Abstract
Brain Angiogenesis Inhibitor 1 (BAI1) is a putative G protein-coupled receptor with potent anti-angiogenic and anti-tumorigenic properties that is mutated in certain cancers. BAI1 is expressed in normal human brain, but it is frequently silenced in glioblastoma multiforme (GBM). In this study we show this silencing event is regulated by overexpression of methyl-CpG-binding domain protein 2 (MBD2), a key mediator of epigenetic gene regulation, which binds to the hypermethylated BAI1 gene promoter. In glioma cells, treatment with the DNA demethylating agent 5-aza-2′-deoxycytidine (5-Aza-dC) was sufficient to reactivate BAI1 expression. Chromatin immunoprecipitation (ChIP) showed that MBD2 was enriched at the promoter of silenced BAI1 in glioma cells and that MBD2 binding was released by 5-Aza-dC treatment. RNAi-mediated knockdown of MBD2 expression led to reactivation of BAI1 gene expression and restoration of BAI1 functional activity, as indicated by increased anti-angiogenic activity in vitro and in vivo. Taken together, our results suggest that MBD2 overexpression during gliomagenesis may drive tumor growth by suppressing the anti-angiogenic activity of a key tumor suppressor. These findings have therapeutic implications since inhibiting MBD2 could offer a strategy to reactivate BAI1 and suppress glioma pathobiology.
Keywords: MBD2, BAI1, glioblastoma, epigenetic, overexpression
Introduction
DNA methylation is a naturally occurring event that consists of the addition of a methyl group to the fifth carbon position of the cytosine pyrimidine ring by DNA methyltransferases. Alterations in the patterns of DNA methylation are widespread in human cancers and include genome-wide hypomethylation and the hypermethylation of CpG island-associated gene promoters, the latter of which represents one mechanism leading to the epigenetic silencing of genes in human cancers (1, 2). DNA methylation alterations have been widely reported in human glioblastoma (GBM), a highly vascularized and aggressive primary intracranial tumor (3–7). A distinct subgroup of primary GBM displays concordant hypermethylation at a large number of loci, indicating the existence of a glioma CpG island methylator phenotype (gCIMP) (8). Interestingly, the subset of GBM exhibiting gCIMP is associated with isocitrate dehydrogenase (IDH) mutations providing a link to an altered metabolic profile (9).
Methyl-CpG-binding domain (MBD) proteins interpret the DNA methylation marks and thus are critical mediators of many epigenetic processes (10–12). The MBD family comprises five members; MBD1-4 and MeCP2. MBD1, MBD2 and MeCP2 bind selectively to methylated CpGs and repress transcription from methylated promoters in vitro and in vivo. By contrast, MBD3 binding is not dependent on DNA methylation and MBD4, while selective for methylated DNA, has been primarily characterized as a thymine DNA glycosylase with little role in transcriptional repression (10–12). However, the expression pattern and functional roles of MBDs in glioblastoma pathogenesis remain yet unidentified.
BAI1 is an orphan GPCR-like receptor abundantly expressed in normal brain with potent anti-angiogenic and anti-tumorigenic properties that was initially identified in a screen for p53-regulated genes (13–18). Importantly, BAI1 and its related family members BAI2 and BAI3 were recently found to undergo somatic mutation in several cancers, including lung, breast and ovarian cancers (19). BAI1 contains several well-defined protein modules in the N-terminus such as an integrin binding RGD motif followed by five thrombospondin type 1 repeats (TSRs), a hormone binding domain and a G-protein-coupled receptor proteolytic cleavage site (GPS) (16). The TSRs within the extracellular region of BAI1 mediate direct binding to phosphatidylserine on apoptotic cells and BAI1 can cooperate with the engulfment and cell motility 1 (ELMO1)/dedicator of cytokinesis 1 (Dock180)/Rac to promote maximal engulfment of apoptotic cells (20). Interestingly, the ELMO1/Dock180 association is also involved in the invasive phenotype of glioma cells (21). The C-terminus is less well characterized and has a QTEV motif that mediates binding to PDZ domain-containing proteins. The BAI1 N-terminal extracellular domain can be cleaved at the GPS site and the resulting 120 kDa fragment known as vasculostatin (Vstat120), is able to inhibit angiogenesis in vitro and suppress intracranial tumor growth in vivo (14, 15). A second N-terminal cleavage site was recently identified, generating a smaller vasculostatin (Vstat40) (17).
Our previous results demonstrated that BAI1 expression was absent in most human glioma cell lines and primary glioblastoma samples examined (22), but the underlying mechanisms remain unknown. In the present study, we provide evidence that MBD2 is upregulated in glioblastomas and that it plays a central role in the epigenetic silencing of BAI1 gene expression, thereby suppressing the anti-angiogenic activity of BAI1.
Materials and Methods
Primary tumors and cell lines
The primary GBM tumor samples were obtained from Emory University Hospital and were reviewed by neuropathologists (Dr. Daniel J. Brat and Dr. Stephen B. Hunter) for histological confirmation of GBM before being included in this study. Human GBM cell lines LN71, SF188 and LN443 were originally established in our lab (22). Human GBM cell lines U87MG, LN229 and U251MG were obtained from the American Type Culture Collection (ATCC) and maintained as described (23). All cell lines were authenticated by the ATCC for viability, morphology and isoenzymology. Human brain microvascular endothelial cells (HBVECs) were purchased from Cell Systems Corp (Kirkland, WA). For chemical treatment, glioma cells were plated (3 × 105 cells/l00-mm dish) and treated 24 h later with 5-Aza-dC (5 μM, Sigma) for 1 to 5 days.
DNA methylation analysis of the BAI1 gene exon 1
We determined CpG island methylation status by bisulfite-sequencing and methylation-specific PCR (MS-PCR) as previously described (3, 24). Additional details, including primer sequences, are provided in Supplementary Materials and Methods.
Reverse Transcription-PCR (RT-PCR)
To determine the messenger RNA (mRNA) levels of the BAI1 gene, RT-PCR was performed on the total RNA extracted from the cells or GBM samples as described (13). Additional details are provided in Supplementary Materials and Methods.
Western blot
Western blot was performed as described (15). The antibodies used were mouse anti-MBD2 (Abcam, cat# ab45027), rabbit anti-MeCP2 (Abcam, cat# ab2828), goat anti-actin (Santa Cruz Biotechnology) and rabbit anti-BAI1 (22). The HRP-conjugated secondary antibodies and enhanced chemiluminescence were from Thermo.
Immunohistochemical Analysis
Immunohistochemistry was performed on archived formalin-fixed and paraffin-embedded human GBM resection specimens. For the tissue array study, 5 non-neoplastic brain and 54 GBM tumor specimens were sectioned and mounted on 2 slides. Sections were deparaffinized and subjected to antigen retrieval by boiling (20 minutes, 100°C) in 0.01 M Tris HCL (pH 10). Slides were then incubated with a 1:200 dilution of MBD2 antibody. Immunostaining was detected with the avidin-biotin complex method, using diaminobenzidine as the chromogen (Abcam). Slides were scanned at 40x resolution with a Nanozoomer 2.0 HT (Hamamatsu) and staining intensity (Five fields/tumor ) was quantified by the MetaMorph Premier software; MBD2 status was assessed based on relative staining intensity unit [absent (0), weak (1) (units, 1–75), moderate (2) (units, 76–150), strong (3) (units, 151–225)] and percentage of positive tumor cells [0% (0), <10% (1), 10–50% (2), 51–80% (3), 81–100% (4)]. Immunoreactivity scores (IHC scores) were determined through multiplying the staining score by the percentage score to give a maximum of 12 (25).
