Abstract
Transcription by RNA polymerase III (Pol III) requires multiple general initiation factors that, in isolated form, assemble onto the promoter in an ordered fashion. Here, it is shown that all components required for transcription of the VA1 and tRNA genes, including TFIIIB, TFIIIC, and RNA Pol III, can be coimmunopurified from a HeLa cell line that constantly expresses a FLAG epitope-tagged subunit of human RNA Pol III. This finding of an RNA Pol III “holoenzyme” suggests similarities between transcription initiation by RNA Pol II and RNA Pol III and has led to the identification of a novel general initiation factor (TDF, translation dependent factor) that is present within the holoenzyme. TDF is selectively inactivated during protein synthesis inhibition by cycloheximide and at a late stage of adenovirus infection, thus accounting for the loss of RNA Pol III-mediated transcription of the tRNA and VA RNA genes under these conditions. On the basis of these observations, possible mechanisms for the global regulation of transcription by RNA Pol III and for disassembly of RNA Pol III initiation complexes are proposed.
Keywords: Transcription, holoenzyme, RNA polymerase III
Eukaryotic RNA polymerases I, II, and III (Pol I, Pol II, Pol III) are structurally distinct, transcribe distinct sets of genes in conjunction with distinct sets of general initiation factors, and respond to largely distinct gene-specific activators (for review, see Zawel and Reinberg 1995; Roeder 1996a). Despite these differences, some mechanistic similarities are apparent. The three RNA polymerases share five common subunits and other subunits that are highly related (Woychik et al. 1990; McKune et al. 1995; Shpakovski et al. 1995), whereas the TATA-binding protein (TBP) is shared by three RNA polymerase type-specific accessory factors (Struhl 1994). Studies with isolated components have shown ordered pathways for the assembly of RNA polymerases and cognate initiation factors into active preinitiation complexes (for review, see Zawel and Reinberg 1995; Roeder 1996a). In addition, there is a parallel between RNA Pol II recruitment by interaction with the TFIIB component of a TFIIB–TFIID–promoter complex (for review, see Roeder 1996b) and RNA Pol III recruitment by interaction with a TFIIB-related component of a TFIIIB–TFIIIC–promoter complex (Werner et al. 1993; Wang and Roeder 1997).
Although the stepwise assembly of functional preinitiation complexes is readily demonstrated in vitro, recent studies have identified large complexes (RNA Pol II “holoenzymes”) that contain RNA Pol II, some or all of the general initiation factors, and various cofactors (Kim et al. 1994; Koleske and Young 1994; Barberis et al. 1995; Ossipow et al. 1995; Chao et al. 1996; Maldonado et al. 1996; for review, see Koleske and Young 1995; Björklund and Kim 1996; Halle and Meisterernst 1996). These holoenzymes (along with the missing general initiation factors) mediate both basal and activator-dependent transcription and suggest a simplified promoter activation model that involves concomitant recruitment of multiple (preassembled) components to the template. The observation that various SRB proteins are both integral components of the biochemically defined RNA Pol II holoenzyme (Kim et al. 1994; Koleske and Young 1994) and essential for transcription in vivo (Thompson and Young 1995) has suggested that the holoenzyme may be the active form of the enzyme in vivo. Because the carboxy-terminal domain (CTD) of the largest subunit plays a key role in formation of the RNA Pol II holoenzyme (Kim et al. 1994; Koleske and Young 1994), the absence of a similar structure in RNA polymerases I and III suggests that any corresponding holoenzymes would depend on novel RNA polymerase-factor interactions.
In the case of RNA Pol III, transcription of corresponding genes (encoding small structural RNAs) requires a number of accessory factors (Gabrielsen and Sentenac 1991; Geiduschek and Kassavetis 1992; White 1994; Roeder 1996a). These include the “common” factors TFIIIC and TFIIIB, which suffice for transcription of tRNA and VA RNA genes, and, in some cases, various gene-specific factors (e.g., TFIIIA for 5S genes and a PSE-binding factor, PTF, for mammalian U6 and 7SK genes). In the best studied cases, preinitiation complex assembly involves promoter recognition by TFIIIC (A and B boxes in tRNA and VA RNA genes) or by TFIIIC plus TFIIIA (A and C boxes in 5S RNA genes), TFIIIB recruitment through interactions with TFIIIC, and RNA Pol III recruitment through interactions with TFIIIB. TFIIIC from yeast consists of a six-subunit complex that binds strongly to the A and B boxes (for review, see Gabrielsen and Sentenac 1991; Geiduschek and Kassavetis 1992), whereas human TFIIIC can be separated into a five-subunit TFIIIC2 that alone binds weakly to the B box and a large less-well-characterized TFIIIC1 complex that together with TFIIIC2 binds strongly to the complete promoter (Yoshinaga et al. 1987; Dean and Berk 1988; Wang and Roeder 1996). Human TFIIIB contains, minimally, TBP and a 90-kD polypeptide (TFIIIB90) that has sequence similarity to both TFIIB and the TFIIB-related subunit of yeast TFIIIB (Wang and Roeder 1995). Like its yeast homolog (Werner et al. 1993), human TFIIIB90 interacts with a conserved subunit of human RNA Pol III (Wang and Roeder 1997). The most purified human RNA Pol III reported to date contains 16 subunits (Wang and Roeder 1997) and is similar in complexity to the yeast RNA Pol III (Gabrielsen and Sentenac 1991).
Studies of these factors in various regulatory responses have shown that serum factors and early stage adenovirus infection stimulate RNA Pol III transcription by increasing the levels of the active form of TFIIIC2 (Sinn et al. 1995), whereas transcription has been reported to be down-regulated through TFIIIB in differentiating mouse F9 embryonic carcinoma (EC) cells and during mitosis (Scott et al. 1983; White et al. 1989; Gottesfeld et al. 1994). Transcription by RNA Pol III is also down-regulated in other situations, including growth into stationary phase (Tower and Sollner-Webb 1988), growth arrest induced by protein synthesis inhibitors (Gokal et al. 1986), starvation for an essential nutrient (Hoeffler and Roeder 1985), and late-stage virus infection (Söderlund et al. 1976; Weinmann 1976). However, the detailed mechanisms involved in these events are poorly understood.
To further investigate the possibility of an RNA Pol III holoenzyme, as well as the initiation mechanism, we created previously a cell line (BN51) that constitutively expresses an epitope-tagged subunit of human RNA Pol III (Wang and Roeder 1997). We have used this cell line to affinity-purify an RNA Pol III complex (tentatively designated holoenzyme) that contains all the essential factors and to identify a novel general initiation factor that is inactivated during translation inhibition and adenovirus infection.
Results
TFIIIB and TFIIIC are coimmunoprecipitated with RNA polymerase III from the cell line BN51
The cell line BN51 constitutively expresses a FLAG epitope-tagged form of the RPC53 subunit that is specific to RNA Pol III (Wang and Roeder 1997). Nuclear extract from BN51 cells was subjected to immunoprecipitation with immobilized anti-FLAG monoclonal antibody (M2 agarose) in BC buffer containing 100 mm KCl and 0.05% NP-40. RNA Pol III was eluted from the M2 agarose with FLAG peptide and analyzed by SDS-PAGE. In addition to the 16 tightly associated subunits of human RNA Pol III (Wang and Roeder 1997), several additional polypeptides specifically copurified in the immunoprecipitate from BN51 nuclear extract but not in the control immunoprecipitate from HeLa cell nuclear extract (Fig. 1A, lane 3 vs. lane 2). The positions and sizes of the 16 RNA Pol III subunits (Pol III) and the other major BN51 immunoprecipitate-specific polypeptides (“unidentified”) are indicated on the right side of Figure 1A.
Figure 1.
