Abstract
Canonical Hedgehog (HH) signaling is characterized by Smoothened (Smo)-dependent activation of the transcription factors Gli1 and Gli2, which regulate HH target genes. In human colon carcinoma cells, treatment with the Gli small molecule inhibitor GANT61 induces extensive cell death, in contrast to the Smo inhibitor cyclopamine. Here we elucidate cellular events upstream of cell death elicited by GANT61, which reveal the basis for its unique cytotoxic activity in colon carcinoma cells. Unlike cyclopamine, GANT61 induced transient cellular accumulation at G1/S (24 hr) and in early S-phase (32 hr), with elevated p21Cip1, cyclin E and cyclin A in HT29 cells. GANT61 induced DNA damage within 24 hr, with the appearance of p-ATM and p-Chk2. Pharmacologic inhibition of Gli1 and Gli2 by GANT61 or genetic inhibition by transient transfection of the Gli3 repressor (Gli3R), downregulated Gli1 and Gli2 expression, induced γH2AX, PARP cleavage, caspase-3 activation and cell death. GANT61 induced γH2AX nuclear foci, while transient transfection of Gli3R demonstrated expression of Gli3R and γH2AX foci within the same nuclei in HT29, SW480 and HCT116. GANT61 specifically targeted Gli1 and Gli2 substantiated by specific inhibition of 1) direct binding of Gli1 and Gli2 to the promoters of target genes HIP1 and BCL-2, 2) Gli-luciferase activity, and 3) transcriptional activation of BCL-2. Taken together, these findings establish that inhibition of HH signaling at the level of the GLI genes downstream of Smo is critical in the induction of DNA damage in early S-phase, leading to cell death in human colon carcinoma cells.
INTRODUCTION
Binding of the secretory HH ligands to their transmembrane receptor Patched (Ptch1) initiates the classical HH signaling pathway, by releasing Smo from Ptch1-dependent suppression. Smo activates the final arbiters of HH signaling, the Gli transcription factors, that regulate HH target genes (1, 2). Aberrantly activated HH signaling has been identified in the malignant phenotype of several types of human cancers (reviewed in (3)), involving amplification of GLI1 or GLI2 and mutations in PTCH1 or SMO (4, 5). There is emerging evidence that the HH pathway progresses during colon carcinogenesis (6, 7) and in metastatic disease (8), whereas in normal colonic tissue, HH signaling is involved in differentiation (9–11). Canonical HH signaling genes are expressed in primary colon cancers, metastatic disease, human carcinoma xenografts (8) and in human colon carcinoma cell lines (12, 13). In several studies, HH signaling molecules have been linked to genomic instability, involving inactivation of homologous recombination (HR) or non-homologous end joining (NHEJ), defects in checkpoint activation, and predisposition to development of cancers (14–16). However, little is known functionally about this signaling pathway and how it affects the survival and pathogenesis of colon cancer.
The majority of studies that determine the effects of inhibiting the HH signaling pathway have utilized the classic Smo inhibitor, cyclopamine, which crosslinks Smo (17). Cyclopamine has demonstrated variable activity in several different types of cancer cells (18). Oncogene-driven signaling pathways converge downstream of Smo on the Gli transcription factors providing non-canonical regulation of HH signaling (5, 19–22). Such non-canonical activation of the Gli proteins can therefore circumvent the inhibition of Smo resulting in reduced efficacy of or resistance to Smo inhibitors. The Gli family of transcription factors is comprised of Gli1, Gli2 and Gli3 that regulate HH-target gene expression (3). Gli2 appears to be the primary activator of HH signaling, with Gli1 as a transcriptional target of Gli2, which may amplify HH-induced target gene expression (4, 23–25). Gli1 and Gli2 induce transcription of overlapping and distinct sets of target genes (4, 23–25). Full length Gli3 has activator functions while a C-terminus cleaved form mediates repressor activity (3, 5). Expression of the repressor form of Gli3 (Gli3R) inhibited proliferation and induced cell death in primary cultures of human colon cancers and metastases. Further, human colon carcinoma cells transduced with Gli3R failed to form xenografts in nude mice (8), indicating the importance of Gli1 and Gli2 and corroborating the role of the HH signaling pathway in colon cancer cell survival.
To identify the mechanisms that regulate HH-driven cellular survival in the context of colon cancer, we employed cyclopamine to target Smo. In addition, to target the Gli proteins downstream of Smo, we employed a small molecule inhibitor of both Gli1 and Gli2, GANT61, identified in a cell-based small molecule screen for inhibitors of Gli1-mediated transcription (26). We have previously demonstrated that GANT61 reduced GLI1, GLI2 and PTCH1 mRNA expression in human colon carcinoma cell lines, and significantly modulated cDNA microarray gene expression profiles downstream of Gli1/Gli2 function (12). Further, inhibition of HH signaling by GANT61 induced greater cytotoxicity in human colon carcinoma cell lines than targeting Smo using cyclopamine (13). To elucidate the mechanisms regulating this differential response, studies were conducted in the human colon carcinoma cell line HT29, which is mutant for p53. Cells treated with GANT61 (20 µM) accumulated at G1/S and in early S at 24 hr and 32 hr, respectively, and by 48 hr underwent cell death. In contrast, cyclopamine (20 µM)-treated HT29 cells demonstrated minimal effects on cell cycle distribution or cell death. GANT61-treated cells accumulated p21Cip1, cyclin E and cyclin A in G1- and S- phase cells at 24 hr–40 hr, in contrast to cyclopamine-treated cells, and stable knockdown of p21Cip1 did not influence sensitivity to GANT61. GANT61 but not cyclopamine induced DNA damage by 24 hr with the appearance of γH2AX nuclear foci at the sites of DNA strand breaks. Similar to the cellular effects induced by pharmacologic inhibition of Gli1 and Gli2 by GANT61, transient transfection of the Gli3R mutant, which inhibits the activating functions of Gli1 and Gli2 (3), downregulated expression of Gli1 and Gli2, induced expression of γH2AX, cleavage of PARP and caspase-3, and cell death. GANT61 activated ATM and Chk2 (but not ATR and Chk1) by 4 hr that was sustainable. Transient expression of Gli3R in HT29, SW480 and HCT116 cells induced nuclear localization of the Gli3R protein and induced formation of γH2AX nuclear foci within the same cells. Data suggest that exogenously expressed Gli3R is functional and suppression of HH/Gli signaling results in DNA damage in multiple human colon carcinoma cell lines that demonstrate active HH signaling. This phenomenon is p53-independent, since HT29 and SW480 cells express a mutant form of p53 while HCT116 harbor a wild-type p53 gene. Within 1 hr of exposure, GANT61 1) reduced the binding of Gli1 and Gli2 to the promoter regions of the Gli target genes HIP1 and BCL-2 but not FAS, which is not a direct Gli target, and 2) inhibited the transcriptional regulation of BCL-2. Further, GANT61 specifically inhibited Gli-luciferase activity in contrast to NF-κB- or AP1- luciferase activities indicating its specificity for Gli1 and Gli2. These findings emphasize the importance of targeting the Gli proteins to functionally inhibit HH signaling, and their critical role in the cellular survival of human colon carcinoma cells.
MATERIALS AND METHODS
Cell culture and reagents
HT29 cells were obtained from ATCC and routinely verified by morphology, growth characteristics, response to cytotoxic agents (Annexin V/PI staining). cDNA microarray gene profiles were also characteristic. Cells were verified biannually to be mycoplasma-free. Cells were maintained in folate-free RPMI 1640 medium containing 10% dFBS and 80 nM [6RS]5-methyltetrahydrofolate. The cells were trypsinized and counted using a Z2 Coulter particle count and size analyzer (Beckman Coulter, Inc., CA). For Western analysis, antibodies against p21Cip1, β-actin and HSP90α/β were purchased from Santa Cruz Biotechnology (CA), anti-Gli1 antibody was from Novus Biologicals (CO) and anti-Gli2 antibody was from Cell Signaling Technology (MA). Anti-c-myc antibody (9E10) was obtained from the Hybridoma Core, Lerner Research Institute. Anti-p21Cip1, anti-cyclin E, and anti-cyclin A antibodies used for bivariate flow cytometry were purchased from BD Biosciences (CA). For Western analysis and confocal microscopy, antibodies against γH2AX, p-Chk1, Chk-1, p-ATR, ATR, p-Chk2, Chk-2 and ATM were purchased from Cell Signaling Technology (MA); the p-ATM antibody was from Rockland Immunochemicals Inc. (PA). AlexaFluor 488 goat anti-rabbit, and AlexaFluor 633 goat anti-mouse secondary antibodies were obtained from Invitrogen (CA). GANT61 was purchased from Alexis Biochemicals (CA), and cyclopamine from Toronto Research Chemicals, Canada.