Chromatin Immunoprecipitation Assay (ChIP)
ChIP was performed using a commercial kit (Cell Signaling, cat# 9003) with some modifications. After cross-linking, the cells were lysed and sonicated using a Misonix sonicator MX2020 (settting 15, 15 seconds for 3 times). Sonicated lysates were centrifuged at 14,000 rpm at 4°C for 15 minutes to get rid of insoluble fractions. An aliquot of the chromatin preparation was set aside and designated as input fraction. The cleared chromatin (100 μg) was immunoprecipitated with 2 μg of either anti-MBD2 or anti-MeCP2 antibody and incubated overnight at 4°C with rotation. The second day, salmon sperm DNA/Protein A/G agarose slurry was added to these samples and rocked for 4 hours at 4°C. Protein A/G immune complexes were collected and washed. Immune complexes were eluted and DNA was recovered by DNA purification columns, and analyzed by PCR. The primers used were 5′-GCT CAC TCT GAC CCT CTG CTC TTTC-3′ (forward) and 5′-AGT AGC CGA AGA ACT TTC CCT GC-3′ (reverse) for BAI1 promoter, the primers used for MGMT promoter were described previously (26). Acetyl-Histone H3 (Lys9) antibody was from Cell Signaling (cat# 9649S) and Histone H3 (tri-methyl-K9) antibody was from Abcam (cat# 8898).
Construction of short hairpin RNA (shRNA) vectors and transient transfection
Constructs for shRNA were generated with the BLOCK-iTU6 RNAi Entry Vector kit (Invitrogen) as described (27), and primer sequences are provided in Supplementary Materials and Methods. Transient transfection of glioma cells with plasmid DNA (2 μg/60 mm dish) was performed with lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen) with minor modification (27).
Scratch-wound endothelial cell migration assay
This assay was performed as previously described (14). In brief, conditioned medium (CM) from glioma cells transfected with shRNAs was collected and concentrated 100x using an UltraCel filter (Amicon). Confluent HBVECs were incubated in 1% serum medium overnight in 12-well plates, then wounded with a 10 μL pipette tip and detached cells were removed by PBS washes. The cells were then treated with CM collected as above and diluted to 10x in endothelial cell culture to induce cell migration. Initial wound width was measured, and the cells were allowed to migrate for 8 hours, and wound width was measured again. The experiment was repeated independently 3 times and the significance was determined by Student’s t test.
In vivo angiogenesis assay
Quantification of the anti-angiogenic responses was performed utilizing the directed in vivo angiogenesis assay (DIVAA) as previously described (28) with the DIVAA Inhibition Assay Kit (Trevigen, cat# 3450-096-IK). Collection of CM from glioma cells transfected with shRNAs was described as in scratch-wound endothelial cell migration assay. 2 μL of 100x concentrated CM was mixed with 18 μL Matrigel containing growth factors and filled into sterile surgical silicone tubing (“angioreactors”). These angioreactors were incubated at 37°C for 1 hour to allow for gel formation, before subcutaneous implantation into the dorsal flank of athymic nude mice (females, 6 to 8 weeks of age; National Cancer Institute, Frederick, MD). 2 weeks later, angioreactors were harvested and the Matrigel was removed and digested. Cell pellets and insoluble fractions were collected by centrifugation at 5000 × g for 2 minutes. The cell pellets were washed and incubated at 4°C overnight in 200 μl of 25 μg/ml of FITC-labeled Griffonia lectin (FITC-lectin), an endothelial cell selective reagent. The relative fluorescence was measured in 96-well plates using a Molecular Device spectrofluorometer (excitation 485 nm, emission 510 nm; Sunnyvale, CA). The mean relative fluorescence ±SD for eight replicate assays was determined.
Results
Down-regulation of BAI1 gene expression in glioblastoma is correlated with aberrant DNA methylation of exon 1
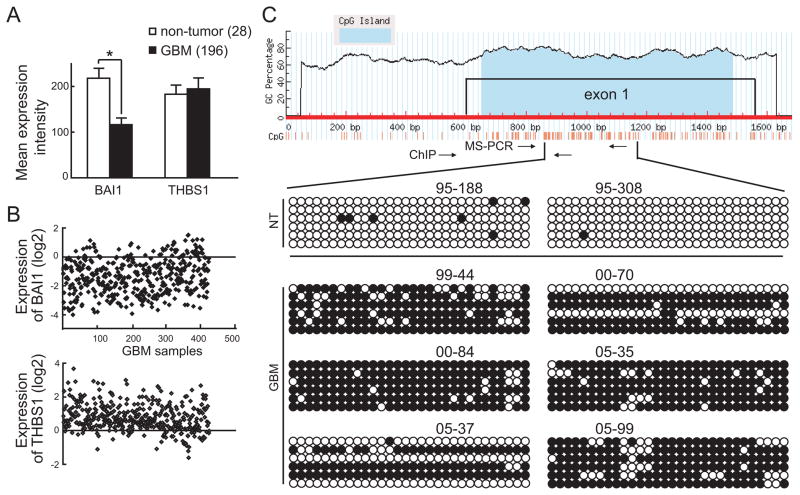
Our previous studies on a limited sample set suggested loss of expression of the BAI1 tumor suppressor in human GBM specimens and cell lines (22), although no mechanism was identified. To independently confirm and investigate the extent of BAI1 loss in GBM, we analyzed large datasets from two brain tumor databases, namely the NCI Repository of Molecular Brain Neoplasia Data (Rembrandt) and The Cancer Genome Atlas (TCGA). The expression of BAI1 was first determined in 28 non-tumor brain tissues and 196 GBM samples (institutional diagnosis) in the Rembrandt dataset. As shown in Fig. 1A, the levels of BAI1 gene expression were significantly decreased (p<0.01) in GBM samples compared with the non-tumor tissues. In contrast, the expression of THBS1, which encodes angiogenesis inhibitor thrombospondin 1 harboring 3 TSRs, showed no change (Fig. 1A). In the TCGA dataset, the expression of BAI1 was available in a total of 424 GBM samples. Analysis of this dataset also showed a consistent and dramatic loss of BAI1 expression, with 250 samples (59%) showing more than a 2-fold decrease (Fig. 1B). In contrast, the expression of THBS1 increased in 374 samples (88%) (Fig. 1B). The variation in relative THBS1 expression between the Rembrandt and TCGA databases may be due to the different probe sets and array platforms used. Taken together, these data suggest that a large fraction of primary GBM exhibit a significant loss or down-regulation of BAI1 mRNA expression.
Figure 1.
Aberrant DNA methylation at BAI1 exon 1 in GBM. A, down-regulation of BAI1 gene expression in human glioblastomas. Raw data from Rembrandt Affymetrix Human Genome HTS U133A 2.0 Array for BAI1 and THBS1 expression in 28 non-tumor and 196 GBM samples was analyzed and quantified. *, p<0.01. B, expression of BAI1 and THBS1 in 424 GBM samples from the TCGA database. The normalized expression values are expressed as log ratios (base 2) for the y-axis, each spot representing one sample (x-axis). C, analysis of CpG island methylation in the BAI1 exon 1 in human glioblastomas by bisulfite-sequencing. Top, schematic diagram indicates the structure of the BAI1 gene promoter showing the CpG island located in exon 1. The CpG island was identified by the MethyPrimer software. Bottom, bisulfite-sequencing results on 2 brain samples from non-tumor (NT) patients and 6 samples from human GBM patients are included. 6 sequenced clones per sample are shown. The methylation status of the individual CpG dinucleotides is shown by unmethylated (empty) or methylated (filled) circles.