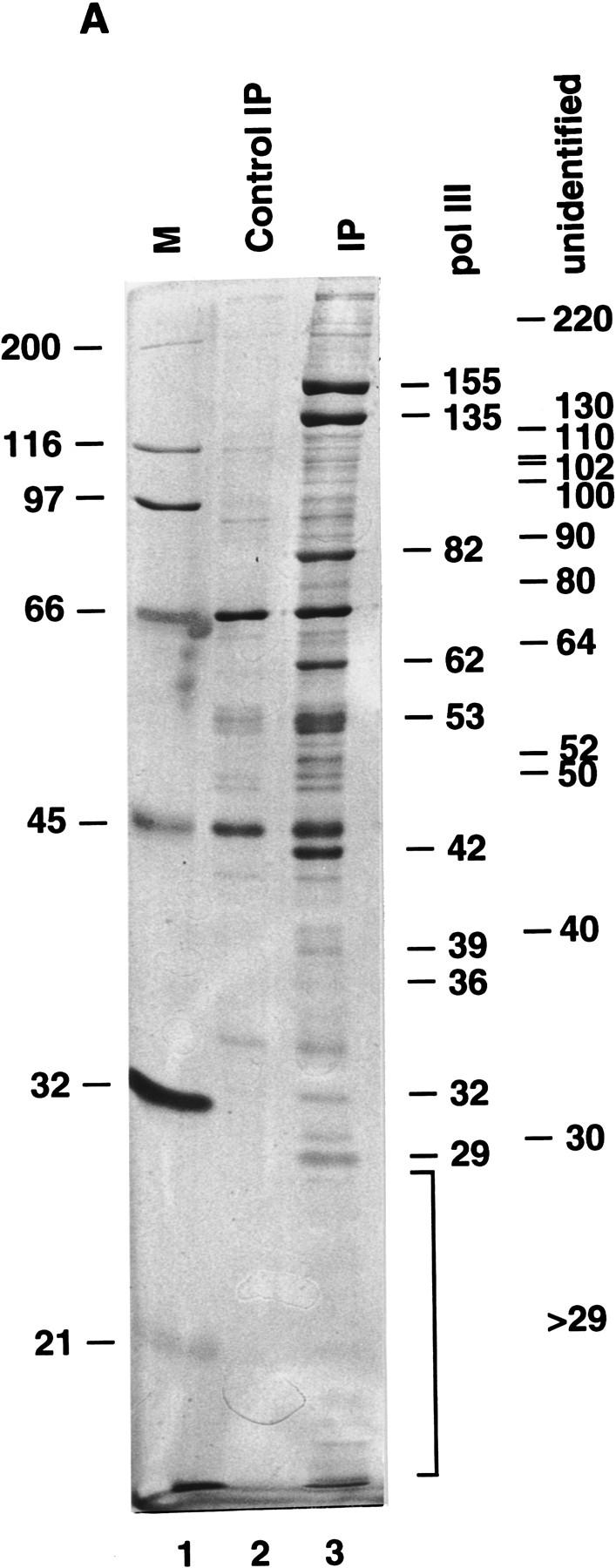
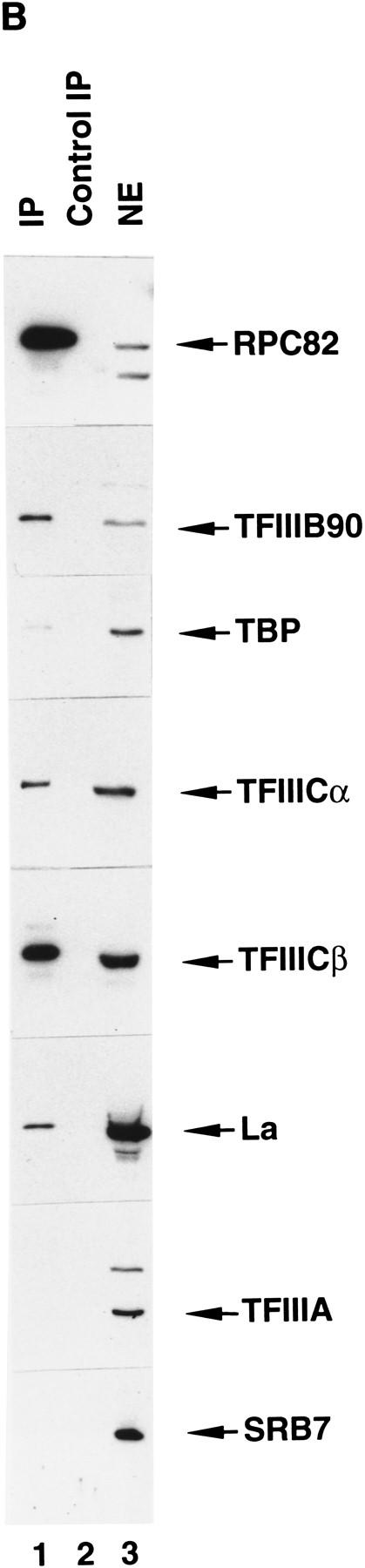
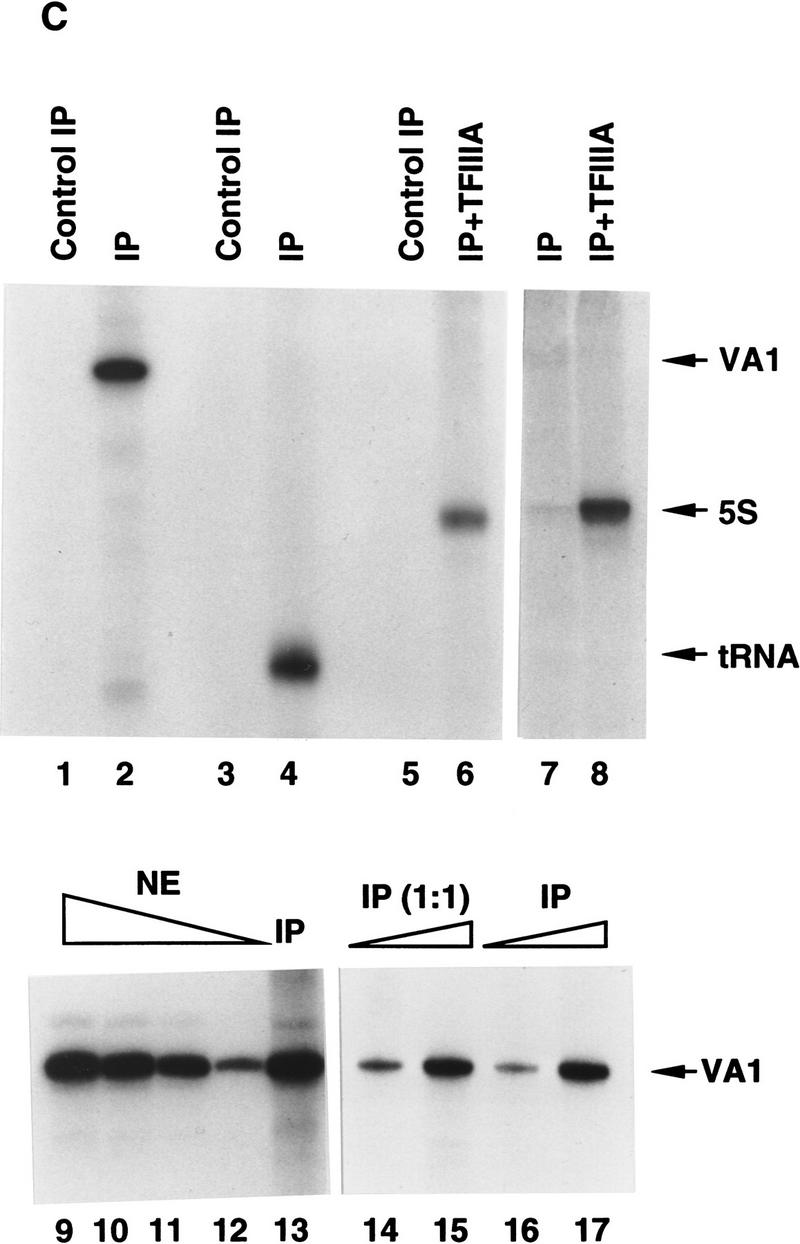
An anti-RNA Pol III immunoprecipitate contains all general components required for transcription by human RNA Pol III. (A) Ten percent SDS-PAGE analysis of the FLAG-eluted immunoprecipitate. (Lane 1) Standard molecular weight markers (Bio-Rad); (lane 2) 5 μl of the FLAG-eluted immunoprecipitate from 270 μg of a control HeLa cell nuclear extract; (lane 3) 5 μl of the FLAG-eluted immunoprecipitate from 270 μg of the nuclear extract made from the cell line BN51. Positions of molecular weight markers are indicated at left. Subunits of RNA Pol III are marked at right. Major unidentified polypetides specifically coimmunoprecipitated with RNA Pol III are marked at right. (B) Western blot analysis of the FLAG-eluted immunoprecipitate. Five microliters of the BN51 immunoprecipitate (lane 1), 5 μl of the control HeLa immunoprecipitate (lane 2), or 27 μg of HeLa nuclear extract (lane 3) was analyzed by Western blot analysis with polyclonal antibodies against proteins indicated at right. (C) Transcription of VA1 RNA (lanes 1,2,9–17), tRNA (lanes 3,4), and 5S RNA (lanes 5–8) genes with 3 μl of FLAG-eluted immunoprecipitate from HeLa (lanes 1,3,5) or BN51 (lanes 2,4,6–8,13) cells or with 10.8 μg (lane 9), 8.1 μg (lane 10), 5.4 μg (lane 11), or 2.7 μg (lane 12) HeLa nuclear extract. Purified TFIIIA was added in lanes 6 and 8. Two microliters (lanes 14,16) or 4 μl (lanes 15,17) of the FLAG-eluted immunoprecipitate from the two-fold diluted BN51 cell nuclear extract (lanes 14,15) or from the normal BN51 cell nuclear extract (lanes 16,17) were used in lanes 14–17.
At present, it is known that transcription factors TFIIIB, TFIIIC2, TFIIIC1, and TFIIIA (5S gene only) are involved in transcription of tRNA, VA RNA, and 5S RNA genes by human RNA Pol III. To determine whether any of these factors were associated with RNA Pol III in the immunoprecipitate, Western blot analysis was performed with available antibodies against corresponding recombinant subunits of these factors. In addition to RNA Pol III subunit RPC82, the BN51 immunoprecipitate contained the 38-kD TBP and 90-kD TFIIIB90 components of TFIIIB and the 220-kD TFIIICα and 110-kD TFIIICβ subunits of TFIIIC2, whereas the normal HeLa immunoprecipitate did not (Fig. 1B). A more quantitative analysis of the stoichiometry of the immunoprecipitated factors is presented below, but it is noted here that ∼60% of the TFIIIB and TFIIIC in the nuclear extract was recovered in the immunoprecipitate. This reflects the fact that a significant fraction of the total RNA Pol III contains the natural (non-FLAG-tagged) RPC53 subunit and is not immunoprecipitated by M2 agarose.
Footprint analyses on the VA1 and tRNA templates revealed that TFIIIC1, essential for extended footprints over the internal promoter (Wang and Roeder 1996), is also in the BN51 immunoprecipitate (data not shown). The 50-kD autoantigen La, which was previously reported to be involved in RNA Pol III transcription termination (Maraia 1996), was also present in the immunoprecipitate, but the 5S RNA gene-specific TFIIIA was not (Fig. 1B). Several specific components (SRB7, cyclin C/SRB10, and CDK8/SRB11) that are diagnostic of yeast and human RNA Pol II holoenzyme complexes (Koleske and Young 1994; Chao et al. 1996; Maldonado et al. 1996) were not found in the immunoprecipitate (Fig. 1B; data not shown). Similarly, two subunits (MAT1 and the 62-kD polypeptide) of TFIIH, one of the general initiation factors found in yeast and human RNA Pol II holoenzyme preparations, also were not detected in the BN51 immunoprecipitate (data not shown).
