by the presenilin γ-secretase complex at the S3 site located in the transmembrane region of the receptor. Upon γ-secretase cleavage, the intracellular portion of NOTCH1 (ICN1) is released from the membrane, translocates to the nucleus and triggers the expression of target genes in association with the RBPJ/CSL DNA binding protein and members of the mastermind-like family of transcriptional co-activators. (5)
Activating mutations in NOTCH1 are present in over 50% of human T-ALL cases.(6) These mutations typically result in increased processing of the receptor at the membrane or in increased stability of the activated intracellular form of NOTCH1 in the nucleus. Mutations that induce increased activation of NOTCH1 at the membrane encompass several mechanisms of action. NOTCH1 class I HD mutations usually consist of point mutations or small in-frame insertions that cause alterations in the conformation of the HD-LNR domains.(7) NOTCH1 class II HD mutations are larger insertions located in the distal part of the HD domain which displace the S2 site outside the reach of the protective HD-LNR complex.(7) Finally, NOTCH1 JME mutations consist of insertions in the extracellular juxtamembrane region of the receptor, which displace the HD-LNR complex, and the S2 site within it, away from the plasma membrane.(8) Given the strict requirement of the release of NOTCH1 from the plasma membrane for activation of the receptor, small molecule inhibitors of the γ-secretase complex, which block S3 processing, effectively block NOTCH1 signaling and have been shown to impair the growth and proliferation of some T-ALL cell lines harboring activating mutations in NOTCH1.(6, 9) Importantly, NOTCH1 processing at the S2 site is similarly required for activation of the receptor, suggesting that inhibition of S2 cleavage could be exploited as therapeutic target for the treatment of TALL. Two closely related ADAM metalloproteases, ADAM10 and ADAM17, have been implicated in the S2 processing of NOTCH receptors in different organisms.(10–14) Genetic studies have demonstrated that the ADAM10 orthologs Kuzbanian and sup-17 are responsible for NOTCH processing in Drosophila and in C. elegans, respectively.(10–12) Similarly, analysis of mouse knockout models has shown that Adam10-deficient mice are defective in the processing of NOTCH receptors and show NOTCH-related phenotypes.(15) Moreover, selective ablation of Adam10 in T-cells using Adam10 conditional knockout resulted in developmental defects similar to those observed in Notch1 deficient thymocytes.(16, 17) However, the specific mechanism of metalloproteinase processing involved in NOTCH signaling remains controversial. First ADAM17 was identified as an alternative protease capable of processing NOTCH1 in vitro. (13) Moreover, a recent report showed that ADAM10 but not ADAM17 is essential in executing ligand-induced extracellular cleavage at site 2 (S2) and suggested the presence of unknown proteases with the ability to process NOTCH signaling.(18) In contrast Bozkulak and coworkers have shown that oncogenic forms of NOTCH1 can be a substrate for both ADAM10 and ADAM17.(19)
Here we further explored the differential role of the ADAM proteases in the activation of oncogenic forms of NOTCH1 in T-ALL. Specifically, we asked what is the proteolytic machinery responsible for NOTCH1 S2 cleavage in T-ALL? Are different oncogenic forms of NOTCH1 processed in the same way? Can inhibition of the enzymes mediating S2 cleavage effectively abrogate oncogenic NOTCH1 signaling in T-ALL?
Materials and methods
Cells and cell culture
HeLa cells and MEFs were grown in DMEM supplemented with 10% fetal bovine serum, 100 U/mL penicillin G and 100 μg/mL streptomycin at 37°C in a humidified atmosphere under 5% CO2. Wild type and Adam10 deficient fibroblasts were a gift from Dr. Carl Blobel (Hospital for Special Surgery, Cornell University, New York, USA). Adam17 null cells were a gift from Dr. Paul Saftig (Christian-Albrechts Universität Kiel, Kiel, Germany). T-ALL cell lines were cultured in RPMI1640 media supplemented with 10% fetalbovine serum, 100 U/mL penicillin G, and 100 μg/mL streptomycin at 37°C in a humidified atmosphere under 5% CO2.
