Abstract
Host genetic variation, particularly within the human leukocyte antigen (HLA) loci, reportedly mediates heterogeneity in immune response to certain vaccines; however, no large study of genetic determinants of anthrax vaccine response has been described. We searched for associations between the IgG antibody to protective antigen (AbPA) response to Anthrax Vaccine Adsorbed (AVA) in humans and polymorphisms at HLA class I (HLA-A, -B, and -C) and class II (HLA-DRB1, -DQA1, -DQB1, -DPB1) loci. The study included 794 European-Americans and 200 African-Americans participating in a 43-month, double-blind, placebo-controlled, clinical trial of AVA (clinicaltrials.gov identifier NCT00119067). Among European-Americans, genes from tightly linked HLA-DRB1-DQA1-DQB1 haplotypes displayed significant overall associations with longitudinal variation in AbPA levels at 4, 8, 26, and 30 weeks from baseline in response to vaccination with 3 or 4 doses of AVA (global p=6.53×10−4). In particular, carriage of the DRB1-DQA1-DQB1 haplotypes *1501-*0102-*0602 (p=1.17×10−5), *0101-*0101-*0501 (p=0.009), and *0102-*0101-*0501 (p=0.006) was associated with significantlylower AbPA levels. In carriers of two copies of these haplotypes, lower AbPA levels persisted following subsequent vaccinations. No significant associations were observed amongst African-Americans or for any HLA class I allele/haplotype. Further studies will be required to replicate these findings and to explore the role of host genetic variation outside of the HLA region.
Keywords: Anthrax vaccines, Bacillus anthracis, Bacterial vaccines, Vaccination, HLA Antigens
INTRODUCTION
As an agent of bioterrorism, Bacillus anthracis poses a threat to U.S. military personnel and the civilian population.1-3 Anthrax remains a prevalent public health concern with recent outbreaks among heroin users in Europe and false alarms in Alabama (January 2010).4 B.anthracis, a Centers for Disease Control and Prevention (CDC) Category A agent,5 represents a high priority threat because it can be distributed into large air volumes over long distances and can result in inhalation anthrax with a case-fatality rate of 86-89%.6 A vaccine was proven effective for the prevention of anthrax after a single human field trial, leading to licensure in the US in 1970 7. Since then, Anthrax Vaccine Adsorbed (AVA, Biothrax™, Bioport Corporation, Lansing MI), a cell-free filtrate prepared from an avirulent strain of B. anthracis adjuvanted with aluminum hydroxide, has been available for the pre-exposure prevention of anthrax with only minor modifications.8 Since its initial licensure, more than two million individuals have received over eight million doses of AVA, and vaccination remains mandatory for all U.S. military personnel.9,10
Anthrax toxin is composed of three proteins: the binding component, protective antigen (PA), and two catalytic components, lethal factor and edema factor. Vaccine-induced antibody responses to PA (AbPA) correlate with survival following lethal spore challenge in animal models.11, 12 AbPA levels as a correlate of protective immunity in humans have been inferred from these animal models, and are generally used as the relevant measure of AVA immunogenicity. Given the rarity of anthrax in humans, it is unlikely that true vaccine efficacy and a direct correlate of protection based on levels of AbPA in humans can be determined. While lethal toxin neutralizing antibodies (TNA) may also be relevant, studies in humans 13 and mice 14 have generally indicated a strong correlation between AbPA and TNA levels. Levels of AbPA in response to priming doses of AVA vary greatly among individuals, with 100- to 1000-fold differences in peak AbPA response following at least two doses of AVA.15-19 However, different numbers of received doses and marked differences in assay techniques could have accounted in large part for that variability. In the recent tightly controlled route- and dose-altering trial of the licensed product, the proportion of responders (e.g., > 4-fold increase over pre-vaccination levels) was very high, but the wide range of variation resembled that described above.19 Without serologic measurement in the context of an efficacy trial, however, it is currently unknown how this variability in AbPA response may influence protection from infection.
Recent research has highlighted the role that host genetic variation can play in the mechanisms and dynamics of the human immune response to vaccines.20, 21 In the case of vaccines for viral infections such as hepatitis B, measles, rubella, and smallpox, heterogeneity in immune response due to host genetic variation has been attributed to variation within the classical human leukocyte antigen (HLA) genes, cytokine and cytokine receptors, and the chemokine and chemokine receptors amongst others.22 Further support for the contribution of host genetic variation has been enhanced by the identification of associations between polymorphisms in other innate immunity genes and immune responses to vaccines.23, 24 To our knowledge, there have been no studies of the relationship between genetic polymorphisms and the humoral immune response to anthrax vaccine at the population level. Although we cannot directly assess the impact of human genetic variation on protective immunity induced by AVA, we can document the occurrence and magnitude of associations between genetic polymorphisms and heterogeneity in AbPA response to AVA. That was the major objective of our investigation into the role of polymorphisms at HLA class I (HLA-A,-B,-C) and II loci (HLA-DRB1, -DQB1,-DQA1, and -DPB1) in the AbPA response to AVA administered in a Centers for Disease Control and Prevention (CDC)-sponsored clinical trial of alternative regimens.19
The licensed AVA regimen previous to 2008 consisted of subcutaneous (SQ) vaccine administration in 0.5 ml doses at weeks 0, 2, and 4 and months 6, 12, and 18 with subsequent yearly booster doses as long as an individual remains at risk of infection. In 2002, a pilot study of 173 volunteers demonstrated that AVA could be given as an intramuscular (IM) injection and that eliminating the 2 week injection could lead to fewer local reactions without significant effect on AVA immunogenicity.16 Shortly thereafter the U.S. Congress funded the CDC to study the safety and efficacy of modified regimens of AVA administration under the Anthrax Vaccine Research Program. In response, the CDC initiated AVA000 (clinicaltrials.gov identifier NCT00119067)— a 43-month prospective, randomized, double-blinded, placebo controlled comparison of AVA given by either SQ or IM administration in as many as 8 doses (licensed regimen) or as few as 4 doses. Investigators at 5 clinical sites in the U.S. enrolled 1,563 healthy adults that provided informed consent. Subjects were randomized into one of seven study groups: group 1 received the licensed regimen (8 doses, SQ), groups 2-5 received between 4 and 8 doses IM, and groups 6a (SQ) and 6b (IM) received saline placebo. Further details of NCT00119067 can be found in Marano et al.,19 which details an interim analysis that led to a change in administration in the currently licensed product.25
RESULTS
Of the 1,563 NCT00119067 participants, 1,303 were randomized to groups receiving AVA, and 1,064 individuals had available typing at HLA class I and II loci as well as genome-wide single nucleotide polymorphism (SNP) data (Affymetrix Genome-Wide 6.0 array). Based on a principal components analysis of the genome-wide SNP data, we further reduced this population to subsets of 794 European-Americans and 200 African-Americans (Table I). This subpopulation was not significantly different from the group of participants without genetic information (of those randomized to groups receiving AVA) in terms of gender (p=0.350) or vaccination group (p=0.342, data not shown). However, participants included in genetic analyses were older (mean age ± standard error: 40.1±0.34 years versus 34.1±0.71 years, p<0.001) than those that were not included from the full NCT00119067 cohort. While the European- and African-American sub-cohorts were generally balanced according to sex and vaccination schedule, African-American participants were somewhat younger. In addition, similar to the overall trial recruitment, the majority of African-American participants in the sub-cohort were recruited from two centers (Washington, DC and Birmingham, AL).