Factors contained in the BN51 immunoprecipitate complex are sufficient for transcription of the VA1, tRNA, and 5S RNA genes
Because the BN51 immunoprecipitate complex contained RNA Pol III and corresponding essential general transcription initiation factors, we tested whether the affinity-purified (peptide-eluted) complex was indeed capable of accurate transcription initiation. Figure 1C demonstrates that the derived affinity-purified complex alone was sufficient for transcription of the VA1 (Fig. 1C, lane 2) and tRNA (lane 4) genes and, in conjunction with the gene-specific activator TFIIIA, 5S RNA genes (lanes 6,8). As expected on the basis of the immunoblot results described in Figure 1B (lane 2), no transcripts could be discerned in the transcription reaction with the control HeLa immunoprecipitate (Fig. 1C, lanes 1,3,5). A dose-response analysis (Fig. 1C, lanes 9–13) further showed that the transcription signal obtained with 3 μl of the BN51 immunoprecipitate was comparable with that obtained with 10.8 μg of nuclear extract protein. Because the immunoblot analysis of Figure 1B showed that 5 μl of the BN51 immunoprecipitate contains levels of TFIIIB and TFIIIC comparable with those in 27 μg of nuclear extract protein, but an ∼10-fold higher level of RNA Pol III, it appears that TFIIIB and TFIIIC are the limiting components in the immunoprecipitate. It was also found that comparable amounts of holoenzyme, assayed by transcription activities, were immunoprecipitated even when the BN51 cell nuclear extract was diluted twofold before immunoprecipitation (Fig. 1C, lanes 14 and 15 vs. lanes 16 and 17). These results indicate that the holoenzyme complex is stable to dilution and unlikely to result from a concentration-dependent nonspecific aggregation of the RNA Pol III components.
The results presented here mirror in part those observed for both yeast and human RNA Pol II holoenzyme preparations that were reported to contain all or part of the general transcription factors but no gene-specific activators (Koleske and Young 1994; Ossipow et al. 1995; Maldonado et al. 1996).
The components commonly required for transcription by human RNA polymerase III form an RNA polymerase III holoenzyme complex
To provide additional evidence that the components present within and released from the immunoprecipitate form a stable complex, the material eluted from the immunoprecipitate was further purified by gel-filtration chromatography on Superose 6. Western blot analysis revealed that the majority of RNA Pol III eluted in fractions 12 and 13, corresponding to a size roughly equivalent to that (∼700 kD) of the free 16-subunit form (Wang and Roeder 1997) of RNA Pol III (Fig. 2A, top panel). However, small amounts of RNA Pol III were coeluted with TFIIIB90 (Fig. 2A, second panel), TBP (third panel), TFIIIC63 (fourth panel), and TDF (see below for definition; data not shown) in fractions (10–11) corresponding to a native size of ∼1500 kD (assuming that the molecular mass of blue Dextran 2000 is 2000 kD). Fractions (10 and 11) containing all these components also mediated transcription of the VA1 RNA gene in the absence of other components, whereas those fractions (13 and 14) containing only RNA Pol III did not (bottom panel). When analyzed independently under the same conditions, individual TFIIIB (∼200 kD), TFIIIC (∼500 kD), and RNA Pol III complexes eluted in fractions 15 and 16, 13 and 14, and 12 and 13, respectively (data not shown). One explanation for these results is that fractions 10 and 11 contain a complex composed of RNA Pol III, TFIIIC, and TFIIIB, and this possibility is favored by the apparent size of the complexes responsible for the transcription activity. Another possibility is that fractions 10 and 11 contain both RNA Pol III–TFIIIC and RNA Pol III–TFIIIB complexes; this possibility is consistent with the ability of TFIIIB subunits to interact directly with RNA Pol III (Wang and Roeder 1997), although comparable interactions of TFIIIC with RNA Pol III have not been reported.
Figure 2.
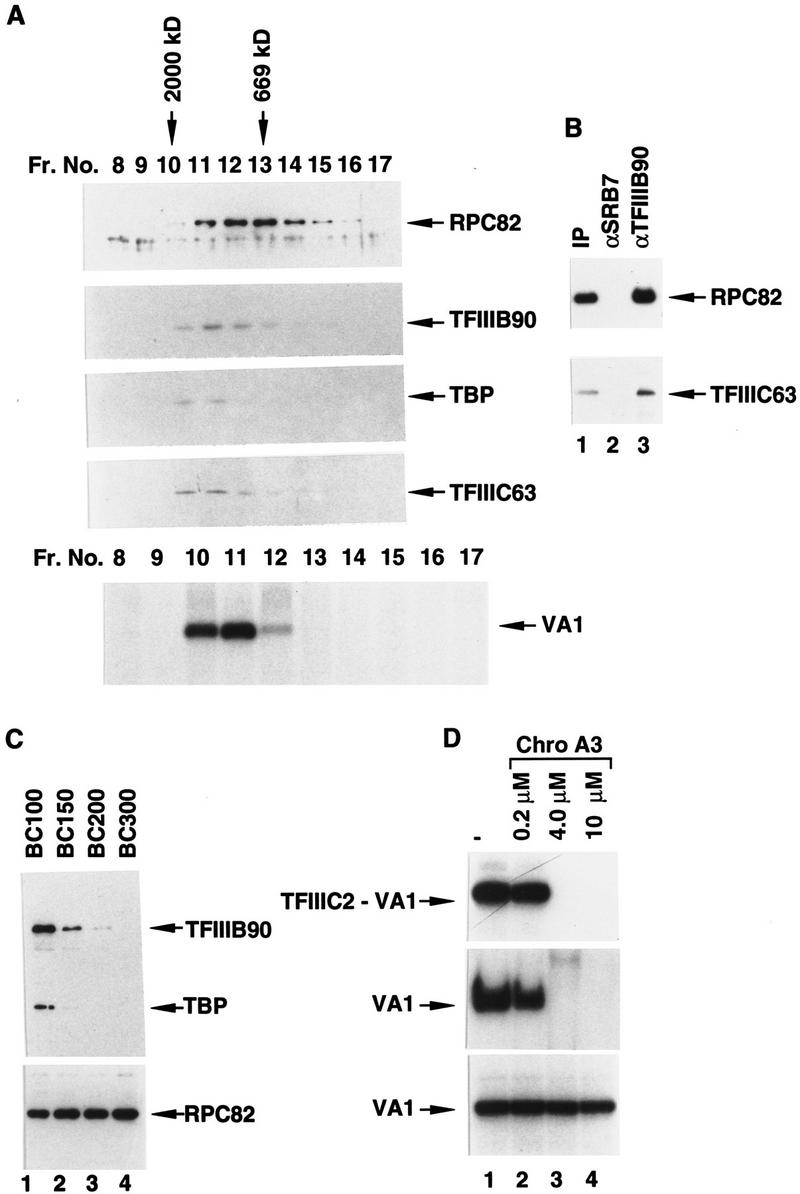
Human RNA Pol III, TFIIIB, TFIIIC, and other essential components can be isolated as a holoenzyme complex. (A) RNA Pol III, TFIIIB90, TBP, and TFIIIC63 coelute on a Superose 6 column. Fifty microliters of the FLAG-eluted immunoprecipitate from 2.7 mg of BN51 nuclear extract was chromatographed on a Superose 6 column in BC100–0.01% NP-40. Five microliters (top) or 20-μl (middle) aliquots of each fraction were analyzed by immunoblot with antibodies against the RPC82 subunit of RNA Pol III, TFIIIB90, TBP, and TFIIIC63 as indicated. The elution peaks of blue Dextran 2000 and thyroglobulin (669 kD) were in fractions 10 and 13, respectively. The bottom panel shows transcription from the VA1 gene with 5-μl aliquots of the same fractions. (B) Coimmunoprecipitation of RNA Pol III and TFIIIC with TFIIIB from the FLAG-eluted immunoprecipitate. Ten microliters of antigen-affinity-purified-immobilized anti-SRB7 (lane 2) or anti-TFIIIB90 (lane 3) protein A–Sepharose 4B was incubated at 4°C with 0.5 ml of BC100 containing 2 mg/ml of BSA and 0.05% NP-40 for 1 hr. The beads were washed with BC100 containing 0.05% NP-40 twice and incubated at 4°C with 100 μl of the FLAG-eluted immunoprecipitate containing 0.5 mg/ml of BSA for 2 hr. The beads were washed five times with BC100–0.05% NP-40, and then boiled in the SDS sample buffer for Western blot analysis with antibodies against RPC82 (top) or TFIIIC63 (bottom). IP (lane 1) denotes 10% of material used for the immunoprecipitation. (C) Salt sensitivity of the RNA Pol III holoenzyme. BN51 nuclear extract was subjected to immunoprecipitation in BC100 (lane 1), BC150 (lane 2), BC200 (lane 3), or BC300 (lane 4), and corresponding FLAG eluates (5 μl) were analyzed by Western blot analysis with the antibodies against TFIIIB90 (top), TBP (middle) and RPC82 (bottom) as indicated. (D) The RNA Pol III holoenzyme is not sensitive to Chromomycin A3. (Top) Mobility retardation assays of human TFIIIC2 (5 ng) and a VA1 promoter in the presence of variable levels of Chromomycin A3. Only the region containing the TFIIIC2–DNA complex is shown. (Middle) In vitro transcription of the VA1 template in 10.8 μg of BN51 nuclear extract in the presence of variable levels of Chromomycin A3. (Bottom) In vitro transcription of the VA1 template with RNA Pol III holoenzyme preparations (FLAG eluates) immunoprecipitated in the presence of variable levels of Chromomycin A3.
To provide evidence for the first possibility—that TFIIIB, TFIIIC, and RNA Pol III can form a single complex—the initial FLAG-eluted immunoprecipitate was subjected to immunoprecipitation with immobilized antigen-purified anti-TFIIIB90 antibodies. Figure 2B shows that both RNA Pol III (detected by anti-RPC82 antibodies; top panel) and TFIIIC (detected by anti-TFIIIC63 antibodies; bottom panel) were coimmunoprecipitated. The immunoprecipitate contained almost all of the input TFIIIB, ∼10% of the RNA Pol III, and ∼20% of the TFIIIC. The partial immunoprecipitation of TFIIIC may reflect either antibody-mediated disruption of an RNA Pol III–TFIIIC–TFIIIB complex or the presence of natural subcomplexes (RNA Pol III–TFIIIB and RNA Pol III–TFIIIC) in addition to the complete RNA Pol III–TFIIIB–TFIIIC complex in the original (FLAG-eluted) immunoprecipitate. As a negative control, immobilized antigen-purified anti-SRB7 antibodies did not bring down RNA Pol III or TFIIIC under the same conditions (lane 2). Overall, these results indicate that TFIIIB, TFIIIC, and RNA Pol III form a complex that is stable to immunoprecipitation and peptide elution and that we tentatively, and for convenience, refer to as an RNA Pol III holoenzyme. However, they do not exclude the presence of partial complexes that could also represent assembly intermediates.