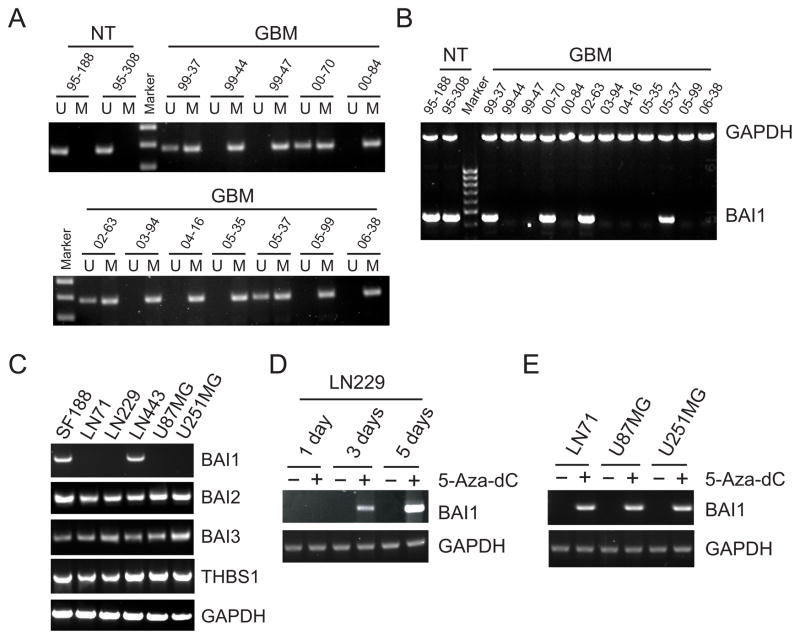
To identify the mechanism underlying BAI1 downregulation, we first considered whether BAI1 might be located in a region of genomic loss in GBM. The BAI1 gene is located on chromosome 8q24, a region that is not reported to exhibit loss of heterozygosity in gliomas, and a fact we confirmed by analyzing the TCGA genomic dataset (results not shown). We considered next the possibility of epigenetic mechanisms of silencing of the BAI1 gene. Using the MethPrimer software, we identified a CpG island in the first exon of the BAI1 gene (Fig. 1C). Bisulfite-sequencing was used to determine the methylation pattern in exon 1 in two non-tumor brain (NT) and six independent GBM samples; six clones per sample were sequenced. While the non-tumor samples were mostly methylation-free, extensive methylation was detected in the GBM samples, with most CpG sites found methylated in 31 of the 36 clones sequenced (Fig. 1C). The few clones that showed minimal or no methylation could have been derived from stromal tissue within the tumor or represent tumor heterogeneity. To further confirm the bisulfite-sequencing result, the DNA methylation status of the six tumors above plus an additional six tumors and two control samples from normal human brain white matter were analyzed by MS-PCR. Eight of 12 GBM samples exhibited prominent PCR products with the methylated primer set, but no products with the unmethylated primer set (Fig. 2A). In contrast, the two non-tumoral brain samples exhibited detectable PCR bands only from the unmethylated primer set. Examination of BAI1 expression in the same tumors showed an inverse correlation between methylation and gene expression in that expression of BAI1 was only observed in those samples with some degree of unmethylated DNA, while the GBM samples that were completely methylated lacked BAI1 expression altogether (Fig. 2B). These data indicate that an aberrant methylation pattern in exon 1 is associated with the silencing of BAI1 expression in a subset of GBM. The glioma-associated silencing appeared to be specific for BAI1 as the expression of two homologs, BAI2 and BAI3, and THBS1 did not vary among human glioma cell lines (Fig. 2C). We next examined the effect of a demethylating agent 5-Aza-dC on BAI1 gene expression. Treatment with 5 μM 5-Aza-dC for 5 days restored BAI1 mRNA expression in LN229 cells in a time-dependent manner (Fig. 2D). Similar reactivation was observed in three other BAI1-silent glioma cell lines (Fig. 2E). Since 5-Aza-dC is known to be highly effective at inducing the expression of genes inappropriately silenced by de novo methylation (29), these results suggest that DNA methylation of the BAI1 gene is likely involved in the gene silencing.
Figure 2.
Treatment of demethylating agent reactivates BAI1 gene expression. A, analysis of methylation pattern at the BAI1 exon 1 by methylation-specific PCR (MS-PCR). Normal tissue was found to be unmethylated (U), while methylation (M) was found in 8 of 12 GBM samples. B, RT-PCR analysis of BAI1 mRNA expression in normal brain and GBM samples, GAPDH mRNA was amplified as a loading control. C, BAI1 mRNA expression in human glioma cell lines analyzed by RT-PCR. Note BAI1 mRNA can only be detected in SF188 and LN443 cells, while, BAI2, BAI3 and THBS1 mRNAs can be detected in all six glioma cell lines. D, reactivation of BAI1 mRNA expression in a BAI1-silent glioma cell line using demethylation agent 5-Aza-dC (5 μM). LN229 cells were treated for 1, 3, and 5 days, the medium was changed every day with fresh chemicals. Cells mock-treated with the same volume of dimethyl sulfoxide (DMSO) were used as negative control. RNA was then isolated and RT-PCR was performed. E, reactivation of BAI1 gene expression by 5-Aza-dC (5 μM) in glioma cell lines LN71, U87MG and U251MG after 3 days of treatment.
The methyl-CpG binding protein MBD2 is selectively overexpressed in GBM
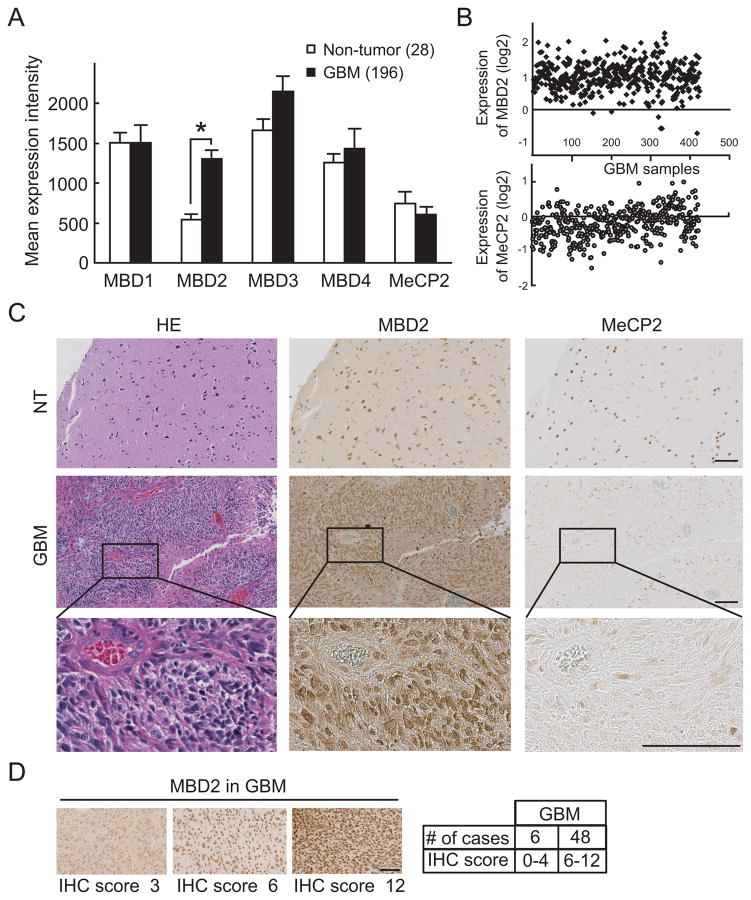
Since the impact of DNA methylation on gene silencing is often mediated through the binding of MBDs (30, 31), we then analyzed the expression of various MBD proteins in GBM by mining the expression data in the Rembrandt database. The expression of MBD1, MBD3, MBD4 and MeCP2 showed no significant difference between non-tumor control and glioblastomas (Fig. 3A). In contrast, we found that MBD2 is significantly overexpressed in GBM with a more than 2-fold greater mean expression in tumors as compared to non-tumor brain tissues. Comparison of the expression of MBD2 and MeCP2 in 424 GBM samples from the TCGA database also showed that MBD2 expression was markedly increased in a significant fraction of GBM (Fig 3B), while in a similar comparison the mean gene expression of MeCP2 was not significantly different. To determine whether MBD2 was also overexpressed at the protein level, we applied immunohistochemistry (IHC) on two randomly selected GBM specimens and found dramatically increased MBD2 immuno-positivity in tumor (GBM) vs. adjacent non-tumor (NT) areas (Fig 3C), consistent with the gene expression data. In contrast, MeCP2 exhibited only low to background levels of staining and most GBM tumor cells were negative for MeCP2 staining. The analysis of MBD2 protein expression in GBM was expanded in a tissue array containing 5 non-tumor brain samples and 54 GBM (Fig 3D). Weak to moderate MBD2 expression was detected in less than 50% of the tumor cells in 6/54 specimens (11%); these tumors were grouped as low-expressing tumors (IHC score 0–4). The remaining 48/54 tumors (89%) exhibiting moderate or strong MBD2 expression in >50% of tumor cells were included in a high-expressing group (IHC score 6–12). Taken together, these data suggest that MBD2 is significantly overexpressed in GBM.