Table I.
Characteristics of healthy volunteers participating in the NCT00119067 trial and available for ancillary study on genetic correlates.
| European- Americans (N=794) |
African-Americans (N=200) |
||
|---|---|---|---|
| Frequency (%) | Frequency (%) | p-valuea | |
| Sex: Female/Male (Ratio) | 409/385 (1.06) | 112/88 (1.27) | 0.291 |
| AGE AT ENROLLMENT | 0.006 | ||
| < 30 years | 176 (22.2) | 50 (25.0) | |
| 30 to 39 years | 172 (21.7) | 56 (28.0) | |
| 40 to 49 years | 250 (31.5) | 67 (33.5) | |
| 50 to 61 years | 196 (24.7) | 27 (13.5) | |
| VACCINATION GROUP | 0.703 | ||
| Group 1 | 160 (20.2) | 32 (16.0) | |
| Group 2 | 165 (20.8) | 40 (20.0) | |
| Group 3 | 151 (19.0) | 41 (20.5) | |
| Group 4 | 147 (18.5) | 39 (19.5) | |
| Group 5 | 171 (21.5) | 48 (24.0) | |
| CLINICAL SITE | <1.00E-15 | ||
| Rochester, MN | 208 (26.2) | 0 (0.0) | |
| Atlanta, GA | 130 (16.4) | 18 (9.0) | |
| Washington, DC | 125 (15.7) | 88 (44.0) | |
| Houston, TX | 149 (18.8) | 24 (12.0) | |
| Birmingham, AL | 182 (22.9) | 70 (35.0) | |
p-value from Chi-Square test. Group 1 received subcutaneous administration at 0, 2, 4, and 26 weeks. Group 2 received intramuscular administration at 0, 2, 4, and 26 weeks. Groups 3-5 received intramuscular administration at 0, 4, and 26 weeks.
We modeled the relationship between HLA class I and II polymorphisms and the AbPA response to the priming regimen of 3 or 4 doses of AVA. Due to the high degree of polymorphism at each of the HLA loci, we first conducted global hypothesis tests for an association with each locus and AbPA response (Table II). No locus showed a significant (p<0.05) global association among African-Americans. Among European-Americans, no association of the class I loci was globally significant, nor was any association with an individual HLA-A allele group or HLA-B-HLA-C haplotype (Supplementary Table 1). In contrast, the locus represented by HLA-DR-DQ haplotypes showed a highly significant global association with AbPA response (p=6.53×10−4) in European-Americans. In univariate analysis, four individual HLA-DR-DQ haplotypes were significantly (p<0.05) associated with either higher or lower AbPA levels (Table III). Models of associations with the individual allelic components of those haplotypes demonstrated no significant and distinct relationships beyond those presented in Table III (data not shown). Full results of univariate analysis for all common alleles/haplotypes in European- and African-Americans are displayed in Supplementary Tables 1 and 2.
Table II.
Global tests of association for HLA class I and II Loci with IgG antibody to protective antigen (AbPA) response
| Locus | European-Americans (N=794) p-valuea |
African-Americans (N=200) p-valuea |
|---|---|---|
| HLA-A | 0.713 | 0.859 |
| HLA-B-C b | 0.480 | 0.607 |
| HLA-C | 0.350 | 0.711 |
| HLA-B | 0.529 | 0.738 |
| HLA-DRB1-DQA1-DQB1b Haplotype | 6.53×10−4 | 0.207 |
| HLA-DRB1 | 2.18×10−4 | 0.115 |
| HLA-DQA1 | 0.001 | 0.540 |
| HLA-DQB1 | 0.002 | 0.219 |
| HLA-DPB1 | 0.150 | 0.168 |
p-value from global F-test of no association for locus of interest based on longitudinal model fit to log10[AbPA] measurements at 4, 8, 26, and 30 weeks assuming constant additive allele/haplotype effects across time on mean log10[AbPA] response.
Global tests performed using common haplotypes formed by particular loci as the unit of measurement. All models adjusted for sex, age group, study site, route of administration, time between vaccination and blood draw, and cumulative number of AVA doses (See Methods).
Table III.