To test the salt sensitivity of the RNA Pol III holoenzyme, immunoprecipitation from BN51 nuclear extracts was performed at different salt conditions (Fig. 2C, lanes 1–4). The amount of the RPC82 subunit of RNA Pol III in the immunoprecipitate was essentially unchanged at all salt conditions (Fig. 2C, bottom panel, lanes 1–4), consistent with our previous demonstration that RNA Pol III is a highly stable multisubunit complex (Wang and Roeder 1997). High levels of TFIIIB90 (Fig. 2C, top panel), TBP (middle panel), and other factors (TFIIIC2, TFIIIC1, and TDF) mentioned above (data not shown) were immunoprecipitated at 100 mm KCl (Fig. 2C, lane 1). However, these levels were greatly reduced, by similar proportions, at 150 mm KCl (lane 2) and at 200 mm KCl (lane 3). When the KCl concentration was increased to 300 mm KCl, no TFIIIB90 or TBP could be detected in the immunoprecipitate (Fig. 2C, lane 4). Thus, association between RNA Pol III and other components is easily disrupted by high salt under the specific buffer conditions used. The holoenzyme was also disrupted by ion exchange chromatography on phosphocellulose because it could not be detected by immunoprecipitation in any of the conventional P11 fractions (0.1, 0.35, and 0.6 m KCl eluates) (data not shown). These results may help to explain why the RNA Pol III holoenzyme could not be unambiguously identified previously through traditional chromatographic methods (Wingender et al. 1986).
Because transcription initiation factors can form preinitiation complexes on the DNA template and interact nonspecifically with DNA at low ionic strength, an important issue is whether components of the RNA Pol III holoenzyme may be tethered together by DNA present in nuclear extract. Chromomycin A3, an antibiotic with high affinity to double-stranded DNA, strongly inhibits DNA-directed RNA synthesis both in vivo and in vitro (Keniry et al. 1993). As shown in Figure 2D (top panel, lanes 2–4), Chromomycin A3, at concentrations of 4 and 10 μm, severely interfered with the formation of a specific VA1 promoter complex with TFIIIC2 (the primary promoter recognition factor). Consistent with this observation, in vitro transcription from the VA1 template was completely abolished at Chromomycin A3 concentrations of 4 and 10 μm (middle panel, lanes 2–4). However, similar levels of VA1 transcripts were synthesized by the RNA Pol III holoenzyme isolated in the absence (Fig. 2D, bottom panel, lane 1) or presence (Fig. 2D, lanes 2–4, bottom panel) of Chromomycin A3. Similar results (persistence of the holoenzyme) were also observed in the presence of ethidium bromide (data not shown), another reagent more commonly used to distinguish DNA-dependent versus DNA-independent protein associations (Lai and Herr 1992). The presence of DNA was also undetectable by an assay that involved treatment of the holoenzyme with phosphatase followed by incubation with [γ32P]ATP and T4 polynucleotide kinase (data not shown). Therefore, DNA is unlikely to be responsible, either through specific or nonspecific factor interactions, for the association of the complex.
We then quantitated by immunoblot the levels of coimmunoprecipitated RPC82, TFIIIB90, TBP, and TFIIIC63 that were released by peptide elution of antibody-bound RNA Pol III. Two different amounts of the immunoprecipitate complex were analyzed, along with variable amounts of recombinant RPC82, TFIIIB90, TBP, and TFIIIC63 as standards, and quantitated by the Storm Imager System (Molecular Dynamics) (Bai et al. 1996). Based on the standard curves, 1 μl of the immunoprecipitate complex was shown to contain 2.7 fmoles of TFIIIB90, 2.9 fmoles of TBP, 2.2 fmoles of TFIIIC63, and 24 fmoles of RPC82. These data indicate the presence of similar molar amounts of TFIIIB (TFIIIB90 and TBP) and TFIIIC (TFIIIC63) components and an ∼10-fold higher amount of RNA Pol III (RPC82 subunit) in the immunoprecipitate. These results are consistent with the results of the gel-filtration analysis of Figure 2A (showing mostly free RNA Pol III) and with studies in yeast showing ∼10% of the total RNA Pol II in the holoenzyme and 90% in a free form (Koleske and Young 1995).
Although an early study (Wingender et al. 1986) also reported cosedimentation of human RNA Pol III and cognate factors, appropriate controls to eliminate the possibility of nonspecific association (especially with nucleic acids) were not provided and the aberrant sizes and poor separation of the individual components also rendered the results inconclusive. Moreover, the extensive salt resistance of the reported RNA Pol III complex contrasts markedly with the moderate salt sensitivity of the complex described here.
Loss of a novel transcription factor found within the RNA Pol III holoenzyme accounts for transcription inactivation during translation inhibition
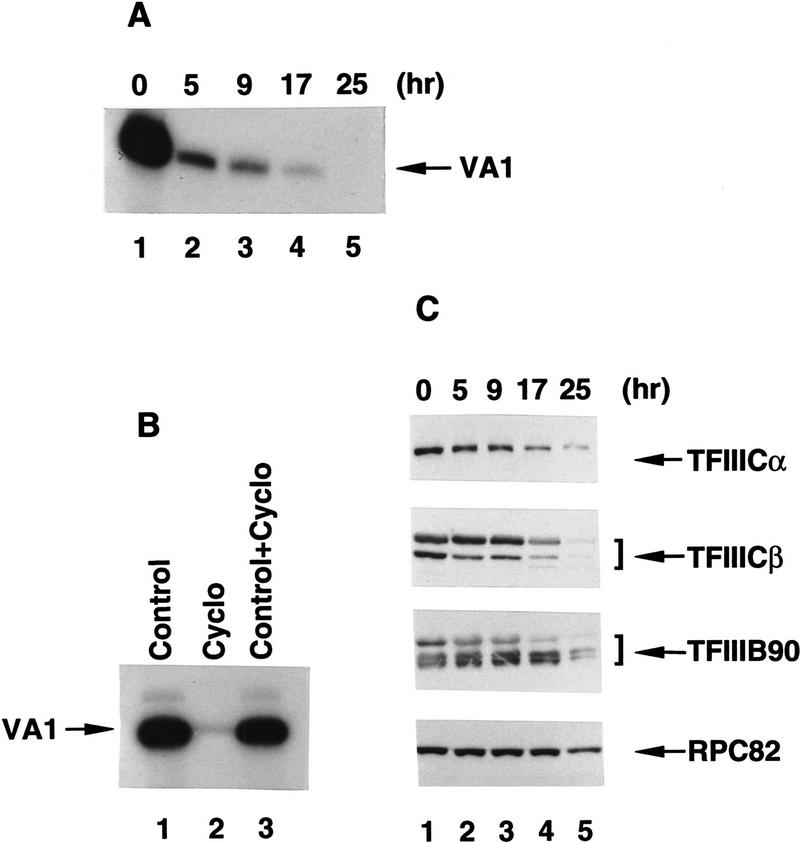
We examined the effect of cycloheximide, a drug that inhibits protein synthesis, on VA1 gene transcription in HeLa cells. Extracts prepared from the same number of mock or cycloheximide-treated cells contained nearly identical amounts of total protein. Parallel transcription assays were performed with the same amounts of nuclear extract on the VA1 template. Consistent with previous results from studies of murine lymphosarcoma and yeast cells (Gokal et al. 1986; Dieci et al. 1995), extracts of cells harvested at different times after cycloheximide addition became increasingly less competent for VA1 gene transcription—showing 90% inactivation at 5 hr and 9 hr (Fig. 3A, lanes 2 and 3), 95% inactivation at 17 hr (lane 4), and complete inactivation at 25 hr (lane 5). After 17 hr of drug treatment, cells were still viable as determined by trypan blue staining; but after 25 hr, >50% of the cells were dead. As shown in Figure 3B, extract derived from 17-hr cycloheximide-treated cells (lane 2) did not inhibit transcription by the control extract (lane 1) when the two were mixed together before assay (lane 3). This result indicates that transcriptional impairment in the nuclear extract derived from cycloheximide-treated cells is not attributable to the presence of a diffusible inhibitor.
Figure 3.
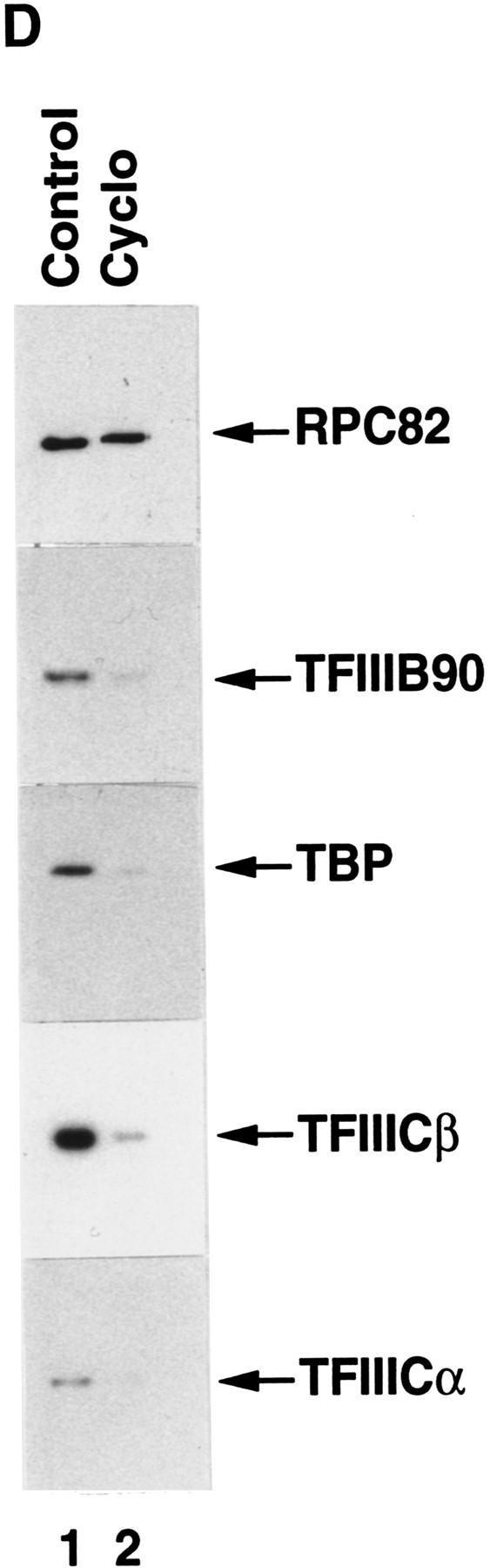
Cycloheximide-mediated translation inhibition represses RNA Pol III transcription. (A) Transcription of the VA1 gene was analyzed in 10.8 μg of nuclear extract from BN51 cells treated with cycloheximide for the indicated times. (B) Transcription of the VA1 gene with 10.8 μg of nuclear extract from untreated (lane 1) or cycloheximide-treated (lane 2) BN51 cells or with a mixture (10.8 μg each) of the two extracts. (C) Western blot analysis of nuclear extract (27 μg) from cells treated with cycloheximide for the times indicated. Extracts (from the same analyses as A) were probed with antibodies against the RPC82 subunit of RNA polymerase III, TFIIIB90, TBP, TFIIICα, and TFIIICβ as indicated. (D) Cycloheximide treatment prevents formation of the RNA Pol III holoenzyme. Western blot analysis of the FLAG-eluted immunoprecipitates derived from nuclear extracts prepared from nontreated (lane 1) or cycloheximide-treated (lane 2) BN51 cells. Blots were probed with polyclonal antibodies against RPC82, TFIIIB90, TBP, TFIIICα, and TFIIICβ as indicated.