Plasmid constructs
The pcDNA3 NOTCH1 L1601P-ΔPEST encodes a double HD (substitution of L to P at position 1601) plus ΔPEST (truncation at position 2472) mutant form of NOTCH1 tagged with a FLAG tag epitope in the C-terminus. The pcDNA3 NOTCH1 L1601P-PEST construct was a gift from Dr. Iannis Aifantis (New York University, New York, US). The pcDNA3 NOTCH1 Jurkat JME17 mutant was generated by cloning a partial NOTCH1 transcript (exons 19 to 29) amplified by PCR from Jurkat cells, which contains an internal tandem duplication of 51 bases within exon 28 of the NOTCH1 gene, in the unique BamH1 and NotI restriction sites of pcDNA3 NOTCH1.(8) The pcDNA3 NOTCH1 P12 mutant was generated by cloning a partial NOTCH1 transcript (exons 19 to 29) amplified by PCR from P12-ICHIKAWA cells, which harbor an internal tandem duplication of 42 bases within exon 27 of the NOTCH1 gene, in the unique BamH1 and NotI restriction sites of pcDNA3 NOTCH1. The pcDNA3.1 TACE vector, encoding the full length wild type mouse Adam17 tagged with a myc tag epitope at the C-terminus, was a gift from Dr. Joaquin Arribas (Vall d’Hebron University Hospital, Barcelona, Spain). The pcDNA3 ADAM10 construct was a gift from Dr. Falk Fahrenholz (Johannes Gutenberg-University, Institute of Biochemistry, Mainz, Mainz, Germany) and encodes the full length bovine ADAM10 with a HA tag at the C-terminus.
Drugs and Inhibitors
The recombinant protein inhibitors of metalloproteases TIMP1 (Calbiochem PF019), TIMP2 (Calbiochem PF021) and TIMP3 (SIGMA T9197) were used at 125 nM. Compound E (Alexis Biochemicals) was used at 10 nM concentration. The cysteine protease inhibitor E-64 (Sigma E3132) was used at 100 μM and the serine protease inhibitor PMSF (Sigma P7626) at 1 mM.
Luciferase reporter assays
NOTCH1 expression plasmids (pcDNA3) were transiently transfected using FuGene (Roche) transfection reagent together with the pGaLUC artificial luciferase reporter construct (a gift from Dr. Tasuku Honjo at Kyoto University, Japan), which contains six tandem RBPJ/CSL binding sites. The pRL plasmid, a vector which expresses the Renilla luciferase gene under the control of the CMV promoter was used as internal control. Total DNA was kept constant by adding empty vector as needed. All transfections were carried out in triplicate. Cell lysates were harvested 48 hours post-transfection and luciferase assays were carried out using the Dual Luciferase Assay System (Promega, Madison, WI) on a LUMAT LB 9507 luminometer (Berthold Technologies, Oak Ridge, TN).
Western blot
Antibodies against activated NOTCH1 (NOTCH1 Val 1774; Cell Signaling, Beverly, MA), Renilla luciferase (Chemicon 4410; Chemicon, Temecula, CA), beta tubulin (sc-8035; Santa Cruz Biotechnology, Santa Cruz, CA), alpha actin (sc-1615 Santa Cruz Biotechnology, Santa Cruz, CA), ADAM17 (SC-6416; Santa Cruz Biotechnology, Santa Cruz, CA), and ADAM10 (Chemicon 19026; Chemicon, Temecula, CA) were used in immunoblotassays following the manufacturers’ instructions.
RNA interference
A lentiviral vector expressing a shRNA targeting human ADAM10 was purchased from Open Biosystems (pGIPZ V2LHS-94294; Open Biosystems, Huntsville, AL). Lentiviral vectors expressing shRNAs targeting mouse Adam10 and mouse Adam17 were generated by cloning corresponding long oligonucleotides with a stem loop structure in the pLKO-puro vector (gift from Dr. William Hahn, Dana Farber Cancer Institute, Boston, MA). Targeted sequences were as follows: mouse Adam10, 5′ CCGGCCTGCCATTTCACTCTGTC 3′; mouse Adam17, 5′ CCGGGACTTCTTCAGTGGTCATGT 3′. Corresponding controls contained the following scrambled stem sequences: mouse Adam10 scrambled, 5′ CCGGGGTATATGCGCCATACACTACCC 3′; mouse Adam17 scrambled, 5′ CCGGATCTTCTACGTGGGTTAGCT 3′. Lentivirus production and infection were performed as previously described.(30)
Quantitative real time PCR
Total RNA from T-ALL cell lines was extracted with RNAqueous kit (Ambion) following the manufacturer’s instructions. cDNAs were generated with the SuperScript RT-PCR system (Invitrogen) and analyzed by quantitative real-time PCR using SYBRGreen RT-PCR Core Reagents kit and the 7300 Real-Time PCR System, both from Applied Biosystems. Relative expression levels were based on GAPDH levels as reference control. Primer sequences were as follows: GAPDH Fw: 5′ GAA GGT GAA GGT CGG AGT 3′; GAPDH Rv: 5′ GAA GAT GGT GAT GGG ATT TC 3′; DELTEX 1 Fw: 5′ AAG AAG TTC ACC GCA AGA GGA TT 3′; DELTEX 1 Rv: 5′ CTA GGT AGC TAG CGT CCG GGT AG 3′
Cell growth assay
Cell viability was determined by MTT assay in triplicate using the Cell Proliferation Kit I (Roche).