Annexin V-FITC/PI staining and flow cytometric analysis
These were as described previously (27).
Cell cycle distribution, Bivariate Flow Cytometric Analysis and BrdU incorporation
For cell cycle distribution and Bivariate flow cytometry, cells were analyzed as previously described (28). For analysis of BrdU incorporation, cells were plated (50,000 cells/well) in a 6-well format, and treated with GANT61 (20µM) or cyclopamine (20µM) for up to 48 hr. Cells were pulsed with BrdU (10 µM; BD Biosciences, MD) for 30–45 min and analyzed by flow cytometry for distribution within the cell cycle as per the manufacturer protocol.
Western analysis
This was performed as previously described (27).
RNA interference studies
HT29 cells stably expressing p21Cip1shRNA were generated by transducing HT29 cells with scrambled shRNA or the gene specific shRNA-expressing retroviruses. Details are provided in Supplementary Materials and Methods.
COMET Assay
Cells were processed, and electrophoresed in agarose gels as described (29). Tail Moment (TM) and Tail Length (TL) were used to characterize the DNA damage within individual cells. Image analysis and quantification were conducted using the NIH imageJ software. TM = %DNA in the tail × TL; where % DNA in the tail = tail area (TA) × tail area intensity (TAI) × 100/(TA × TAI) +[head area (HA) × head area intensity (HAI)].
Confocal microscopy
Cells were plated (50,000/well) on coverslips in 6-well plates. The cells were treated with GANT61 (20µM) or cyclopamine (20µM) for up to 48 hr and processed for microscopy. Details of microscopy are described in the Supplementary Materials and Methods.
Gli3R and transient transfections
The myc-tagged c-terminus deleted construct Gli3R (gift of Dr. Ariel Ruiz i Altaba, University of Geneva Medical School) has been previously described (3). HT29 cells were transiently transfected using Lipofectamine 2000 (Invitrogen) with Gli3R or the empty vector pCS2-MT (gifted by Dr. David Turner, University of Michigan MBNI). Cells were used for experiments 24 hr, 48 hr or 72 hr post-transfection.
ChIP analysis
HT29 cells were treated with GANT61 (20 µM) for 1 hr or 24 hr and ChIP analysis was conducted using Gli1 or Gli2 antibodies and Abcam ChIP kit (MA), according to the manufacturer’s protocol. Details are provided in Supplementary Materials and Methods.
Luciferase reporter assays
The Gli-luciferase reporter construct (kindly provided by Dr. Rune Toftgård, Karolinska Institutet) has been previously described (30). The NF-κB-luciferase plasmid p5XIP10κB was previously reported (27). The AP1-Luciferase (kindly provided by Dr. Philip H. Howe, Cleveland Clinic) contains a basic promoter element (TATA box) joined to tandem repeats of the AP1 binding element (Stratagene, La Jolla, CA, USA). Transient transfection for 24 hr with luciferase reporters was performed 24 hr after GANT61 (20 µM) treatment, as described (13).
RT-PCR
HT29 cell were treated with GANT61 (20 µM) for 1 hr followed by RNA isolation for up to a further 4 hr. Following conversion into cDNA, samples were used for qPCR as described previously (13). Primers:
BCL-2-Forward: 5’- GGAGGATTGTGGCCTTCTTT-3’
BCL-2-Reverse: 5’- GCCGTACAGTTCCACAAAGG-3’
RESULTS
GANT61 and cyclopamine exhibit differences in cell cycle regulation and induction of cell death
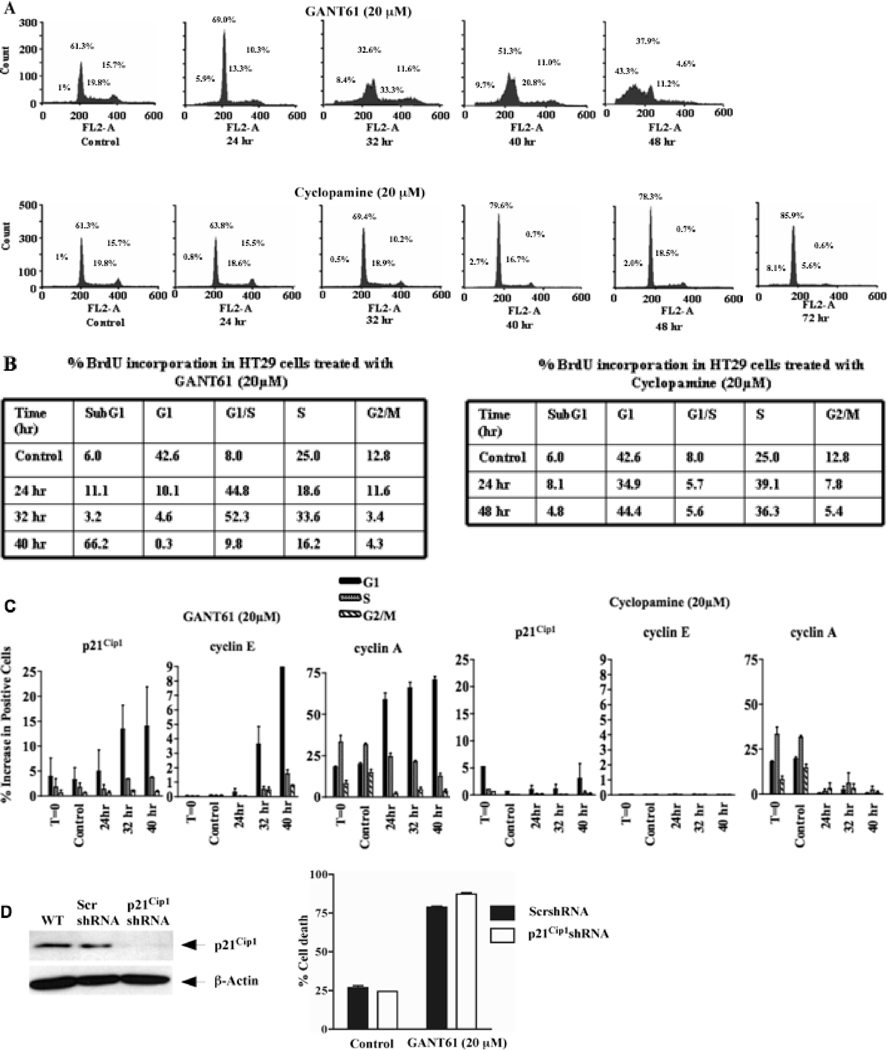
We have demonstrated previously in a panel of six human colon carcinoma cell lines, that at equimolar concentrations (10 µM–30 µM), GANT61 induced > 80% cell death by 72 hr of treatment, in contrast to cyclopamine (13). These concentrations and time frames for the induction of cellular effects are similar to those determined in other model systems for inhibitors of HH signaling (8, 26, 31, 32). A more detailed study of the mechanisms regulating the differential effects between GANT61 and cyclopamine was conducted in HT29 cells, which express mutant p53. Cells were treated with GANT61 (20 µM) or cyclopamine (20 µM) followed by PI staining and flow cytometric analysis of cell cycle distribution. GANT61-treated cells accumulated at G1/S by 24 hr, moving into early S-phase by 32 hr, and subsequently becoming subG1 by 48 hr (Figure 1A). In contrast, treatment with cyclopamine resulted in a modest increase in G1/S-phase cells by 48 hr; by 72 hr cells had not progressed either into S-phase, or into subG1 (Figure 1A).
Figure 1.
