Abstract
Analysis and quantification of analytes in biological systems is a critical component of metabolomic investigations of cell function. The most widely used methods employ chromatographic separation followed by mass spectrometric analysis, which requires significant time for sample preparation and sequential chromatography. We introduce a novel high-throughput, separation-free methodology based on MALDI mass spectrometry that allows for the parallel analysis of targeted metabolomes. Proof-of-concept is demonstrated by analysis of prostaglandins and glyceryl prostaglandins. Derivatization to incorporate a charged moiety into ketone-containing prostaglandins dramatically increases the signal-to-noise ratio relative to underivatized samples. This resulted in an increased dynamic range (15 fmol – 2000 fmol on plate) and improved linearity (r2= 0.99). The method was adapted for high-throughput screening methods for enzymology and drug discovery. Application to cellular metabolomics was also demonstrated.
Introduction
Quantification of individual or multiple analytes in biological systems is a critical element of enzymology, metabolomics and biomonitoring. The most commonly used methods employ chromatographic separation followed by mass spectrometric analysis. Quantification is achieved by stable isotope dilution methods using isotopically labeled internal standards. These methods are highly sensitive and specific, but they require considerable time for sample preparation and chromatographic separation and utilize serial rather than parallel sample handling. Matrix-assisted laser desorption/ionization mass spectrometry1 (MALDI MS) is a powerful analytical technique capable of parallel processing of hundreds of samples without the need for prior separation2. MALDI MS is extremely sensitive (low attomole – femtomole sensitivity) and fast, with analysis times in large part dependent on the frequency of the irradiating laser. Solid-state lasers with repetition rates of 1 kHz allow multiple samples to be analyzed in under one second (since typically many fewer than 1000 laser shots are needed)3. The fast analytical capability of MALDI MS is ideally suited for fast serial analysis of large numbers of samples. MALDI MS is widely used for characterization of protein samples but is not routinely employed in quantitative analyses. Some reports have described the use of MALDI for the analysis of endogenous metabolites4, 5, but no reports have described its use for the routine analysis of prostaglandins.
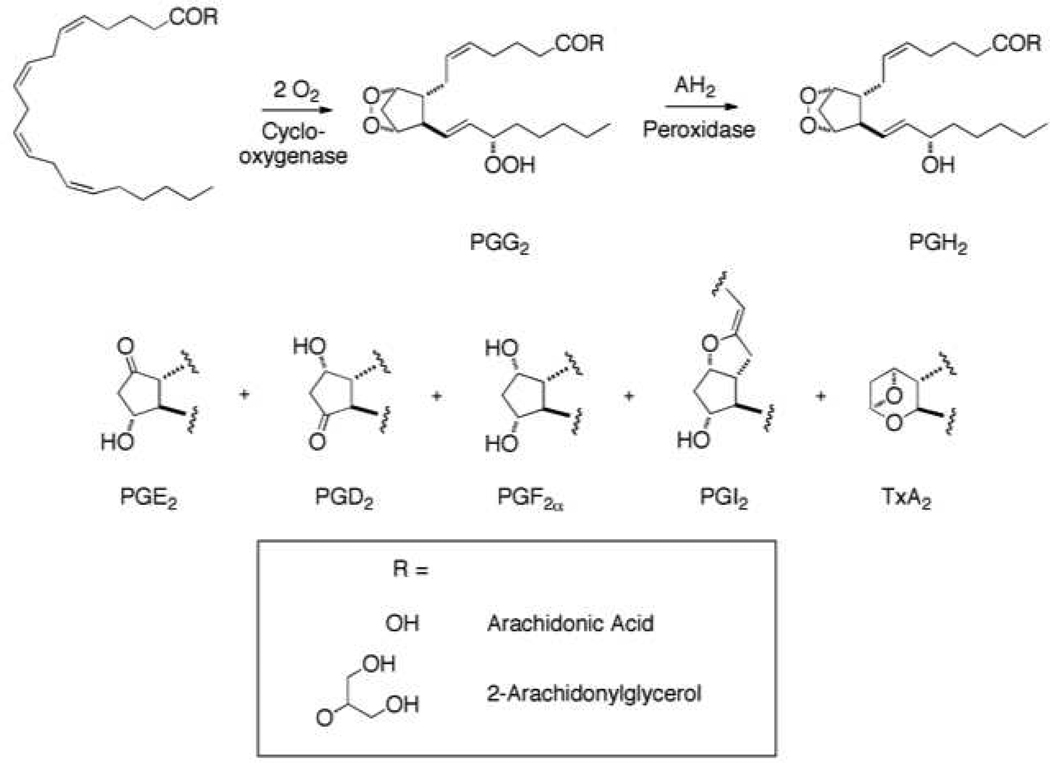
We describe herein the development of a robust MALDI MS-based approach for the high-throughput analysis of an important class of bioactive lipids. Selective derivatization of ketone-containing prostaglandins (PGs) with positively charged hydrazines converted them to charged hydrazones that were readily quantified by MALDI MS. PGs are products of the cyclooxygenase (COX) pathway of arachidonic acid (AA) metabolism and PG-glycerol esters (PG-Gs) are products of oxygenation of the endocannabinoid, 2-arachidonoylglcyerol (2-AG) (Figure 1)6. PGs and PG-Gs are potent lipid mediators that exert a broad range of biological responses through a series of membrane-bound G-protein-coupled receptors7. PGs have been implicated in diverse physiological and pathophysiological responses such as platelet aggregation, gastrointestinal integrity, wound healing, inflammation, hyperalgesia, and fever8, 9. Many analytical methods have been described for their quantification based on mass spectrometry and they are widely applied in clinical and preclinical studies10–13. All of the methods require extensive workup prior to analysis and/or time consuming HPLC separations followed by mass spectrometry. Because of the clinical importance of PG measurement, we developed a rapid high-throughput analytical method based on the derivatization of ketone-containing PGs followed by MALDI MS analysis. By using stable isotopically labeled internal standards, it was possible to develop quantitative mass spectrometric assays that displayed a large dynamic range. The assay was fully automated which enabled rapid liquid handling, high-throughput assay execution, and simultaneous deposition on a 384 well MALDI target. Multiple reaction monitoring for a particular PG and its internal standard allowed parallel quantification of samples with high sensitivity, specificity, and speed. This method should be applicable to an extraordinarily wide-range of biomolecules and to applications ranging from high-throughput enzyme assays to targeted metabolomics.