Statistical analysis
Statistical significance was assessed using Student’s t-test.
Results
Inhibition of ADAM proteases impairs the activity of NOTCH1 mutant alleles
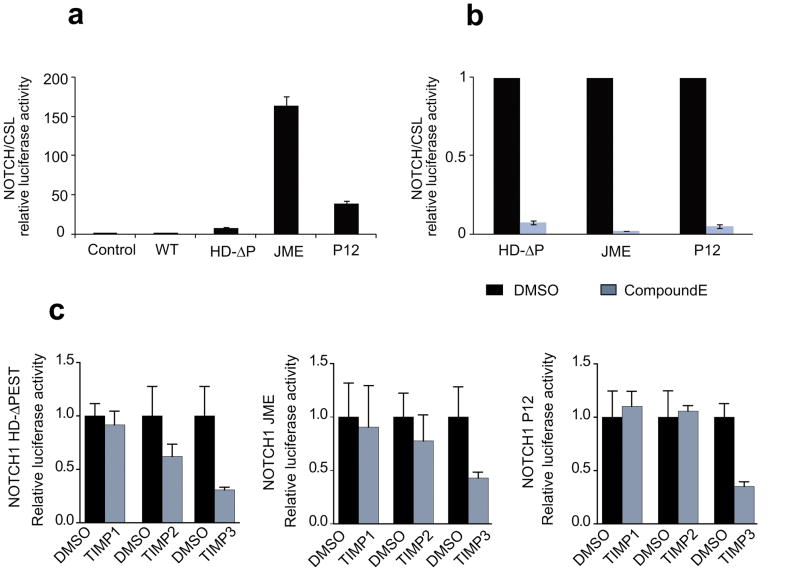
To address the role of ADAM proteases in the shedding and activation of NOTCH1 in human cells we selected highly active NOTCH1 mutants processed at the canonical S2 site such as the NOTCH1 L1601P ΔPEST allele, the NOTCH1 JME17 Jurkat allele and the NOTCH1 P12 allele.(6–8) NOTCH1 L1601P belongs to class I HD mutants which disrupt the HD LNR repeat complex facilitating S2 cleavage. This allele was combined with the truncation of the C-terminal PEST domain to increase its intrinsic activity via increased ICN1 stability.(7) The recently described NOTCH1 JME17 allele is representative of highly active juxtamembrane expansion mutations and consists of the insertion of 17 amino acids in the extracellular juxtamembrane region of the receptor and requires integrity of the S2 cleavage site for activation.(8) The NOTCH1 P12 allele belongs to the class II HD mutants and it’s characterized by an in frame insertion which displaces the C-terminus portion of the HD domain outside the LNR complex generating an alternative S2 cleavage site.(7) Luciferase reporter assay shows that expression of NOTCH1 L1601P ΔPEST, NOTCH1 JME17 and NOTCH1 P12 mutant allele leads to approximately 8, 38 and 160 fold activation of NOTCH1 signaling compared to basal levels (Figure 1a). The activity of the three NOTCH1 mutants is also dependent on the processing of the receptor at the S3 site, therefore inhibition of the γ-secretase complex by Compound E, a highly selective γ-secretase inhibitor, dramatically reduces NOTCH1 activity (Figure 1b).