GANT61 induces accumulation of HT29 cells at G1/S and early S in contrast to cyclopamine. HT29 cells were treated with A: GANT61 (20 µM) or cyclopamine (20 µM) for up to 72 hr. DNA was extracted, stained and cell cycle distribution was analyzed using flow cytometry. Data are representative of 3 independent experiments. B: BrdU incorporation demonstrates accumulation of GANT61-treated cells at the G1/S boundary and in early S. HT29 cells were treated with GANT61 (20 µM) or cyclopamine (20 µM) for up to 48 hr. % BrdU incorporation was determined using flow cytometry. Data are representative of duplicate determinations. C. Bivariate flow cytometric analysis of p21Cip1, cyclin E and cyclin A expression in different phases of the cell cycle. HT29 cells were treated with GANT61 (20 µM) or cyclopamine (20 µM) for up to 40 hr, followed by staining and cell cycle analysis. Data are representative of duplicate determinations. D: Influence of stable p21Cip1shRNA knockdown on sensitivity of HT29 cells to GANT61. HT29 cells stably expressing p21Cip1- or scrambled-shRNA were treated for 72 hr with GANT61 (20 µM). Cell death was analyzed by Annexin V/PI staining and flow cytometry. Data represent the mean ± SD of duplicate determinations. Reduced expression of p21Cip1 was confirmed by western analysis.
BrdU incorporation analysis
In GANT61-treated cells, cellular accumulation at the G1/S boundary was evident by 24 hr, as demonstrated by a 37% increase in BrdU incorporation, which increased to 52% by 32 hr, and an 8% increase in S-phase cells at this time. By 40 hr, there was a decrease in cells in G1/S (> 40% decrease in BrdU incorporation), in S-phase (17%), and an increase (60%) in cells within the subG1 compartment (Figure 1B and Supplementary Figure 1). These effects were consistent with decreased BrdU-labeled cells in G2/M. In contrast, following cyclopamine treatment, the appearance of a stronger G1/S peak by 48 hr observed in cell cycle analysis (Figure 1A), was paralleled by an 11%–14% increase in BrdU-labeled cells in the S-phase (Figure 1B), clearly demonstrating differences in cell cycle regulation between GANT61- and cyclopamine- treated HT29 cells.
Bivariate flow cytometric analysis of the distribution of p21Cip1, cyclin E and cyclin A within the cell cycle
Cell cycle progression is regulated by different cyclin-cdk complexes. Cyclin E, which regulates Cdk2, is expressed in late G1 and early S-phase (33). Cyclin A, expressed in late G1, begins to accumulate in S-phase and is rapidly destroyed at the onset of mitosis (34). Further, p21Cip1 could have a potential role at the GI/S boundary. Expression of these proteins was analyzed by bivariate flow cytometric analysis, simultaneously with DNA content. In GANT61-treated cells, p21Cip1 was induced and continued to be elevated in G1-phase cells over a period of 24 hr–40 hr (Figure 1C). Similarly, Cyclin E appeared at 24 hr in G1-phase cells, and in S-phase cells at 32 hr–40 hr; the largest accumulation of cyclin E occurred in G1-phase cells where most remained accumulated at 40 hr. Cyclin A accumulated significantly in the G1-phase following GANT61 treatment, while the percentage of cells expressing cyclin A in S-phase as well as G2/M-phase cells declined. In cyclopamine-treated cells, p21Cip1 and cyclin E remained at low levels in all cell cycle phases for up to 40 hr. Cyclin A was expressed in untreated cells in G1, S and G2/M, but decreased in all phases by 24 hr following cyclopamine treatment (Figure 1C and Supplementary Figure 2). Data are consistent with cellular accumulation at the G1/S boundary and in early S-phase in GANT61-treated HT29 cells with accumulation of p21Cip1, cyclin E and cyclin A mostly in G1- and partially in S-phase cells. In contrast, no effects on p21Cip1 or cyclin E distribution, or sustained accumulation of cyclin A were evident in cyclopamine-treated cells, consistent with lack of significant cell cycle perturbation, or induction of cell death.
GANT61-induced cell death is independent of p21Cip1
HT29 cells stably transduced with p21Cip1shRNA or scrambled-shRNA (control) were treated with GANT61 (20 µM) for 72 hr, followed by by Annexin V/PI staining and flow cytometric analysis (Figure 1D). GANT61 induced similar levels of cell death (75–80%) in scrambled-shRNA- or p21Cip1shRNA- transduced cells, indicating the lack of a functional role for p21Cip1, as well as p53, in the mechanism of GANT61-induced cell death.
GANT61 induces DNA damage
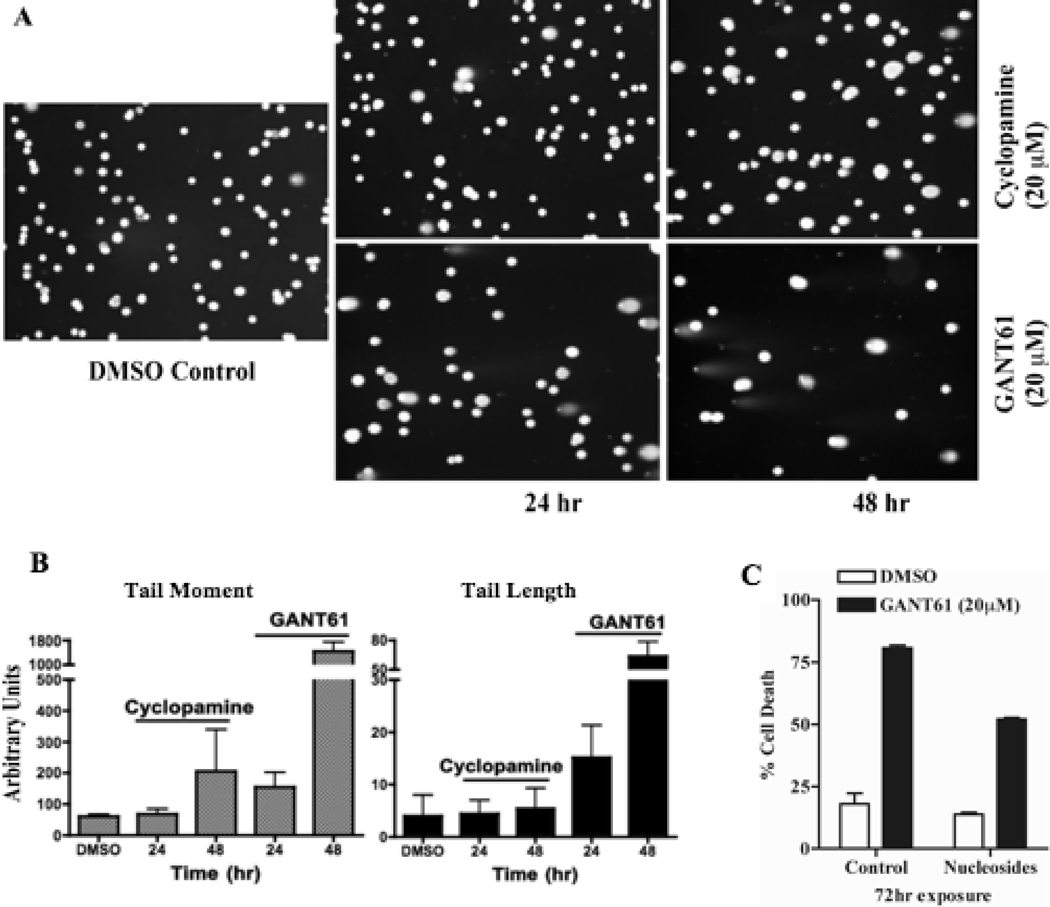
To determine whether GANT61 induces DNA damage following cellular accumulation at G1/S and early S, HT29 cells were treated with GANT61 (20 µM) or cyclopamine (20 µM) for 24 hr or 48 hr. Single cells were analyzed by the COMET assay, which detects DNA damage by alteration in the pattern of cellular elution through agarose gels (Figure 2A). Significant changes in elution profiles were detected in GANT61-treated cells by fluorescence miscroscopy, Tail Moment and Tail Length (Figure 2B). In contrast, cyclopamine-treated cells demonstrated an increase in Tail Moment but not Tail Length at 48 hr. HT29 cells were also exposed to GANT61 (20 µM) or DMSO (control) in the absence or presence of nucleosides (thymidine, adenosine, cytidine, and guanosine; 20 µM each). Supplementation with nucleosides conferred partial protection (~ 50% cell death) from GANT61-induced cytotoxicity (~ 80% cell death; Figure 2C), indicating a role of DNA damage signaling in GANT61-induced cytotoxicity.
Figure 2.