Univariate and multivariable associations of HLA-DR-DQ (DRB1-DQA1-DQB1) haplotypes with IgG antibody to protective antigen (AbPA) response in European-Americans
| Univariate | Full Multivariable | Reduced Multivariable | |||||
|---|---|---|---|---|---|---|---|
| Frequency | Mean Change (SE) | p-value | Mean Change (SE) | p-value | Mean Change (SE) | p-value | |
| HLA DR-DQ Haplotypes | |||||||
| *0101-*0101-*0501 | 0.164 | −0.075 (0.035) | 0.033 | −0.09 (0.036) | 0.011 | −0.093 (0.036) | 0.009 |
| *0102-*0101-*0501 | 0.032 | −0.171 (0.067) | 0.010 | −0.182 (0.067) | 0.007 | −0.186 (0.067) | 0.006 |
| *1401-*0104-*0503 | 0.037 | 0.148 (0.069) | 0.032 | 0.115 (0.070) | 0.101 | - | - |
| *1501-*0102-*0602 | 0.232 | −0.116 (0.029) | 6.52×10−5 | −0.125 (0.029) | 2.16×10−5 | −0.128 (0.029) | 1.17×10−5 |
| Sex, Male (Female) | −0.064 (0.028) | 0.022 | −0.065 (0.028) | 0.019 | |||
| Study Site (Atlanta, GA) | |||||||
| Birmingham, AL | −0.074 (0.043) | 0.084 | −0.077 (0.042) | 0.070 | |||
| Houston, TX | −0.134 (0.046) | 0.004 | −0.137 (0.046) | 0.003 | |||
| Washington, DC | −0.085 (0.053) | 0.109 | −0.087 (0.053) | 0.100 | |||
| Rochester, MN | −0.027 (0.042) | 0.510 | −0.023 (0.041) | 0.577 | |||
| Age (< 30 years) | |||||||
| 30 to 39 years | −0.144 (0.040) | 2.97×10−4 | −0.144 (0.040) | 2.98×10−4 | |||
| 40 to 49 years | −0.239 (0.039) | 8.81×10−10 | −0.241 (0.039) | 5.73×10−10 | |||
| 50 to 61 years | −0.323 (0.044) | 2.11×10−13 | −0.323 (0.044) | 1.73×10−13 | |||
( ) denotes the reference category where applicable. Frequency denotes carriage frequency of particular haplotype based on N chromosomes. Mean change denotes the estimated regression coefficient representing the mean change in log10[AbPA] relative to the reference category based on a multivariable longitudinal model fit to measurements at 4, 8, 26, and 30 weeks. Regression coefficients for haplotype effects assume an additive effect on log10[AbPA] response. (SE) Standard error. All models also adjusted for route of administration, cumulative number of AVA doses, and time since last vaccination (See Methods).
Age, gender, and the vaccine regimen affected the AbPA response in the trial.19 In multivariable analyses including those three non-genetic variables, carriage of three of the four HLA-DR-DQ haplotypes highlighted in univariate analysis remained significantly associated with lower mean AbPA levels (Table III). The HLA-DR15 haplotype (DRB1*1501-DQA1*0102-DQB1*0602), carried by 23.2% of European-Americans, was associated with lower mean AbPA response (25.5% [95% confidence interval (CI): 15.1-34.7%] and 44.5% [95% CI: 27.9-57.3% for heterozygous and homozygous carriers, respectively). (Percent reductions calculated based on a linear scale for AbPA in μg/mL). The DRB1*0101-DQA1*0101-DQB1*0501 haplotype, carried by 16.4% of European-Americans, was associated with a 19.3% lower mean AbPA level (95% CI: 5.0-31.4%) for heterozygous carriers and 34.8% (95% CI: 9.8-53.0%) for homozygous carriers. The relatively uncommon DRB1*0102-DQA1*0101-DQB1*0501 haplotype (carriage frequency=3.2%) showed a similar association, with a 34.8% (95% CI: 11.8-51.9%) and a 57.5% (95% CI: 22.2-76.8%) lower mean AbPA level for heterozygous and homozygous carriers, respectively.
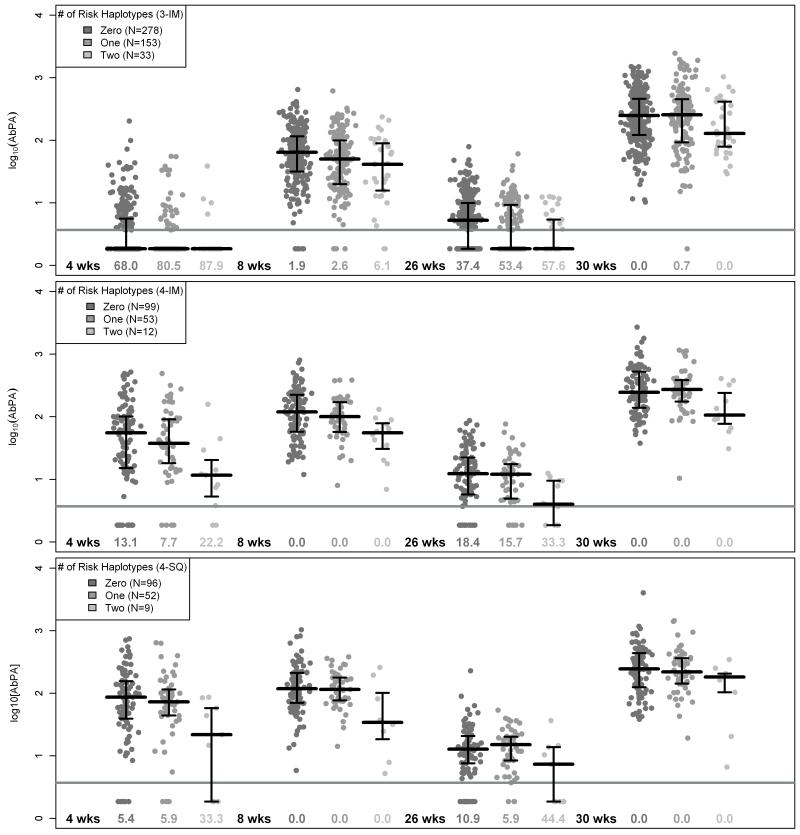
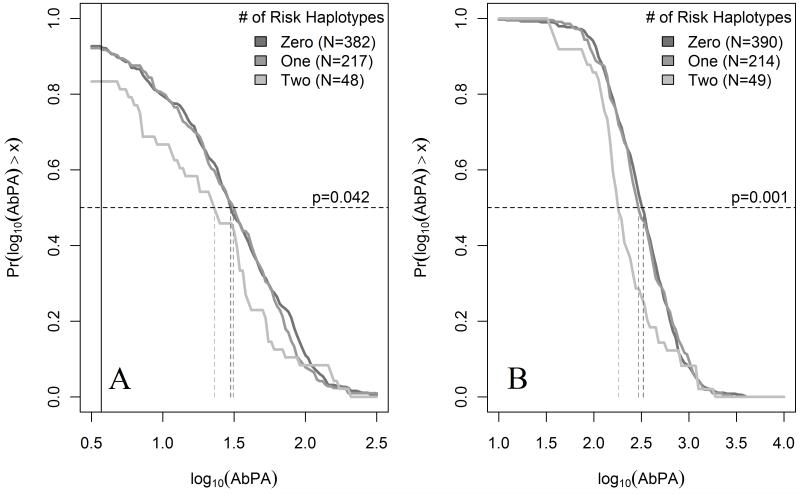
The pattern of AbPA response (unadjusted for age, study site, and sex) for carriers of the identified haplotypes involving DRB1*0101, DRB1*0102, and DRB1*1501 is displayed in Figure 1 across each measurement time point. In general, individuals with one copy had decreased AbPA levels initially compared to those with no copies (4 and 8 weeks, and 26 weeks for 3-IM) but similar AbPA levels at 30 weeks following the vaccination at 6 months. However, individuals with two copies of the identified haplotypes had consistently lower AbPA levels that persisted after the 6 month vaccination. Figure 2 displays the reverse cumulative distribution function for AbPA measured immediately preceding the 42 month booster dose and approximately one month following that dose (43 months), similarly stratified according to the number of risk haplotypes carried. These plots were restricted to data from European-Americans who did not deviate from the study protocol (See methods). Both before and after the 42 month vaccination, there appears to be little difference in the AbPA distributions between individuals who carry either no copy and those who carry only one copy of these haplotypes. Similar to the pattern observed at 30 weeks in Figure 1, the distribution of AbPA levels among carriers of two haplotypes is shifted towards lower values both before the 42 month vaccination dose (p=0.042 compared to 0/1 copies) and after it (p=0.007 compared to 0/1 copies).