Figure 1.
Conversion of AA and 2-AG to PGs and PG-Gs by COX-2. PGI2 and TxA2 are unstable and rapidly hydrolyze to 6-keto-PGF1α and TxA2, respectively.
Materials and Methods
Reagents
AA was purchased from Nu-Chek Prep, Inc. (Elysian, MN, USA) Prostaglandin E2, prostaglandin D2, prostaglandin E2-d4, prostaglandin E2-1-glyceryl ester, 6-keto-prostaglandin F1α and (R)-flurbiprofen ((R)-(-)-2-fluoro-α-methyl-4-biphenylacetic acid) were purchased from Cayman chemicals (Ann Arbor, MI, USA). α-Cyano-4-hydroxy-cinnamic acid (CHCA), Girard’s reagent T, dimethyl sulfoxide (DMSO, Biotechnology performance certified), sulindac sulfide ((Z)-5-fluoro-2-methyl-1-[p-(methylthio)benzylidene]indene-3-acetic acid), indomethacin (1-(4-chlorobenzoyl)-5-methoxy-2-methyl-3-indoleacetic acid) and (S)-flurbiprofen ((S)-(+)-2-fluoro-α-methyl-4-biphenylacetic acid) were purchased from Sigma-Aldrich (St. Louis, MO, USA). All organic solvents were HPLC grade. Water and ethyl acetate were purchased from Fisher Scientific (Pittsburgh, PA, USA). Acetonitrile was purchased from Acros (Morris Plains, NJ, USA). Ethanol was purchased from Pharmco-AAPER (Shelbyville, KY, USA). RAW264.7 (RAW) cells were obtained from the American Type Culture Collection (ATCC) and were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with GlutaMax, high glucose, sodium pyruvate, and pyridoxine-HCl purchased from Gibco (Grand Island, NY, USA). Heat-inactivated fetal bovine serum (hi-FBS) was purchased from Summit Biotechnologies (Fort Collins, CO, USA). Lipopolysaccharide (LPS) was purchased from Calbiochem (San Diego, CA, USA).
Enzymes
The expression and purification of recombinant murine COX-2 (mCOX-2) from SF-9 cells was performed as previously described14.
High-Throughput Screening Materials
Labcyte (Sunnyvale, CA, USA) 384-well flat bottom polypropylene plates were used for the stock and reaction plates. Tips for the 384-tip head of the Agilent (Santa Clara, CA, USA) Bravo used in the study were purchased from Biotix (San Diego, CA, USA).
Derivatization and Analysis of Prostaglandins
Prostaglandin standards were dissolved in a solution of 1:1-ethanol:water to a final concentration of 20 µM. Aliquots of this solution were mixed with an equivalent volume of Girard’s reagent T (10 mg/mL solution in 20% acetic acid). The mixture was allowed to react for 10 min at room temperature then added to an equivalent volume of a solution of 20 mg/mL CHCA in 60% acetonitrile in water. A one µL aliquot was spotted on a MALDI plate (5 pmol analyte on-plate) and analyzed with a Bruker UltrafleXtreme TOF/TOF MS, Bruker Autoflex Speed TOF MS (Billerica, MA, USA) or on a Thermo LTQ-XL linear ion trap equipped with a MALDI source (San Jose, CA, USA) for MS/MS analysis of derivatized prostaglandins (PGE2 transition m/z 466.3→389.3, PGE2-d4 m/z 470.3→393.3).
Mass Spectrometry Parameters
All mass spectra were analyzed in positive ion mode. Both the Bruker UltrafleXtreme and Autoflex Speed TOF MS instruments are equipped with a Nd:YAG (solid state) laser at 355 nm utilizing SmartBeam™ modulation. Both instruments were operated in reflector mode. The UltrafleXtreme was operated with the following settings: extraction voltage, 10 kV; delay time, 70 ns; mass range, 20–1000 m/z. The Autoflex Speed was operated with the following settings: extraction voltage, 19 kV; delay time, 40 ns; mass range, 160–1000 m/z. The LTQ-XL is equipped with a N2 laser operating at 337 nm. MS/MS experiments for individual analytes utilized a collision energy of 50, activation Q of 0.300, activation time of 50 ms and a product ion mass range of 150–600 m/z.
COX-2 inhibitor assay
The ability of COX-2 to oxygenate AA or 2-AG was quantified by monitoring conversion to PGE2 or PGE2-G. A solution of COX-2 protein (200 nM) was made in 100 mM Tris/ 500 µM phenol solution, with a 2x molar equivalent of hematin, and was preincubated for 5 min at room temperature. A concentration range of inhibitors was added to aliquots of the enzyme solution and allowed to incubate at 37°C for 20 min. AA was added to this solution (50 µM) and reaction allowed to proceed for 30 sec. The solution was quenched with an equal volume of ethyl acetate (containing PGE2-d4 internal standard), the organic layer was collected and placed in a clean glass tube, then dried down under nitrogen. Samples were reconstituted in 1:1-ethanol:water and derivatized with an equivalent volume of Girard’s reagent T (10 mg/mL solution in 20% acetic acid) and analyzed by MALDI as described above. All inhibitor concentrations for 50% enzyme activity (IC50) were calculated by nonlinear regression analysis using GraphPad Prism software.