GANT61 induces DNA damage in HT29 cells. A: COMET assay for determination of DNA damage in single cells following treatment with GANT61 or cyclopamine. HT29 cells were treated with GANT61 (20 µM) or cyclopamine (20 µM) for up to 48 hr, harvested, and analyzed by COMET assay. B: Graphical representation of DNA damage by Tail Length and Tail Moment, calculated as described in Materials and Methods. C: Cell death analysis of HT29 cells treated with either DMSO or GANT61 in the presence and absence of nucleosides. HT29 cells were simultaneously exposed to nucleosides (thymidine, adenosine, cytidine, and guanosine; each 20 µM) and/or GANT61 (20 µM) for 72 hr. Cell death was determined by flow cytometry following Annexin V-FITC/PI staining.
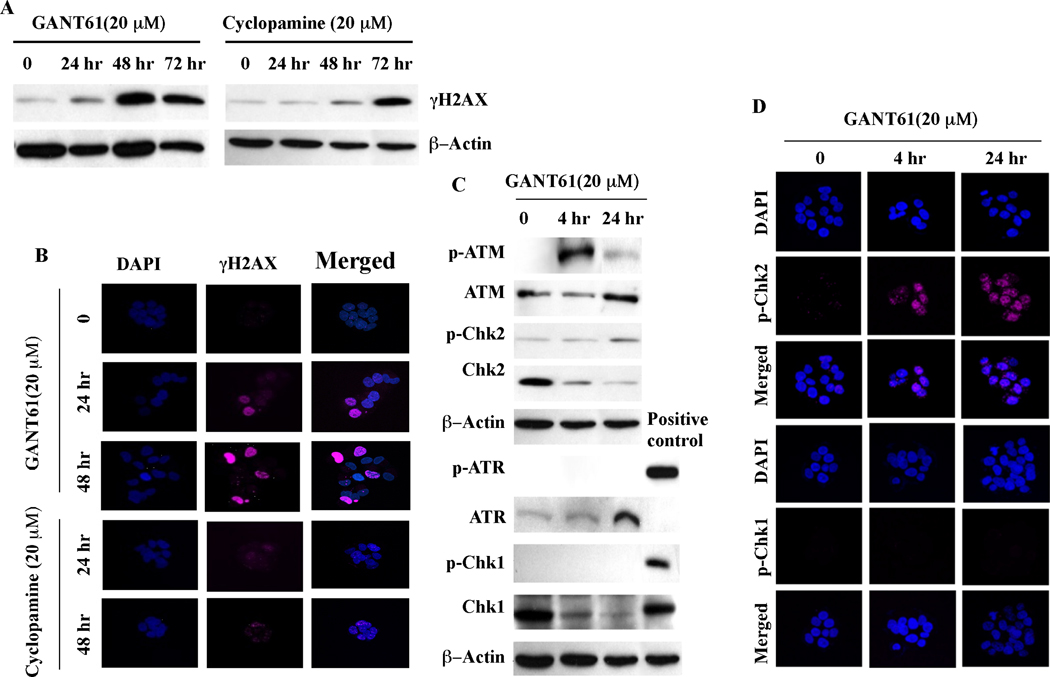
To further characterize the DNA damage response, expression of γH2AX, a marker of double strand breaks (35), was determined by western analysis in HT29 cells treated for up to 72 hr with GANT61 (20 µM) or cyclopamine (20 µM; Figure 3A). Appearance of γH2AX was detected at 24 hr after GANT61 treatment upstream of cell death, and was strongly expressed at 48 hr, when cells were undergoing apoptosis. In contrast, γH2AX was barely detectable in cyclopamine-treated cells at 24 hr by western analysis, and only slightly increased at 48 hr. Further evaluation of γH2AX expression by confocal microscopy is shown in Figure 3B. Following treatment of HT29 cells with GANT61, γH2AX was strongly detected at 24 hr along with a change in cellular morphology comprising cellular dissociation, in the absence of cell death. Changes in cellular morphology by confocal microscopy and γH2AX foci were not detectable within 48 hr of cyclopamine exposure (Figure 3B).
Figure 3.
DNA damage response in GANT61- or cyclopamine-treated cells. A: Western analysis of γH2AX in HT29 cells treated with GANT61 (20 µM) or cyclopamine (20 µM) for up to 72 hr. B: Confocal microscopy of HT29 cells treated with GANT61 (20 µM) or cyclopamine (20 µM) for up to 48 hr. Cells were fixed, permeabilized, and stained for nuclei (DAPI) or γH2AX (DNA damage). C: Expression of phosphorylated or total ATM, Chk2, ATR or Chk1 by western analysis. D: Confocal microscopy to determine p-Chk2 or p-Chk1 nuclear foci following GANT61 (20 µM) treatment.
GANT61 activates ATM and Chk2 in HT29 cells
To determine the molecular mechanism underlying GANT61-induced DNA damage signaling, HT29 cells were treated with GANT61 (20 µM) or cyclopamine (20 µM) for up to 24 hr, and expression of the phosphorylated (active) forms of ATM, ATR, Chk1 and Chk2 were examined by Western analysis (Figure 3C), and p-Chk1 and p-Chk2 by confocal microscopy (Figure 3D). In GANT61-treated cells, p-ATM and p-Chk2 were detected as early as 4 hr, and their expression was sustained for 24 hr. In contrast, p-ATR and p-Chk1 expression remained undetectable. Further, p-Chk2 but not p-Chk1 nuclear foci were detected by confocal microscopy in GANT61-treated cells, indicating an active ATM-Chk2 axis in the GANT61-induced DNA damage response.
Genetic downregulation of Gli1 and Gli2 by Gli3R induces DNA damage and cell death
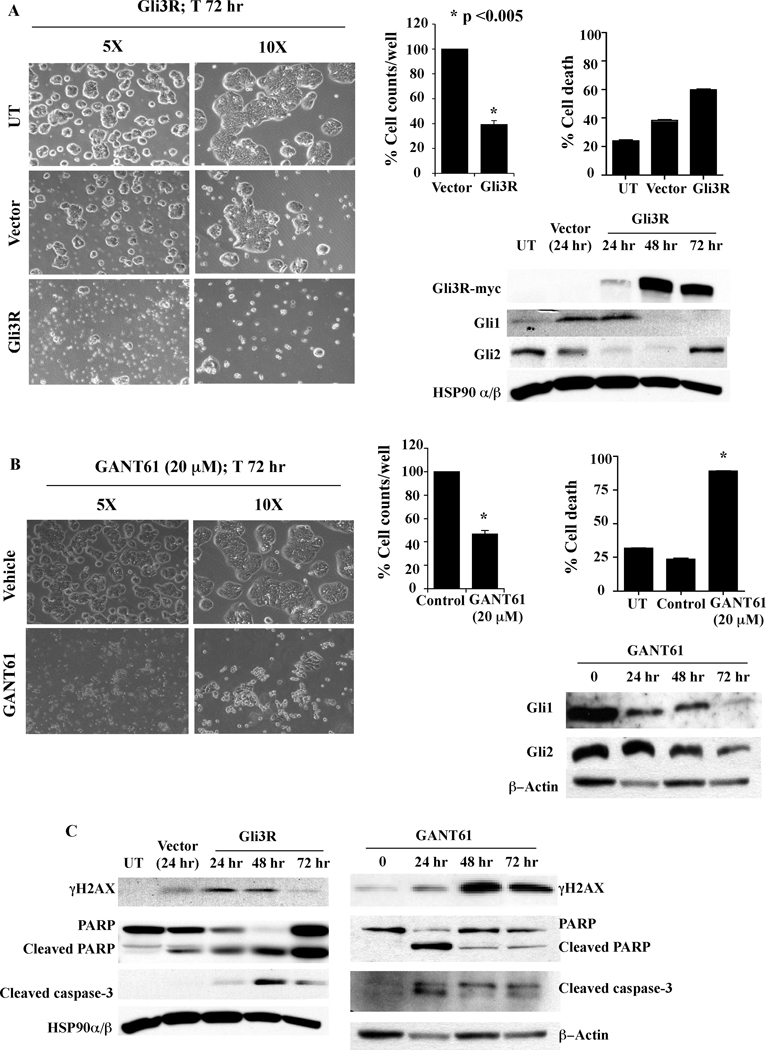
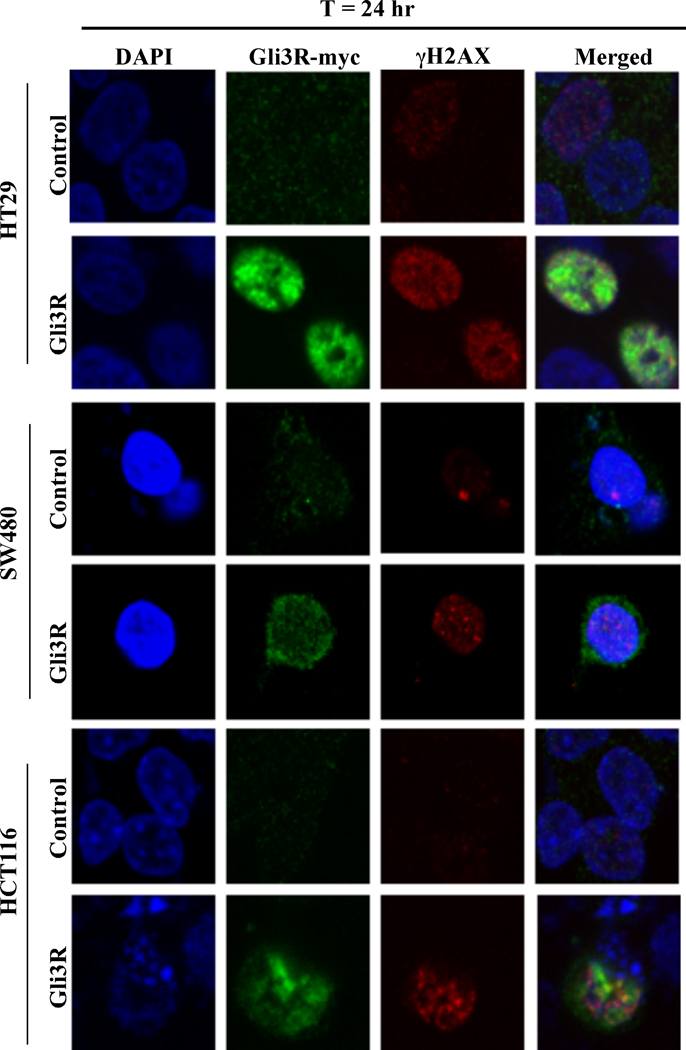
The critical role of Gli1 and Gli2 function in cellular survival in colon carcinoma cells was further investigated by genetic downregulation of Gli1 and Gli2. A c-terminus deleted mutant form of Gli3 (Gli3R, Gli3C’ΔCla1 with a myc-tag (3, 8)) was employed, which contains the N-terminus region that determines nuclear localization and repressor activity. Transient transfection of HT29 cells with Gli3R-pCS2-MT reduced cell growth by 60% over a period of 72 hr (microscopy, cell number), induced cell death (microscopy, annexin V/PI staining), and decreased Gli1 and Gli2 protein expression (western). By 72 hr post-transfection Gli2 protein was re-expressed while decreased Gli1 protein was sustained (Figure 4A). Gli3R was determined by expression of the myc-tag, which was detected by 24 hr and was highest at 48 hr