Figure 1.
Distribution of IgG antibody to protective antigen (AbPA) response for European-Americans according to number of HLA-DR-DQ risk haplotypes
(3-IM) Intramuscular injection at 0, 4, and 26 weeks (wks). (4-IM) Intramuscular injection at 0, 2, 4, and 26 weeks. (4-SQ) Subcutaneous injection at 0, 2, 4, and 26 weeks. Bold black lines denote sample median of unadjusted log10[AbPA] concentration (along with the inter-quartile range) computed by substituting 1/2 the empirical reactivity threshold (3.7 μ g / mL) of the ELISA assay (gray line) for observations below the limit. Numbers below data points denote the proportion of individuals below the reactivity threshold at a particular measurement time point. HLA-DR-DQ risk haplotypes were defined as the number of copies of the following haplotypes: HLA-DRB1-DQA1-DQB1 *0101-*0101-*0501, *0102-*0101-*0501, or *1501-*0102-*0602
Figure 2.
Reverse cumulative distribution plots of IgG antibody to protective antigen (AbPA) for European-Americans prior to and following the 42 month vaccination, stratified according to number of HLA-DR-DQ risk haplotypes
Panel A displays the distribution of log10(AbPA) pre-vaccination at 42 months stratified according to the number of HLA DR-DQ risk haplotypes. Panel B displays the log10(AbPA) distribution measured approximately 1 month post-vaccination. HLA-DR-DQ risk haplotypes were defined as the number of copies of the following haplotypes: HLA-DRB1-DQA1-DQB1 *0101-*0101-*0501, *0102-*0101-*0501, or *1501-*0102-*0602. The solid black line denotes the empirical reactivity threshold of the ELISA assay (3.7 μg/dL). Dashed lines correspond to the sample median for each HLA defined subgroup. Displayed p-values compare the AbPA distribution for individuals with two risk haplotypes versus zero/one haplotypes.
DISCUSSION
This study represents the first to explore the role of host HLA genetic variation in AbPA response to AVA, the only licensed vaccine against anthrax and one of the few licensed vaccines available to counter bioterrorist threats. In our study, at least two relatively common HLA DR-DQ haplotypes or their constituent alleles were associated with a decreased AbPA response to AVA within European-Americans. The HLA-DR15 haplotype (DRB1*1501-DQA1*0101-DQB1-*0602) has been consistently associated with susceptibility to occurrence or pathogenesis of multiple sclerosis and systemic lupus erythematosus along with protection against type I diabetes.26-28 With regard to vaccines, HLA-DRB1 alleles have been most consistently associated with aberrant response to hepatitis B vaccination as measured by anti-HBsAg levels: DRB1*0301 and DRB1*0701 with non-response and DRB1*0804 less consistently with high antibody response.29, 30 A role for the DRB1*15-DQB1*06 haplotype in a relatively lower vaccine response has been suggested only once, in a study of children receiving at least two doses of the measles-mumps-rubella vaccine, where it was associated with lower levels of IgG antibody to rubella virus.31 The DRB1*0101-DQA1*0101-DQB1*0501 and DRB1*0102-DQA1*0101-DQB1*0501 haplotypes have also been implicated in certain vaccine responses, in autoimmune or immunodeficiency diseases such as rheumatoid arthritis,32 in the absence of virus RNA (DRB1*0101) in hepatitis C infection,33 and immunoglobulin A deficiency (DRB1*0102).34 In a study of measles-mumps-rubella vaccine response, Ovsyannikova et al. demonstrated an association of DRB1*0101 with increased mumps-specific lymphoproliferation among a cohort of schoolchildren in Minnesota.35
We observed the strongest class II associations with lower mean AbPA levels. Other investigators have begun to explore how HLA-DRB1-mediated antigen presentation might contribute to variably immunogenic AVA response, but they reported no direct evidence for differential contributions by the specific alleles highlighted in our study.36 With a relatively low likelihood that the vaccine regimen will again be administered to large numbers of subjects in a clinical trial, there is a more compelling need for confirmation of the biological effect of the associated human class II alleles on antibody response in ex vivo laboratory experiments and of similar relationships with homologous leukocyte antigen alleles in primate anthrax models, where the correlate of protection from disease by AbPA can be closely approximated.
Virtually all AVA vaccinees displayed a significant rise in AbPA concentration after at least 2 doses of AVA. While vaccinees carrying two copies of the highlighted alleles/haplotypes more frequently had AbPA levels below the ELISA assay reactivity threshold prior to the 6 month dose, the effect of carrying a single copy of one of these alleles/haplotypes appeared to dissipate following the 6 month dose. However, even the difference in initial response could be meaningful in the context of an anthrax outbreak within civilian populations. In a setting where a regimen of vaccine plus antibiotic treatment might be used for prophylaxis at the population level,37 the decreased response associated with carrying a single haplotype copy could influence the efficacy of the early vaccine doses.