High-Throughput Analysis
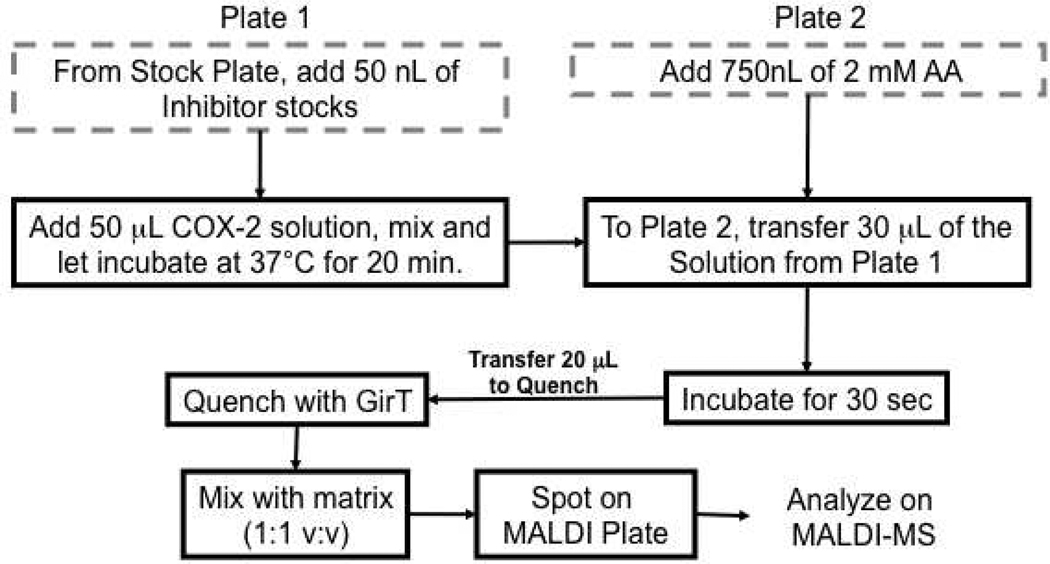
Modifications to the COX-2 inhibitor assay were made to adapt the standard assay to a high-throughput format. Two 384-well stock plates were prepared with AA and inhibitors in DMSO. Using a Labcyte Echo® Liquid Handling system, 50 nL of inhibitor was dispensed into a waiting 384-well reaction plate (DMSO was dispensed for no inhibitor control). Each inhibitor concentration was tested in quadruplicate. A solution of COX-2 (50 µL), containing 200 nM enzyme with appropriate cofactors as described above, was added to the reaction plate and allowed to incubate at 37°C for 20 min. Using the Echo® system again, 750 nL of AA was added to another 384-well plate and placed on an Agilent Bravo 384-well liquid handler. To the waiting 750 nL of AA, 29.25 µL of the inhibitor/enzyme solution was added; samples were mixed using the Bravo and allowed to react for 30 sec. Thirty µL of the reaction solution was aspirated and dispensed into a pre-dispensed 30 µL of quench solution (Girard’s reagent T in 20% acetic acid in ethanol solution with PGE2-d4 internal standard). These samples were allowed to react as described above. Ten µL of the derivatized solutions was added to 10 µL of a 20 mg/mL solution of CHCA and 1 µL of the final solution was spotted on a MALDI plate using the Bravo. All samples were analyzed using the Thermo LTQ ion trap mass spectrometer as described above (Chart 1).
Chart 1.
Flow chart representing the protocol used for the high throughput analysis. Liquid handling with the Bravo is represented in black boxes ( ) whereas liquid handling with the Echo is indicated in grey, dashed boxes (
) whereas liquid handling with the Echo is indicated in grey, dashed boxes ( ).
).
Activated Macrophage Eicosanoid Analysis
RAW cells were plated at 1.5 × 106 cells in a 30 mm dish in DMEM with 10% hi-FBS and 20 ng/mL of GM-CSF and allowed to incubate overnight. After the overnight incubation, the medium was removed and fresh DMEM with 10% hi-FBS was added. One set of cells was treated with only medium, the other set was treated with medium containing LPS (200 ng/mL) and IFNγ (20 U/mL) for 6 hr followed by addition of AA (20 µM). After 6 hr incubation, the medium was collected. A 2:1-ethyl acetate:0.1% acetic acid solution was added to the collected medium. Samples were vortexed, centrifuged (1000Xg for 5 min) and the organic layer was collected in clean glass tubes and dried down under nitrogen. Samples were reconstituted in ethanol, derivatized as described above and analyzed using the Bruker Autoflex Speed TOF-MS.
