Abstract
There is a growing need both clinically and experimentally to improve the characterization of blood lipids. A liquid chromatography-mass spectrometry (LC-MS) method, developed for the qualitative and semi-quantitative detection of lipids in biological samples and previously validated in mitochondrial samples, was now evaluated for the profiling of serum lipids. Data were acquired using high resolution full scan MS and high energy collisional dissociation (HCD) all ion fragmentation. The method was designed for efficient separation and detection in both positive and negative ionization mode and evaluated using standards spanning 7 lipid classes. Platform performance, related to the identification and characterization of serum triglycerides (TGs) was assessed using extracted ion chromatograms with mass tolerance windows of 5 ppm or less from full scan exact mass measurements determined using SIEVE non-differential LC-MS analysis software. The platform showed retention time coefficients of variation (CV) < 0.3%, mass accuracy values < 2 ppm error and peak area CV < 13%, with the majority of that error coming from sample preparation and extraction rather than the LC-MS analysis and linearity was shown to be over four orders of magnitude (r2=0.999) for the standard TG (15:0)3 spiked into serum. Instrument mass accuracy and precision were critical to the identification of unknown TG species, in part because these parameters enabled us to reduce false positives. In addition to detection and relative quantitation of TGs in serum, TG structures were characterized through the use of alternating HCD scans at different energies to produce diagnostic fragmentations on all ions in the analysis. The lipidomics method was applied to serum samples from 192 rats maintained on diets differing in macronutrient composition. The analysis identified 86 TG species with 81 unique masses that varied over 3.5 orders of magnitude and showed diet-dependency - consistent with TGs linking diet and disease risk.
Keywords: Serum, triglyceride, LC-MS, HCD, dietary macronutrients, lipidomics, profiling
INTRODUCTION
Blood serum (plasma) is a readily accessible and widely studied homeostatic biofluid that provides information regarding an individual’s physiological status. In both research and the clinic, most serum (plasma) analyses focus on a limited number of species or groups of species with known biological importance. The analysis of serum lipids, particularly of cholesterol and triglycerides (TGs), is standard in medicine. These tests are part of the routine assessment for heart disease and metabolic disease risk in middle-aged men and post-menopausal women, and are increasingly used in younger individuals considered at high risk for these conditions. Total TG concentration serves as a basis for these health recommendations because it is often linked to increased insulin resistance and the onset of cardiovascular disease.1, 2 It is a growing view that modern -omics profiling approaches will change this standard, by taking advantage of advanced analytical technology and developing sensitive platforms that better capture the overall physiological status of an individual.
As noted, current clinical analyses consider only the aggregate TG concentration level, and thus implicitly define all TG species as equally important. The limitation to this method lies in the possibility that individual TG species may be differentially associated with the condition of interest. Indeed, there is a growing understanding that there are TG species-specific implications for health. Most notably is the association between saturated TGs with short carbon chains and insulin resistance.3 Even in these studies the TGs of interest are only defined by their total acyl carbons and double bonds, but not the specific acyl chain composition. It is reasonable to consider that acyl chain makeup may provide greater insight to the relationship between these species and overall disease risk; however, it is not readily being used
Evaluation of an individual’s TGs in the appropriate context of other plasma lipids may further increase the ability to accurately evaluate that individual’s health/disease status. The diversity and potential importance of plasma lipids has recently been exploited by Quehenberger et al.2 in a quantitative assessment of the six main lipid categories found in plasma: fatty acyls (FA), glycerolipids, glycerophospholipids, sphingolipids, sterols and prenols. In this study, each lipid category was analyzed using targeted approaches developed for the optimum extraction, separation and MS detection of relevant lipids. A variety of extraction techniques, as well as both normal and reversed phased LC, gas chromatography-MS, sample derivitization, and three different types of MS instrumentation, were used to assess the levels of over 500 distinct molecular species that were distributed among the major plasma lipid categories. Overall, 18 unique triglycerides (TGs) were identified, although the method used did not allow the acyl side chains of these species to be fully characterized. The authors speculate that by using calculations to take into account non-isomeric and isobaric species, it would be possible to detect over 200 individual TGs in human plasma. These efforts highlight the complexity of the human plasma lipidome and establish a quantitative baseline that researchers undertaking non-targeted profiling experiments can reference when establishing changes related to individual physiology, nutrition or disease.
To evaluate the chemical and biological diversity of lipids in biological samples, mass spectrometry (MS) based analytical methods ideally should be highly sensitive, with a large dynamic range, excellent mass accuracy and high mass resolution. Lipidomic strategies vary in the degree of analytical bias associated with the method based on the goal of the study4. A variety of targeted approaches—in which particular lipids are analyzed—are available, which allow researchers to concentrate on anything from one or a few specific lipids for absolute quantitation in a biological sample to one specific group or subgroup of lipid species. Although these techniques can be quite sensitive, a potential drawback is that they can only measure the specific predetermined metabolite(s) that are of known importance to the study.5
In a study where all lipid species are of potential biological importance, the most comprehensive measurement of a lipidome from a single analysis would require a non-targeted profiling approach, in which no deliberate bias is given toward any of the 8 lipid categories established and defined by LIPID MAPS (www.lipidmaps.org).6, 7 This type of broad coverage can be achieved using high resolution (HR) MS instrumentation and mass fragmentation in one of two ways. (i) First, shotgun approaches,8–13 which do not involve any chromatographic separation before MS detection, can characterize and quantify over 340 lipid molecular species. Such an approach has been successfully used to establish the lipid profile of the entire yeast lipidome9. (ii) Alternatively, lipids can be separated before detection using liquid chromatography (LC)-MS approaches, which have achieved differential lipid identification and characterization in mouse plasma14, human plasma15 and rat mitochondria.16 Both approaches are relatively comprehensive in terms of lipid coverage; however, using LC-MS with full scan MS acquisitions and all ion fragmentation16 allows for chromatograms to be retroactively searched and new lipids to be identified. This feature is advantageous because in many cases neither the biological importance nor the compositional complexities of all lipid species is completely understood prior to analysis.
This current report uses an HR LC-MS full scan profiling method with all ion high-energy collisional dissociation (HCD) for semi-quantitative lipidomics, applied to profile the serum lipids from rats undergoing a dietary study. The method is presented in the context of general serum lipidome coverage with a specific focus on the characterization and relative quantitation of TGs, highlighting its robustness and analytical range. A total of 86 TG molecules, representing 81 unique masses, were detected in sera samples from 192 rats; trends relating to the major dietary fat constituent and glycemic index were also determined. This method was effectively used to study the significance of nutritional macronutrient diversity (in the form of diets that differed in fat and carbohydrate composition) on the serum lipidome.
MATERIALS AND METHODS
Chemicals
LC-MS grade acetonitrile (ACN), methanol (MeOH), and isopropanol (IPA), as well as highperformance liquid chromatography (HPLC) grade dichloromethane (DCM) and dimethyl sulfoxide were purchased from Fisher Scientific (Pittsburg, PA) and ammonium formate was purchased from Sigma-Aldrich (St. Louis, MO). A detailed list of all lipid standards purchased for LC-MS as well as their abbreviations and sources are in the supplemental information (Tables S1 and S2).