Our results also suggest that the lower response by individuals carrying two copies of the identified haplotypes persists beyond the earlier vaccinations. These individuals retained a capacity for response but exhibited smaller increases in AbPA levels following vaccination. This persistence of decreased AbPA levels may be particularly important in view of the effort to reduce the frequency of booster doses from the currently licensed yearly interval to an every third year regimen based on the results of NCT0019067. However, without a direct estimate of minimal AbPA levels necessary to confer protection in humans, the risk of vaccine failure in the interval(s) between vaccine doses is not clear. Ongoing studies with the CDC Anthrax Vaccine Research Program (AVRP) are building models to establish a correlate of immune protection by AVA against aerosolized B. anthracis in rhesus macaques and applying these models to estimate vaccine effectiveness in humans (C. Quinn and D. Madigan, personal communication). Unfortunately, the reliability of these estimates rests on the intrinsically un-testable a priori assumption of cross-species equivalence in associations of measures of immune response and survival. If lower AbPA levels do correlate with reduced likelihood of protection, human subpopulations with sub-optimal response to AVA may benefit from additional doses of AVA (similar to vaccines for hepatitis B 38 and measles 39) and/or alternative methods to prevent B. anthracis related disease. In these circumstances knowledge of the factors that predict sub-optimal response may be advantageous.
The observed effects of HLA class I and II markers on AbPA response among the African-Americans in the trial were not significant, most likely because this smaller group provided less statistical power. This underscores the importance of including larger, more ethnically diverse populations in future studies of host genetic variation in vaccine response. The strong linkage disequilibrium between alleles of HLA-DRB1, -DQA1, and -DQB1 precluded discrimination of their individual contributions to the diminished AbPA response. With much larger cohorts unlikely to be available to reveal allele-specific effects at the population level, studies carefully designed to assess functions such as peptide binding and presentation by individual alleles will be needed to tease apart both the separate contributions of alleles at the individual loci and epistatic effects of these and other genetic markers.
We did not explicitly evaluate the potential effects of class I alleles at full resolution. Higher resolution might have detected a relationship with HLA-A and -B supertype groupings of alleles that bind overlapping peptides40 or a relationship with one or more lower frequency alleles that were missed by their two-digit grouping. However, several factors make it unlikely that a class I relationship was missed: global tests for class I loci were far less indicative of involvement than for class II, no trend suggestive of an association was seen with any cluster of alleles resolved totwo-digit specificity, and the role for class I molecules in presenting antigenic peptides from a bacterial protein is less firmly established. Despite these limitations, the implementation of this study in the context of the clinical trial was a major advantage in establishing uniformity in administration of vaccine and measurement of AbPA immune response. The trial setting provided an unprecedented opportunity in vaccine immunogenetics to investigate host genetic correlates under rigorous conditions seldom achieved in observational studies.
Our modeling approach assumed constant genetic effects across time. Because the first AbPA measurements were taken at 4 weeks following initial immunization, they probably reflect immune status at the early stages of the adaptive response. We are therefore unlikely to have observed temporal heterogeneity that might have occurred during a transition from an innate to an adaptive response. Finally, previous work on derangements of adaptive immunity in general and on vaccine responses in particular focused our initial attention on HLA genes, but ongoing studies in AVA vaccinees at the genomic and functional levels will hopefully provide a broader perspective on the host determinants of AbPA levels along with other features of the response to anthrax vaccine, consistent with a vaccinomics/systems biology approach.41
MATERIALS AND METHODS
POPULATION GENETICS ANALYSIS PROGRAM (POPGEN) STUDY POPULATION
NCT00119067 was designed to enroll 1560 participants, and approximately 5 out of 6 of those enrolled (1303) were randomized to receive AVA. Because our genetic study began approximately two years after the initiation of the trial,; only 1,064 individuals were available for typing at HLA class I and II loci or for a genome-wide association study of individual SNPs (Affymetrix Genome-Wide 6.0 array). The trial included individuals of European, African, Hispanic, and Asian ancestries; therefore, a principal components analysis (PCA) based on the genome-wide SNP data was used to define two major racial/ethnic groups representing European- and African-Americans 42. Individuals who did not cluster among the top two principal components representing these ethnicities were excluded from further analysis (N=65). After excluding five additional vaccinees with missing immunogenicity measurements, we analyzed 794 European-Americans and 200 African-Americans. This study of genetic correlates was approved by the institutional review board at the University of Alabama at Birmingham.
MEASUREMENT OF ANTIBODY TO PROTECTIVE ANTIGEN
According to the NCT00119067 protocol, serum samples were collected from all participants at 0, 4, 8, 26, and 30 weeks and 12, 13, 18, 19, 30, 31, 42, and 43 months. At each time point, AbPA was measured as anti-PA specific IgG with comparison to reference samples of known AbPA concentrations in a quantitative enzyme-linked immunosorbent assay (ELISA) with an empirical reactivity threshold of 3.7 μg/ml. We have focused this analysis on AbPA response during the initial vaccination series (AVA administered at 0, 2, 4, and 26 weeks or 0, 4, and 26 weeks) because nearly all vaccinees reach peak AbPA levels after 3-4 doses of AVA, and later measurements are largely predictable from the initial response profile and subsequent vaccination schedule.43 In addition, 3 doses of vaccine plus antibiotic treatment is currently recommended for the optimal protection of previously unvaccinated persons after potential exposure.37 Following Marano et al.,19 groups 1 (4-SQ) and 2 (4-IM) received the originally licensed regimen (0, 2, 4, and 26 weeks) within the first 7 months and were evaluated separately. For groups 3-5, the vaccination schedules (0, 4, and 26 weeks) did not differ within the first 7 months and were aggregated for certain analyses (3-IM).