post-transfection (Figure 4A). Similar effects on cell growth, cell death and Gli1 and Gli2 protein expression were induced by GANT61 (20 µM; Figure 4B). Further, induction of DNA damage was detected following transient transfection with Gli3R, marked by elevated expression of γH2AX, detected within 24 hr. This was associated with cleavage of full length PARP and caspase-3, also determined in GANT61-treated cells (Figure 4C). To ensure that the effects of Gli3R were not specific to HT29 only, Gli3R was transiently expressed in SW480 and HCT116 cells and γH2AX expression determined by confocal microscopy. The expression of Gli3R was visualized using an anti-myc antibody. Within 24 hr of transient transfection, myc-tagged Gli3R was detected in nuclei of cells that also expressed γH2AX nuclear foci in all 3 cell lines, HT29, SW480 and HCT116 (Figure 5). Collectively, data demonstrate that both pharmacologic and genetic inhibition of HH signaling result in reduced cell growth, increased cell death and induction of DNA damage that is associated with reduced Gli1 and Gli2 protein levels. These findings emphasize the significance of the HH signaling pathway in human colon cancer cell growth and survival, regulated at the level of the GLI genes.
Figure 4.
Gli3R and GANT61 produce similar cellular responses in HT29 cells. A: Transient transfection with Gli3R for 72 hr, or B: Treatment with GANT61 (20 µM) for 72 hr. Untransfected (UT), empty vector transfected (Vector), GLI3R transfected (Gli3R) cells, or untreated (UT), DMSO- or GANT61-treated cells, were examined under an inverted microscope at 72 hr to determine changes in cellular morphology (left panels). Cells were also counted using a coulter counter to monitor cell growth per well. Cell death was determined at 72 hr by flow cytometry following Annexin V-FITC/PI staining. Total cell lysates were extracted from treated cells at 0 hr, 24 hr, 48 hr and 72 hr; 60 µg of total protein was analyzed by western blot using antibodies specific for the myc tag, Gli1, Gli2 and HSP90α/β as the loading control. C: Expression of γH2AX, cleaved PARP and cleaved caspase-3 determined by western analysis.
Figure 5.
Gli3R localizes to the nucleus and induces DNA damage in three human colon carcinoma cell lines. HT29, SW480 and HCT116 cells were transiently transfected with Gli3R for 24 hr and examined by confocal microscopy. Empty vector transfected (Control), or GLI3R transfected (Gli3R) cells were fixed, permeabilized, and stained for nuclei (DAPI), myc expression (myc-tagged Gli3R) or γH2AX (DNA damage).
GANT61 inhibits specific binding of Gli1 and Gli2 to Gli target genes and blocks transcriptional activity
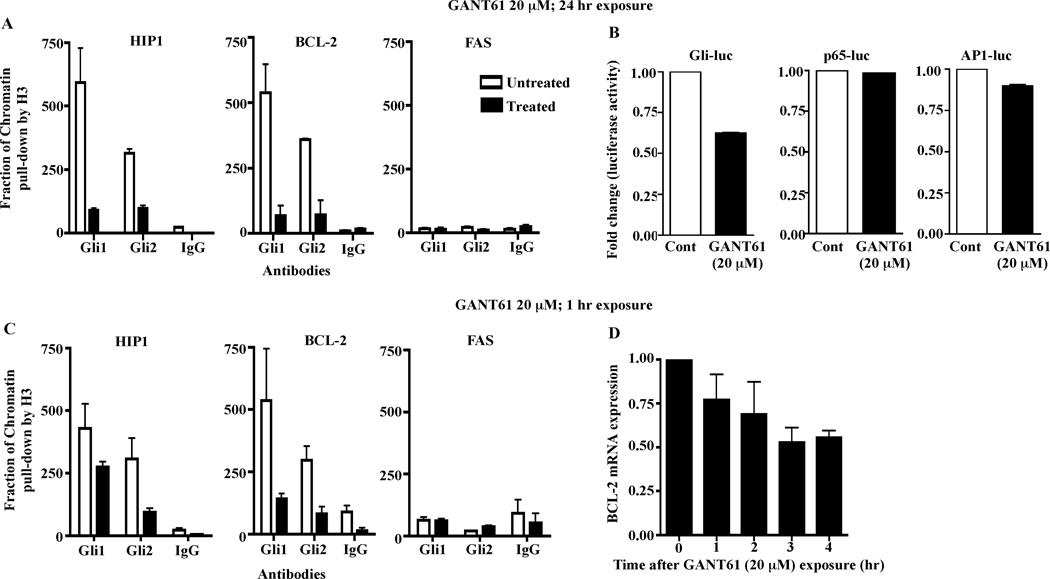
To further determine the specificity of GANT61 for targeting the function of Gli1 and Gli2, ChIP analysis was conducted using GANT61-treated HT29 cells (20 µM; 24 hr). Following isolation of chromatin, immunoprecipitation with Gli1- or Gli2- specific antibodies, or antibodies against IgG (negative control) or histone H3 (positive control) were performed. Subsequent qPCR using primers that flanked the promoter regions of the Gli target genes HIP1 and BCL-2, or FAS (negative control), revealed inhibition of binding by GANT61 of both Gli1 and Gli2 to their target gene promoters of HIP1 and BCL-2 but not to FAS, where no Gli binding sites were detected (Figure 6A). GANT61 (20 µM; 24 hr) specifically inhibited Gli-luciferase reporter activity in transiently transfected HT29 cells. In contrast, neither p65-luciferase nor AP1-luciferase activities were affected (Figure 6B). In addition using short, 1 hr GANT61 (20 µM) exposure, 1) inhibition of Gli1 and Gli2 binding to the promoter regions of HIP1 and BCL-2 was determined at this early time point, similar to data derived after 24 hr drug exposure (Figure 6C), and 2) inhibition of the transcriptional regulation of BCL-2 for up to a further 4 hr examined (Figure 6D). Collectively these data substantiate the specificity of GANT61 in targeting Gli transcriptional activity.