Results
Method Development and Validation
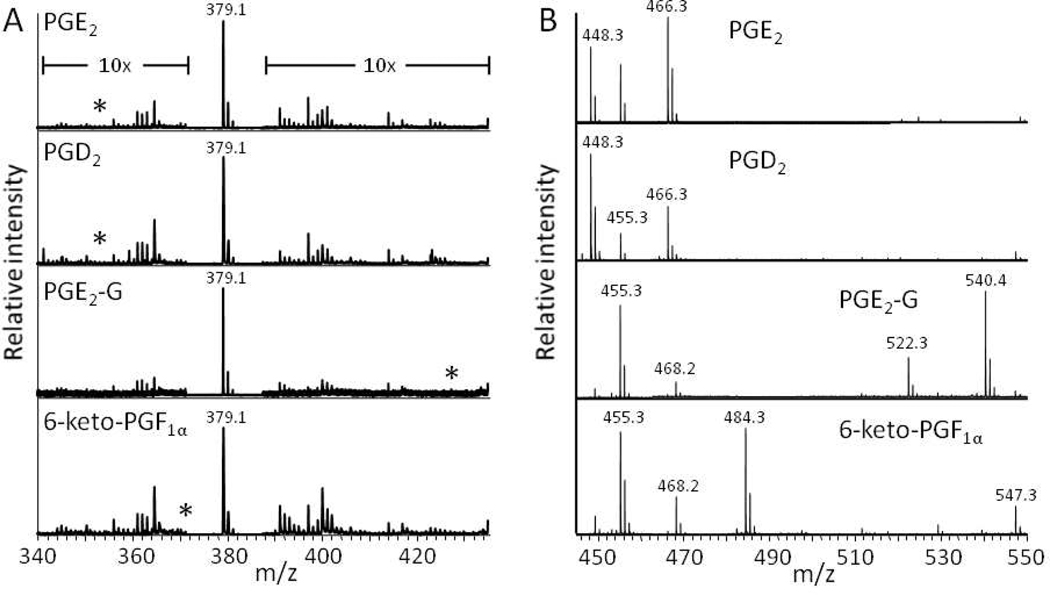
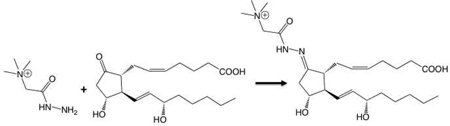
The ability to directly analyze prostaglandins using standard MALDI protocols and a number of different matrices in either positive or negative mode was assessed. Figure 2a shows the mass spectrum for 3 PGs and a PG-G. All spectra were dominated by peaks from the matrix (CHCA). It was determined that a charged moiety could be added to the analytes in order to improve detection by pre-ionizing the compounds. Girard’s T reagent reacts with ketones to give hydrazone derivatives bearing charged moieties15. The given equation displays the chemical reaction between PGE2 and Girard’s T reagent. Figure 2b represents the mass spectra obtained from the same prostaglandins derivatized with the Girard’s T reagent. The mass spectra are dominated by the expected masses of the derivatized prostaglandins (PGE2 m/z = 466.3, PGD2 m/z = 466.3, PGE2-G m/z = 540.3 and 6-keto-PGF1α m/z = 484.3) and have a much higher signal-to-noise ratio than their underivatized counterparts (Fig. 2a). Ions were observed at m/z = 448 in both PGE2 and PGD2 spectra corresponding to dehydration products likely formed by the acidic Girard’s reagent T. (A contaminant ion was observed in all four spectra at m/z = 455.)
Figure 2.
MALDI mass spectra of PGE2, PGD2, PGE2-G, and 6-keto-PGF1α before (A) and after (B) derivatization with Girard’s T reagent. All mass spectra were obtained with CHCA matrix in the positive ion mode. The spectra in (A) are dominated by a protonated CHCA-dimer signal at m/z 379.1. The area surrounding this signal was increased 10x for all four spectra. The expected [M+H]+ ions are indicated with asterisks in (A) at m/z 353.2, 353.2, 427.3, and 371.2 for PGE2, PGD2, PGE2-G, and 6-keto-PGF1α, respectively. The expected Girard’s T products are shown in (B) at m/z 466.3, 466.3, 540.4, and 484.3, respectively.
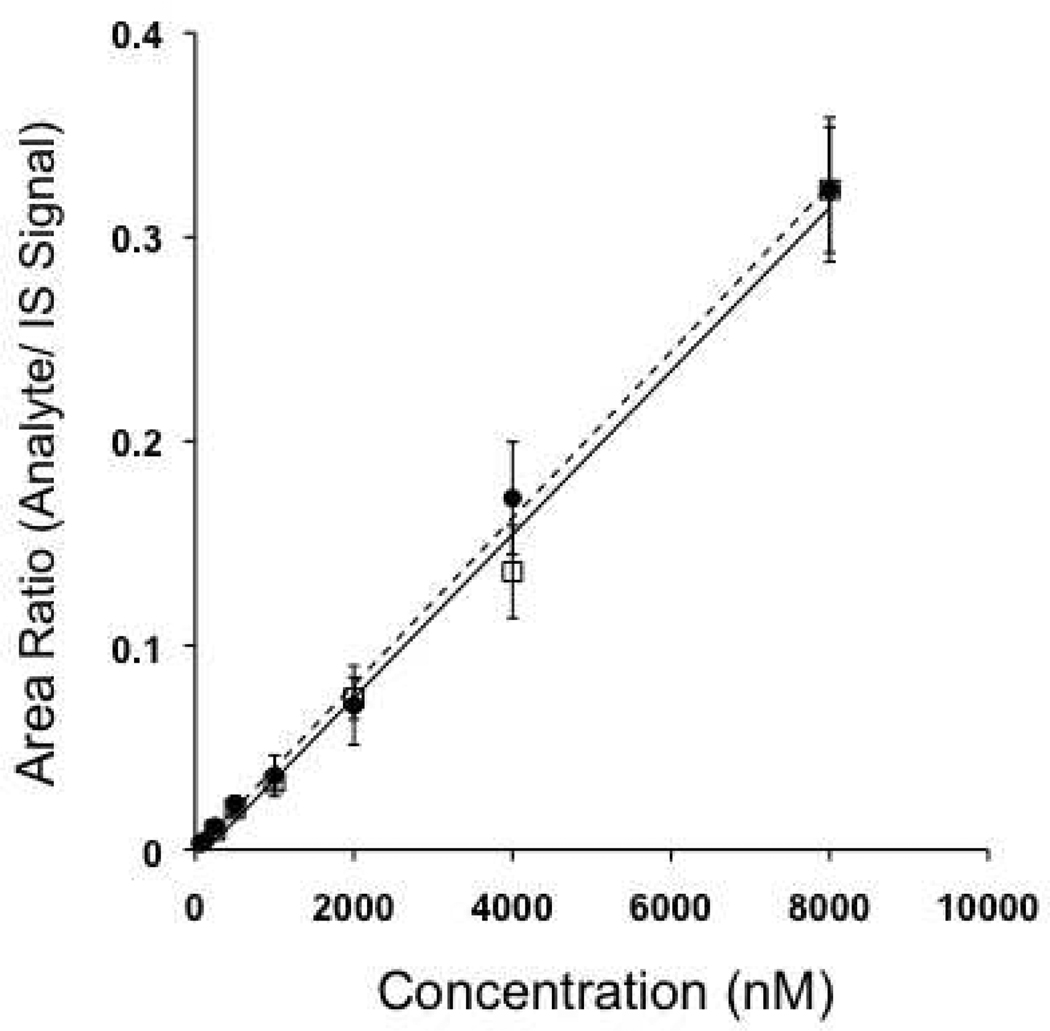
To assess linearity, the lower limit of detection, and the potential for signal interference, an 8-point concentration curve of PGE2 and PGE2 mixed with PGE2-G (PGE2-d4 was added as internal standard) was constructed in triplicate from 15 fmol on-plate to 2000 fmol on-plate. Quantification of PGE2 and PGE2-G was based on the MS2 transitions: m/z 466.3→389.3 for PGE2 and m/z 540.3→463.3 for PGE2-G (see supplemental Fig. 1). Figure 3 displays the concentration-response curves for PGE2 alone and PGE2 in solution with PGE2-G. The regression of the lines for PGE2 alone and PGE2 mixed with PGE2-G are similar and neither slope differs significantly from unity (r2=0.99). No signal interference was observed between the two curves demonstrating that neither prostaglandin affected the analysis of the other on-plate. This signal response and sensitivity is comparable to that obtained from LC-MS/MS (data not shown).