Figure 3.
Overexpression of MBD2 mRNA and protein in GBM. A, expression of MBD family members in the Rembrandt brain tumor database. Expression data of 28 non-tumor and 196 GBM samples was analyzed and quantified from Rembrandt Affymetrix Human Genome HTS U133A 2.0 Array. Note a statistically significant increase in MBD2 mRNA expression in GBM as compared to non-tumoral brain *, p<0.01. B, expression data of MBD2 and MeCP2 from TCGA database in 424 GBM samples. The normalized expression values are expressed as log ratios (base 2) for the y-axis, each spot representing one sample (x-axis). C, IHC results of MBD2 and MeCP2 staining in non-tumor tissue (NT, upper panel) and in GBM tissue (middle and lower panel). Scale bar, 100 μm. D, representative images show the quantification of MBD2 staining intensity in GBM tissue arrays. Representative images from low-expressing (IHC score 0–4; 6 cases) and high-expressing GBMs (IHC score 6–12; 48 cases) are shown with the respective scores indicated. Scale bar, 100 μm.
MBD2 is necessary to maintain the silencing of BAI1
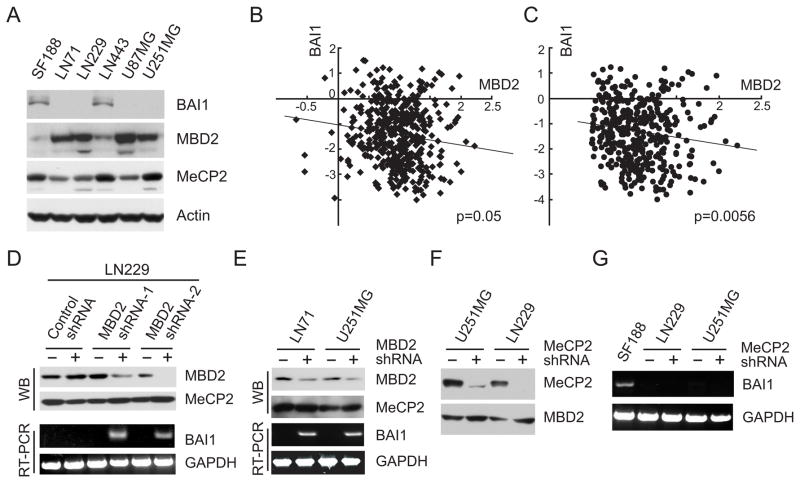
We next sought to determine whether there was any relationship between the levels of MBD2 and expression of BAI1 in glioma cell lines. The protein levels of MBD2 and BAI1 were determined and our data suggested a correlation between lack of BAI1 protein expression and elevated MBD2 protein levels (Fig. 4A). We next examined the relationship between the expression of MBD2 and BAI1 in primary GBM. Among the 424 primary GBM for which gene expression data were available from TCGA, there was a statistically significant negative correlation between the expression of MDB2 and BAI1 (Spearman correlation coefficient of −0.095; p=0.05; n=424) (Fig 4B). If one considers only those 373 tumors for which MBD2 was overexpressed by >= 1.4-fold relative to normal tissues (log2 >= 0.5), this association was even more significant (Spearman correlation coefficient of −0.14329; p=0.0056; n=373) (Fig 4C). Taken together, these data support a negative correlation between MBD2 and BAI1. To determine whether MBD2 played a direct role in BAI1 gene silencing, the endogenous levels of MBD2 mRNA were knocked down by transient transfection of specific shRNA expression vectors. We tested the effect of two MBD2 shRNAs first in LN229 cells, a glioma cell line that exhibits abundant MBD2 expression and is silent for BAI1 expression (Fig. 4A). Transfection with either shRNA exhibited a significant reduction in MBD2 protein expression (Fig. 4D), but had no effect on MeCP2 protein levels, indicating specificity of these shRNAs. Transfection of LN229 cells with the MBD2-specific shRNA expression vector led to a reactivation of BAI1 mRNA expression as compared with the non-specific control shRNA (Fig. 4D). Similar results were observed in two other BAI1-silent glioma cell lines (Fig. 4E). Since MeCP2 is known to be involved in transcriptional repression of multiple genes, we also designed MeCP2-specific shRNA vectors and determined their effects on BAI1 mRNA expression. While these vectors potently downregulated MeCP2 protein levels (Fig. 4F), they failed to reactivate BAI1 gene expression (Fig. 4G). Together, these results show that MBD2 contributes to BAI1 silencing.
Figure 4.
Reactivation of BAI1 by MBD2 knockdown. A, determination of protein expression of MBD family members in human glioma cell lines by Western blotting. Note low MBD2 levels in SF188 and LN443 cells, the only cell lines in which BAI1 protein is detected. B and C, correlation between the expression of MBD2 and BAI1 in TCGA GBM samples. The relative expression level of MBD2 mRNA is plotted against that of BAI1 mRNA in 424 total available TCGA GBM samples and in 373 GBM samples in which MBD2 is overexpressed (C). Expression values are expressed as log2 ratio of tumor/normal. D, knockdown of MBD2 reactivates BAI1 gene expression. LN229 cells were transiently transfected with two different shRNA expression vectors against human MBD2 or control scrambled shRNA. Two days later western blot (WB) was performed to determine the reduction in MBD2 levels and reactivation of BAI1 gene expression by RT-PCR. E, reactivation of BAI1 by knockdown of MBD2 using one shRNA expression vector in glioma cell lines LN71 and U251MG. Procedures as in B. F, the MeCP2 shRNA expression vector suppresses expression of MeCP2, but not MBD2. LN229 and U251MG cells were transfected with MeCP-shRNA or control shRNA. Lysates of transfected cells were subjected to immunoblotting using indicated antibodies after 48 hours. G, knockdown of MeCP2 expression is not able to reactivate BAI1 gene expression. BAI1 gene expression was examined by RT-PCR on total mRNAs extracted from the cells transfected in panel D. Expression of BAI1 mRNA in SF188 was used as a positive control.