To monitor possible modifications or changes in concentration of known transcription factors or RNA Pol III, nuclear extracts from control and cycloheximide-treated cells were analyzed by Western blot. As shown in Figure 3C, there were no detectable changes in the levels of TFIIICα, TFIIICβ, TFIIIB90, and the RPC82 subunit of RNA Pol III at times (5 hr and 9 hr) when transcription activity was already dramatically (>90%) reduced. Mild decreases (∼50%) were apparent by 17 hr but did not correlate with the very severe (95%) reduction in transcription at that time. Consistent with significant cell death at 25 hr, there was significant degradation of TFIIIC, TFIIIB, and (to a lesser extent) RNA Pol III components at 25 hr. There were no apparent changes in electrophoretic mobility that would indicate post-translational modifications, although more discriminating analyses are necessary to rule out this possibility.
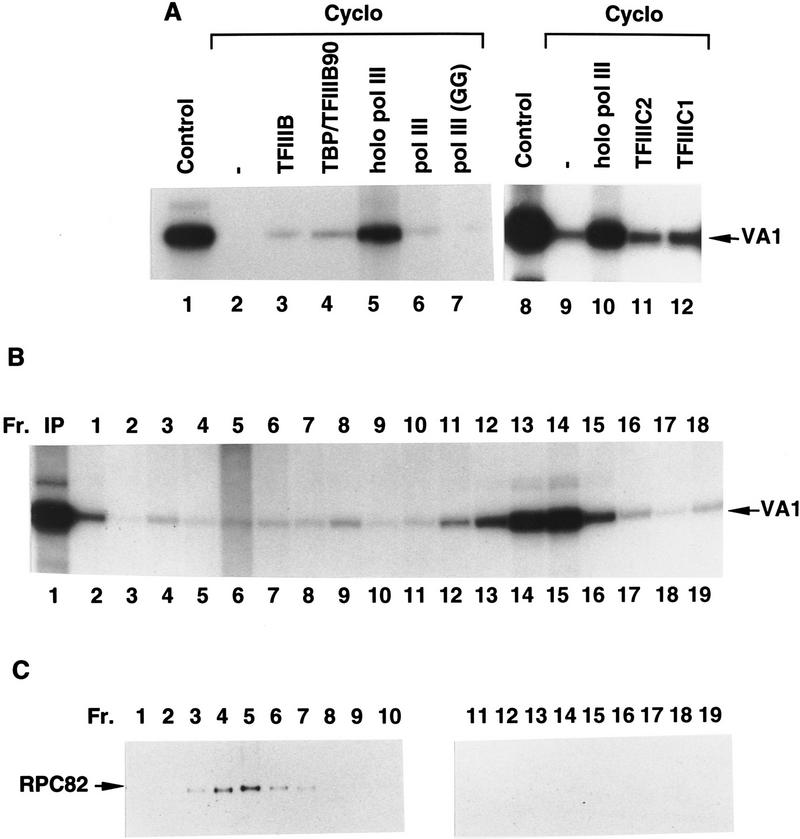
To identify the components responsible for the cycloheximide-induced down-regulation of RNA Pol III-mediated transcription, we first tried to restore transcription in nuclear extract from cycloheximide-treated cells by addition of individual transcription factors isolated from control cells. As shown in Figure 4A, insignificant to marginal levels of activity were restored by addition of natural TFIIIB (lane 3), recombinant core TFIIIB (lane 4), immunopurified core (16-subunit) RNA Pol III (lane 6), conventional chromatographically purified RNA Pol III (lane 7), TFIIIC2 (lane 11), or TFIIIC1 (lane 12). In contrast, addition of the transcription-competent RNA Pol III holoenzyme to the inactive extract resulted in a high level of transcription activity (Fig. 4A, lane 5 vs. lane 1 and lane 10 vs. lane 8). This latter result again indicates that transcriptional impairment in the nuclear extract derived from cycloheximide-treated cells is not attributable to the presence of a diffusible inhibitor.
Figure 4.
The RNA polymerase III holoenzyme contains a novel factor that is inactivated by cycloheximide treatment of cells. (A) Transcription of the VA1 RNA gene was analyzed in nuclear extract (10.8 μg) from untreated (lanes 1,8) or 17-hr cycloheximide-treated (lanes 2–7 and 9–12) BN51 cells. Extracts from the cycloheximide-treated BN51 cells were complemented with no additional factor (lanes 2,9) or with 5 μg of the native TFIIIB (lane 3), 5 ng of the recombinant TFIIIB (lane 4), 2 μl of RNA polymerase holoenzyme (lanes 5,10), 2 μl of the highly purified core RNA Pol III isolated by affinity (lane 6) or conventional (lane 7) methods, 2 μl of TFIIIC2 (lane 11), and 2 μl of TFIIIC1 (lane 12). The extracts used in lanes 1–7 and 8–12 were from separate experiments. (B) The novel cycloheximide-sensitive factor disassociates from RNA Pol III during sucrose gradient sedimentation at high salt. The FLAG-eluted immunoprecipitate from BN51 cells was subjected to sucrose centrifugation at 500 mm KCl and both input (IP) and 3-μl aliquots of gradient fractions were analyzed for effects on VA1 gene transcription in 10.8 μg of nuclear extract from cycloheximide-treated BN51 cells. (C) Western blot analysis of the sucrose gradient fractions (2 μl) from panel B using polyclonal antibodies against the RPC82 subunit of RNA Pol III.
In view of these results and because the holoenzyme represented a highly purified source of a complete set of transcription factors, it was used to search for the cycloheximide-sensitive factor. To this end the holoenzyme preparation was subjected to sucrose gradient analysis at high salt (500 mm KCl). As shown in Figure 4B, fractions 13 and 14 fully restored the transcription activity of the cycloheximide-treated nuclear extract. When the same fractions were analyzed by Western blot (Fig. 4C) and SDS-PAGE with silver staining (data not shown), RNA Pol III (RPC82 subunit) was found in fractions 3–6 but not in fractions 13 and 14. Altogether these results provide direct evidence that the RNA Pol III holoenzyme contains a novel factor, distinct from TFIIIB, TFIIIC1, TFIIIC2, and RNA Pol III, that is responsible for restoring the inactivated transcription activity. Because the novel factor described here is functionally inactivated, relative to other RNA Pol III factors, during inhibition of protein synthesis, it is tentatively named translation dependent factor (TDF).
As shown above, the concentrations and functional activities of TFIIIB, TFIIIC, and RNA Pol III in cycloheximide-treated cells remain relatively unchanged. These results raised the interesting question as to whether these remaining components could still assemble into a stable complex. To test this possibility, nuclear extracts made from control and cycloheximide-treated (17 hr) BN51 cells were subjected to immunoprecipitation on M2 agarose. A Western blot analysis revealed that the immunoprecipitates from control and cycloheximide-treated BN51 cells contained the same amounts of RNA Pol III (detected by anti-RPC82 antibodies), whereas the immunoprecipitates from cycloheximide-treated BN51 cells contained much lower amounts of TFIIIB90, TBP, TFIIICα, and TFIIICβ (Fig. 3D, lane 2 vs. lane 1). These results suggested that holoenzyme assembly can be affected either by a novel factor, such as TDF, that is selectively inactivated by protein synthesis inhibition or by as yet undetected modifications of RNA Pol III or other general factors.
TDF is inactivated at a late stage of adenovirus infection
As shown in Figure 5A, the transcription activity of nuclear extracts prepared from adenovirus type 2 (Ad2)-infected cells dramatically decreases at later stages (21 hr and 31 hr) of infection. This result is consistent with the previously published results of Hoeffler and Roeder (1985) analyzing similarly prepared nuclear extracts. Consistent with the analysis of nuclear extracts from cycloheximide-treated cells, Western blot analyses revealed that the concentrations of TFIIICα, TFIIICβ, TFIIIB90, and the RPC82 subunit of RNA Pol III were unaffected and that there were no apparent modifications (data not shown). Moreover, in direct parallel to the situation with nuclear extract from cycloheximide-treated (17 hr) cells (Fig. 5B, top panel and above), readdition of sucrose gradient-purified TDF (lane 3) but not TFIIIB (lane 5), RNA Pol III (lane 4), or any other RNA Pol III transcription factors (data not shown) fully restored the transcription activity of the nuclear extract made from 21-hr virus-infected cells (Fig. 5B, bottom panel). Because the transcription activity could be restored only by purified TDF, transcription inactivation by late-stage virus infection probably involves the same mechanism as translation inhibition. A previous report that virus infection at late stage inhibits host cell translation also supports this conclusion (O’Malley et al. 1986; Zhang et al. 1994; Schneider 1995).
Figure 5.