Figure 3.
Representation of standard curves for both PGE2 in solution alone (●) and PGE2 in solution with PGE2-G ( ). The equations representing the depicted regression lines are: for PGE2 alone, y = 4e-5x - 0.0008 and for PGE2 mixed with PGE2-G, y = 4e-5x - 0.005. Neither regression line differed significantly from unity (r2=0.99).
). The equations representing the depicted regression lines are: for PGE2 alone, y = 4e-5x - 0.0008 and for PGE2 mixed with PGE2-G, y = 4e-5x - 0.005. Neither regression line differed significantly from unity (r2=0.99).
COX-2 Inhibitor Assay: 14C-TLC or Oxygen Consumption vs. MALDI MS/MS
Analysis of COX activity and inhibition is routinely assessed by monitoring AA metabolism, prostaglandin formation or oxygen consumption utilizing either a 14C-thin layer chromatography (TLC) method or a polarographic electrode to monitor oxygen consumption16, 17. Though both methods give accurate assessments for the analysis of COX activity, each has disadvantages. MALDI MS/MS analysis has the ability to analyze COX activity by using non-radioactively labeled material and has a much faster analysis time compared to either 14C-TLC or oxygen electrode-based analysis. In addition, the large dynamic range of the MALDI detection method enables the use of a wide range of enzyme, substrate, and inhibitor concentrations.
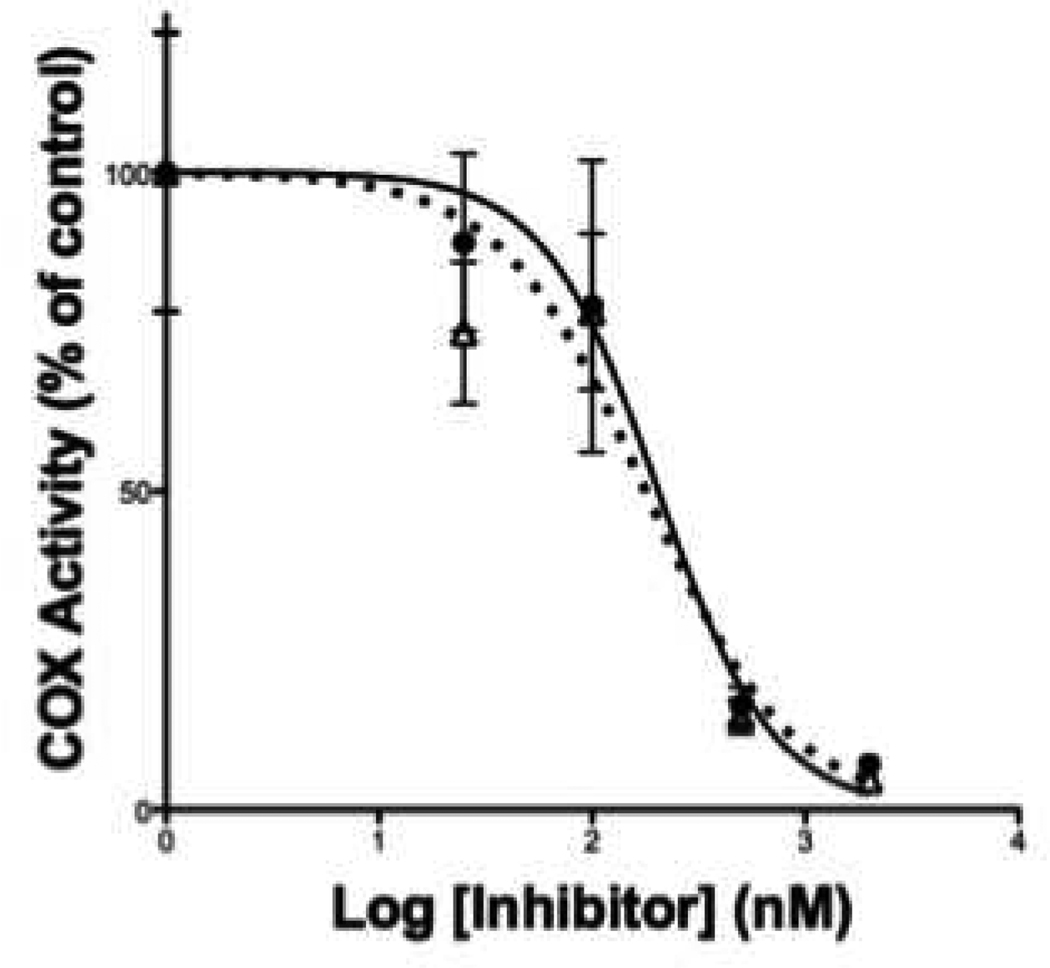
MALDI MS analysis was used to determine the IC50 value for inhibition of COX-2 by the known non-selective inhibitor, indomethacin. The results were compared with the IC50 values obtained from either 14C-TLC or oxygen electrode assays. Figure 4 displays the IC50 curves obtained from MALDI MS analysis for indomethacin. The IC50 value for indomethacin obtained from the 14C-TLC method was 210 nM18 which is comparable to the IC50 of 200 nM obtained from MALDI MS/MS analysis. The IC50 value obtained using an oxygen consumption assay was 900 nM19 (Figure 4). The oxygen electrode used for the oxygen uptake method is relatively insensitive, which requires higher enzyme and substrate concentrations, which may account for the higher IC50 determined by this method. It is clear that by using MALDI MS analysis it is possible to obtain comparable results to either the 14C-TLC or the oxygen uptake methods in dramatically shortened times.