Figure 6.
Specificity of GANT61 for targeting Gli1 and Gli2 in human colon cancer cells. Binding between Gli1 or Gli2 and the promoter regions of HIP1 and BCL-2 but not FAS, was inhibited in GANT61-treated cells. HT29 cells treated with GANT61 (20 µM) for 24 hr (A) or 1 hr (C) were employed for chromatin immunoprecipitation (ChIP) analysis using antibodies specific for Gli1, Gli2, IgG (negative control), or histone H3 (positive control, used for normalization). Subsequent qPCR used primers that flanked the promoter regions of the Gli target genes, HIP1 and BCL-2 or FAS (negative control and not a direct Gli target). B. GANT61 treatment (20 µM; 24 hr) of HT29 cells demonstrated specific decreased reporter activity of a Gli-luciferase reporter. In contrast, exposure to GANT61 did not affect luciferase activity in HT29 cells transfected with p65 (NFκB)- or AP1- luciferase reporters. D. GANT61 treatment (20 µM; 1hr) of HT29 cells inhibits the transcriptional regulation of BCL-2.
DISCUSSION
Regulated function of the HH signaling pathway is critical during embryonic development, while deregulated HH signaling is documented in a variety of human cancers (1, 3–5). Although recent literature suggests the involvement of HH signaling in colon carcinogenesis, progression (6, 7), and metastatic disease (8), there is limited understanding regarding its mechanistic role in colon cancer. Primary human colon carcinomas, liver metastases, xenografts (8), and human colon carcinoma cell lines (12), express HH signaling pathway components. siRNA-mediated downregulation of Gli1 or Gli2 has reduced proliferation and induced apoptosis in primary cultures of human colon cancers and liver metastatses, with the Gli3R acting with greater potency. Further, human colon carcinoma cells transduced with Gli3R have failed to grow as xenografts in nude mice (8).
In the context of colon cancer, previous attempts to block HH signaling at the level of Smo, has induced only moderate cytotoxicity in these cells (36). A recent study reported GANT61, a small molecule inhibitor of both Gli1 and Gli2, which effectively blocks Gli function (26). We have demonstrated that GANT61 induced considerably greater cytotoxicity in six human colon carcinoma cell lines (60%–90% cell death) than that induced by cyclopamine (≈ 30% cell death; (13)). These findings indicate that direct targeting of the Gli transcription factors downstream of Smo is more efficient in interrupting HH signaling, likely due to non-canonical activation of Gli proteins independent of Smo (5, 19–22).
Limited information exists regarding the genes or mechanisms involved in the inhibition of the HH signaling response in cancer cells. In an oral squamous carcinoma cell line, cyclopamine induced a modest (10%) increase in cells at the G1/S boundary (37). Knockdown of Smo using siRNA reduced proliferation of two human colon cancer cell lines, with decreased expression of cyclin E and increased expression of p21Cip1, consistent with accumulation at G1/S (38). However no cytotoxic effects were described in either study. These results are consistent with only modest cytotoxicity demonstrated by cyclopamine in human colon carcinoma cell lines. In detailed studies conducted in HT29 cells, inhibition of Smo by cyclopamine induced a modest increase in cells at the G1/S boundary and modest perturbations in cell cycle distribution, with minimal entry of cells into the subG1 compartment, even after 72 hr of exposure. In contrast, inhibition of Gli1/Gli2 downstream of Smo by GANT61, induced transient accumulation of cells at the G1/S boundary and in early S, followed by entry into the subG1 compartment. Previously, cDNA microarray gene profiling demonstrated up-regulated expression of p21Cip1 mRNA and downregulated expression of genes involved in the G1/S (CYCLIN E, CYCLIN A, CDK2, CDC25A) transition in HT29 (and GC3/c1) human colon carcinoma cell lines treated with GANT61 for 24 hr (12). Using bivariate flow cytometric analysis, accumulation of p21Cip1, cyclin E and cyclin A was observed in the G1- and S- phases in contrast to cyclopamine-treated cells. Incorporation of BrdU also demonstrated accumulation of GANT61-treated cells at the G1/S boundary and delay in early S without further progression. Previously it was demonstrated in GANT61-treated HT29 cells that the mRNAs of genes involved in DNA replication, including thymidylate synthase, thymidine kinase, topoisomerase2, E2F and DNA polymerases, were also downregulated at this time (12), supporting the lack of progression of cells through S-phase. p21Cip1 binds to and inhibits cyclin/cdk complexes with a preference for those containing cdk2 (39), and plays an essential role in growth arrest after DNA damage (40, 41). Over-expression of p21Cip1 can lead to G1 and G2 (42) or S-phase (43) arrest. However, stable p21Cip1 knockdown had no effect on GANT61-induced cell death in HT29 cells supporting a p21Cip1 –independent mechanism.
We have previously reported that GANT61-treated cells demonstrated modifications in genes involved in DNA damage response signaling, including H2AFX, MDC1, BRCA1, FANCD2, CDC45L, the DDI and RAD genes (12). The present study characterized the DNA damage response elicited by GANT61-mediated inhibition of HH signaling activity in human colon cancer cells. In mammalian cells there are two parallel pathways that respond to stress-induced DNA damage: the ATM pathway, which responds to double strand breaks (DSBs), and ATR, which responds to DSBs and to agents that interfere with replication forks (44, 45). Both ATM and ATR are kinases that phosphorylate several target proteins, are early transducers of the DNA damage response (reviewed in (46–48)), and are recruited to DNA break sites following activation (49). Checkpoint functions of ATM are primarily mediated by the effector kinase Chk2, and of ATR by Chk1, following phosphorylation (reviewed in (46, 49)). Efficient transduction of DNA damage signals downstream of ATM and ATR also requires a class of checkpoint mediators and adaptors, whose mechanisms are not yet completely defined (50). One of the earliest modifications of chromatin in the DNA damage response is phosphorylation of H2AX (γH2AX), a direct phosphorylation target of ATM and ATR (46, 49), located at the sites of DNA strand breaks as immunoreactive foci. Expression of γH2AX was detected by both western analysis and confocal microscopy by 24 hr in GANT61-treated cells upstream of cell death. This was not observed in cyclopamine-treated cells. A differential DNA damage response evaluated in single cells in GANT61-treated vs. cyclopamine-treated cells was also determined by COMET assay. The involvement of DNA damage in GANT61-induced cytotoxicity was further substantiated from the protective effect of nucleoside supplementation during exposure of HT29 cells to GANT61, that would elevate the pool of dATP, dGTP, dCTP and dTTP required for DNA replication. Subsequent examination of the early response genes, the activated forms of ATM and Chk2, demonstrated the appearance of p-ATM and p-Chk2 (but not p-ATR or p-Chk1) at 4 hr following GANT61 treatment, that was sustained; p-Chk2 nuclear foci were also determined in individual cells by confocal microscopy (summarized in Figure 7). No activation of ATM, ATR, Chk1 or Chk2 was detected in cyclopamine-treated cells.