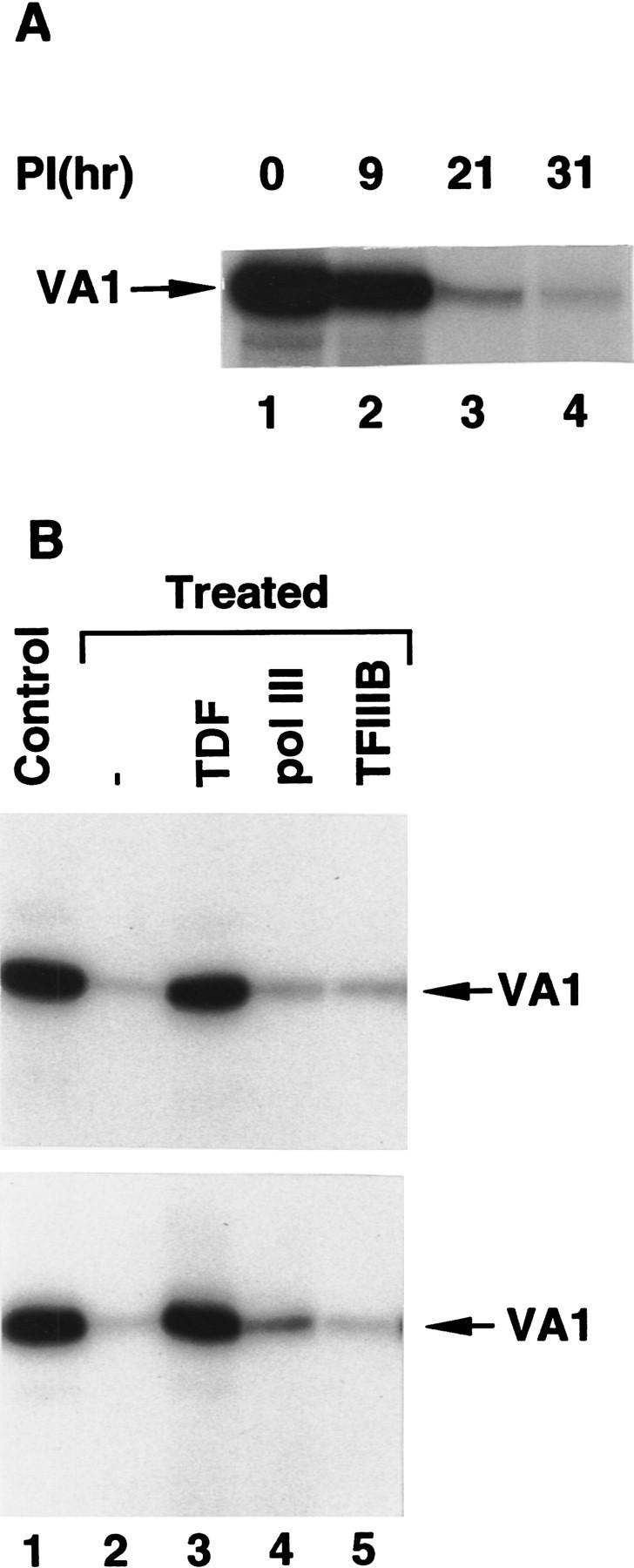
Late-stage adenovirus infection inhibits RNA Pol III transcription through inactivation of TDF. (A) VA1 gene transcription in nuclear extracts (10.8 μg) from BN51 cells infected with adenovirus and harvested at the indicated times postinfection (PI). (B) TDF restores transcription to extracts from both adenovirus-infected and cycloheximide-treated cells. VA1 gene transcription was analyzed in nuclear extracts from untreated cells (lane 1, top and bottom) or in nuclear extracts (10.8 μg) from 17-hr cycloheximide-treated (top) or adenovirus-infected (bottom) BN51 cells in the presence of no additional factors (lane 2), 3 μl of TDF (sucrose gradient fraction 13) (lane 3), 2 μl of core RNA Pol III (lane 4), or 2 μl of native TFIIIB (lane 5).
TDF is an essential basal factor that is broadly required for transcription by RNA Pol III
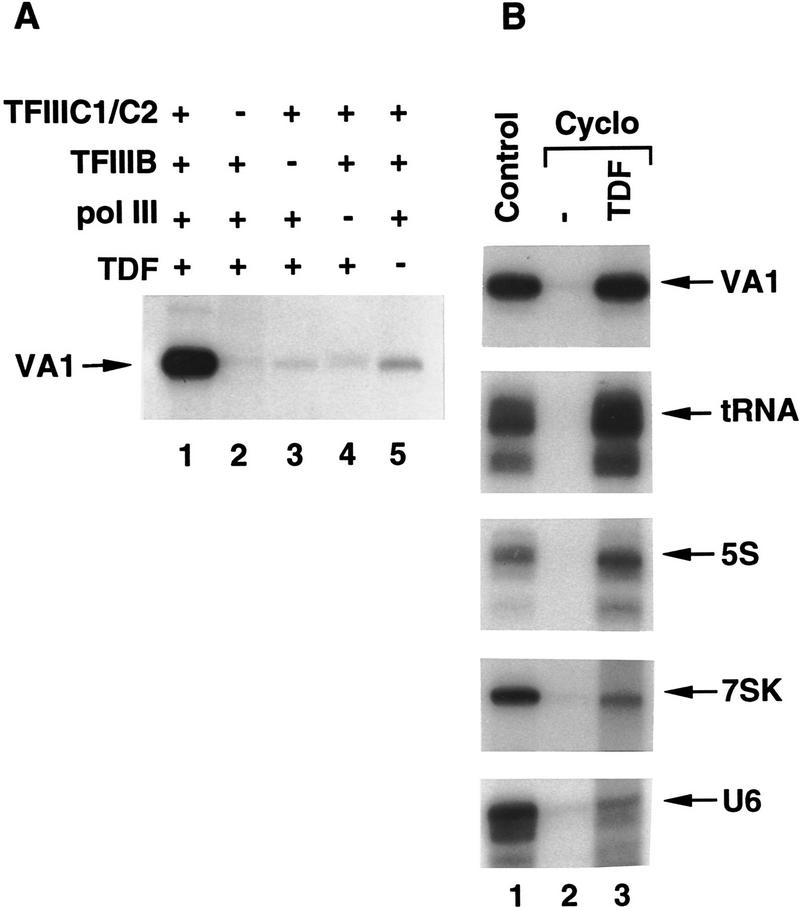
TDF is defined by its ability to restore transcription activity to nuclear extracts from cycloheximide-treated cells. Because these extracts contain negative cofactors that could potentially suppress RNA Pol III transcription as well as RNA Pol II transcription (White et al. 1994), certain opposing factors could be required conditionally (in crude systems) for transcription. Hence, it was important to investigate this possibility for TDF, especially because previous studies had indicated that TFIIIB, TFIIIC1, and TFIIIC2 are sufficient for transcription of VA and tRNA genes by RNA Pol III in purified systems (Wang and Roeder 1996). The assay system used here contained highly purified preparations of TFIIIB, TFIIIC, (containing both TFIIIC2 and TFIIIC1), and core (16-subunit) RNA Pol III, in addition to a partially purified TDF preparation [fast protein liquid chromatography (FPLC) Superose 6 fraction; see Materials and Methods] that was uncontaminated by other factors (TFIIIB, TFIIIC1, TFIIIC2, and RNA Pol III) on the basis of Western blot and transcription assays (data not shown). All fractions were from untreated cells. As shown in Figure 6A, all of these factors are essential for optimal transcription because the absence of any single factor resulted in <10% of the maximal transcription activity with all present. From this result, it is concluded that TDF functions as an essential basal transcription factor in the highly purified reconstituted system. Our previous results (Wang and Roeder 1996) can be explained by the fact that the TFIIIC1 preparation used (FPLC Mono Q fraction) was contaminated with TDF activity because of similar chromatographic behavior between TFIIIC1 and TDF on P11 and Mono Q columns (see Materials and Methods). In the present analysis, TDF was separated from TFIIIC1 on an high-pressure liquid chromatography (HPLC) heparin–5PW column.
Figure 6.
TDF is a basal factor that is generally required for transcription by human RNA Pol III. (A) In vitro transcription of the VA1 gene in a reconstituted system containing the indicated combinations of 1 μl of highly purified TFIIIC containing TFIIIC2 and TFIIIC1, 5 μg of purified native TFIIIB, 0.5 μl of purified core RNA Pol III, and 2 μl of the partial purified TDF (Superose 6 fraction). (B) TDF-dependent transcription of different class III genes. Transcription of VA1, tRNA, 5S, U6, and 7SK genes was analyzed independently in control BN51 nuclear extracts (lane 1) and in nuclear extracts made from cycloheximide-treated BN51 cells analyzed alone (lane 2) or complemented with 3 μl of TDF (sucrose gradient fraction 13) (lane 3).
As a further test of the specificity of TDF, we analyzed transcription of subclass 1 (5S RNA) and subclass 3 (U6 and 7SK RNA) genes, in addition to other (tRNA and VA RNA) subclass 2 genes, in nuclear extracts from normal versus cycloheximide-treated cells. As shown in Figure 6B, transcription from each of these genes was active in control extract (lane 1), inhibited in extract from cycloheximide-treated cells (lane 2), and active in extract from cycloheximide-treated cells supplemented with partially purified TDF (lane 3). However, whereas purified TDF completely restored transcription of the subclass 1 (5S RNA) and subclass 2 (tRNA and VA RNA) genes, it only partially restored transcription of subclass 3 (U6 and 7SK RNA) genes. The latter result suggests that additional subclass 3-specific factors also are partially inactivated by cycloheximide treatment and are consistent with previous demonstrations (for review, see Yoon et al. 1995) of such factors. Nevertheless, these results indicate that TDF is a general transcription factor essential for transcription of all three subclasses of class III genes.
Discussion
An affinity purification technique, in conjunction with a cell line expressing an epitope-tagged subunit specific to RNA Pol III, has allowed us to isolate and characterize a human RNA Pol III holoenzyme that contains all of the basal factors essential for transcription of the VA1 and tRNA genes. These include previously characterized factors, as well as a novel translation-dependent factor (TDF) that appears to be essential for transcription of all class III genes. These results have important implications both for the mechanism of transcription initiation by RNA Pol III and for the regulation of class III genes in various physiological processes.