Figure 4.
IC50 values for COX-2 inhibitor, indomethacin, determined by MALDI MS/MS; validation of Girard’s reagent T as quench. The IC50 value of indomethacin inhibition of COX-2 was determined using either ethyl acetate as a quench (●) followed by Girard’s T reagent or direct addition of Girard’s reagent T in 20% acetic acid in ethanol as a quench (△),with similar IC50 values of 200 nM and 180 nM, respectively.
COX-2 Inhibitor Assay - High Throughput Screening (HTS) Method Development
The rapidity of analysis by MALDI MS/MS makes it attractive to combine it with high-throughput liquid handling. We proceeded to develop a high-throughput assay for the analysis of inhibitors against either isoform of COX by analysis of PGE2 formation. This was accomplished through a combination of ultra-low volume acoustic liquid handling using a Labcyte Echo to produce the test compound plates and a Bravo liquid handler controlled by VWorks (Agilent, Santa Clara, CA, USA) using a 384-tip pipetting head. The highly accurate and rapid pipetting and mixing with the Bravo together with the precise timing of assay steps afforded by using VWorks allowed facile, rapid, and reproducible execution of 384 parallel enzyme reactions, derivatizations and spotting operations. We found that reactions could be quenched with Girard’s T reagent, which eliminated the liquid-liquid extraction and sample evaporation steps. Figure 4 presents a comparison of the concentration-dependent inhibition curves obtained with indomethacin using ethyl acetate quench or direct addition of Girard’s T as a quench. Both methods gave comparable results so it was determined that a high-throughput assay could be developed without the need for a liquid-liquid extraction.
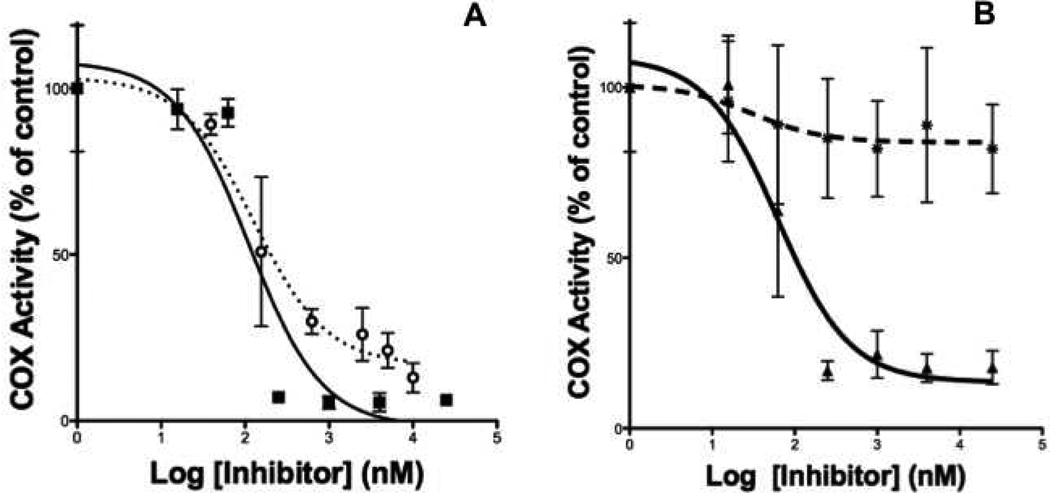
Once the method was developed, we validated the assay by assessing the IC50 values obtained from four known COX-2 inhibitors. Figure 5A displays the results obtained from the analysis of the inhibition of COX-2 to metabolize AA by indomethacin and sulindac sulfide. The measured IC50 values (indomethacin IC50 = 110 nM, sulindac sulfide IC50 = 290 nM) are comparable to previously reported values. Although there is some difference between the value of 110 nM for indomethacin measured with a high-throughput assay and the value of 200 nM measured with manual pipetting and sample handling, the difference was not statistically significant (students t-test, p = 0.4)16,17, 20. In addition to potent COX-2 inhibitors, the ability to distinguish an inhibitor from a non-inhibitor was determined by assessing COX-2 inhibition by stereoisomers flurbiprofen. Figure 5B displays the results obtained from the analysis of inhibition of COX-2 to metabolize AA by (S)-flurbiprofen and (R)-flurbiprofen. The measured IC50 values ((S)-flurbiprofen, IC50 = 120 nM; and (R)-flurbiprofen, no inhibition) were comparable to previously reported values18, 19, 21. The HTS method is capable of assessing COX-2 inhibition of a number of different known COX-2 inhibitors as well as distinguishing between compounds that do and do not inhibit COX-2.
Figure 5.
IC50 values determined for COX-2 inhibitors using the high throughput assay. (A) COX-2 inhibitors indomethacin (■) and sulindac sulfide( ) were assessed against COX-2 metabolism of AA. IC50 values determined were 110 nM and 290 nM, respectively. (B) Sterioisomers R-flurbiprofen (✱) and S- flurbiprofen (▲) were assessed against COX-2 metabolism of AA. IC50 values determined were no inhibition and 120 nM, respectively.
) were assessed against COX-2 metabolism of AA. IC50 values determined were 110 nM and 290 nM, respectively. (B) Sterioisomers R-flurbiprofen (✱) and S- flurbiprofen (▲) were assessed against COX-2 metabolism of AA. IC50 values determined were no inhibition and 120 nM, respectively.