Preparation of Lipid Standards
Stock lipid samples were prepared by dissolving lipid standards in DCM:MeOH (2:1 v/v) at concentrations ranging from 1–10 mg/mL and were stored at −20°C. The internal standard (IS) mixture spiked into each sample before extraction consisted of 25 μg/mL of TG (15:0)3, PC (17:0)2, PG (14:0)2, lysoPC (20:0), PS (16:0)2 and FA (18:1) (Table S1, panel A). A second set of standards with varying concentrations, used to asses method linearity, limit of detection and extraction efficiency, are shown in Table S1, panel B. The biological standard (BIOSTD), which was used to assess the method’s ability to separate and detect multiple lipid species, as well as the HCD fragmentations of those species, contained the 37 standards in Table S2, all at a working concentration of ~5 μg/mL.
Rat Dietary Experiments
Sets of male Fisher 344 × Brown Norway F1 (FBNF1) rats (n=8 per set), aged 7–9 weeks, were fed ad libitum one of 24 isocaloric diets that differed in fat and carbohydrate composition (total n=192). The diets were comprised of six different fat groups, with the major constituent of each being either saturated fats (SFAs), trans fats (Trans), monounsaturated fats (MUFAs), or one of 3 groups of polyunsaturated fats (PUFAs), which vary in the ω-6/ω-3 PUFA ratios. Each type of fat was combined with one of 4 carbohydrate groups that varied based on sucrose content. The fat, carbohydrate and protein percentages in all diets were held consistent at 5, 66, and 20 (w/w), equivalent to 12, 68, and 21 (kcal %). Body weights and food composition were measured twice a week and rats were sacrificed after 8 weeks, collecting the blood serum. Greater details of their husbandry and diets are not critical for the current report and will be presented elsewhere (Greenberg, Stavrovskaya, Baranov, Kristal, manuscripts in preparation).
Lipid Extraction, LC-MS Conditions and Experiments
Lipids were extracted from 30 μL serum aliquots of all individual rat samples and total study pool samples, created by taking aliquots from each of the 192 samples, according to the method of Bligh and Dyer17, substituting DCM for chloroform16, 18. First, 30 μL of IS was added to each 30 μL sample, followed by 190 μL of MeOH. Samples were then vortexed for 20 seconds. Next, 380 μL of DCM was added, the sample was again vortexed for 20 seconds and 120 μL of water was added to induce phase separation. The samples were then vortexed for 10 seconds and allowed to equilibrate at room temperature for 10 minutes before centrifugation at 8000 g for 10 minutes at 10 °C. A total of 370 μL of the lower lipid-rich DCM layer was then collected and the solvent evaporated to dryness under vacuum. Samples were reconstituted in 300 μL of ACN/IPA/H2O (65:30:5 v/v/v) containing PG (17:0)2 at a concentration of 5 μg/mL before LCMS analysis. Ten μL of sample was injected onto the LC-MS system.
Details of the LC-MS method and SIEVE analysis have been described previously.16 Lipid extracts were separated on an Ascentis Express C18 2.1 × 150 mm 2.7μm column (Sigma- Aldrich, St. Louis, MO) connected to a Thermo Fisher Scientific PAL autosampler, Accela quaternary HPLC pump and an Exactive benchtop orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with a heated electrospray ionization (HESI) probe. Mobile phase A in the chromatographic method consisted of 60:40 Water:ACN in 10 mM ammonium formate and 0.1% formic acid and mobile phase B consisted of 90:10 IPA:ACN also with 10 mM ammonium formate and 0.1% formic acid. The spray voltage was set to 3.5 kV, whereas the heated capillary and the HESI probe were held at 250 °C and 350 °C, respectively. The sheath gas flow was set to 25 units and the auxiliary gas set to 15 units. These conditions were held constant for both positive and negative ionization mode acquisitions. The instrument was tuned by direct infusion of PG (17:0)2 in both positive and negative mode and external mass calibration was performed using the standard calibration mixture approximately every five days.
All rat samples were analyzed, in randomized order, by both positive and negative ionization mode in separate experiments acquiring full scan MS only, with resolution set to 60,000 (or 2 Hz) and the scan range between m/z 120–2000. Pool samples and BIOSTDs, used for identification studies, underwent LC-MS experiments in both positive and negative ionization mode that alternated full scan MS acquisitions with HCD scans at 30, 60 or 100 eV. Lipid extraction efficiencies were done by comparing pool samples spiked with the IS mixture before extraction to those spiked with the IS mixture after extraction. Each extraction was performed in triplicate and 4 injections of each sample were done. These experiments were also run in both positive and negative mode. The same chromatographic conditions and buffers were used for all LC-MS experiments.
Data Analysis and Lipid Identification
Results from all LC-MS profiling experiments were analyzed using the MS label free differential analysis software package SIEVE v 1.3 (Thermo Fisher Scientific and Vast Scientific, Cambridge, MA). The chromatograms were time-aligned, referencing a pool sample acquired in the middle of our sequence, and 10k frames were built off of this same reference file. The framing parameters in these experiments were set at 0.01 Daltons for the m/z window and 1.00 minute for the RT window; 1000 was used at the intensity threshold. The frames built off the reference were then applied to all samples in the experiment and the resultant information, which corresponded to m/z, RT, and intensity, were then used to differentiate between the diets via statistical analysis and to identify unknown lipid species in the samples.
For identification, the frame m/z values were used to do batch searches on the Metlin database19, the human metabolome database20 (HMDB) and the LIPID MAPS database7. The frame m/z and RT information was then used to do extracted ion chromatograms (XICs) in the HCD experiments of the pool, using QualBrowser software (Thermo Fisher, San Jose, CA), both to confirm the SIEVE analysis and use the HCD fragmentations to supplement exact match database searches for lipid identification and characterization.
RESULTS AND DISCUSSION
Rat Serum Lipidome Coverage
The diverse nature of the serum lipidome, which includes species with a range of polarities, ionizable moieties, efficiencies and preferences, suggests that comprehensive LC-MS profiling methods should utilize both positive and negative ionization mode. The LC-MS method being used here previously exploited this concept when it was applied to profiling rat mitochondria 16 where multiple categories and subclasses of lipids, in addition to the mitochondria-specific lipid relating to mechanism, cardiolipin, were detected and identified using two profiling experiments run in positive and negative mode separately.
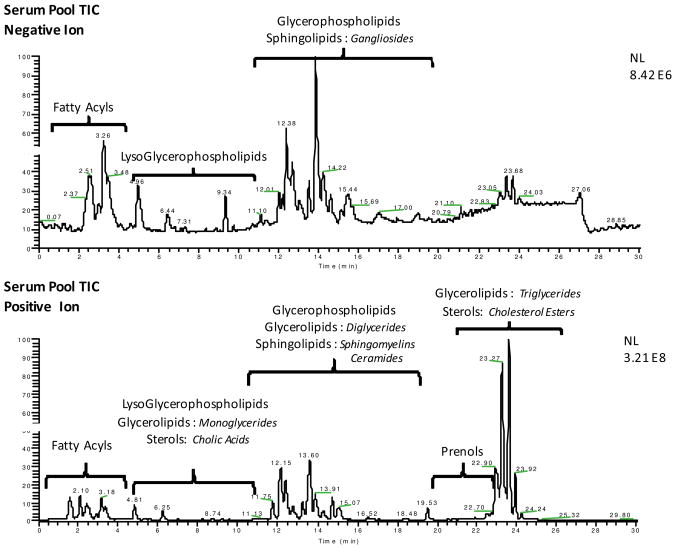
When this methodology was directly applied to serum lipids, similar results were expected and obtained. Figure 1 shows two serum pool total ion chromatograms (TICs) that highlight not only the regions where certain lipid class signals are absent or enhanced based on the ionization mode, but also the dramatic change to the overall chromatogram intensity. The base peak intensity of the negative ion TIC (panel A) was 8.42 × 106, whereas the intensity of the positive ion TIC (panel B) was nearly two orders of magnitude higher at 3.21 × 108. This discrepancy in signal is a result of the highly abundant TGs in serum and their subsequently sensitive detection as NH4+ adducts in positive ionization mode.