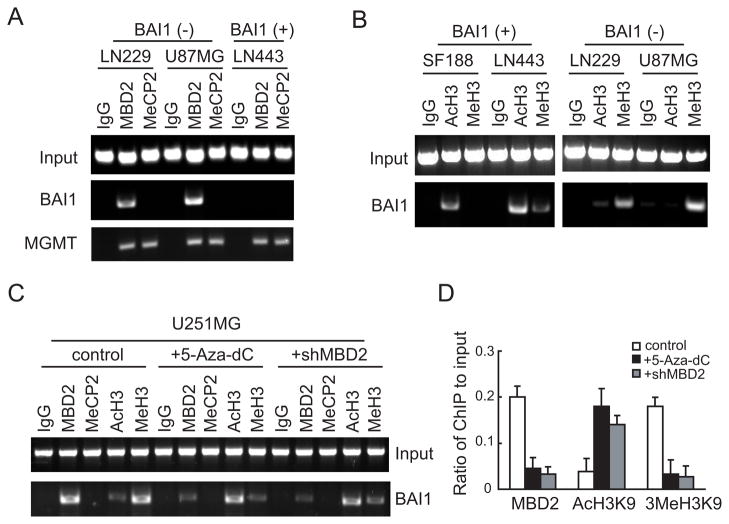
MBD2 is recruited to the BAI1 gene promoter in glioma cells
To further examine the role of MBD2 in the BAI1 gene regulation, we performed ChIP using antibodies against MeCP2 or MBD2. ChIP assays showed that MBD2 was enriched at the BAI1 promoter in the BAI1-silent cell lines (LN229, U87MG), whereas it was not associated with the locus in BAI1-expressing cells (LN443) (Fig. 5A). There was no evidence for MeCP2 association with the BAI1 promoter in any of the three cell lines (Fig. 5A). The inability to detect MeCP2 is not due to a lack of expression or technical aspects as these cells express high levels of MeCP2. Moreover, MeCP2 was efficiently recruited to the promoter of O6-methylguanine-DNA methyltransferase (MGMT), a gene to which MeCP2 has been shown to bind in a methylation-dependent manner in all three cell lines (Fig. 5A) (26, 32). MBDs, and in particular MBD2, are components of NURD/Mi2 corepressor complexes and are thought to direct transcriptional silencing of methylated CpG islands through the recruitment of histone deacetylase activity (10, 11). Consistent with this, aberrant DNA methylation of CpG island promoters is associated with histone hypoacetylation and the acquisition of H3K9 methylation (1, 33). We therefore examined the status of acetylation of lysine 9 (AcH3K9) and tri-methylation of lysine 9 (3MeH3K9) on histone H3 by ChIP in two BAI1-expressing (SF188 and LN443) and two BAI1-silent (LN229 and U87MG) cell lines. 3MeH3K9, a marker of condensed chromatin, was enriched at the BAI1 promoter in BAI1-silent cells, but was absent in BAI1-expressing cells (Fig. 5B). In contrast, AcH3K9, a marker for transcriptionally-active chromatin, was enriched at the BAI1 promoter in BAI1-expressing cells, but was greatly reduced in BAI1-silent cells (Fig. 5B). These results demonstrate a correlation between BAI1 expression and changes in histone modification.
Figure 5.
ChIP assay demonstrates the specific binding of MBD2 to the BAI1 promoter. A, Crosslinked chromatin was prepared from BAI1-silent (LN229, U87MG) and BAI1-expressing (LN443) glioma cell lines, sonicated to shear DNA fragments and immunoprecipitated with anti-MBD2 or MeCP2 antibodies or control immunoglobulins (IgG). The immunoprecipitates were then subjected to PCR analysis using primer pairs spanning the BAI1 or MGMT promoters. Note that binding of MBD2 to the BAI1 promoter was only found in BAI1-silent cells (LN229, U87MG), while MeCP2 did not bind altogether. Binding to the MGMT promoter was used as a positive control for both proteins. B, the presence of acetylation (AcH3K9) and tri-methylation (3MeH3K9) on lysine 9 of histone H3 associates with the activation status of the BAI1 promoter. ChIPs were performed in BAI1-expressing (SF188, LN443) and in BAI1-silent (LN229, U87MG) cells. Note that the mark of active chromatin (AcH3) is associated with BAI1-expressing cells, while that of inactive chromatin (MeH3) with silent cells. C, release of MBD2 from the BAI1 promoter in the presence of demethylation agent 5-Aza-dC and MBD2 knockdown. U251MG cells were treated with either 5-Aza-dC (5 μM) for 5 days or transfected with MBD2 shRNA or non specific shRNA control for 72 hrs before ChIP assays. Note that both 5-Aza-dC and shMBD2 treatments led to decrease of MBD2 binding and concomitant switch from inactive to active chromatin markers on the BAI1 promoter. D, quantification of the MBD2, AcH3K9 and 3MeH3K9 ChIP fractions on the BAI1 promoter from C. ChIP quantification was plotted as a ratio of bound MBD2, AcH3K9 or 3MeH3K9 to input signal and expressed on the y-axis.
Next we investigated the influence of 5-Aza-dC on the association of MBD2 with the BAI1 CpG island. Treatment of the BAI1-silent cell line U251MG (Fig. 2C and Fig. 4A) with 5-Aza-dC caused a significant reduction in MBD2 occupancy at the BAI1 promoter as determined by ChIP (Fig. 5C and D). Concomitant with the loss of MBD2, 5-Aza-dC treatment also induced hyperacetylation of histone H3 in the BAI1 promoter region and a reduction of H3K9 tri-methylation (Fig. 5C and D). Similar results were obtained when a shRNA against MBD2 was employed (Fig. 5C and D). Interestingly, the depletion of MBD2 did not allow for the binding of another methyl-CpG binding protein MeCP2 (Fig. 5C). Taken together, these findings support the conclusion that MBD2 selectively binds to the BAI1 promoter, and that its presence is necessary to maintain characteristics of closed chromatin and transcriptional silencing at the BAI1 locus. These data also suggest that the targeting of MBD2 may be as effective as DNA demethylating agents in restoring chromatin conformation and prompting the re-activation of BAI1 gene expression.