MALDI TOF analysis of Activated Macrophages
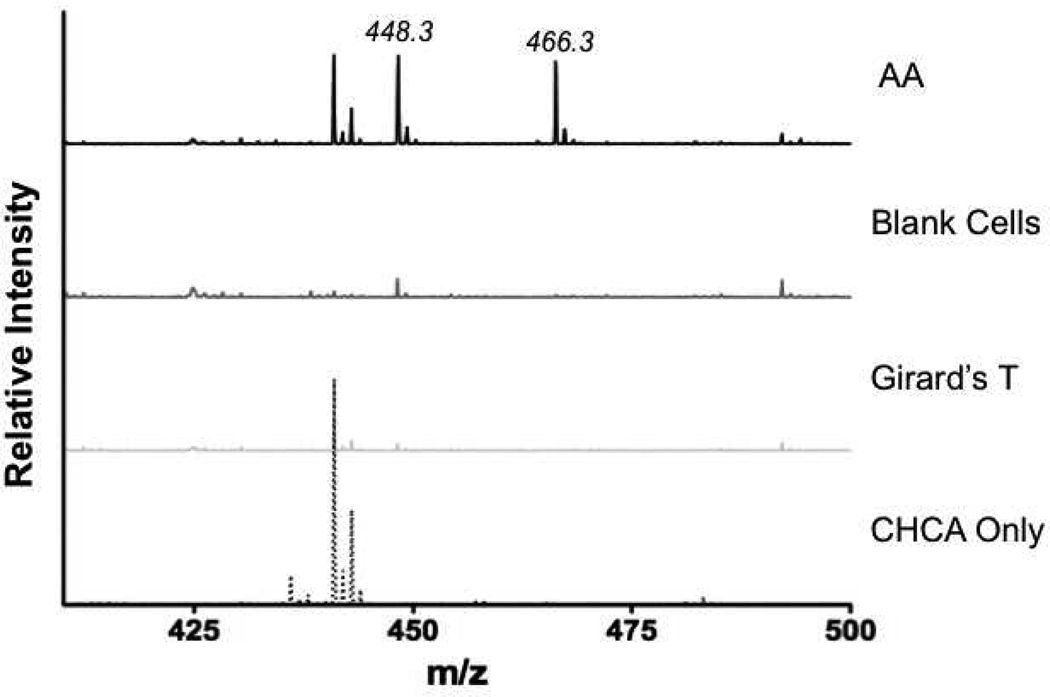
Analysis of AA metabolism in cultured cells has been performed mainly by LC-MS analysis22. These methods have demonstrated the formation of a number of different prostaglandins, including PGE2/D2 and 15-keto-13,14-dihydro-PGE2. Previous work also has shown that RAW cells treated with LPS/INF-γ have increased levels of COX-2, and addition of AA leads to increased production of PGD211,23–25. To assess whether this derivatization method could be applied to the analysis of cellular metabolism of AA, RAW cells were treated as described in Materials and Methods. Medium from these cells was extracted and analyzed on a MALDI TOF MS. Figure 6 is a comparison between blank CHCA, Girard’s T reagent only, untreated RAW cells and RAW cells treated with AA. When cells were treated with AA, increased levels of PGE2/PGD2 (m/z = 466.3) were detected which were similar to published studies of LPS/INF-γ treated RAW cells23. Previous investigations have shown that the ratio of PGD2-to-PGE2 produced by RAW cells is greater than 10-to-123. Previous work from our lab has shown that activation of RAW cells with LPS treatment stimulated the production of PGD2 compared to untreated samples23. The utilization of the MALDI MS method allows us to quantify prostaglandins at levels similar to those quantified by LC-MS/MS analysis.
Figure 6.
MALDI MS of media extracted from LPS-activated RAW cells. Cells dosed with AA (AA) showed increases in the formation of PGE2/PGD2 (m/z = 466.3)
Discussion
We have successfully analyzed ketone-containing prostaglandins after derivatization with Girard’s T reagent by MALDI MS. This procedure will not distinguish between isomeric species (such as PGE2 and PGD2 or 6-oxo-PGF1-alpha and thromboxane B2), however additional analytical procedures such as tandem MS can be utilized. We assessed the suitability of using MALDI to detect the production of prostaglandins in an endpoint assay for COX-2 inhibition. Initially MALDI MS was used as the analytical tool to detect derivatized prostaglandins, however we wanted to assess its suitability for high-throughput analyses. We determined that Girard’s T reagent solution could be added as the quench for the assay, making it suitable for the automated process. We utilized state-of-the art liquid handling robotics to assay the biological activity of COX-2 in the presence of several inhibitors. The entire process was automated from substrate aliquoting, enzyme addition, reaction quenching, matrix mixing, application to a 384-well MALDI plate and automated MS analysis. Three hundred eighty-four independent biochemical assays can be completed from initial incubation to final analysis in approximately 6 hr. This is a drastic improvement in time and number of samples analyzed compared to other analytical techniques. For example, a similar number of assays would take at least a week to manually perform the incubation, liquid-liquid extraction, and pre-concentration steps, as well as a further 18 hr for the HPLC-MS/MS analysis.
Additionally, we wanted to assess the capability of this newly developed MALDI method to analyze more complex cellular samples. We showed that we are able to obtain comparable results to LC-MS, again in a fraction of the time. This was demonstrated from the analysis of extracted media from activated macrophages where we found increased production of PG’s in AA-treated RAW cells compared to untreated cells. Individual data points were acquired in less than a second using no more than 1 µL of samples compared to ~15 min for a complete LC separation13.
In the quarter of a century since MALDI was first described, it has been one of the prime technologies that has revolutionized the analysis of biological samples, especially the study of peptides and proteins. As the speed and resolution of mass spectrometry technology has advanced, it now holds great potential for high-throughput analysis. This report has demonstrated the efficacy of MALDI MS for the high-throughput, sensitive, and quantitative analysis of prostaglandins in complex samples including those extracted from whole cell media. This technology has wide application in many areas of biological and chemical analysis.