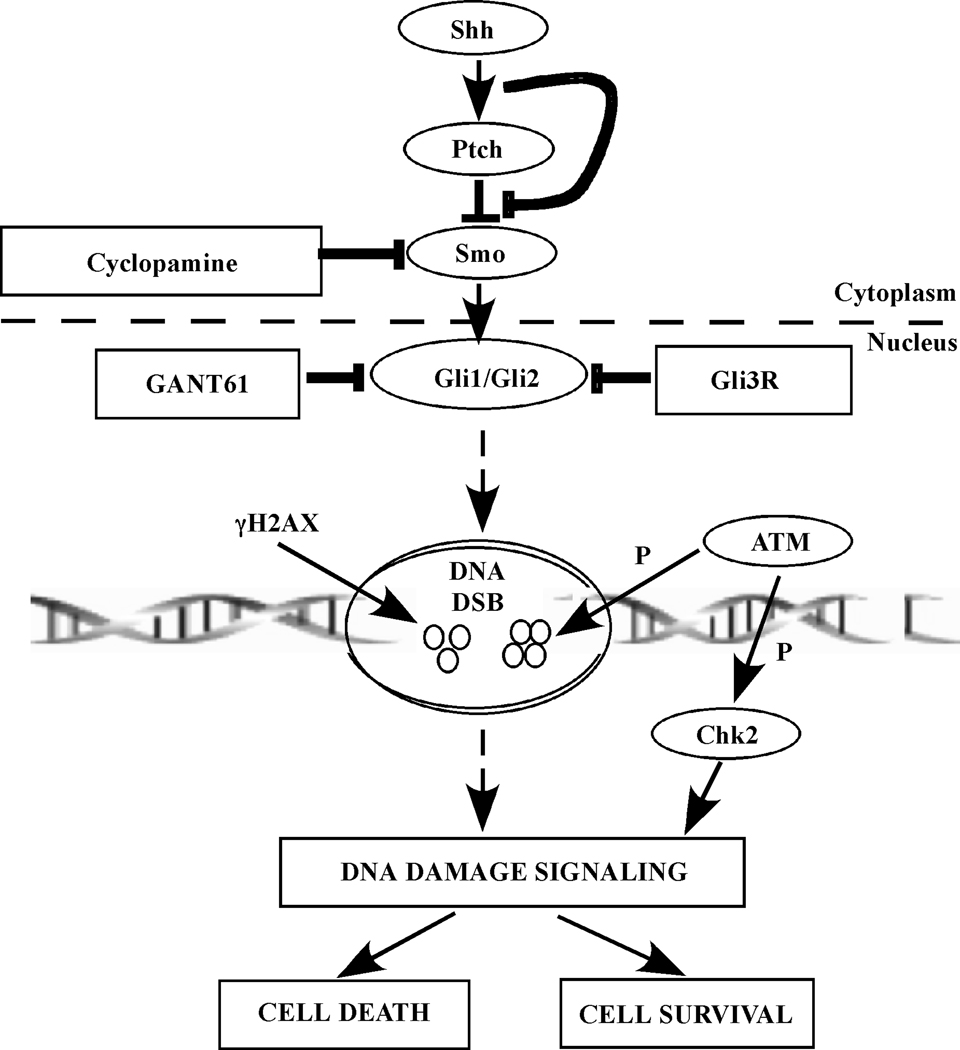
Figure 7.
Schematic representation of the mechanisms underlying inhibition of HH/Gli signaling that result in cell death. Targeting Smo by cyclopamine is upstream and less effective than targeting Gli1/Gli2 by either GANT61 or Gli3R in eliciting cytotoxicity in human colon carcinoma cells. Blocking Gli1/Gli2 function with GANT61 or Gli3R induces DNA damage marked by formation of γH2AX foci at DNA break sites. An ATM/Chk2 axis is involved in DNA damage signaling downstream of inhibiting Gli1/Gli2 activity. P represents phosphorylation and DSB is double strand break.
The role of the Gli proteins in colon cancer cell survival was further confirmed using the c-terminus deleted repressor Gli3R, to inhibit Gli1 and Gli2 activity. Transient expression of Gli3R over a period of 72 hr paralleled the effects of GANT61 by decreasing growth and expression of Gli1 and Gli2 in HT29 cells, inducing cell death, γH2AX expression, cleavage of PARP and caspase-3. Multiple human colon carcinoma cell lines (HT29, SW480, HCT116) respond to the exogenous expression of Gli3R (a C-terminus truncated mutant of the Gli3 protein which localizes to the nucleus (3)), by induction of immunoreactive γH2AX nuclear foci in the same cells expressing nuclear Gli3R. These data demonstrate the far-reaching consequences of Gli3R expression in human colon carcinoma cell lines that express active HH signaling. The GANT61- or Gli3R- induced DNA damage response is also independent of p53, since expression HT29 and SW480 express mutant p53, whereas HCT116 is p53 wild-type.
GANT61 a) functions in the nucleus to abrogate Gli function, b) blocks both Gli1- and Gli2-mediated transcription, c) reduces expression of GLI1 and HIP1 mRNA (qRT-PCR) in contrast to cyclopamine in SUFU-null MEFS, and d) inhibits Gli1 DNA binding activity (EMSA; (26)). Further confirmation of the specificity of Gli1 and Gli2 as targets for GANT61 is provided by ChIP analysis, luciferase reporter assays, and inhibition of the transcriptional regulation of BCL-2. In ChIP analysis GANT61 specifically inhibited the binding of Gli1 and Gli2 transcription factors to promoter regions of the Gli target genes HIP1 and BCL-2 in contrast to that of FAS, which is not a direct target of the Gli proteins, as early as 1 hr following GANT61 exposure. Treatment with GANT61 specifically inhibited Gli-luciferase but not the activity of NF-κB (p65) or AP1 transcription factors in luciferase reporter assays. Inhibition of BCL-2 transcriptional regulation was also determined after 1 hr GANT61 exposure. These findings further substantiate the specificity of GANT61 in targeting Gli transcriptional activity in human colon carcinoma cells.
In summary, inhibition of the HH signaling pathway by targeting the transcription factors Gli1 and Gli2 is highly effective at inducing cell death in human colon carcinoma cells in contrast to targeting Smo upstream of Gli. Inhibition of Gli1 and Gli2 by GANT61 induced inhibition of DNA replication in early S-phase leading to DNA damage signaling involving an ATM/Chk2 axis and induction of cell death. Both pharmacologic (GANT61) and genetic (Gli3R) downregulation of Gli1 and Gli2 by Gli3R reduced Gli1 and Gli2 expression, cell proliferation, and induced changes in cellular morphology, DNA damage, γH2AX nuclear foci, cleavage of PARP and caspase-3, and cell death (schematically represented in Figure 7). The mechanisms underlying the induction of Gli1/Gli2-regulated DNA damage, the significance of an early S-phase response, and the inability to repair damaged DNA, are currently under investigation.
Supplementary Material
Acknowledgements
The authors would like to acknowledge financial support from NCI awards RO1 CA 32613 (JAH), NCI RO1 108929 (JAH), and from the Cleveland Clinic.