Figure 1.
Panels A and B show the total ion chromatogram (TIC) separation of the same serum pool sample using LC-MS performed in negative and positive ionization modes, respectively. Sections of the chromatograms are labeled with the lipid categories detected, indicating the regions where each will elute using the LC-MS method. Highlighted in italics are some lipid classes from those categories that are observed in the serum pool samples. A more detailed list is in Table 1.
In addition to the clinically relevant TGs, other lipid categories are detected more readily by the positive ion chromatogram than the negative ionization TIC; this observation is highlighted by both the labels on the chromatogram and the list of these detectable species in Table 1. There is some cross-over in the ability of the two ionization techniques to detect certain classes of lipids, such as phosphocholines (PCs), free fatty acids (FFAs) and sphingomyelins (SMs), but overall both are needed for the most comprehensive lipid class coverage.
Table 1.
is a breakdown of the lipid categories and classes observed in the serum pools using the LC-MS system described herein. The ionization and adduct preference of each class are defined by the major adduct observed upon the analysis of standards. Observations of these classes in serum samples are also indicated.
| Lipid Class | − Ionization | + Ionization | Found in Serum |
|---|---|---|---|
| Glycerophospholipids (GP) | |||
| Phosphocholine (PC) | [M+FormAcid] − | [M+H]+ | + |
| LysoPhosphocholine (Lyso -PC) | [M+FormAcid] − | [M+H]+ | + |
| Phosphoethanolamine (PE) | [M−H] − | + | |
| Phosphoserine (PS) | [M−H] − | + | |
| Phosphoinositol (PI) | [M−H] − | + | |
| Phosphoglycerol (PG) | M−H] − | [M+NH4]+ | + |
| Phosphatidic Acid (PA) | [M−H] − | [M+NH4]+ | + |
| Cardiolipin (CL) | [M−H] − | [M+NH4]+ | + |
| Fatty Acyls (FA) | + | ||
| Free Fatty acids (FFA) | [M−H] − | [M+NH4]+ | + |
| Eicosanoids and Metabolites | [M−H] − | [M+NH4]+ | + |
| Acylcarnitines (AcCar) | [M+H]+ | + | |
| Sphingolipids (SL) | |||
| Sphingomyelins (SM) | [M+FormAcid] − | [M+H]+ | + |
| Ceramides (Cer) | [M+H]+ | + | |
| Gangliosides (Gan) | [M−H] −/[M−H20−H] − | + | |
| Sterol Lipids (ST) | |||
| Cholic Acids (CA) | [M+H]+ | + | |
| Cholesterol Esters (CE) | [M+NH4]+ | + | |
| Glycerolipids (GL) | |||
| Monoacylglycerol (MG) | [M+NH4]+ | + | |
| Diacylglycerol (DG) | [M+NH4]+ | + | |
| Triacylglycerol (TG) | [M+NH4]+ | + | |
| Prenol Lipids (PL) | |||
| coenzyme -Q (CoQ) | [M+H]+/[M+NH4]+ | + | |
Because many lipid species are isobaric, an understanding of the chromatographic retention time pattern can be used for the characterization or identification of unknown lipids. By using reversed phase LC-MS, lipid separation is being achieved by differences in both head group polarity and FA side chain composition. Lipid standards spanning 7 lipid classes (detailed in Figure S2) were used to determine the elution profile of those species commonly found in biological samples. This information is valuable not only for identifying lipids between classes but also those which fall within the same class and elute over a short retention time (RT) window such as TGs. In these cases, RT, exact mass, and all ion fragmentation, can all be used together to eliminate false positive identifications and make more explicit characterizations.
Serum Triglyceride Identification and Characterization
In order to effectively highlight our LC-MS profiling approach to total serum lipidome coverage, the manuscript concentration will now change to give a systematic account of the identification, and characterization of unknown TGs found in rat serum. This shift in focus is a result of the sheer abundance of TGs in the biofluid, as well as the potential importance of these molecules in the areas of disease diagnosis, therapeutics and risk assessment.
Using the LC-MS profiling method described above, the six TG compounds in the BIOSTD and IS (Tables S1 and S2)—TG (8:0)3, TG (10:0)3, TG (12:0)3, TG (14:0)3, TG (15:0)3 and TG (16:0)3—eluted between 10 and 24 minutes or between 66 – 97% mobile phase B in the gradient. The TGs were detected in positive mode as predominately [M+NH4] + ions with [M+Na] +, [2M+NH4] + and [2M+Na] + ions also being observed at less than 10% of the relative abundance of the [M+NH4] + signal. The TG standards elution profiles, which are based off of acyl chain length and degree of unsaturation, are used when identifying unknown TG species in serum samples.
The BIOSTD was injected a total of 5 times over the course of a 5 day period; during this time, the TG standards were associated with retention time (RT) coefficients of variance (CV) of less than 0.3% when compared across all 5 injections. The TG standards that eluted between 19 and 25 minutes had similar relative intensities, suggesting similar ionization efficiency at this point in the chromatogram. TG (15:0)3 eluted at 23 minutes in the chromatogram and represents the region where the majority of the unknown TG species are to be found. Assessment of TG platform linearity, in relationship to TG (15:0)3 spiked into rat serum, was linear over 4 orders of magnitude with an R2 value of 0.999.
Identification of unknown TGs in serum samples was achieved by first sorting the full scan exact mass measurements and RT pairs determined by SIEVE (termed frames) into chromatographic regions that could contain TGs. The data reducing constraints were determined by the TG standards previously discussed. Four regions of the chromatogram were searched: between RTs of 9.50–14.50 minutes and m/z 450–575, 14.50–19.50 minutes and m/z 575–675, 19.50–22.45 minutes and m/z 675–750, and 22.45–27 minutes and m/z 750–1150. Online databases were then searched for hits that matched frames falling into any of these regions within 0.01 Daltons and those hits were verified via extracted ion chromatograms (XICs) of < 5 ppm which confirmed the monoisoptic peak selection performed by SIEVE.
The Metlin19 and Human Metabolome Database20 (HMDB) were searched for all lipid identifications, not just those that were TG specific, and the LIPID MAPS online MS tool was used to search its virtual glycerolipids database. Results from these databases were at times complementary to each other, and it was necessary to search all of them to increase the absolute number of possible hits as well as the number of high probability hits. The Metlin and HMDB databases allow the user to search by ionization mode, in a batch setting, in which multiple possible exact mass adducts, such as [M+NH4] +, are calculated and included as possible identification options. LIPID MAPS searches were restricted to specific user-defined adducts only. These characteristics were important because the LC-MS method not only yielded [M−H] or [M+H] ions from serum lipids, but also multiple adduct variations depending on the lipid species. Therefore, these databases can still be effectively used even when a search is not focusing on TG identification.