Figure 1. Expression of different NOTCH1 mutant alleles induces constitutive NOTCH signaling.
a. NOTCH/CSL luciferase reporter assays in HeLa cells expressing a class I HD NOTCH1 mutant (HD-ΔP), a class II HD NOTCH1 mutant (P12) and a NOTCH1 JME allele (JME). b. Effects of the γ-secretase inhibitor CompE in NOTCH1 signaling induced by expression class I HD NOTCH1 mutant (HD-ΔP), a class II HD NOTCH1 mutant (P12) and a NOTCH1 JME allele (JME) in HeLa cells. c. Effects of the TIMP1, TIMP2 and TIMP3 metalloprotease inhibitors in NOTCH1 signaling induced by expression of class I HD, a class II HD and NOTCH1 JME alleles in HeLa cells.
To address the role of metalloproteases and other proteolytic systems in the shedding and activation of NOTCH1 we transfected HeLa cells with expression vectors containing NOTCH1 activated alleles and treated with a panel of selective and broad protease inhibitors including TIMP1, a protein that binds to and inhibits soluble metalloproteases; TIMP2, an inhibitor of both soluble and membrane-bound metalloproteases and TIMP3, a broad metalloprotease inhibitor capable of inhibiting the activity of soluble, membrane-bound and ADAM metalloproteases. In these experiments, treatment with TIMP1 had no effect on the processing of NOTCH1 HD class I, HD class II and JME alleles (Figure 1c). TIMP2 decreased the activity of our NOTCH1 HD class I mutant and to some extent the activity of the NOTCH1 JME allele, but not that of the HD class II construct (Figure 1c). In contrast, the activity of all three mutants was consistently and significantly inhibited in the presence of TIMP3 (Figure 1c). Finally, inhibition of cysteine proteases with E64 and serine proteases with PMSF did not affect the activity of NOTCH1 mutant alleles (data not shown). These finding suggests that ADAM proteases are the main enzymatic machinery responsible for S2 processing of oncogenic forms of NOTCH1.
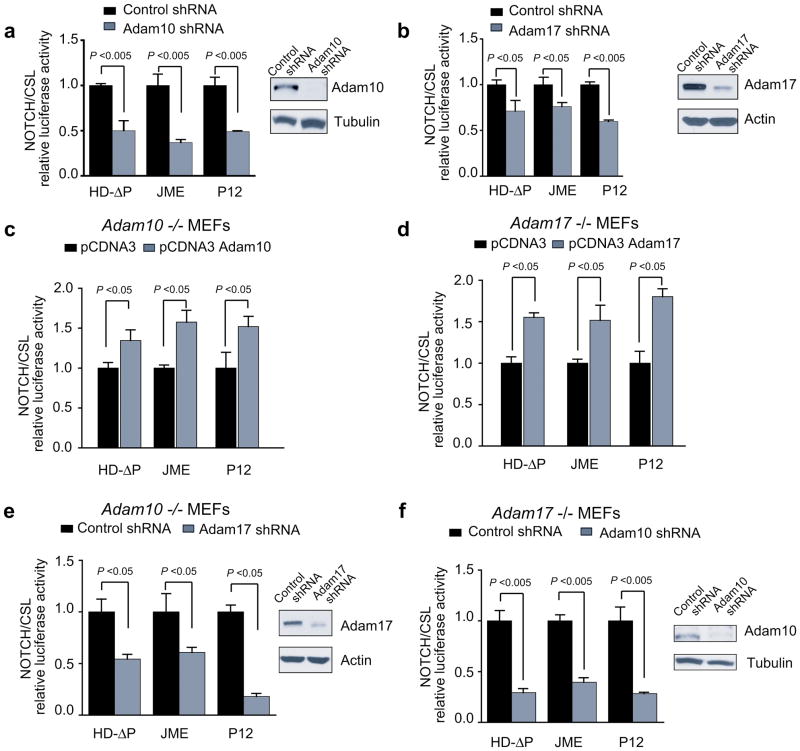
To characterize the requirement of specific ADAM proteases for the activity of NOTCH1 mutants, we first tested the activity of oncogenic NOTCH1 alleles upon shRNA knockdown of Adam10. Consistent with the well established role of Adam10 in the processing and activation of wild type NOTCH1, shRNA inactivation of Adam10 in mouse fibroblasts resulted in a marked decrease in the activity of NOTCH1 HD class I, class II and JME NOTCH1 alleles expressed (Figure 2a). Similarly, shRNA knockdown of Adam17 also impaired the activity of all three NOTCH1 mutants (Figure 2b). Similarly, expression of exogenous Adam10 or Adam17 in mouse embryonic fibroblasts (MEFs) derived from Adam10 and Adam17 knockout animals, effectively increased activation of NOTCH1 signaling upon expression of activated forms of NOTCH1 in these cells (Figure 2c–d). To further analyze the possible overlapping roles of Adam10 and Adam17 in the regulation of NOTCH1 activity, we tested the effects of inactivating both proteases simultaneously by knocking down Adam17 in Adam10 deficient cells and vice versa, by knocking down Adam10 in Adam17 knockout MEFs. These experiments showed that simultaneous inactivation of Adam10 and Adam17 resulted in more pronounced decreases in the activity of oncogenic NOTCH1 mutants (Figure 2e,f). All together these results suggest that although both Adam10 and Adam17 are competent in NOTCH1 processing, these proteases play a non redundant role in the processing of NOTCH1 mutant alleles.