REFERENCES
- 1.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 2.Lum L, Beachy PA. The Hedgehog response network: sensors, switches, and routers. Science. 2004;304:1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- 3.Ruiz i Altaba A. Gli proteins encode context-dependent positive and negative functions: implications for development and disease. Development. 1999;126:3205–3216. doi: 10.1242/dev.126.14.3205. [DOI] [PubMed] [Google Scholar]
- 4.Katoh Y, Katoh M. Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr Mol Med. 2009;9:873–886. doi: 10.2174/156652409789105570. [DOI] [PubMed] [Google Scholar]
- 5.Ruiz i Altaba A, Mas C, Stecca B. The Gli code: an information nexus regulating cell fate, stemness and cancer. Trends Cell Biol. 2007;17:438–447. doi: 10.1016/j.tcb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshikawa K, Shimada M, Miyamoto H, et al. Sonic hedgehog relates to colorectal carcinogenesis. J Gastroenterol. 2009;44:1113–1117. doi: 10.1007/s00535-009-0110-2. [DOI] [PubMed] [Google Scholar]
- 7.Bian YH, Huang SH, Yang L, Ma XL, Xie JW, Zhang HW. Sonic hedgehog-Gli1 pathway in colorectal adenocarcinomas. World J Gastroenterol. 2007;13:1659–1665. doi: 10.3748/wjg.v13.i11.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varnat F, Duquet A, Malerba M, et al. Human colon cancer epithelial cells harbour active HEDGEHOG-GLI signalling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. EMBO Mol Med. 2009;1:338–351. doi: 10.1002/emmm.200900039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varnat F, Zacchetti G, Ruiz i Altaba A. Hedgehog pathway activity is required for the lethality and intestinal phenotypes of mice with hyperactive Wnt signaling. Mech Dev. 127:73–81. doi: 10.1016/j.mod.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Alinger B, Kiesslich T, Datz C, et al. Hedgehog signaling is involved in differentiation of normal colonic tissue rather than in tumor proliferation. Virchows Arch. 2009;454:369–379. doi: 10.1007/s00428-009-0753-7. [DOI] [PubMed] [Google Scholar]
- 11.van den Brink GR, Bleuming SA, Hardwick JC, et al. Indian Hedgehog is an antagonist of Wnt signaling in colonic epithelial cell differentiation. Nat Genet. 2004;36:277–282. doi: 10.1038/ng1304. [DOI] [PubMed] [Google Scholar]
- 12.Shi T, Mazumdar T, Devecchio J, et al. cDNA microarray gene expression profiling of hedgehog signaling pathway inhibition in human colon cancer cells. PLoS One. 5 doi: 10.1371/journal.pone.0013054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazumdar T, DeVecchio J, Shi T, Jones J, Agyeman A, Houghton JA. Hedgehog signaling drives cellular survival in human colon carcinoma cells. Cancer Res. 71:1092–1102. doi: 10.1158/0008-5472.CAN-10-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leonard JM, Ye H, Wetmore C, Karnitz LM. Sonic Hedgehog signaling impairs ionizing radiation-induced checkpoint activation and induces genomic instability. J Cell Biol. 2008;183:385–391. doi: 10.1083/jcb.200804042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frappart PO, Lee Y, Russell HR, et al. Recurrent genomic alterations characterize medulloblastoma arising from DNA double-strand break repair deficiency. Proc Natl Acad Sci U S A. 2009;106:1880–1885. doi: 10.1073/pnas.0806882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snijders AM, Huey B, Connelly ST, et al. Stromal control of oncogenic traits expressed in response to the overexpression of GLI2, a pleiotropic oncogene. Oncogene. 2009;28:625–637. doi: 10.1038/onc.2008.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez P, Hernandez AM, Stecca B, et al. Inhibition of prostate cancer proliferation by interference with SONIC HEDGEHOG-GLI1 signaling. Proc Natl Acad Sci U S A. 2004;101:12561–12566. doi: 10.1073/pnas.0404956101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasper M, Schnidar H, Neill GW, et al. Selective modulation of Hedgehog/GLI target gene expression by epidermal growth factor signaling in human keratinocytes. Mol Cell Biol. 2006;26:6283–6298. doi: 10.1128/MCB.02317-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schnidar H, Eberl M, Klingler S, et al. Epidermal growth factor receptor signaling synergizes with Hedgehog/GLI in oncogenic transformation via activation of the MEK/ERK/JUN pathway. Cancer Res. 2009;69:1284–1292. doi: 10.1158/0008-5472.CAN-08-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji Z, Mei FC, Xie J, Cheng X. Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. J Biol Chem. 2007;282:14048–14055. doi: 10.1074/jbc.M611089200. [DOI] [PubMed] [Google Scholar]
- 22.Yoon JW, Kita Y, Frank DJ, et al. Gene expression profiling leads to identification of GLI1-binding elements in target genes and a role for multiple downstream pathways in GLI1-induced cell transformation. J Biol Chem. 2002;277:5548–5555. doi: 10.1074/jbc.M105708200. [DOI] [PubMed] [Google Scholar]
- 23.Thiyagarajan S, Bhatia N, Reagan-Shaw S, et al. Role of GLI2 transcription factor in growth and tumorigenicity of prostate cells. Cancer Res. 2007;67:10642–10646. doi: 10.1158/0008-5472.CAN-07-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikram MS, Neill GW, Regl G, et al. GLI2 is expressed in normal human epidermis and BCC and induces GLI1 expression by binding to its promoter. J Invest Dermatol. 2004;122:1503–1509. doi: 10.1111/j.0022-202X.2004.22612.x. [DOI] [PubMed] [Google Scholar]
- 25.Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- 26.Lauth M, Bergstrom A, Shimokawa T, Toftgard R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci U S A. 2007;104:8455–8460. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jani TS, DeVecchio J, Mazumdar T, Agyeman A, Houghton JA. Inhibition of NF-kappaB signaling by quinacrine is cytotoxic to human colon carcinoma cell lines and is synergistic in combination with tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) or oxaliplatin. J Biol Chem. 285:19162–19172. doi: 10.1074/jbc.M109.091645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juan G, Darzynkiewicz Z. Bivariate analysis of DNA content and expression of cyclin proteins. Chapter 7. Curr Protoc Cytom. 2001;(Unit 7):9. doi: 10.1002/0471142956.cy0709s04. [DOI] [PubMed] [Google Scholar]
- 29.Plesca D, Crosby ME, Gupta D, Almasan A. E2F4 function in G2: maintaining G2-arrest to prevent mitotic entry with damaged DNA. Cell Cycle. 2007;6:1147–1152. doi: 10.4161/cc.6.10.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kogerman P, Grimm T, Kogerman L, et al. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat Cell Biol. 1999;1:312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- 31.Stecca B, Mas C, Clement V, et al. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci U S A. 2007;104:5895–5900. doi: 10.1073/pnas.0700776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dormoy V, Danilin S, Lindner V, et al. The sonic hedgehog signaling pathway is reactivated in human renal cell carcinoma and plays orchestral role in tumor growth. Mol Cancer. 2009;8:123. doi: 10.1186/1476-4598-8-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ekholm SV, Reed SI. Regulation of G(1) cyclin-dependent kinases in the mammalian cell cycle. Curr Opin Cell Biol. 2000;12:676–684. doi: 10.1016/s0955-0674(00)00151-4. [DOI] [PubMed] [Google Scholar]
- 34.Yam CH, Fung TK, Poon RY. Cyclin A in cell cycle control and cancer. CELL Mol Life Sci. 2002;59:1317–1326. doi: 10.1007/s00018-002-8510-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lobrich M, Shibata A, Beucher A, et al. gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle. 9:662–669. doi: 10.4161/cc.9.4.10764. [DOI] [PubMed] [Google Scholar]
- 36.Qualtrough D, Buda A, Gaffield W, Williams AC, Paraskeva C. Hedgehog signalling in colorectal tumour cells: induction of apoptosis with cyclopamine treatment. Int J Cancer. 2004;110:831–837. doi: 10.1002/ijc.20227. [DOI] [PubMed] [Google Scholar]
- 37.Nishimaki H, Kasai K, Kozaki K, et al. A role of activated Sonic hedgehog signaling for the cellular proliferation of oral squamous cell carcinoma cell line. Biochem Biophys Res Commun. 2004;314:313–320. doi: 10.1016/j.bbrc.2003.12.097. [DOI] [PubMed] [Google Scholar]
- 38.Arimura S, Matsunaga A, Kitamura T, Aoki K, Aoki M, Taketo MM. Reduced level of smoothened suppresses intestinal tumorigenesis by down-regulation of Wnt signaling. Gastroenterology. 2009;137:629–638. doi: 10.1053/j.gastro.2009.04.059. [DOI] [PubMed] [Google Scholar]
- 39.Harper JW, Elledge SJ, Keyomarsi K, et al. Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dulic V, Kaufmann WK, Wilson SJ, et al. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 41.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 42.Niculescu AB, 3rd, Chen X, Smeets M, Hengst L, Prives C, Reed SI. Effects of p21(Cip1/Waf1) at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Mol Cell Biol. 1998;18:629–643. doi: 10.1128/mcb.18.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogryzko VV, Wong P, Howard BH. WAF1 retards S-phase progression primarily by inhibition of cyclin-dependent kinases. Mol Cell Biol. 1997;17:4877–4882. doi: 10.1128/mcb.17.8.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 45.Osborn AJ, Elledge SJ, Zou L. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 2002;12:509–516. doi: 10.1016/s0962-8924(02)02380-2. [DOI] [PubMed] [Google Scholar]
- 46.Nojima H. G1 and S-phase checkpoints, chromosome instability, and cancer. Methods Mol Biol. 2004;280:3–49. doi: 10.1385/1-59259-788-2:003. [DOI] [PubMed] [Google Scholar]
- 47.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 48.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 49.Shimada M, Nakanishi M. DNA damage checkpoints and cancer. J Mol Histol. 2006;37:253–260. doi: 10.1007/s10735-006-9039-4. [DOI] [PubMed] [Google Scholar]
- 50.Liu WF, Yu SS, Chen GJ, Li YZ. DNA damage checkpoint, damage repair, and genome stability. Yi Chuan Xue Bao. 2006;33:381–390. doi: 10.1016/S0379-4172(06)60064-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.