Supplementary Material
Acknowledgements
This work was supported by research grants (GM15431, GM58008, and CA89450) and a training grant (GM65086) from the National Institutes of Health and a center grant from the National Foundation for Cancer Research. We are grateful to Dan Dorset in the Vanderbilt High-Throughput Screening Facility for assistance with the liquid-handling equipment.
References
- 1.Karas M, Bachmann D, Bahr U, Hillenkamp F. Int. J. Mass Spectrom. Ion Proc. 1987;78:53–68. [Google Scholar]
- 2.Miura D, Fujimura Y, Tachibana H, Wariishi H. Anal. Chem. 2009;82:498–504. doi: 10.1021/ac901083a. [DOI] [PubMed] [Google Scholar]
- 3.Holle A, Haase A, Kayser M, Höhndorf J. J. Mass Spectrom. 2006;41:705–716. doi: 10.1002/jms.1041. [DOI] [PubMed] [Google Scholar]
- 4.Nordstrom A, Want E, Northen T, Lehtio J, Siuzdak G. Anal. Chem. 2008;80:421–429. doi: 10.1021/ac701982e. [DOI] [PubMed] [Google Scholar]
- 5.Kubo A, Ohmura M, Wakui M, Harada T, Kajihara S, Ogawa K, Suemizu H, Nakamura M, Setou M, Suematsu M. Anal. Bioanal. Chem. 2011;400:1895–1904. doi: 10.1007/s00216-011-4895-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rouzer CA, Marnett LJ. J. Biol. Chem. 2008;283:8065–8069. doi: 10.1074/jbc.R800005200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nirodi CS, Crews BC, Kozak KR, Morrow JD, Marnett LJ. Proc. Natl. Acad. Sci. USA. 2004;101:1840–1845. doi: 10.1073/pnas.0303950101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narumiya S, Toda N. Br. J. Pharmacol. 1985;85:367–375. doi: 10.1111/j.1476-5381.1985.tb08870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe T, Narumiya S, Shimizu T, Hayaishi O. J. Biol. Chem. 1982;257:14847–14853. [PubMed] [Google Scholar]
- 10.Murphy RC, Barkley RM, Zemski Berry K, Hankin J, Harrison K, Johnson C, Krank J, McAnoy A, Uhlson C, Zarini S. Anal. Biochem. 2005;346:1–42. doi: 10.1016/j.ab.2005.04.042. [DOI] [PubMed] [Google Scholar]
- 11.Blair IA, Barrow SE, Waddell KA, Lewis PJ, Dollery CT. Adv. Prostag. Thromb. L. 1983;11:197–205. [PubMed] [Google Scholar]
- 12.Barrow SE, Heavey DJ, Ennis M, Chappell CG, Blair IA, Dollery CT. Prostaglandins. 1984;28:743–754. doi: 10.1016/0090-6980(84)90032-7. [DOI] [PubMed] [Google Scholar]
- 13.Kingsley PJ, Rouzer CA, Saleh S, Marnett LJ. Anal. Biochem. 2005;343:203–211. doi: 10.1016/j.ab.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Rowlinson SW, Crews BC, Lanzo CA, Marnett LJ. J. Biol. Chem. 1999;274:23305–23310. doi: 10.1074/jbc.274.33.23305. [DOI] [PubMed] [Google Scholar]
- 15.Hong HH, Wang YS. Anal. Chem. 2007;79:322–326. doi: 10.1021/ac061465w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalgutkar AS, Kozak KR, Crews BC, Hochgesang GP, Jr, Marnett LJ. J. Med. Chem. 1998;41:4800–4818. doi: 10.1021/jm980303s. [DOI] [PubMed] [Google Scholar]
- 17.Kulmacz RJ, Miller JF, Jr, Pendleton RB, Lands WE. Methods Enzymol. 1990;186:431–438. doi: 10.1016/0076-6879(90)86136-j. [DOI] [PubMed] [Google Scholar]
- 18.Kalgutkar AS, Crews BC, Rowlinson SW, Marnett AB, Kozak KR, Remmel RP, Marnett LJ. Proc. Natl. Acad. Sci. USA. 2000;97:925–930. doi: 10.1073/pnas.97.2.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gierse JK, Hauser SD, Creely DP, Koboldt C, Rangwala SH, Isakson PC, Seibert K. Biochem. J. 1995;305:479–484. doi: 10.1042/bj3050479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walters MJ, Blobaum AL, Kingsley PJ, Felts AS, Sulikowski GA, Marnett LJ. Bioorg. Med. Chem. Lett. 2009;19:3271–3274. doi: 10.1016/j.bmcl.2009.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnett J, Chow J, Ives D, Chiou M, Mackenzie R, Osen E, Nguyen B, Tsing S, Bach C, Freire J, et al. Biochim. Biophys. Acta. 1994;1209:130–139. doi: 10.1016/0167-4838(94)90148-1. [DOI] [PubMed] [Google Scholar]
- 22.Yang P, Felix E, Madden T, Fischer SM, Newman RA. Anal. Biochem. 2002;308:168–177. doi: 10.1016/s0003-2697(02)00218-x. [DOI] [PubMed] [Google Scholar]
- 23.Rouzer CA, Jacobs AT, Nirodi CS, Kingsley PJ, Morrow JD, Marnett LJ. J. Lipid Res. 2005;46:1027–1037. doi: 10.1194/jlr.M500006-JLR200. [DOI] [PubMed] [Google Scholar]
- 24.Mesaros C, Lee SH, Blair IA. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2009;877:2736–2745. doi: 10.1016/j.jchromb.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kempen EC, Yang P, Felix E, Madden T, Newman RA. Anal. Biochem. 2001;297:183–190. doi: 10.1006/abio.2001.5325. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.