GENOTYPING OF HLA CLASS I AND II GENES IN THE HUMAN MHC
Genotyping was performed with methods similar to those applied in our previous studies of European, African, and North American populations.30, 44, 45 Briefly, HLA class I genotyping was performed using a combination of PCR-based techniques, including PCR with sequence-specific primers (Dynal/Invitrogen), automated reference-strand conformation analysis (RSCA) (Dynal/Invitrogen), sequence-specific oligonucleotide (SSO) probe hybridization automated by the Auto LiPA 30 system (Innogenetics Inc., Alpharetta, GA), and automated sequencing-based typing with the ABI 3130xl DNA Analyzer (Abbott Molecular Inc., Des Plaines, IL). Resolution of alleles to 4-digits was performed selectively when individual alleles were expected to occur frequently enough for meaningful analysis or when an association at the allele level was likely (e.g. for B*27, B*35, and B*58). HLA-DQA1 and –DQB1 alleles were resolved to 4-digit specificity with automated hybridization of 35 and 37 SSO probes, respectively, to gene-specific PCR amplicons (Innogenetics). Homozygosity was confirmed and ambiguities with SSO results were resolved by sequencing-based typing (SBT), using commercial kits from Abbott Molecular Inc. HLA-DRB1 genotyping depended primarily on SBT (Abbott Molecular Inc.) accompanied by SSO assays with 57 probes (Innogenetics) to resolve ambiguities.
STATISTICAL METHODS
HLA class I loci were analyzed according to allele groups defined by their first two digits. HLA class II loci analyzed either by two-digit allele groups or by four-digit individual allele designations, which define functionally distinct amino acid sequences and protein structures. Because of the extensive linkage disequilibrium (LD) between HLA-B and -C and HLA-DRB1, -DQA1, and -DQB1, haplotypes were inferred using an expectation-maximization (EM) algorithm (SAS v9.1.3, SAS, Cary, NC) and taken as the primary unit of analysis for those sets of loci. Haplotype assignments with posterior probabilities less than 0.95 were treated as ambiguous and excluded from subsequent association testing.
To account for the longitudinal measurements of AbPA and left-censoring due to the reactivity threshold of the ELISA assay, parametric survival models including subject-specific random effects were used to model associations between HLA alleles/haplotypes and log10 transformed AbPA.46 All models assumed a logistic error distribution and used robust sandwich variance estimates for model parameters. Age at enrollment (<30, 30-39, 40-49, and 50-61 years of age), sex, study site, cumulative number of AVA doses (1, 2, 3, and 4) at a particular vaccination, time between vaccination and blood draw, and route of administration (SQ vs IM) were included as covariates in all models. The effect of route of administration was allowed to vary at each measurement time point, allowing for a difference between the effects of SQ and IM administration at 4, 8, 26, and 30 weeks. In addition, the time lag between vaccination and blood sampling was modeled as a piecewise linear function of the square root of time in days, with a single breakpoint in the rate of AbPA decay at 42 days. If blood sampling was within 42 days of vaccination, there was indication of a subtle correlation between AbPA level and the time since vaccination, with an expected large decrease in AbPA for measurements taken at longer intervals (e.g., at 26 weeks, roughly 5 months following vaccination at 4 weeks). We also considered alternatives to the model described above, with further adjustment for the top 10 principal components based on the GWAS data in order to control for latent population structure or cryptic relatedness. Analyses incorporating this additional adjustment produced results almost identical to the original, and they are not presented here.
All models assumed an additive dose-response coding of allele/haplotype effects, where the effect of either was assumed to be constant across time. To maintain control of the overall type I error rate, global tests were first conducted within each of the racially/ethnically distinct populations to test for effects of any of the loci individually, as well as for HLA-B-C and HLA-DRB1-DQA1-DQB1 haplotypes 47. For the global tests, haplotypes/alleles that were observed less than 15 times within a particular racial/ethnic group were collapsed into a single reference category.
We have also presented plots of the reverse cumulative distribution48 for AbPA (equivalent to 1 minus the empirical cumulative distribution function) measured at 42 and 43 months in the NCT00119067 trial. We restricted this analysis to European-American participants who did not deviate from the trial protocol in order to remove the effects of missed vaccinations or time inconsistencies in AbPA measurement. For AbPA measured at 42 months, this was defined as receiving the specified number of AVA doses, with AbPA measured within 1 year ± 28 days (groups 1, 2, or 3), 2 years ± 28 days (group 4), and 3 years ± 28 days (group 5). For AbPA measured at 43 months, AbPA was required to be measured within 42 days of the specified final AVA vaccination. Differences in the distributions across HLA defined subgroups were tested using the Peto and Peto modification of the Gehan-Wilcoxon test in the presence of left-censoring49, or with the Kolmogorov-Smirnov test otherwise. All analyses were performed in the R statistical computing environment.50
Supplementary Material
ACKNOWLEDGMENTS
Samples used in this study were made available through a clinical trial funded by the Centers for Disease Control and Prevention, including a supplemental grant to G.A.P. This work was supported by the National Institute of Allergy and Infectious Diseases through contract N01-AI40068 (to R.A.K.). N.M.P. was supported by post-doctoral training grant T32 HL072757 from the National Heart, Lung, and Blood Institute. The authors would like to thank Conrad Quinn, Ph.D., Charles Rose, Ph.D. and Brian Plikaytis, M.S. for helpful conversations concerning the NCT00119067 trial data and Jelai Wang, M.S. and Vinodh Srinivasasianagendra, M.S. for informatics and data management assistance. They also gratefully acknowledge the other site principal investigators from the NCT00119067 trial for their efforts in data collection: Janiine Babcock, M.D. (Walter Reed Army Institute of Research), Wendy Keitel, M.D. (Baylor College of Medicine), and Harry Keyserling, M.D. (Emory University School of Medicine).
Footnotes
Conflicts of interest: G.A.P has served as a consultant to Emergent Biosolutions, Inc. All other authors declare no conflicts of interest or competing financial interests.