Figure 2. ADAM10 and ADAM17 mediate the activation of oncogenic NOTCH1 mutant alleles.
a. NOTCH/CSL luciferase reporter assays in mouse fibroblasts expressing a class I HD NOTCH1 mutant (HD-ΔP), a class II HD NOTCH1 mutant (P12) and a NOTCH1 JME allele (JME) in the presence a shRNA targeting Adam10 (Adam10 shRNA) or a control inactive shRNA (control). Western blot analysis demonstrates effective Adam10 knockdown in cells expressing the Adam10 shRNA. b. NOTCH/CSL luciferase reporter assays in mouse fibroblasts expressing activated NOTCH1 mutants in the presence a shRNA targeting Adam17 (Adam17 shRNA) or a control inactive shRNA (control). Western blot analysis demonstrates effective Adam17 knockdown in cells expressing the Adam10 shRNA. c. NOTCH/CSL reporter assays in Adam10 deficient mouse fibroblasts expressing oncogenic NOTCH1 alleles upon reexpression of Adam10. d. NOTCH/CSL reporter assays in Adam17 deficient mouse fibroblasts expressing oncogenic NOTCH1 alleles upon reexpression of Adam17. e. NOTCH/CSL reporter assays in Adam10 deficient mouse fibroblasts expressing oncogenic NOTCH1 alleles upon shRNA knockdown of ADAM17. f. NOTCH/CSL reporter assays in Adam17 deficient mouse fibroblasts expressing oncogenic NOTCH1 alleles upon shRNA knockdown of ADAM10. Experiments show mean and standard deviation of triplicate replicas from representative experiments. Each experiment was repeated at least three times.
Inhibition of ADAM10 abrogates NOTCH1 signaling in human T-ALL
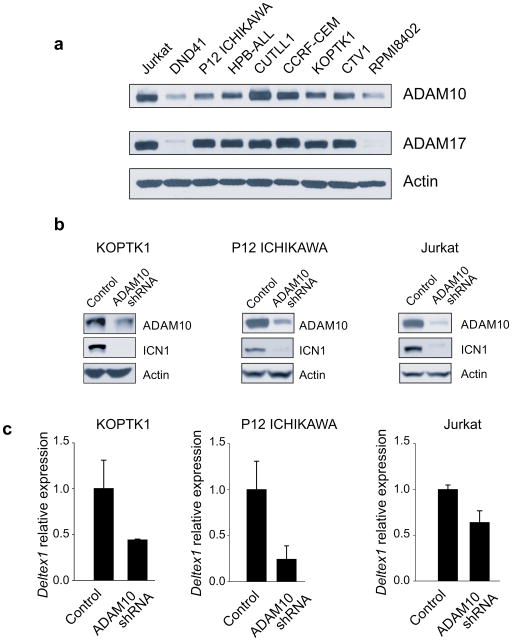
Recently, biochemical analysis of NOTCH1 mutant proteins expressed in the C2C12 mouse myoblast cell line showed that both ADAM10 and ADAM17 can effectively process oncogenic forms of NOTCH1.(18) However, genetic data from Adam10 conditional knockout mice has shown that Adam10 is strictly required for the processing and activation of Notch1 in T-cells(16, 17) raising the possibility that T-cell lymphoblasts could be more dependent on ADAM10 processing than predicted based on analysis of NOTCH1 mutants in an heterologous system. To investigate the specific requirement of ADAM10 in the cleavage and activation of NOTCH1 in human T-ALL cells we first analyzed the expression of these proteases in a panel of T-ALL cell lines. Western blot analysis showed variable levels of expression of ADAM10 and ADAM17 across different T-ALL cell lines harboring activating mutations in NOTCH1, with CUTLL1, CEM and Jurkat cells showing high levels and RPMI8402 and DND41 showing relatively low levels of expression (Figure 3a). ADAM17 was homogeneously expressed at high levels in most T-ALL cell lines with the exception of DND41 and RPMI8402 which showed very low and undetectable levels of ADAM17 expression, respectively (Figure 3a).