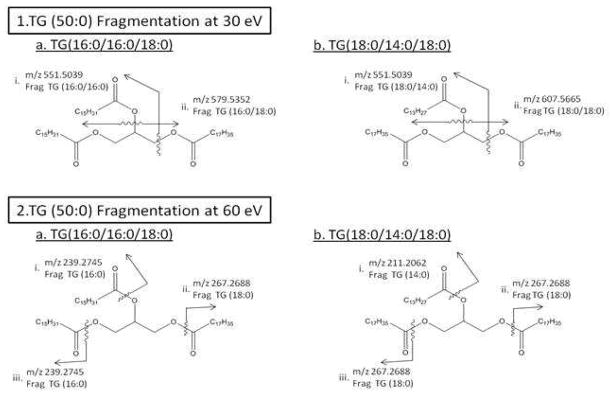
Given that each TG molecule contains 3 FA side chains, each database search could yield multiple possible TG variations for a given exact mass. For example, during a search for the identity of a lipid with m/z 852.8015 found at RT 24.06 minutes, the HMDB provided two possible TG isomers, both with the composition of TG (50:0), indicating the total number of acyl carbons followed by the total number of double bonds based on the exact mass of [M+NH4]+ ions. To differentiate these isobars, the HCD capabilities of the Exactive was used to fragment all ions in the spectrum, using multiple energies, and reconstruct TG molecules based on their individual diagnostic fragmentations. These data are all acquired in one MS scan event, rather than in several isolation and fragmentation steps that can only focus on one ion at a time. This process has the ability to eliminate many false positives and to refine the list of exact mass hits.
HCD fragmentation was performed on the serum pools at three different energies to provide differential fragmentation patterns that can more effectively define each structure. The two HMDB structures of TG (50:0) are shown in Figure 2 with each diagnostic 30eV HCD and 60 eV HCD fragmentation indicated. At 30 eV, each TG is broken into representative diacylglycerol groups that yield one unique fragment for each molecule, labeled as fragment (ii) on each structure in Figure 2 panel 1, and one shared fragment mass, m/z 551.5039, labeled as fragment (i) on each structure in Figure 2 panel 1. At 60 eV each TG is broken into distinct FA groups labeled on each structure individually in Figure 2 panel 2.
Figure 2.
shows the two possible structures of TG (50:0) found in the HMDB, [M+NH4] + m/z 852.8015, as a. and b. of both panels 1 and 2. Panel 1 indicates HCD fragmentation at 30 eV to the unique and shared diacylglycerol fragments on each structure and panel 2 shows the FA fragments from HCD fragmentation at 60 eV.
The TG (50:0) diacylglycerol and FA fragmentation m/z values are theoretically calculated based on positive ionization and HCD fragmentation. Possible exact masses of FA and diacylglycerol fragments were based on the common TGs found in each database and XICs of the theoretically calculated masses were used to search the serum pool samples. Observation of a fragment peak at a given RT that matched a full MS measurement indicates a TG species that contained a diagnostic fragment. That information was used to characterize unknown TG molecules.
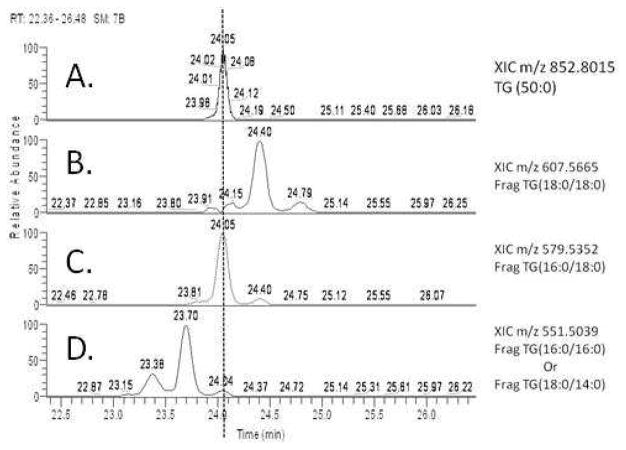
The characterization of TG (50:0) was done using HCD all ion fragmentation with subsequent XIC searches to chromatographically align parent ion masses with fragment masses. Figure 3 shows the XIC alignment of the parent ion m/z 852.8015 and the three possible 30 eV HCD diacylglycerol fragmentations from Figure 2 panel 1.
Figure 3.
shows the XIC of parent ion m/z 852.8015 in panel A, with panels B–D showing XICs of the 3 possible 30 eV diacylglycerol fragments shown in Figure 2. Chromatographic alignment of the parent ion peak at 24.05 minutes can be seen with fragments m/z 579 and m/z 551 in panels C and D.
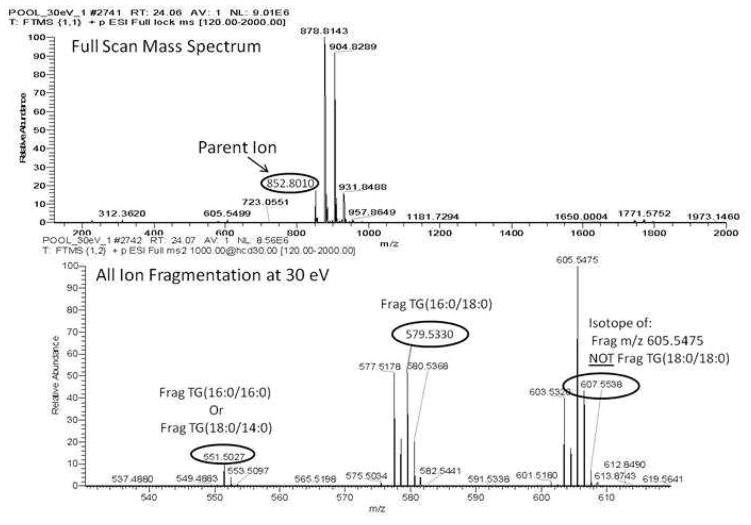
There is a very clear chromatographic alignment of the parent ion peak in Figure 3 panel A with both m/z 579 and m/z 551 (Figure 3, panels C and D), indicating that TG (50:0) is most likely TG (16:0/16:0/18:0). However, it is not clear if panel B, corresponding to m/z 607.5665, shows a peak at 24.05 minutes or not. From the inspection of the full scan mass spectrum at 24.05 minutes and the HCD spectrum at 24.06 minutes, as seen in Figure 4, it is clear that any appreciable signal from m/z 607 is resulting from the [M+2] isotope of m/z 605.5475. This phenomenon is a result of the all ion fragmentation method being used, and shows how by using chromatographic alignment and HR MS data, the ambiguous result can be resolved.
Figure 4.
the top panel, shows the full mass spectrum of the species with an RT of 24.05 minutes from the chromatogram in Figure 3, indicating the parent ion peak of interest at m/z 852.8010. The bottom panel shows the all ion fragmentation mass spectrum at RT 24.06 minutes between m/z 530 - m/z 620. The fragmentations resulting from the parent ion are labeled at m/z 551 and m/z 579, with fragment m/z 607 clearly marked as an M+2 isotope of m/z 605, not a unique fragment from the parent ion of interest.