RNA Pol III holoenzyme
As originally defined in prokaryotes, an RNA polymerase holoenzyme reflects an autonomously initiating entity composed of a catalytically active RNA polymerase core and an initiation-specific σ-factor subunit (for review, see McClure 1985). In more recent studies in eukaryotes, the term holoenzyme has been used to describe complexes containing a catalytically active 12-subunit RNA Pol II, cofactors important for regulatory factor function, and variable complements of the general initiation factors (introductory section). Partly because of variations in isolation procedures and partly because of different criteria for distinguishing complexes that arise from adventitious factor interactions, there is considerable disagreement over what constitutes the most physiologically relevant holoenzyme forms (Koleske and Young 1995; Björklund and Kim 1996; Halle and Meisterernst 1996; Orphanides et al. 1996). One difficulty in this regard is the possibility that factor interactions that normally take place only on the promoter might also occur, under certain in vitro conditions, even in the absence of DNA. Clearly, what remains to be established is whether partial or completely preassembled holoenzyme complexes, akin to those isolated biochemically, actually initiate transcription, as such, either in vivo or in vitro. In any case, however, the preassembly of partial or complete complexes may have mechanistic and regulatory consequences for the overall pathway of preinitiation complex assembly and function.
The number and sizes of the polypeptides present in the RNA Pol III holoenzyme described here (Fig. 1) are consistent with those of the 16 known subunits in RNA Pol III, the known subunits in TFIIIC2 (220, 110, 102, 90, and 63 kD) and TFIIIB (38 and 90 kD), several candidate subunits in TFIIIC1 (Z. Wang and R.G. Roeder, unpubl.), and an additional candidate subunit in TFIIIB (Y. Hsieh, Z. Wang, and R.G. Roeder, unpubl.). The RNA Pol III preparation described here fits the strict holoenzyme concept in that it contains all components necessary for transcription initiation. Other important criteria include near stoichiometric levels of the associated general initiation factors, an apparent native size (∼1500 kD) consistent with that predicted from estimates of the sizes of the constituents, and coassociation of the components during an RNA Pol III-specific selection and subsequent size fractionation. The RNA Pol III holoenzyme is also deficient in the one class III gene-specific activator tested (TFIIIA), consistent with the lack of gene-specific activators in RNA Pol II holoenzymes. In contrast to those RNA Pol II holoenzyme complexes that remain intact through multiple chromatographic steps, but that lack complete complements of the general initiation factors (Kim et al. 1994; Koleske and Young 1994; Chao et al. 1996; Maldonado et al. 1996), the RNA Pol III holoenzyme described here appears rather sensitive both to moderate KCl concentrations and to ion-exchange chromatography. In this regard, the only RNA Pol II holoenzyme preparations reported to be self-sufficient for initiation are these isolated from cellular extracts by single-step affinity purification methods at low salt (Ossipow et al. 1995; McCracken et al. 1997). As an additional argument against adventitious in vitro association of RNA Pol III and general initiation factors TFIIIB and TFIIIC, the formation of the RNA Pol III holoenzyme described here correlates with the presence of the newly described TDF. In this regard, the role of TDF could be superficially analogous to that of the SRBs in the RNA Pol II holoenzyme (Kim et al. 1994; Koleske and Young 1994), although the basis for general factor interactions (as opposed to cofactor interactions) is not yet established in the latter case.
The isolation of an autonomously initiating complex of RNA Pol III and associated factors extends the list (see introductory section) of similarities between transcription initiation by RNA Pol II and III and indicates a more unified mechanism. Given increasing evidence for eukaryotic holoenzymes, but bearing in mind the cautionary points raised above, we tentatively designate the complex described here as an RNA Pol III holoenzyme.
A novel accessory factor for initiation by RNA Pol III
Here, we have taken advantage of both the affinity-purified RNA Pol III holoenzyme and the selective sensitivity of specific factors to protein synthesis inhibition to identify and partially purify a novel RNA Pol III transcription factor (TDF). Given the complexity and incomplete purification of other general factors required by RNA Pol III, it is important to consider their potential relationships to TDF. The well-characterized human factors include TFIIIC2, which contains five subunits and binds to the B box, and a core TFIIIB, which contains TBP and a TFIIB-related polypeptide that, like its yeast homolog (Werner et al. 1993; Chaussivert et al. 1995), interacts both with a component of TFIIIC2 (Y. Hsieh, R. Kovelman, Z. Wang, and R.G. Roeder, unpubl.) and with a subunit of RNA Pol III (Wang and Roeder 1997). Preliminary data (Y. Hsieh, Z. Wang, and R.G. Roeder, unpubl.) also indicate that human TFIIIB contains a third subunit with weak sequence relationships to the third (largest) subunit of yeast TFIIIB. Human TFIIIC1, which has no known counterpart in yeast, is less well characterized but stabilizes and extends promoter interactions of TFIIIC2 (for review, see Wang and Roeder 1996) and can interact with TFIIIC2 both in the absence and presence of promoter DNA (Sinn et al. 1995; Z. Wang, and R.G. Roeder, unpubl.).
Several observations presented here indicate that TDF is an essential basal factor distinct from these factors. First, transcription capability was restored to a cycloheximide-treated extract by RNA Pol III holoenzyme-derived TDF but not by purified preparations of natural TFIIIB, TFIIIC2, TFIIIC1, or core RNA Pol III. Second, in a system reconstituted solely with more purified components, transcription was dependent on TDF plus all the previously described components. Third, Western blot analyses with antibodies to the various TFIIIC2 and TFIIIB subunits failed to show any relationship with TDF (Z. Wang, Y. Hsieh, and R.G. Roeder, unpubl.). Fourth, neither RNA Pol III nor the previously characterized factors showed the same sensitivity to translation inhibition as did TDF activity. Nonetheless, although TDF is apparently distinct, we cannot exclude the formal possibility that it is a loosely associated subunit of one of the other factors. It might also be related to yeast transcription factor TFIIIE, which is reported to be required for transcription by highly purified yeast RNA Pol III, TFIIIC, and TFIIIB but not yet purified (Dieci et al. 1993; Ruth et al. 1996).
Regulation of human RNA Pol III-mediated transcription through TDF
Cycloheximide down-regulates both RNA Pol I- and RNA Pol III-mediated transcription (Gokal et al. 1986) and could involve regulatory mechanisms that are also used in more physiological situations (see introductory section). The down-regulation of RNA Pol III-mediated transcription by cycloheximide has been reported to be mediated by selective inactivation of TFIIIB (both TFIIIB70 and TFIIIB90 subunits) in yeast (Dieci et al. 1995), whereas the present data suggest that in human this is mediated through the novel TDF factor, rather than TFIIIB (at least not hTBP and hTFIIIB90). The inactivation of TDF as a consequence of protein synthesis inhibition suggests that it could serve as a point of regulation in more physiological processes. The present study indicates that TDF is inactivated in nuclear extracts from adenovirus-infected cells late in the lytic cycle, at which time there is a general decline in both host 5S and tRNA and viral VA RNA synthesis (Söderlund et al. 1976; Weinmann 1976). The observation that late-stage viral infection results in an inhibition of protein synthesis (O’Malley et al. 1986; Zhang et al. 1994; Schneider 1995) provides a simple explanation, namely TDF inactivation in the absence of continued protein synthesis, for the mechanism involved. Although the basis for the preferential loss of TDF activity during protein synthesis inhibition is not understood, this could reflect covalent modifications and/or proteolytic degradation; and preliminary studies have shown that the cycloheximide-mediated inactivation of TDF activity is largely prevented in the presence of an inhibitor (LLnL) of the ubiquitin-dependent proteasome pathway (Z. Wang, T. Luo, and R.G. Roeder, unpubl.). Should TDF prove to be critical for formation of holoenzyme and corresponding preinitiation complexes and should it be directly targeted within these complexes by the inactivation/degradation pathway, then our results would have significant implications for the unresolved problem of how the class III preinitiation complexes that are so stable in vitro (Bogenhagen et al. 1982; Lassar et al. 1983; Kassavetis et al. 1990; Wang and Roeder 1996) are inactivated (disassembled) in vivo.
Materials and methods
Extract preparation, in vitro transcription, and mobility retardation assays
Growth of BN51 cells (Wang and Roeder 1997), nuclear extract preparation (Dignam et al. 1983), and in vitro transcription assay conditions (Wang and Roeder 1995, 1997) with pVA1, pH2D (Santos and Zasloff 1981), pH5S8544, U6, and 7SK templates have been described. In transcription assays with Chromomycin A3, 0.2 μg of the pVA1 template was first incubated with 2 μl of nuclear extract (10.8 μg of protein) at 30°C for 30 min to form a preinitiation complex. This was followed by a 10-min incubation with Chromomycin A3 and a subsequent 30-min incubation in the presence of nucleoside triphosphates to allow transcription. In mobility retardation assays with Chromomycin A3, the TFIIIC2–DNA complex was formed by incubating the VA1 probe with TFIIIC2 for 30 min at 30°C (Wang and Roeder 1996) and was subsequently challenged with Chromomycin for 10 min.
Immunopurification of RNA Pol III holoenzyme
One milliliter of nuclear extract (5.4 mg of protein), prepared either from normal HeLa cells or BN51 cells, was adjusted to 100 mm KCl–0.05% NP-40 in BC buffer (Wang and Roeder 1995) by addition of 2.5 m KCl and 10% NP-40 and incubated with 50 μl of immobilized anti-FLAG antibody (M2 agarose) at 4°C for 3–6 hr with rotation. After washing five times with 1 ml of BC100–0.05% NP-40, proteins were eluted from the M2 agarose by incubation with 50 μl of BC100–0.1% NP-40 plus 0.2 μg/ml of FLAG peptide at 4°C for 30 min.