Figure 3. ADAM10 inactivation impairs NOTCH1 signaling in T-ALL.
a. Western blot analysis of ADAM10 and ADAM17 expression in T-ALL cell lines. b. Western blot analysis of activated NOTCH1 protein (ICN1) in T-ALL cell lines expressing a shRNA construct targeting ADAM10 or an inactive control shRNA. c. RT-PCR analysis of the Deltex1 NOTCH1 target gene in T-ALL cell lines expressing a shRNA construct targeting ADAM10 or an inactive control shRNA.
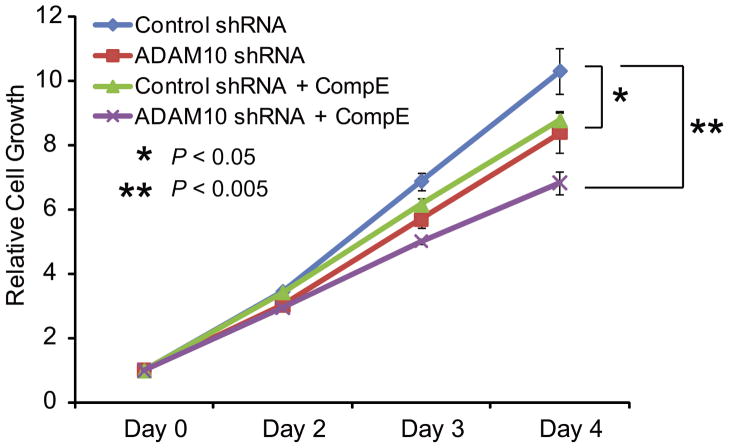
Next, we performed RNA knock down of ADAM10 across different T-ALL cell lines harboring NOTCH1 HD class I mutations (KOPTK1), NOTCH1 HD class II mutations (P12 ICHIKAWA) and NOTCH1 JME mutations (Jurkat). Strikingly, even though ADAM10 shRNA expression resulted in only partial downregulation of ADAM10, this translated into effective inhibition of NOTCH1 activation (Figure 3b) and transcriptional downregulation of the NOTCH1 target gene Deltex1 (Figure 3c). These results demonstrate that selective inactivation of ADAM10 can effectively downregulate oncogenic NOTCH1 in human T-ALL lymphoblasts. Consistent with this thesis, downregulation of ADAM10 via shRNA knockdown impaired the growth of KOPTK1 cells, a T-ALL cell line sensitive to inhibition of NOTCH1 signaling with GSIs (6) (Figure 4). Moreover, ADAM10 knockdown sensitized KOPTK1 cells to low concentrations (10 nM) of Compound E, a highly active GSI (Figure 4). Altogether these experiments identify NOTCH1 S2 processing and ADAM10 as targets for inhibition of NOTCH1 signaling in T-ALL and suggests that combined inhibition of ADAM and γ-secretase NOTCH1 cleavage may be exploited for the treatment of T-ALL.
Figure 4. Effects of ADAM10 inactivation in T-ALL cell growth.
Analysis of cell growth in KOPTK1 cells expressing a control inactive shRNA or a shRNA targeting ADAM10 in the presence or absence of low (10nM) concentrations of CompE, a highly active γ-secretase inhibitor. Bars represent average relative cell numbers and standard deviation as determined using a colorimetric MTT assay.