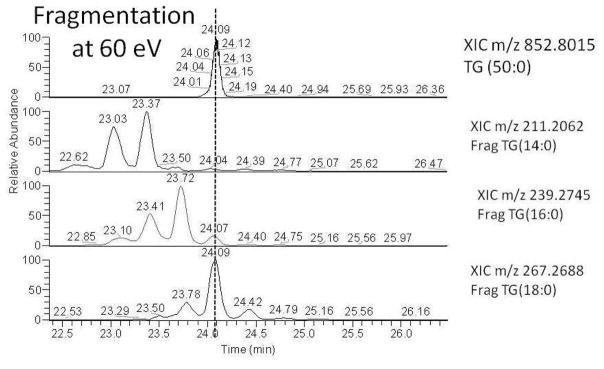
Using HCD at 60 eV further confirms the characterization of TG (50:0) by aligning the parent ion peak at 24.05 minutes in Figure 5 with the 3 possible FA mass XICs from Figure 2 panel 2. The parent ion mass at 24.09 minutes is aligned with m/z 239.2745 and m/z 267.2688, which correspond to FA 16:0 and FA 18:0, respectively. From this experiment, it is apparent that the unknown structure with two possible HMDB hits has been characterized to be TG (16:0/16:0/18:0) (i.e., structure 1 from Figure 2). To be clear, we are characterizing the fatty acyl composition of the TG molecule; however, we cannot tell at which position on the glycerol backbone the FA chains are located nor can we distinguish possible positional isomers from fragmentation alone.
Figure 5.
shows the XIC of parent ion m/z 852.8015 in panel A, with panels B–D showing XICs of the 3 possible 60 eV FA fragments shown in Figure 2. Chromatographic alignment of the parent ion peak at 24.05 minutes can be seen with fragments m/z 239 and m/z 267 in panels C and D, confirming the unknown TG (50:0) to be TG (16:0/16:0/18:0).
This type of analysis—using all ion fragmentation, XIC chromatograms and chromatographic alignment—was used to characterize the TG ions identified by using SIEVE and exact mass searches on the aforementioned databases. With this method, out of 81 unique masses detected from the total of 86 TG species found in rat serum, 62 TG molecules were structurally characterized, as indicated in Table 2.
Table 2.
Chart 2 shows the 86 unique TG species detected in the serum pool samples organized by the C:DB of each TG. The species in italics represent an identified isomer of the previous TG.
| RT | MZ | New Error | TG (C:DB) | ID | % composition | CV (n=18) | |
|---|---|---|---|---|---|---|---|
| 1 | 22.51 | 740.6742 | −0.37 | TG(42:0)* | TG(14:0/14:0/14:0) | 0.128 | 5.35 |
| 2 | 21.99 | 738.6590 | 0.25 | TG(42:1)* | TG(12:0/12:0/18:1) | 0.065 | 28.87 |
| 3 | 22.52 | 766.6905 | 2.10 | TG(44:1)* | 0.138 | 5.13 | |
| 4 | 22.94 | 768.7067 | 1.26 | TG(44:0)* | 0.189 | 7.61 | |
| 5 | 22.26 | 726.6601 | 1.67 | TG(41:0)* | 0.007 | 8.50 | |
| 6 | 21.96 | 712.6437 | 0.69 | TG(40:0)* | 0.093 | 7.49 | |
| 7 | 21.36 | 736.6438 | 0.89 | TG(42:2)* | 0.029 | 7.73 | |
| 8 | 21.16 | 684.6123 | 0.69 | TG(38:0)* | 0.062 | 11.50 | |
| 9 | 22.56 | 792.7061 | 0.36 | TG(46:2) | TG(16:1/14:0/16:1) | 0.273 | 6.02 |
| 10 | 22.95 | 794.7220 | 0.74 | TG(46:1) | TG(16:0/14:0/16:1) | 0.817 | 2.73 |
| 11 | 23.32 | 796.7384 | 1.71 | TG(46:0) | TG(16:0/14:0/16:0) | 0.273 | 14.41 |
| 12 | 22.62 | 818.7216 | 0.22 | TG(48:3) | TG(16:1/14:0/18:2) | 0.373 | 7.13 |
| 13 | 22.95 | 818.7229 | 1.79 | TG(48:3)^ | 0.065 | 10.78 | |
| 14 | 22.97 | 820.7372 | 0.21 | TG(48:2) | TG(16:0/16:1/16:1) | 3.304 | 1.81 |
| 15 | 23.32 | 822.7532 | 0.57 | TG(48:1) | TG(16:0/16:0/16:1) | 3.037 | 6.68 |
| 16 | 23.68 | 824.7693 | 1.18 | TG(48:0) | TG(16:0/16:0/16:0) | 0.507 | 27.79 |
| 17 | 23.15 | 834.7535 | 0.86 | TG (49:1) | TG(15:0/18:1/16:1) | 0.441 | 4.00 |
| 18 | 23.50 | 836.7691 | 0.84 | TG (49:2) | TG(15:0/18:1/16:0) | 0.501 | 4.95 |
| 19 | 22.25 | 840.7067 | 1.04 | TG(50:6) | TG(18:3/14:0/18:3) | 0.012 | 5.53 |
| 20 | 22.48 | 842.7223 | 1.05 | TG(50:5) | TG(18:2/14:0/18:3) | 0.065 | 5.72 |
| 21 | 22.68 | 844.7372 | 0.21 | TG(50:4) | TG(16:1/16:2/18:1) | 0.513 | 8.20 |
| 22 | 23.01 | 846.7524 | −0.40 | TG(50:3) | TG(16:0/16:1/18:2) | 4.278 | 2.62 |
| 23 | 23.32 | 848.7681 | −0.26 | TG(50:2) | TG(16:1/16:0/18:1) | 11.622 | 4.14 |
| 24 | 23.65 | 850.7843 | 0.23 | TG(50:1) | TG(16:0/16:0/18:1) | 6.431 | 6.23 |
| 25 | 24.02 | 852.8005 | 0.82 | TG(50:0) | TG(16:0/16:0/18:0) | 0.270 | 43.15 |
| 26 | 22.90 | 858.7536 | 0.98 | TG(51:4) | TG(15:0/18:2/18:2) | 0.090 | 5.10 |
| 27 | 23.19 | 860.7688 | 0.56 | TG(51:3)^ | 0.427 | 5.06 | |
| 28 | 23.50 | 862.7840 | −0.04 | TG(51:2) | TG(15:0/18:1/18:1) | 1.106 | 6.30 |
| 29 | 22.28 | 866.7221 | 0.78 | TG(52:7) | TG(20:4/14:0/18:3) | 0.014 | 6.12 |
| 30 | 22.59 | 868.7374 | 0.38 | TG(52:6) | TG(16:1/16:1/20:4) | 0.103 | 7.25 |
| 31 | 22.92 | 870.7524 | −0.42 | TG(52:5) | TG(18:1/14:0/20:4) | 0.657 | 8.20 |
| 32 | 23.08 | 872.7679 | −0.55 | TG(52:4) | TG(18:1/16:0/18:3) | 4.223 | 3.56 |
| 33 | 23.35 | 874.7831 | −1.14 | TG(52:3) | TG(18:1/16:0/18:2) | 11.400 | 2.78 |
| 34 | 24.02 | 874.7851 | 1.15 | TG(52:3)^ | 0.164 | 16.26 | |
| 35 | 23.65 | 876.7991 | −0.78 | TG(52:2) | TG(18:1/16:0/18:1) | 18.753 | 2.84 |