Reactivated BAI1 expression inhibits endothelial cell migration and in vivo angiogenesis
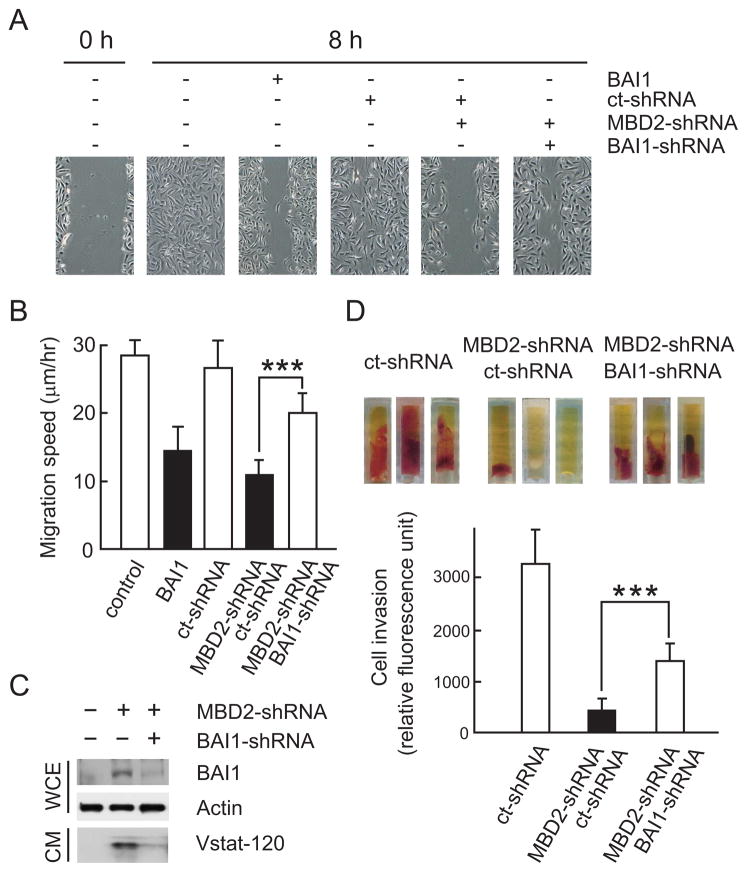
The above data, combined with our previous demonstration that the cleaved 120 kDa N-terminal fragment of BAI1 (vasculostatin or Vstat120) can suppress angiogenesis in vitro and inhibit tumor growth in vivo (14, 15), suggests that reactivation of BAI1 expression by MBD2 knockdown may be a novel therapeutic approach for GBM. However, whether the levels or functionality of reactivated BAI1 are sufficient to restore the anti-angiogenic activity is not known. Therefore, we first determined whether reactivation of BAI1 gene expression by MBD2 silencing could restore BAI1 protein synthesis and anti-angiogenic activity as measured in an in vitro endothelial cell migration assay. BAI1-silent U251MG cells were transiently transfected with control or MBD2-specific shRNA vectors as above and conditioned medium (CM) was collected three days later. Confluent human brain microvascular endothelial cells (HBVECs) were wounded and incubated with CM from transfected U251MG cells. 8 hours later, phase contrast images were captured to monitor the distance traveled by HBVECs from the wound edge to the center of the wound. As a positive control, we used CM from U251MG cells stably transfected with a BAI1-expressing vector. U251MG-BAI1 cell CM dramatically inhibited wound closure as compared to untreated control (Fig. 6A; compare panels 2 and 3), consistent with our previous results (14). Similarly, HBVECs treated with CM from MBD2-shRNAs transfected U251MG cells exhibited clearly reduced migration as compared to control shRNAs transfected cells (Fig. 6A); quantification of the migration speed showed more than 50% reduction in wound closure (Fig. 6B).
Figure 6.
Reactivation of BAI1 expression by MBD2 knockdown restores anti-angiogenic activity of BAI1 in the cell conditioned media (CM). A, CM of U251MG glioma cells with reactivated BAI1 expression inhibits human brain-derived endothelial cell (HBVECs) migration in a scratch-wound assay. Representative pictures of wounded HBVECs before (0 hr) and 8 hrs after treatment with CM are shown. CM from cells stably transfected with a BAI1 expression vector (U251-BAI1; clone B12) inhibited cell migration substantially (compare 2nd and 3rd panels). CM from cells transiently transfected for 72 hrs with shRNAs against MBD2 or BAI1, singly or in combination, inhibited cell migration differentially (panels 4–6). Ct-shRNA, control shRNA. B, Quantification of migration speed in the scratch-wound assay of A. Final wound width was measured after 8 hrs and the distance migrated was calculated. The experiment was repeated three times with similar results. Columns, mean; bars, SD (n = 6 for each condition); *, P < 0.001. C, Western blot demonstrating the levels of BAI1 and secreted Vasculostatin (Vstat-120) 72 hrs after transfection with shRNA expression vectors in U251MG cells used in A, B. WCE, whole cell extract; CM, conditioned medium. D, Quantitative in vivo angiogenesis assay. Angioreactors containing CM from U251MG cells transfected with indicated shRNAs were implanted subcutaneously in athymic nu/nu mice. Representative images show vascularization of angioreactors from each group 2 weeks later (upper panel). The relative mouse endothelial cell invasion in the angioreactor was quantified by dissociating the cells in the angioreactor, incubating them with a fluorescently-labeled plant lectin specific for endothelial cells and measuring the fluorescence of cell pellets derived from each group (n=8, lower panel). *, P < 0.001.
Since MBD2 knock down has the potential to re-activate the expression of other genes that are silenced in a DNA methylation-dependent manner in addition to BAI1, it is possible that the inhibitory effect on wound closure determined above could result from the re-expression of other genes. To address this issue, we designed BAI1 shRNAs to determine whether the effect is BAI1-specific. BAI1-specific shRNAs suppressed BAI1 protein expression substantially, resulting in strongly reduced levels of Vstat120 in the CM (Fig. 6C). We then repeated the scratch wound assay with CM from cells transfected with both MBD2 and BAI1 shRNAs. The migration of HBVECs treated with CM from U251MG cells transfected with both MBD2 and BAI1 shRNAs was more than 50% faster than that of HBVECs treated with CM from cells transfected with both MBD2 and control shRNAs (Fig. 6A and B). These results show that a concomitant reduction in BAI1 expression partially abolished the anti-angiogenic effect of MBD2 knockdown. We further determined the anti-angiogenic activity of reactivated BAI1 in an in vivo angiogenesis assay. CM from U251MG cells transfected with either MBD2-shRNAs alone or in combination with BAI1-shRNAs, were mixed with matrigel plus FGF-2 and VEGF (angioreactor) and subcutaneously implanted into athymic nude mice. Two weeks later, angioreactors were dissected and visually inspected for evidence of angiogenesis. Vascularization was readily observed and the angiogenic response had penetrated deep into the angioreactor with CM from control shRNA transfected U251MG cells (Fig. 6D, upper panel). In contrast, whereas there was an occasional angiogenic response observed in angioreactors exposed to CM from MBD2-shRNA transfected cells, the extent of this response was usually minimal and had only superficially penetrated the angioreactor. To directly measure the angiogenic response, we dissociated the cells in the angioreactor and used a fluorescein-labeled lectin (FITC-lectin) that specifically binds to endothelial cells to quantify the number of murine endothelial cells infiltrated into the angioreactor. A statistically significant difference in endothelial cell content was found between angioreactors exposed to CM from cells transfected with MBD2-shRNA versus MBD2-shRNA in combination with BAI1-shRNA (Fig. 6D, lower panel). Taken together, these results demonstrate that restoration of BAI1 expression by MBD2 knockdown reactivates BAI1 synthesis and anti-angiogenic activity.
Discussion
Here we provide evidence that the BAI1 gene is epigenetically silenced in GBM and that manipulation of the silencing event can reactivate BAI1 expression and anti-angiogenic tumor suppressor activity, which can be exploited for therapeutic means.
In glioma cell lines, BAI1 silencing could be reversed by treatment with the demethylating agent 5-Aza-dC. Therefore, our data support the notion that DNA methylation contributes to inactivation of the BAI1 gene. Transcriptional silencing through promoter DNA methylation has been proposed to occur through several different molecular mechanisms, such as by direct interference with transcription factor binding, by altering the structure of chromatin, and/or by recruiting MBD proteins (10, 11, 30, 31, 34). MBD proteins are critical mediators of many epigenetic processes in that they interpret the methylation marks on DNA and facilitate the establishment of a repressive chromatin environment. ChIP assays demonstrated binding of MBD2 to the CpG island region in the BAI1 promoter specifically in cell lines where the gene was methylated and silenced, whereas there was no association of MeCP2 with the methylated BAI1 promoter. Furthermore, in BAI1-silent glioma cell lines, shRNA-directed knockdown of MBD2 resulted in the local depletion of MBD2 and restored the chromatin state to one similar to that of expressing cell lines (eg. hyperacetylated at H3K9) and re-activated BAI1 expression. Taken together, these results support a mechanism wherein the specific binding of MBD2 to the methylated BAI1 promoter is necessary to maintain transcriptional repression of BAI1.