Discussion
Blocking aberrant NOTCH signaling by inhibition of the proteolytic system responsible for the processing and activation of oncogenic NOTCH1 receptors encoded by NOTCH1 mutant alleles is emerging as a molecularly targeted therapy for the treatment of T-ALL.(20, 21) Thus, inhibition of the NOTCH1 S3 cleavage by the γ-secretase complex has been shown to effectively abrogate NOTCH activation and to impair the growth of T-ALL lymphoblasts.(6, 21, 22) A second strategy towards inhibition of NOTCH1 signaling consists of anti-NOTCH1 inhibitory antibodies capable of blocking the access of the S2 cleavage site to ADAM proteases.(23, 24) Alternatively, the proteolytic machinery responsible for S2 NOTCH1 processing could also be exploited for the development of anti-NOTCH1 therapies. Indeed, genetic models and biochemical studies have established that physiologic activation of NOTCH receptors requires their proteolytic processing at the S2 site by an ADAM protease. Two ADAM proteases, ADAM10 and ADAM17 have been proposed as NOTCH S2 processing enzymes.(10–14) However, data from knockout mice supports that ADAM10 may be a more critical player in regulating Notch function in vertebrates.(15) Moreover, data from transgenic mice expressing a dominant-negative form of Adam10 in thymocytes(16) and recent findings using a conditional knockout to selectively ablate Adam10 function in T-cells(17) have shown that Adam10 inactivation impairs T-cell development due to defective activation of Notch1 signaling in the thymus. However, activating mutations in NOTCH1 found in T-ALL frequently alter the structure of the HD-LNR domains that normally control the access of ADAM10 to the S2 site, raising the possibility that S2 cleavage may be processed by alternative proteases in T-ALL. Thus, analysis of the role of ADAM10 and ADAM17 in the processing of NOTCH1 class I and class II HD mutant proteins expressed in heterologous systems have shown that ADAM10 and ADAM17 seem to be able to process mutant NOTCH1 proteins.(19) Here we validated and extended these results showing that both ADAM10 and ADAM17 can process HD and JME mutant forms of NOTCH1. A corollary of these observations is that single inhibition of ADAM10 or ADAM17 would fail to abrogate NOTCH signaling in leukemic lymphoblasts. Still, selective inactivation of ADAM10 in human T-ALL cells showed that inhibition of this protease is sufficient to effectively impair oncogenic NOTCH1 activation. Notably, the identification of ADAM10 and ADAM17 as essential proteases in the processing and activation of ERBB signaling (25–27) has led to the development of selective inhibitors of ADAM10 and ADAM17 as well as dual inhibitors for the treatment of lung, breast and colon cancer.(28, 29) INCB7839, an ADAM10 and ADAM17 inhibitor, is currently in clinical trials in the United States for the treatment of metastatic HER2+ breast cancers in combination with Trastuzumab alone or Trastuzumab and Vinorelbine. Given that activated NOTCH1 signaling, through either activating mutations of the NOTCH1 receptor or inactivating mutations of the FBXW7 tumor suppressor gene, is almost universal in T-ALL, the combination of an ADAM10 and a gamma-secretase inhibitor represent a rationale and attractive option for targeted therapy. While it is reassuring that inhibition of ADAM10 and ADAM17 activity was well tolerated in patients with solid tumors, it remains to be tested whether and to what extent the use of an ADAM inhibitor for the treatment of TALL will cause the gastrointestinal side effects initially observed with the use of gamma secretase inhibitors. In this context, our demonstration of the requirement of ADAM10 for the activation of oncogenic NOTCH1 in T-cell lymphoblasts provides preliminary evidence supporting a potential role for selective ADAM10 protease inhibitors as anti-NOTCH1 agents in T-ALL.
Acknowledgments
This work was supported by the National Institutes of Health (R01CA120196 to A.F.); the WOLF Foundation (A.F), the Leukemia and Lymphoma Society (grants 1287-08 and 6237-08 to A.F.), the Deutsche Forschungsgemeinschaft Sonderforschungsbereich 415 to P.S. and Interuniversity Attraction Poles Program P5/19 of the Belgian Federal Science Policy Office. Adolfo Ferrando is a Leukemia & Lymphoma Society Scholar.
Footnotes
Conflict of interest
The Ferrando laboratory receives research support from Merck and Pfizer. The authors declare no conflicts of interest for this work.