| 36 | 25.46 | 876.8008 | 1.13 | TG(52:2)^ | 0.025 | 20.59 | |
| 37 | 23.99 | 878.8150 | −0.45 | TG(52:1) | TG(16:0/16:0/20:1) | 2.204 | 5.93 |
| 38 | 22.34 | 892.7376 | 0.61 | TG(54:8) | TG(18:3/18:2/18:3) | 0.018 | 5.24 |
| 39 | 22.73 | 894.7524 | −0.35 | TG(54:7) | TG(18:2/18:2/18:3) | 0.190 | 8.21 |
| 40 | 22.94 | 896.7677 | −0.77 | TG(54:6) | TG(16:0/16:0/22:6) | 1.058 | 5.60 |
| 41 | 23.08 | 898.7831 | −1.10 | TG(54:5) | TG(18:1/18:1/18:3) | 3.395 | 3.54 |
| 42 | 23.78 | 898.7835 | −0.57 | TG(54:5) | TG(16:1/18:0/20:4) | 0.303 | 21.38 |
| 43 | 23.39 | 900.7983 | −1.66 | TG(54:4) | TG(18:2/16:0/20:2) | 5.300 | 2.15 |
| 44 | 24.02 | 900.7989 | −1.00 | TG(54:4) | TG(16:0/18:0/20:4) | 0.632 | 20.54 |
| 45 | 23.65 | 902.8137 | −1.80 | TG(54:3) | TG(18:1/16:0/20:2) | 6.319 | 1.38 |
| 46 | 23.99 | 904.8300 | −1.19 | TG(54:2) | TG(16:0/18:2/20:0) | 2.780 | 9.52 |
| 47 | 24.33 | 906.8464 | −0.28 | TG(54:1) | TG(16:0/18:0/20:1) | 0.249 | 21.63 |
| 48 | 22.40 | 918.7531 | 0.33 | TG (56:9)^ | 0.021 | 5.23 | |
| 49 | 22.79 | 920.7677 | −0.72 | TG (56:8) | TG(18:2/16:0/22:6) | 0.207 | 9.87 |
| 50 | 23.10 | 922.7832 | −0.94 | TG (56:7) | TG(18:1/16:0/22:6) | 0.950 | 4.46 |
| 51 | 23.24 | 924.7987 | −1.10 | TG (56:6) | TG(18:1/16:0/22:5) | 1.262 | 4.52 |
| 52 | 23.51 | 926.8141 | −1.36 | TG (56:5) | TG(18:1/18:1/20:3) | 1.160 | 7.51 |
| 53 | 23.76 | 928.8303 | −0.82 | TG (56:4) | TG(18:1/18:1/20:2) | 0.635 | 9.91 |
| 54 | 23.96 | 930.8452 | −1.56 | TG(56:3) | TG(16:1/20:1/20:1) | 0.774 | 14.87 |
| 55 | 24.31 | 932.8613 | −1.12 | TG (56:2) | TG(16:0/20:1/20:1) | 0.227 | 25.13 |
| 56 | 24.71 | 934.8782 | 0.17 | TG (56:1) | TG(18:0/18:0/20:1) | 0.063 | 21.13 |
| 57 | 22.51 | 944.7683 | −0.10 | TG(58:10) | TG(18:2/20:4/20:4) | 0.029 | 6.60 |
| 58 | 22.81 | 946.7834 | −0.70 | TG(58:9) | TG(18:2/18:1/22:6) | 0.101 | 10.03 |
| 59 | 23.11 | 948.7992 | −0.61 | TG(58:8) | TG(18:1/18:1/22:6) | 0.266 | 6.25 |
| 60 | 23.35 | 950.8150 | −0.41 | TG(58:7) | TG(18:1/18:1/22:5) | 0.239 | 4.22 |
| 61 | 23.54 | 952.8304 | −0.73 | TG(58:6)^ | 0.191 | 9.22 | |
| 62 | 23.77 | 954.8466 | −0.11 | TG(58:5)^ | 0.110 | 14.80 | |
| 63 | 23.99 | 956.8620 | −0.30 | TG(58:4) | TG(18:2/20:1/20:1) | 0.091 | 14.41 |
| 64 | 24.31 | 958.8768 | −1.31 | TG(58:3) | TG(18:2/20:0/20:1) | 0.131 | 23.07 |
| 65 | 24.64 | 960.8930 | −0.59 | TG(58:2) | TG(18:1/20:0/20:1) | 0.108 | 14.13 |
| 66 | 25.08 | 962.9096 | 0.33 | TG(58:1) | TG(18:1/20:0/20:0) | 0.038 | 20.78 |
| 67 | 22.35 | 968.7687 | 0.40 | TG (60:12)^ | 0.009 | 5.43 | |
| 68 | 22.67 | 970.7840 | −0.10 | TG (60:11)^ | 0.018 | 9.27 | |
| 69 | 22.95 | 972.7992 | −0.55 | TG (60:10)^ | 0.033 | 6.28 | |
| 70 | 23.12 | 974.8146 | −0.79 | TG (60:9)^ | 0.039 | 8.14 | |
| 71 | 23.42 | 976.8299 | −1.20 | TG(60:8) | TG(20:0/20:4/20:4) | 0.023 | 10.48 |
| 72 | 24.30 | 980.8125 | 0.14 | TG(60:6) | TG(20:1/20:1/20:4) | 0.007 | 6.77 |
| 73 | 24.02 | 982.8783 | 0.36 | TG(60:5) | TG(20:0/20:1/20:4) | 0.031 | 13.41 |
| 74 | 24.31 | 984.8928 | −0.78 | TG(60:4) | TG(20:0/20:0/20:4) | 0.058 | 23.83 |
| 75 | 24.64 | 986.9082 | −1.03 | TG(60:3) | TG(20:1/20:1/20:1) | 0.087 | 17.13 |
| 76 | 25.01 | 988.9247 | −0.15 | TG(60:2) | TG(20:0/20:1/20:1) | 0.063 | 13.93 |
| 77 | 25.49 | 990.941 | 0.79 | TG(60:1) | TG(20:0/20:0/20:1) | 0.014 | 24.77 |
| 78 | 22.50 | 994.7840 | −0.02 | TG(62:13)^ | 0.004 | 8.33 | |
| 79 | 23.06 | 998.8167 | 1.37 | TG(62:11) | TG(22:5/18:1/22:5) | 0.003 | 20.92 |
| 80 | 24.20 | 1008.8941 | 0.54 | TG(62:6)* | 0.008 | 18.39 | |
| 81 | 24.56 | 1010.9097 | 0.44 | TG(62:5)* | 0.016 | 19.33 | |
| 82 | 24.98 | 1014.9404 | −0.12 | TG(62:3)* | 0.033 | 20.22 | |
| 83 | 25.23 | 1002.9411 | 0.59 | TG(61:2)* | 0.009 | 15.59 | |
| 84 | 25.41 | 1016.9567 | 0.52 | TG(62:2)* | 0.019 | 18.81 | |
| 85 | 25.35 | 1042.9726 | 0.69 | TG(64:3)* | 0.009 | 19.32 | |
| 86 | 25.85 | 1044.9886 | 1.10 | TG(64:2)* | 0.006 | 22.41 |
lipid maps glycerolipid virtual database exact mass,
HMDB exact mass
Rat Serum Triglyceride Relative Quantitation
The exact molecular composition of the TG molecules found in serum can vary greatly. This diversity reflects changes in acyl side chain length, the degree of unsaturation and the location of the FA moieties on the glycerol backbone. The possible biological implications of this diversity (e.g., susceptibility to insulin resistance, contribution to increased incidence of type II diabetes)1, 21 highlight the importance of fully characterizing TG populations, as opposed to only monitoring changes in total TG content.