Supplementary Information accompanies the paper on the Genes and Immunity website (http://www.nature.com/gene)
REFERENCES
- 1.Maillard J, Fischer M, McKee KJ, Turner L, Cline J. First case of bioterrorism-related inhalational anthrax, Florida, 2001: North Carolina investigation. Emerg Infect Dis. 2002;8(10):1035–8. doi: 10.3201/eid0810.020389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jernigan D, Raghunathan P, Bell B, Brechner R, Bresnitz E, Butler J, et al. Investigation of bioterrorism-related anthrax, United States, 2001: epidemiologic findings. Emerg Infect Dis. 2002;8(10):1019–28. doi: 10.3201/eid0810.020353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Traeger M, Wiersma S, Rosenstein N, Malecki J, Shepard C, Raghunathan P, et al. First case of bioterrorism-related inhalational anthrax in the United States, Palm Beach County, Florida, 2001. Emerg Infect Dis. 2002;8(10):1029–34. doi: 10.3201/eid0810.020354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McNeil D., Jr. Anthrax: In Scotland, Six Heroin Users Die of Anthrax Poisoning. The New York Times. 2010 [Google Scholar]
- 5.National Institutes of Health . In: NIAID Biodefense Research Agenda for CDC Category A Agents. National Institute of Allergy and Infectious Diseases (NIAID), editor. Bethesda, MD: 2002. [Google Scholar]
- 6.Meselson M, Guillemin J, Hugh-Jones M, Langmuir A, Popova I, Shelokov A, et al. The Sverdlovsk anthrax outbreak of 1979. Science. 1994;266(5188):1202–8. doi: 10.1126/science.7973702. [DOI] [PubMed] [Google Scholar]
- 7.Brachman P, Gold H, Plotkin S, Fekety F, Werrin M, Ingraham N. Field Evaluation of a Human Anthrax Vaccine. Am J Public Health Nations Health. 1962;52(4):632–45. doi: 10.2105/ajph.52.4.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention Use of anthrax vaccine in response to terrorism: supplemental recommendations of the Advisory Committee on Immunization Practices. MMWR Morb Mortal Wkly Rep. 2002;51:1024–1026. [PubMed] [Google Scholar]
- 9.Bienek D, Loomis L, Biagini R. The anthrax vaccine: no new tricks for an old dog. Hum Vaccin. 2009;5(3):184–9. doi: 10.4161/hv.5.3.7308. [DOI] [PubMed] [Google Scholar]
- 10.Lee C. Mandatory anthrax shots to return. Washington Post. 2006 October 17; [Google Scholar]
- 11.Pitt M, Little S, Ivins B, Fellows P, Boles J, Barth J, et al. In vitro correlate of immunity in an animal model of inhalational anthrax. J Appl Microbiol. 1999;87(2):304. doi: 10.1046/j.1365-2672.1999.00897.x. [DOI] [PubMed] [Google Scholar]
- 12.Fellows P, Linscott M, Ivins B, Pitt M, Rossi C, Gibbs P, et al. Efficacy of a human anthrax vaccine in guinea pigs, rabbits, and rhesus macaques against challenge by Bacillus anthracis isolates of diverse geographical origin. Vaccine. 2001;19(23-24):3241–7. doi: 10.1016/s0264-410x(01)00021-4. [DOI] [PubMed] [Google Scholar]
- 13.Crowe S, Ash L, Engler R, Ballard J, Harley J, Farris A, et al. Select human anthrax protective antigen epitope-specific antibodies provide protection from lethal toxin challenge. J Infect Dis. 2010;202(2):251–60. doi: 10.1086/653495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parreiras P, Sirota L, Wagner L, Menzies S, Arciniega J. Comparability of ELISA and toxin neutralization to measure immunogenicity of Protective Antigen in mice, as part of a potency test for anthrax vaccines. Vaccine. 2009;27(33):4537–42. doi: 10.1016/j.vaccine.2009.05.045. [DOI] [PubMed] [Google Scholar]
- 15.Pittman P, Mangiafico J, Rossi C, Cannon T, Gibbs P, Parker G, et al. Anthrax vaccine: increasing intervals between the first two doses enhances antibody response in humans. Vaccine. 2000;19(2-3):213–6. doi: 10.1016/s0264-410x(00)00174-2. [DOI] [PubMed] [Google Scholar]
- 16.Pittman P, Kim-Ahn G, Pifat D, Coonan K, Gibbs P, Little S, et al. Anthrax vaccine: immunogenicity and safety of a dose-reduction, route-change comparison study in humans. Vaccine. 2002;20(9-10):1412–20. doi: 10.1016/s0264-410x(01)00462-5. [DOI] [PubMed] [Google Scholar]
- 17.Pittman P, Hack D, Mangiafico J, Gibbs P, McKee KJ, Friedlander A, et al. Antibody response to a delayed booster dose of anthrax vaccine and botulinum toxoid. Vaccine. 2002;20(16):2107–15. doi: 10.1016/s0264-410x(02)00058-0. [DOI] [PubMed] [Google Scholar]
- 18.Singer D, Schneerson R, Bautista C, Rubertone M, Robbins J, Taylor D. Serum IgG antibody response to the protective antigen (PA) of Bacillus anthracis induced by anthrax vaccine adsorbed (AVA) among U.S. military personnel. Vaccine. 2008;26(7):869–73. doi: 10.1016/j.vaccine.2007.11.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marano N, Plikaytis B, Martin S, Rose C, Semenova V, Martin S, et al. Effects of a reduced dose schedule and intramuscular administration of anthrax vaccine adsorbed on immunogenicity and safety at 7 months: a randomized trial. JAMA. 2008;300(13):1532–43. doi: 10.1001/jama.300.13.1532. [DOI] [PubMed] [Google Scholar]
- 20.Pulendran B. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat Rev Immunol. 2009;9(10):741–7. doi: 10.1038/nri2629. [DOI] [PubMed] [Google Scholar]
- 21.Seib K, Dougan G, Rappuoli R. The key role of genomics in modern vaccine and drug design for emerging infectious diseases. PLoS Genet. 2009;5(10):e1000612. doi: 10.1371/journal.pgen.1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poland G, Ovsyannikova I, Jacobson R. Vaccine immunogenetics: bedside to bench to population. Vaccine. 2008;26(49):6183–8. doi: 10.1016/j.vaccine.2008.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ovsyannikova I, Dhiman N, Haralambieva I, Vierkant R, O’Byrne M, Jacobson R, et al. Rubella vaccine-induced cellular immunity: evidence of associations with polymorphisms in the Toll-like, vitamin A and D receptors, and innate immune response genes. Hum Genet. 2010;127(2):207–21. doi: 10.1007/s00439-009-0763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ovsyannikova I, Haralambieva I, Dhiman N, O’Byrne M, Pankratz V, Jacobson R, et al. Polymorphisms in the vitamin A receptor and innate immunity genes influence the antibody response to rubella vaccination. J Infect Dis. 2010;201(2):207–13. doi: 10.1086/649588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. [Accessed 3/15/2010];BioThrax - Package Insert. http://www.fda.gov/downloads/BiologicsBloodVaccines/BloodBloodProducts/ApprovedProducts/LicensedProductsBLAs/UCM074923.pdf. In.