References
- 1.Ferrando AA. The role of NOTCH1 signaling in T-ALL. Hematology Am Soc Hematol Educ Program. 2009:353–361. doi: 10.1182/asheducation-2009.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paganin M, Ferrando A. Molecular pathogenesis and targeted therapies for NOTCH1-induced T-cell acute lymphoblastic leukemia. Blood Rev. 2011;25(2):83–90. doi: 10.1016/j.blre.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aster JC, Blacklow SC, Pear WS. Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J Pathol. Jan;223(2):262–273. doi: 10.1002/path.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006 Sep;7(9):678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 6.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004 Oct 8;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 7.Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, et al. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006 Jun;26(12):4642–4651. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sulis ML, Williams O, Palomero T, Tosello V, Pallikuppam S, Real PJ, et al. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood. 2008 Aug 1;112(3):733–740. doi: 10.1182/blood-2007-12-130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palomero T, Ferrando A. Oncogenic NOTCH1 control of MYC and PI3K: challenges and opportunities for anti-NOTCH1 therapy in T-cell acute lymphoblastic leukemias and lymphomas. Clin Cancer Res. 2008 Sep 1;14(17):5314–5317. doi: 10.1158/1078-0432.CCR-07-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan D, Rubin GM. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 1997 Jul 25;90(2):271–280. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- 11.Sotillos S, Roch F, Campuzano S. The metalloprotease-disintegrin Kuzbanian participates in Notch activation during growth and patterning of Drosophila imaginal discs. Development. 1997 Dec;124(23):4769–4779. doi: 10.1242/dev.124.23.4769. [DOI] [PubMed] [Google Scholar]
- 12.Wen C, Metzstein MM, Greenwald I. SUP-17, a Caenorhabditis elegans ADAM protein related to Drosophila KUZBANIAN, and its role in LIN-12/NOTCH signalling. Development. 1997 Dec;124(23):4759–4767. doi: 10.1242/dev.124.23.4759. [DOI] [PubMed] [Google Scholar]
- 13.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, et al. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000 Feb;5(2):207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 14.Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000 Feb;5(2):197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 15.Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, et al. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet. 2002 Oct 1;11(21):2615–2624. doi: 10.1093/hmg/11.21.2615. [DOI] [PubMed] [Google Scholar]
- 16.Manilay JO, Anderson AC, Kang C, Robey EA. Impairment of thymocyte development by dominant-negative Kuzbanian (ADAM-10) is rescued by the Notch ligand, delta-1. J Immunol. 2005 Jun 1;174(11):6732–6741. doi: 10.4049/jimmunol.174.11.6732. [DOI] [PubMed] [Google Scholar]
- 17.Tian L, Wu X, Chi C, Han M, Xu T, Zhuang Y. ADAM10 is essential for proteolytic activation of Notch during thymocyte development. Int Immunol. 2008 Sep;20(9):1181–1187. doi: 10.1093/intimm/dxn076. [DOI] [PubMed] [Google Scholar]
- 18.van Tetering G, van Diest P, Verlaan I, van der Wall E, Kopan R, Vooijs M. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J Biol Chem. 2009 Nov 6;284(45):31018–31027. doi: 10.1074/jbc.M109.006775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bozkulak EC, Weinmaster G. Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol Cell Biol. 2009 Nov;29(21):5679–5695. doi: 10.1128/MCB.00406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008 Mar 22;371(9617):1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 21.Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009 Jan;15(1):50–58. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palomero T, Barnes KC, Real PJ, Glade Bender JL, Sulis ML, Murty VV, et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006 Jul;20(7):1279–1287. doi: 10.1038/sj.leu.2404258. [DOI] [PubMed] [Google Scholar]
- 23.Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. Apr 15;464(7291):1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 24.Aste-Amezaga M, Zhang N, Lineberger JE, Arnold BA, Toner TJ, Gu M, et al. Characterization of Notch1 antibodies that inhibit signaling of both normal and mutated Notch1 receptors. PLoS ONE. 5(2):e9094. doi: 10.1371/journal.pone.0009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy G. The ADAMs: signalling scissors in the tumour microenvironment. Nat Rev Cancer. 2008 Dec;8(12):929–941. doi: 10.1038/nrc2459. [DOI] [PubMed] [Google Scholar]
- 26.Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005 Jan;6(1):32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- 27.Bublil EM, Yarden Y. The EGF receptor family: spearheading a merger of signaling and therapeutics. Curr Opin Cell Biol. 2007 Apr;19(2):124–134. doi: 10.1016/j.ceb.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Zhou BB, Peyton M, He B, Liu C, Girard L, Caudler E, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006 Jul;10(1):39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenny PA, Bissell MJ. Targeting TACE-dependent EGFR ligand shedding in breast cancer. J Clin Invest. 2007 Feb;117(2):337–345. doi: 10.1172/JCI29518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006 Mar 24;124(6):1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]