Although DNA methylation has been extensively studied in GBM, the expression and regulation of MBDs has not. For the first time, we provide evidence that MBD2 is specifically overexpressed in GBM. The MBD2 gene is located on chromosome 18q21 (35); no amplification of this region has been reported in GBM, nor have genomic copy number changes or somatic mutations for MBD2 been observed in the data from the Rembrandt and TCGA glioma databases (data not shown). Therefore, the overexpression of MBD2 observed in a subset of GBM may result from a different mechanism, such as enhanced transcription. MBD2 has the greatest binding affinity for methylated DNA among MBD family proteins in vitro (35) suggesting that MBD2 may be the MBD family member with the greatest effect on gene silencing (36). Accumulating evidence demonstrates that MBD2 is involved in the suppression of aberrantly methylated tumor suppressor genes by binding to methylated promoters (37, 38), including p14/ARF and p16/Ink4A (39, 40), 14-3-3σ (24) and GSTP1 (glutathione S-transferase p1) (41). In all cases, siRNA-mediated knockdown of MBD2 resulted in the reactivation of the corresponding gene target, similar to what we observed for BAI1. Consistent with a role in tumor promotion, previous work has shown that knockout of Mbd2 strongly suppresses intestinal tumorigenesis in ApcMin mice (42). The underlying mechanism may be that deficiency of Mbd2 elevates levels of the Wnt target Lect2, a Wnt pathway repressor (43).
The recent discovery of 5-hydroxymethylcytosine (5-hmC) in mammalian DNA has added a new dimension to the regulation of DNA methylation and may be particularly relevant in the pathogenesis of glioblastomas as the TET enzymes that catalyze the hydroxylation of methyl cytosine residues are among those affected by the accumulation of 2-hydroxyglutarate that accompanies IDH gene mutations (44). Although the precise function of 5-hmC in epigenetic regulation is not yet completely understood, recent work suggests that it may facilitate DNA demethylation through a base excision repair mechanism (45). Furthermore, there is emerging evidence that the binding of some MBD proteins, including MBD2b, to DNA is inhibited by 5-hmC (46, 47), thus the conversion of 5-mC to 5-hmC may play a functional role in the dynamic regulation of gene expression. Future studies are warranted to determine the distribution of 5-hmC in the BAI1 gene regulatory regions and its role in BAI1 gene transcription.
At present, epigenetic approaches in cancer therapy have focused primarily on inhibitors of the DNA methyltransferases and histone modifiers (eg. HDACs). Our data suggest that MBD2 may also represent a promising cancer therapeutic target. MBD2-null mice display a surprisingly weak phenotype, and global DNA methylation levels and genomic imprinting are relatively unaffected by the absence of MBD2 (48). The fact that MBD2 knockout mice are viable and resistant to tumorigenesis, coupled with the finding that down-regulation of MBD2 could restore a functional BAI1 with potent anti-angiogenic activity, makes MBD2 a particularly attractive target for therapeutic intervention for GBM and/or in the prevention of glioma progression. Sequence-specific antisense inhibitors of MBD2 have been shown to inhibit both anchorage-independent growth of human cancer cell lines in vitro and the growth of human tumor xenografts in vivo (49, 50). Validation of MBD2 as a viable target in GBM will require careful examination of the global impact on gene expression, as the potential therapeutic benefit will depend upon the reprogrammed transcriptome tipping the tumor towards an anti- or pro-tumorigenic biological response. Our data show that antagonizing MBD2 elicits a global anti-angiogenic response that is largely dependent upon BAI1 expression and would be expected to suppress tumor growth. In summary, our results demonstrate a functional role of MBD2 in the repression of BAI1 in GBM. This study could lead to new therapeutic prospects for the treatment of patients with brain tumors.
Supplementary Material
Acknowledgments
We thank all the Van Meir lab members for helpful comments, the Emory Biomarker CORE facility for DNA sequencing services, Dr. Daniel J Brat for neuropathology diagnosis of GBM samples, Dr Yuan Liu for help with statistical analysis, Dr. Y. Nakamura for the BAI1 expression vector, and Narra S Devi and Zhaobin Zhang for technical support.
Financial support: NIH R01-CA86335 (EGVM), NIH RO1-CA077337 (PMV), the Southeastern Brain Tumor Foundation (DZ and EGVM) and the University Research Council of Emory University (EGVM).
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
Author contributions: DZ and EGVM conceived experiments, DZ performed all experiments, SBH provided the GBM tissue array and histological confirmation, and PMV provided experimental advice. DZ, PMV and EGVM wrote the manuscript.
References
- 1.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–13. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 3.Stone AR, Bobo W, Brat DJ, Devi NS, Van Meir EG, Vertino PM. Aberrant methylation and down-regulation of TMS1/ASC in human glioblastoma. Am J Pathol. 2004;165:1151–61. doi: 10.1016/S0002-9440(10)63376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim TY, Zhong S, Fields CR, Kim JH, Robertson KD. Epigenomic profiling reveals novel and frequent targets of aberrant DNA methylation-mediated silencing in malignant glioma. Cancer Res. 2006;66:7490–501. doi: 10.1158/0008-5472.CAN-05-4552. [DOI] [PubMed] [Google Scholar]
- 5.Martinez R, Esteller M. The DNA methylome of glioblastoma multiforme. Neurobiol Dis. 2010;39:40–6. doi: 10.1016/j.nbd.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 6.Brat DJ, Castellano-Sanchez A, Kaur B, Van Meir EG. Genetic and biologic progression in astrocytomas and their relation to angiogenic dysregulation. Adv Anat Pathol. 2002;9:24–36. doi: 10.1097/00125480-200201000-00004. [DOI] [PubMed] [Google Scholar]
- 7.Nagarajan RP, Costello JF. Epigenetic Profiling of Gliomas. In: Van Meir Erwin., editor. CNS Cancer. New York: Humana; 2009. pp. 615–650. [Google Scholar]
- 8.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen BC, Smith AA, Zheng S, Koestler DC, Houseman EA, Marsit CJ, et al. DNA methylation, isocitrate dehydrogenase mutation, and survival in glioma. J Natl Cancer Inst. 2011;103:143–53. doi: 10.1093/jnci/djq497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballestar E, Wolffe AP. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur J Biochem. 2001;268:1–6. doi: 10.1046/j.1432-1327.2001.01869.x. [DOI] [PubMed] [Google Scholar]
- 11.Wade PA. Methyl CpG binding proteins: coupling chromatin architecture to gene regulation. Oncogene. 2001;20:3166–73. doi: 10.1038/sj.onc.1204340. [DOI] [PubMed] [Google Scholar]
- 12.Sansom OJ, Maddison K, Clarke AR. Mechanisms of disease: methyl-binding domain proteins as potential therapeutic targets in cancer. Nat Clin Pract Oncol. 2007;4:305–15. doi: 10.1038/ncponc0812. [DOI] [PubMed] [Google Scholar]
- 13.Nishimori H, Shiratsuchi T, Urano T, Kimura Y, Kiyono K, Tatsumi K, et al. A novel brain-specific p53-target gene, BAI1, containing thrombospondin type 1 repeats inhibits experimental angiogenesis. Oncogene. 1997;15:2145–50. doi: 10.1038/sj.onc.1201542. [DOI] [PubMed] [Google Scholar]