The 86 total TG species found in rat serum using the general profiling method outlined above are organized in Table 2. This total number is in comparison to the 18 TG species determined in human plasma by Quehenberger et al.2 using a targeted approach, the 45 species in human plasma determined by Krotronen et al.1 using a non-targeted LC-MS profiling method, and the 65 mouse hepatic TGs found by Hu and colleagues22, also via non-targeted LC-MS profiling. It is important to remember that this manuscript focuses on the characterization of TGs in rat serum; however, the method employed monitors all of the lipid classes and categories shown in Table 1. The increased number of TGs identified here could be a direct result of the dietary study investigated. In this study, the animals were fed diets of varying fat sources, leading to a large pool of FAs available for incorporation during lipid biosynthesis, and therefore a greater diversity to be analyzed. It is especially encouraging, however, that the extensive TG coverage found here can be achieved without sacrificing the detection of other lipids in serum.
The TGs in Table 2 are organized according to the total number of acyl carbons and the number of double bonds (C:DB). The mass error (corrected based on the average TG mass defect), total TG percent composition in the pools and relative quantitative precision for the 18 pool sample injections analyzed over 5 days are also shown. Two CV values are given for each TG: (i) before any normalization and (ii) after normalization to the total TG signal from the pool sample. The median CV from the non-normalized data was ~ 20% (with an average of 23%); after normalization the values were reduced to 8 and 11%, respectively.
These data were acquired using two identical aliquots of the common serum pool, POOL1 and POOL2, each extracted and prepared for LC-MS identically, and injected a total of 9 times. The median TG CV across these nine injections and before any correction was 13% for POOL1 and 3% for POOL2. This difference in CV reflects variations upstream from the LC-MS analysis, i.e., in sample preparation and TG extraction. Therefore, both pool samples were used for all normalization. We are aware that this approach of normalizing to total TGs is useful for assessing the precision of the analytical platform (and contributions to this precision), but has limited applicability to individual samples because of inherent biological differences in total TG levels. The total signal correction method used on both pool samples is essentially equivalent to standardization to an internal standard. Thus, these corrected numbers reflect variation from the LC-MS analysis and data reduction steps, removing the majority of error derived from sample extraction and processing in the overall method. There is a range of TG precision values that varies in relation to the total signal observed. These values increase, as is expected, as the limit of detection of the platform is approached. Note that the method recognizes unknown TGs in rat serum that differ in abundance by over 3.5 orders of magnitude (5000-fold difference in relative abundance).
The mass error of each TG is consistently less than 2 ppm after correction, and this value, together with the relative quantitation CVs over the 18 POOL injections, were used to help eliminate false positive TG identifications. SIEVE values which were included in the first data reduction step but that fell outside the normal error and CV boundaries determined from the pools had their individual XICs and MS spectra specifically scrutinized for TG analysis. Using this approach, approximately 20 TG isotopes were found to be misidentified as unique TG species in the initial analysis, similar to the example in Figures 3 and 4.
Of the 86 total TG species identified, 62 were able to have their individual FA chains characterized. The comprehensive list of TGs in Table 2 is labeled to show those molecules that were identified or characterized by exact mass alone. Because a large number of species elute over a limited time period, it is natural for co-elution of some species to occur, and the method of all ion fragmentation is often unable to clearly distinguish which fragment peaks are generated from which parent ion. In many cases, the fragment and parent ion intensities and RT times can be used to determine the FA side chains that comprise each species; however, if the parent ion intensity is too low and the number of co-eluting species is too high, such characterization is not possible. In these cases, the TGs are identified by their C:DB ratio, which still represents a far more specific and biologically important determination of serum TG levels than solely basing this number on the total TGs found.
Diet Dependent Triglyceride Modulation
This rat serum study, a component of the NIH Genes and Environment Initiative (GEI), tests a hypothesis that intra-class shifts of fats and carbohydrates in the diet will affect the serum biochemical fingerprint in such a way that these changes can serve as a pathogenic link between environmental stresses—in this case diet—and any long-term pathobiological outcomes. This study involved 24 different isocaloric diets that varied in the fat and carbohydrate groups included (see Materials and Methods), with 8 rats held for 8 weeks on each diet for a total sample set of 192 rats/cohort.
The effect of these diets on TG concentrations is of interest because of studies that have shown that in humans SFA intake is often related to insulin resistance23, whereas PUFA intake improves insulin sensitivity24. Additionally, a recent study showed that serum concentrations of specific TGs, such as TG (16:0/16:0/18:1) or TG (16:0/18:1/18:0), show more precise relationships to insulin resistance than does the customary total TG concentration1.
The two TG molecules discussed in that study, TG (16:0/16:0/18:1) and TG (16:0/18:1/18:0), were also identified and characterized in our rat serum dietary study. Using the LC-MS method described, we are unable to directly compare the signals of each TG to each other or yield absolute quantitative values without the use of isotopic correction methods, and internal standards.25 However, we are able to compare the intensity of these species individually across all diets and all rats in the study and interpret a relationship between the animal’s food intake and occurrence of these TGs. In this study, relative quantitation was achieved based off of the monoisotpic [M+NH4]+ m/z intensity value, provided by SIEVE. Comparisons of these values across all rats showed trends linking the relative amount of each of these TG species to the major fat component of each diet, but also showed that the trends were independent of dietary glycemic index.
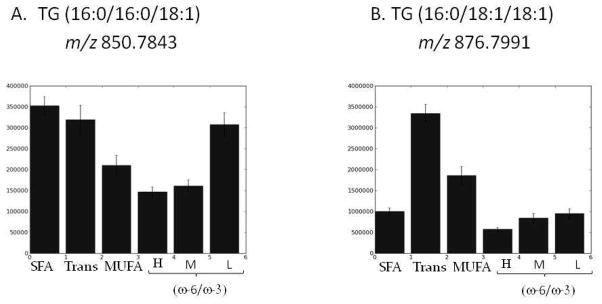
The bar graphs in Figure 6 show how levels of both TG molecules vary depending on which of the six fat groups was present in the diet (with the major fat constituent labeled). Because TG levels were not affected by the carbohydrate in the diet, each bar is comprised of data from all four glycemic index diets associated with a given fat group. TG (16:0/16:0/18:1) shows increases in rats fed the SFA, Trans and low (w-6/w-3) diets. The increase associated with the SFA diets seems natural because the molecule is mostly comprised of saturated fats. TG (16:0/18:1/18:0) showed dramatic increases in rats fed all Trans fat and MUFA diets relative to the other four. Both of these TG species contain an 18:1 FA chain that contains a single double bond in the structure. Two of the fat diets surveyed, Trans and MUFA, contain 18:1 species but with differing stereochemistry (i.e., the Trans diet has the trans isomer and the MUFA diet has the cis isomer). Using our current method, we cannot determine the stereochemistry around that double bond, which could be in either a cis or trans position. However, the observed trend of TG (16:0/18:1/18:0), which increased so dramatically in rats fed the Trans fat diets, suggest that it is mostly the trans isomer that is present. The ability to measure and characterize these TG species will facilitate the studies required to develop a more solid biological interpretation of these results.