- 26.Schmidt H, Williamson D, Ashley-Koch A. HLA-DR15 haplotype and multiple sclerosis: a HuGE review. Am J Epidemiol. 2007;165(10):1097–109. doi: 10.1093/aje/kwk118. [DOI] [PubMed] [Google Scholar]
- 27.Graham R, Ortmann W, Langefeld C, Jawaheer D, Selby S, Rodine P, et al. Visualizing human leukocyte antigen class II risk haplotypes in human systemic lupus erythematosus. Am J Hum Genet. 2002;71(3):543–53. doi: 10.1086/342290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erlich H, Valdes A, Noble J, Carlson J, Varney M, Concannon P, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57(4):1084–92. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C, Tang J, Song W, Lobashevsky E, Wilson C, Kaslow R. HLA and cytokine gene polymorphisms are independently associated with responses to hepatitis B vaccination. Hepatology. 2004;39(4):978–88. doi: 10.1002/hep.20142. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Ni R, Song W, Shao W, Shrestha S, Ahmad S, et al. Clear and independent associations of several HLA-DRB1 alleles with differential antibody responses to hepatitis B vaccination in youth. Hum Genet. 2009;126(5):685–96. doi: 10.1007/s00439-009-0720-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ovsyannikova I, Pankratz V, Vierkant R, Jacobson R, Poland G. Human leukocyte antigen haplotypes in the genetic control of immune response to measles-mumps-rubella vaccine. J Infect Dis. 2006;193(5):655–63. doi: 10.1086/500144. [DOI] [PubMed] [Google Scholar]
- 32.Bridges SJ, Kelley J, Hughes L. The HLA-DRB1 shared epitope in Caucasians with rheumatoid arthritis: a lesson learned from tic-tac-toe. Arthritis Rheum. 2008;58(5):1211–5. doi: 10.1002/art.23433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuniholm M, Kovacs A, Gao X, Xue X, Marti D, Thio C, et al. Specific human leukocyte antigen class I and II alleles associated with hepatitis C virus viremia. Hepatology. 2009 doi: 10.1002/hep.23515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martínez A, Gual L, Fernández-Arquero M, Nogales A, Ferreira A, Garcia-Rodriguez M, et al. Epistatic effects occurring among susceptibility and protective MHC genes in IgA deficiency. Genes Immun. 2003;4(4):316–20. doi: 10.1038/sj.gene.6363955. [DOI] [PubMed] [Google Scholar]
- 35.Ovsyannikova I, Jacobson R, Dhiman N, Vierkant R, Pankratz V, Poland G. Human leukocyte antigen and cytokine receptor gene polymorphisms associated with heterogeneous immune responses to mumps viral vaccine. Pediatrics. 2008;121(5):e1091–9. doi: 10.1542/peds.2007-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laughlin E, Miller J, James E, Fillos D, Ibegbu C, Mittler R, et al. Antigen-specific CD4+ T cells recognize epitopes of protective antigen following vaccination with an anthrax vaccine. Infect Immun. 2007;75(4):1852–60. doi: 10.1128/IAI.01814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright JG, Quinn CP, Shadomy S, Messonnier N, (CDC) CfDCaP Use of anthrax vaccine in the United States: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2009. MMWR Recomm Rep. 2010;59(RR-6):1–30. [PubMed] [Google Scholar]
- 38.Alper C, Kruskall M, Marcus-Bagley D, Craven D, Katz A, Brink S, et al. Genetic prediction of nonresponse to hepatitis B vaccine. N Engl J Med. 1989;321(11):708–12. doi: 10.1056/NEJM198909143211103. [DOI] [PubMed] [Google Scholar]
- 39.Poland G, Jacobson R, Thampy A, Colbourne S, Wollan P, Lipsky J, et al. Measles reimmunization in children seronegative after initial immunization. JAMA. 1997;277(14):1156–8. [PubMed] [Google Scholar]
- 40.Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poland G, Ovsyannikova I, Jacobson R, Smith D. Heterogeneity in vaccine immune response: the role of immunogenetics and the emerging field of vaccinomics. Clin Pharmacol Ther. 2007;82(6):653–64. doi: 10.1038/sj.clpt.6100415. [DOI] [PubMed] [Google Scholar]
- 42.Price A, Patterson N, Plenge R, Weinblatt M, Shadick N, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 43.American Society of Microbiology Anthrax Vaccine Efficacy: A Cross-Species Analysis; ASM Biodefense and Emerging Diseases Research Meeting; Baltimore, MD. 2010. [Google Scholar]
- 44.Shao W, Tang J, Dorak M, Song W, Lobashevsky E, Cobbs C, et al. Molecular typing of human leukocyte antigen and related polymorphisms following whole genome amplification. Tissue Antigens. 2004;64(3):286–92. doi: 10.1111/j.0001-2815.2004.00295.x. [DOI] [PubMed] [Google Scholar]
- 45.Tang J, Shao W, Yoo Y, Brill I, Mulenga J, Allen S, et al. Human leukocyte antigen class I genotypes in relation to heterosexual HIV type 1 transmission within discordant couples. J Immunol. 2008;181(4):2626–35. doi: 10.4049/jimmunol.181.4.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klein J, Moeschberger M. Survival Analysis: Techniques for Censored and Truncated Data. Second edn Springer-Verlag; New York: 2003. [Google Scholar]
- 47.Hothorn T, Bretz F, Westfall P. Simultaneous Inference in General Parametric Models. Biometrical Journal. 2008;50(3):346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 48.Reed G, Meade B, Steinhoff M. The reverse cumulative distribution plot: a graphic method for exploratory analysis of antibody data. Pediatrics. 1995;96(3 Pt 2):600–3. [PubMed] [Google Scholar]
- 49.Peto R, Peto J. Asymptotically efficient rank invariant testing procedures. Journal of the Royal Statistical Society, Series A. 1972;135:185–207. [Google Scholar]
- 50.R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.