- 14.Kaur B, Cork SM, Sandberg EM, Devi NS, Zhang Z, Klenotic PA, et al. Vasculostatin inhibits intracranial glioma growth and negatively regulates in vivo angiogenesis through a CD36-dependent mechanism. Cancer Res. 2009;69:1212–20. doi: 10.1158/0008-5472.CAN-08-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaur B, Brat DJ, Devi NS, Van Meir EG. Vasculostatin, a proteolytic fragment of brain angiogenesis inhibitor 1, is an antiangiogenic and antitumorigenic factor. Oncogene. 2005;24:3632–42. doi: 10.1038/sj.onc.1208317. [DOI] [PubMed] [Google Scholar]
- 16.Cork SM, Van Meir EG. Emerging roles for the BAI1 protein family in the regulation of phagocytosis, synaptogenesis, neurovasculature and tumor development. J Mol Med. 2011 doi: 10.1007/s00109-011-0759-x. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cork SM, Kaur B, Cooper L, Saltz JH, Sandberg EM, Van Meir EG. A furin/MMP-14 proteolytic cascade releases a novel 40 kDa vasculostatin from tumor suppressor BAI1. Manuscript submitted. doi: 10.1038/onc.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogler O, Mikkelsen T. Angiogenesis and apoptosis in glioma: two arenas for promising new therapies. J Cell Biochem. 2005;96:16–24. doi: 10.1002/jcb.20475. [DOI] [PubMed] [Google Scholar]
- 19.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–73. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 20.Park D, Tosello-Trampont AC, Elliott MR, Lu M, Haney LB, Ma Z, et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450:430–4. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- 21.Jarzynka MJ, Hu B, Hui KM, Bar-Joseph I, Gu W, Hirose T, et al. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer Res. 2007;67:7203–11. doi: 10.1158/0008-5472.CAN-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaur B, Brat DJ, Calkins CC, Van Meir EG. Brain angiogenesis inhibitor 1 is differentially expressed in normal brain and glioblastoma independently of p53 expression. Am J Pathol. 2003;162:19–27. doi: 10.1016/S0002-9440(10)63794-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cobbs CS, Whisenhunt TR, Wesemann DR, Harkins LE, Van Meir EG, Samanta M. Inactivation of wild-type p53 protein function by reactive oxygen and nitrogen species in malignant glioma cells. Cancer Res. 2003;63:8670–3. [PubMed] [Google Scholar]
- 24.Pulukuri SM, Rao JS. CpG island promoter methylation and silencing of 14-3-3sigma gene expression in LNCaP and Tramp-C1 prostate cancer cell lines is associated with methyl-CpG-binding protein MBD2. Oncogene. 2006;25:4559–72. doi: 10.1038/sj.onc.1209462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Remmele W, Hildebrand U, Hienz HA, Klein PJ, Vierbuchen M, Behnken LJ, et al. Comparative histological, histochemical, immunohistochemical and biochemical studies on oestrogen receptors, lectin receptors, and Barr bodies in human breast cancer. Virchows Arch A Pathol Anat Histopathol. 1986;409:127–47. doi: 10.1007/BF00708323. [DOI] [PubMed] [Google Scholar]
- 26.Nakagawachi T, Soejima H, Urano T, Zhao W, Higashimoto K, Satoh Y, et al. Silencing effect of CpG island hypermethylation and histone modifications on O6-methylguanine-DNA methyltransferase (MGMT) gene expression in human cancer. Oncogene. 2003;22:8835–44. doi: 10.1038/sj.onc.1207183. [DOI] [PubMed] [Google Scholar]
- 27.Zhu D, Yang Z, Luo Z, Luo S, Xiong WC, Mei L. Muscle-specific receptor tyrosine kinase endocytosis in acetylcholine receptor clustering in response to agrin. J Neurosci. 2008;28:1688–96. doi: 10.1523/JNEUROSCI.4130-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guedez L, Rivera AM, Salloum R, Miller ML, Diegmueller JJ, Bungay PM, et al. Quantitative assessment of angiogenic responses by the directed in vivo angiogenesis assay. Am J Pathol. 2003;162:1431–9. doi: 10.1016/S0002-9440(10)64276-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones PA. Gene activation by 5-azacytidine. In: Razin A, Cedar H, Riggs AD, editors. DNA Methylation: Biochemistry and Biological Significance. New York: Springer; 1984. pp. 165–87. [Google Scholar]
- 30.Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451–4. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 31.Singal R, Ginder GD. DNA methylation. Blood. 1999;93:4059–70. [PubMed] [Google Scholar]
- 32.Patel SA, Graunke DM, Pieper RO. Aberrant silencing of the CpG island-containing human O6-methylguanine DNA methyltransferase gene is associated with the loss of nucleosome-like positioning. Mol Cell Biol. 1997;17:5813–22. doi: 10.1128/mcb.17.10.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–98. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 34.Kass SU, Landsberger N, Wolffe AP. DNA methylation directs a time-dependent repression of transcription initiation. Curr Biol. 1997;7:157–65. doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- 35.Hendrich B, Abbott C, McQueen H, Chambers D, Cross S, Bird A. Genomic structure and chromosomal mapping of the murine and human Mbd1, Mbd2, Mbd3, and Mbd4 genes. Mamm Genome. 1999;10:906–12. doi: 10.1007/s003359901112. [DOI] [PubMed] [Google Scholar]
- 36.Lopez-Serra L, Ballestar E, Ropero S, Setien F, Billard LM, Fraga MF, et al. Unmasking of epigenetically silenced candidate tumor suppressor genes by removal of methyl-CpG-binding domain proteins. Oncogene. 2008;27:3556–66. doi: 10.1038/sj.onc.1211022. [DOI] [PubMed] [Google Scholar]
- 37.To KK, Zhan Z, Bates SE. Aberrant promoter methylation of the ABCG2 gene in renal carcinoma. Mol Cell Biol. 2006;26:8572–85. doi: 10.1128/MCB.00650-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez-Serra L, Ballestar E, Fraga MF, Alaminos M, Setien F, Esteller M. A profile of methyl-CpG binding domain protein occupancy of hypermethylated promoter CpG islands of tumor suppressor genes in human cancer. Cancer Res. 2006;66:8342–6. doi: 10.1158/0008-5472.CAN-06-1932. [DOI] [PubMed] [Google Scholar]
- 39.Magdinier F, Wolffe AP. Selective association of the methyl-CpG binding protein MBD2 with the silent p14/p16 locus in human neoplasia. Proc Natl Acad Sci U S A. 2001;98:4990–5. doi: 10.1073/pnas.101617298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin V, Jorgensen HF, Chaubert AS, Berger J, Barr H, Shaw P, et al. MBD2-mediated transcriptional repression of the p14ARF tumor suppressor gene in human colon cancer cells. Pathobiology. 2008;75:281–7. doi: 10.1159/000151708. [DOI] [PubMed] [Google Scholar]
- 41.Lin X, Nelson WG. Methyl-CpG-binding domain protein-2 mediates transcriptional repression associated with hypermethylated GSTP1 CpG islands in MCF-7 breast cancer cells. Cancer Res. 2003;63:498–504. [PubMed] [Google Scholar]
- 42.Sansom OJ, Berger J, Bishop SM, Hendrich B, Bird A, Clarke AR. Deficiency of Mbd2 suppresses intestinal tumorigenesis. Nat Genet. 2003;34:145–7. doi: 10.1038/ng1155. [DOI] [PubMed] [Google Scholar]
- 43.Phesse TJ, Parry L, Reed KR, Ewan KB, Dale TC, Sansom OJ, et al. Deficiency of Mbd2 attenuates Wnt signaling. Mol Cell Biol. 2008;28:6094–103. doi: 10.1128/MCB.00539-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–8. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–8. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–23. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slack A, Bovenzi V, Bigey P, Ivanov MA, Ramchandani S, Bhattacharya S, et al. Antisense MBD2 gene therapy inhibits tumorigenesis. J Gene Med. 2002;4:381–9. doi: 10.1002/jgm.288. [DOI] [PubMed] [Google Scholar]
- 50.Campbell PM, Bovenzi V, Szyf M. Methylated DNA-binding protein 2 antisense inhibitors suppress tumourigenesis of human cancer cell lines in vitro and in vivo. Carcinogenesis. 2004;25:499–507. doi: 10.1093/carcin/bgh045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.