Figure 6.
shows the bar graphs representing the amount of TG (16:0/16:0/18:1) or TG (16:0/18:1/18:0) (panel A and B respectively) found in serum samples of animals held on each fat diet. In this analysis, the 4 carbohydrate diets did not affect the amount of TGs found; the trends observed were solely dictated by the major fat constituent consumed. Bars show means +/− s.e.m. (standard error of the mean)
CONCLUSIONS
The goal of this study was to use a method recently developed for lipid profiling in all biological fluids or tissues and apply its strengths for surveying the rat serum lipidome. The platform’s breadth of coverage, qualitative information and quantitative precision make this approach a powerful tool for comprehensive LC-MS lipidomic analysis. Although the focus in the current report was on TG detection, characterization and relative quantitation in serum, the HR LC-MS method can be applied not only to other biological samples, such as cerebrospinal fluid or tissue, but it can also be used to quantify and assess the other lipid species that were detected in the analysis. The ability to use HCD scans to fragment all ions in the chromatogram with mass accuracies consistently below 2 ppm allows the data to be retroactively searched and analyzed to characterize classes of lipids that may not have been of initial interest in the study. In addition to TGs, many serum bioactive lipids can be affected by dietary macronutrients and their subsequent variations can often be informative regarding disease risk assessment.
Supplementary Material
Acknowledgments
The studies reported were funded by U01-ES16048 (BSK, PI), a part of the NIH Genes and Environment Initiative (GEI) and RC1ES018411 (BSK, PI), funded through ARRA. The authors also thank ThermoFisher for the loan of an Exactive Benchtop orbitrap for demonstration testing and financial support for scientific meeting attendance.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional information, as noted in the text, is available free of charge at http://pubs.acs.org.
References
- 1.Kotronen A, Velagapudi VR, Yetukuri L, Westerbacka J, Bergholm R, Ekroos K, Makkonen J, Taskinen MR, Oresic M, Yki-Jarvinen H. Diabetologia. 2009;52:684–690. doi: 10.1007/s00125-009-1282-2. [DOI] [PubMed] [Google Scholar]
- 2.Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, Sullards CM, Wang E, Murphy RC, Barkley RM, Leiker TJ, Raetz CR, Guan Z, Laird GM, Six DA, Russell DW, McDonald JG, Subramaniam S, Fahy E, Dennis EA. J Lipid Res. 51:3299–3305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhee EP, Cheng S, Larson MG, Walford GA, Lewis GD, McCabe E, Yang E, Farrell L, Fox CS, O’Donnell CJ, Carr SA, Vasan RS, Florez JC, Clish CB, Wang TJ, Gerszten RE. J Clin Invest. 121:1402–1411. doi: 10.1172/JCI44442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li M, Zhou Z, Nie H, Bai Y, Liu H. Anal Bioanal Chem. 2010 doi: 10.1007/s00216-010-4327-y. [DOI] [PubMed] [Google Scholar]
- 5.Dennis EA. Proc Natl Acad Sci U S A. 2009;106:2089–2090. doi: 10.1073/pnas.0812636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CR, Russell DW, Seyama Y, Shaw W, Shimizu T, Spener F, van Meer G, VanNieuwenhze MS, White SH, Witztum JL, Dennis EA. J Lipid Res. 2005;46:839–861. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 7.Fahy E, Sud M, Cotter D, Subramaniam S. Nucleic Acids Res. 2007;35:W606–612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ejsing CS, Moehring T, Bahr U, Duchoslav E, Karas M, Simons K, Shevchenko A. J Mass Spectrom. 2006;41:372–389. doi: 10.1002/jms.997. [DOI] [PubMed] [Google Scholar]
- 9.Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A. Proc Natl Acad Sci U S A. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han X, Gross RW. J Lipid Res. 2003;44:1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Han X, Gross RW. Mass Spectrom Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 12.Han X, Yang J, Cheng H, Yang K, Abendschein DR, Gross RW. Biochemistry. 2005;44:16684–16694. doi: 10.1021/bi051908a. [DOI] [PubMed] [Google Scholar]
- 13.Han X, Yang K, Yang J, Cheng H, Gross RW. J Lipid Res. 2006;47:864–879. doi: 10.1194/jlr.D500044-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu C, van Dommelen J, van der Heijden R, Spijksma G, Reijmers TH, Wang M, Slee E, Lu X, Xu G, van der Greef J, Hankemeier T. J Proteome Res. 2008;7:4982–4991. doi: 10.1021/pr800373m. [DOI] [PubMed] [Google Scholar]
- 15.Pietilainen KH, Sysi-Aho M, Rissanen A, Seppanen-Laakso T, Yki-Jarvinen H, Kaprio J, Oresic M. PLoS ONE. 2007;2:e218. doi: 10.1371/journal.pone.0000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bird SS, Marur VR, Sniatynski MJ, Greenberg HK, Kristal BS. Anal Chem. 83:940–949. doi: 10.1021/ac102598u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bligh EG, Dyer WJ. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 18.Cequier-Sanchez E, Rodriguez C, Ravelo AG, Zarate R. J Agric Food Chem. 2008;56:4297–4303. doi: 10.1021/jf073471e. [DOI] [PubMed] [Google Scholar]
- 19.Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 20.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. Nucleic Acids Res. 2009;37:D603–610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu FB, van Dam RM, Liu S. Diabetologia. 2001;44:805–817. doi: 10.1007/s001250100547. [DOI] [PubMed] [Google Scholar]
- 22.Hu C, Hoene M, Zhao X, Haring HU, Schleicher E, Lehmann R, Han X, Xu G, Weigert C. PLoS One. 2010;5:e13318. doi: 10.1371/journal.pone.0013318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vessby B, Uusitupa M, Hermansen K, Riccardi G, Rivellese AA, Tapsell LC, Nalsen C, Berglund L, Louheranta A, Rasmussen BM, Calvert GD, Maffetone A, Pedersen E, Gustafsson IB, Storlien LH. Diabetologia. 2001;44:312–319. doi: 10.1007/s001250051620. [DOI] [PubMed] [Google Scholar]
- 24.Corpeleijn E, Feskens EJ, Jansen EH, Mensink M, Saris WH, de Bruin TW, Blaak EE. Diabetologia. 2006;49:2392–2401. doi: 10.1007/s00125-006-0383-4. [DOI] [PubMed] [Google Scholar]
- 25.Han X, Gross RW. Anal Biochem. 2001;295:88–100. doi: 10.1006/abio.2001.5178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.