Gel-filtration chromatography and sucrose gradient centrifugation
The immunoprecipitate (50 μl) was loaded onto a Superose 6 column (3.2 × 300 mm, Pharmacia Smart System), and the column was eluted with BC100–0.01% NP-40 at a flow rate of 40 μl/min. Fractions of 100 μl were collected. High-range gel-filtration molecular weight markers (Pharmacia) were separated on the same column under the same conditions. Sucrose gradient centrifugation was performed as described previously (Wang and Roeder 1997). The immunopurified holoenzyme (0.2 ml) was loaded onto a 4.0-ml, 5%–20% sucrose gradient containing 0.5 m KCl in BC buffer (20 mm HEPES at pH 7.9, 20% glycerol, 0.5 mm EDTA, 1 mm DTT, 0.5 mm PMSF) and centrifuged for 24 hr at 4°C, 56,000 rpm in an SW60 rotor (Beckman). Fractions (0.2 ml) were collected from the bottom of the tube.
Cell treatment with cycloheximide
Exponentially growing normal HeLa cells or BN51 cells were treated for various periods of time with 100 μg/ml of cycloheximide, which reversibly blocks both protein synthesis and cell growth within a couple of hours (Gokal et al. 1986). Unless denoted otherwise, cells were treated with cycloheximide for 17 hr.
Infection of cells with Ad2
Ad2 was titrated on HeLa S3 monolayer cells by plaque assay (Hoeffler and Roeder 1985). Cell cultures were concentrated 10-fold in the absence of serum, and Ad2 was added at 100 plaque-forming units per cell. Virus was absorbed for 1 hr, after which fresh medium was added. Infections were allowed to proceed for different periods of time before the cells were harvested for preparation of nuclear extracts.
RNA Pol III factor isolation and reconstitution
TFIIIA (SDS-PAGE purified), TFIIIB [high-pressure liquid chromatography (HPLC) SP-5W column fraction], TFIIIC2 (B box oligo affinity column fraction), TFIIIC1 (FPLC Mono Q fraction), and chromatographically purified (sucrose gradient fraction) or immunopurified RNA Pol III were isolated as described (Wang and Roeder 1996, 1997). Core TFIIIB was reconstituted from recombinant TBP expressed in bacteria and recombinant TFIIIB90 expressed in Sf9 cells via baculovirus (Wang and Roeder 1995). A highly purified TFIIIC, which contains both TFIIIC1 and TFIIIC2, was isolated by procedures to be described elsewhere (Z. Wang and R.G. Roeder, unpubl.).
TDF activity, detected in a transcription complementation assay with nuclear extract prepared from cycloheximide-treated cells, is present mainly in phosphocellulose (P11) 0.6 m KCl fractions derived from HeLa S100 or nuclear extracts. The P11 0.6 m KCl fraction from S100 was dialyzed against BC100 for 3 hr and loaded onto an FPLC Mono Q column (HR10/10, Pharmacia). The column was eluted with 200 ml of a linear gradient of 100–600 mm KCl in BC buffer. Fractions containing TDF activity (eluted at 230 mm KCl) were pooled and loaded onto an HPLC heparin–5PW column (3.7 × 70 mm, Bio-Rad). The column was eluted with 60 ml of a linear gradient of 100–600 mm KCl in BC buffer. Fractions containing TDF activity were pooled, concentrated, and loaded onto an FPLC Superose 6 column (HR10/30) that was equilibrated and eluted with BC600–0.1% NP-40. The recoveries of TDF activity on Mono Q, heparin–5PW, and Superose 6 columns were 20%, 25%, and 25%, respectively. TDF was partially separated from TFIIIC1 on the Mono Q column (TDF eluted at 230 mm KCl, and TFIIIC1 eluted at 250 mm KCl; Wang and Roeder 1996) and completely separated from TFIIIC1 on an HPLC heparin–5PW column (TDF eluted at 200 mm KCl, and TFIIIC1 at 350 mm KCl). The TFIIIC1 activity was assayed in a reconstituted system containing highly purified TFIIIB, TFIIIC2, and RNA Pol III as described in Wang and Roeder (1996).
Acknowledgments
We thank Y. Hsieh for the recombinant TFIIIC63 protein and corresponding antibodies and Y. Tao for antibodies against human SRB7. T. Luo thanks B. Kaiser and the Rockefeller University Science Outreach Programs. This work was supported by a grant (CA42567) from the National Institutes of Health to R.G.R.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL roeder@rockvax.rockefeller.edu; FAX (212) 327-7949.
References
- Bai L, Wang Z, Yoon JB, Roeder RG. Cloning and characterization of the β subunit of human proximal sequence element-binding transcription factor and its involvement in transcription of small nuclear RNA genes by RNA polymerase II and III. Mol Cell Biol. 1996;16:5419–5426. doi: 10.1128/mcb.16.10.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- Björklund S, Kim YJ. Mediator of transcription regulation. Trends Biochem Sci. 1996;249:335–337. doi: 10.1016/s0968-0004(96)10051-7. [DOI] [PubMed] [Google Scholar]
- Bogenhagen DF, Wormington WM, Brown DD. Stable transcription complexes of Xenopus 5S RNA genes: A means to maintain the differentiated state. Cell. 1982;28:413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- Chao DM, Gadbois EL, Murray PJ, Anderson SF, Sonu MS, Parvin JD, Young RA. A mammalian SRB protein associated with an RNA polymerase II holoenzyme. Nature. 1996;380:82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- Chaussivert N, Conesa C, Shaaban S, Sentenac A. Complex interactions between yeast TFIIIB and TFIIIC. J Biol Chem. 1995;270:15353–15358. doi: 10.1074/jbc.270.25.15353. [DOI] [PubMed] [Google Scholar]
- Dean N, Berk AJ. Ordering promoter binding of class III transcription factors TFIIIC1 and TFIIIC2. Mol Cell Biol. 1988;8:3017–3025. doi: 10.1128/mcb.8.8.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G, Duimio L, Coda-Zabetta F, Sprague KU, Ottonello S. A novel RNA polymerase III transcription factor fraction that is not required for template commitment. J Biol Chem. 1993;268:11199–11207. [PubMed] [Google Scholar]
- Dieci G, Duimio L, Peracchia G, Ottonello S. Selective inactivation of two components of the multiprotein transcription factor TFIIIB in cycloheximide growth-arrested yeast cells. J Biol Chem. 1995;270:13476–13482. doi: 10.1074/jbc.270.22.13476. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielsen OS, Sentenac A. RNA polymerase III(C) and its transcription factors. Trends Biochem Sci. 1991;16:412–416. doi: 10.1016/0968-0004(91)90166-s. [DOI] [PubMed] [Google Scholar]
- Geiduschek EP, Kassavetis GA. Transcription Regulation. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 247–279. [Google Scholar]
- Gokal P, Cavanaugh AH, Thompson EA. The effects of cycloheximide upon transcription of rRNA, 5S RNA, and tRNA genes. J Biol Chem. 1986;261:2536–2541. [PubMed] [Google Scholar]
- Gottesfeld JM, Wolf VJ, Dang T, Forbes DJ, Hartl P. Mitotic repression of RNA polymerase III transcription in vitro mediated by phosphorylation of a TFIIIB component. Science. 1994;263:81–84. doi: 10.1126/science.8272869. [DOI] [PubMed] [Google Scholar]
- Halle JP, Meisterernst M. Gene expression: Increasing evidence for a transcriptosome. Trends Genet. 1996;12:161–163. doi: 10.1016/0168-9525(96)30035-8. [DOI] [PubMed] [Google Scholar]
- Hoeffler W, Roeder RG. Enhancement of RNA polymerase III transcription by E1A gene product of adenovirus. Cell. 1985;41:955–963. doi: 10.1016/s0092-8674(85)80076-3. [DOI] [PubMed] [Google Scholar]
- Kassavetis GA, Braun BR, Nguyen LH, Geiduschek EP. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- Keniry MA, Banville DL, Simmonds PM, Shafer R. Nuclear magnetic resonance comparison of the binding sites of Mithramycin and Chromomycin on the self-complementary oligonucleotide d(ACCCGGGT)2. J Mol Biol. 1993;231:753–767. doi: 10.1006/jmbi.1993.1324. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Björklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Koleske AJ, Young RA. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- ————— The RNA polymerase II holoenzyme and its implications for gene regulation. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- Lai JS, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar AB, Martin PL, Roeder RG. Transcription of class III genes: Formation of preinitiation complexes. Science. 1983;222:740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson CW, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- Maraia RJ. Transcription termination factor La activates RNA polymerase III and controls reinitiation. Proc Natl Acad Sci. 1996;93:3383–3387. doi: 10.1073/pnas.93.8.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure WR. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- McKune K, Moore PA, Hull MW, Woychik NA. Six human RNA polymerase subunits functionally substitute for their yeast counterparts. Mol Cell Biol. 1995;15:6895–6900. doi: 10.1128/mcb.15.12.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RP, Mariano TM, Siekierka J, Mathews MB. A mechanism for the control of protein synthesis by adenovirus VA RNA. Cell. 1986;44:391–400. doi: 10.1016/0092-8674(86)90460-5. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes & Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Ossipow V, Tassan JP, Nigg EA, Schibler U. A mammalian RNA polymerase II holoenzyme containing all components required for promoter-specific transcription initiation. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- Roeder RG. Nuclear RNA polymerase: Role of general initiation factors and cofactors in eukaryotic transcription. Methods Enzymol. 1996a;273:165–171. doi: 10.1016/s0076-6879(96)73016-1. [DOI] [PubMed] [Google Scholar]
- ————— The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996b;21:327–335. [PubMed] [Google Scholar]
- Ruth J, Conesa C, Dieci G, Lefebvre O, Dusterhoft A, Ottonello S, Sentenac A. A suppressor of mutations in the class III transcription system encodes a component of yeast TFIIIB. EMBO J. 1996;15:1941–1949. [PMC free article] [PubMed] [Google Scholar]
- Santos T, Zasloff M. Comparative analysis of human chromosomal segments bearing nonallelic dispersed tRNAimet genes. Cell. 1981;23:699–709. doi: 10.1016/0092-8674(81)90433-5. [DOI] [PubMed] [Google Scholar]
- Schneider RJ. Cap-independent translation in adenovirus infected cells. Curr Top Microbiol Immunol. 1995;203:117–129. doi: 10.1007/978-3-642-79663-0_6. [DOI] [PubMed] [Google Scholar]
- Scott MRD, Westphal KH, Rigby PWJ. Activation of mouse genes in transformed cells. Cell. 1983;34:557–567. doi: 10.1016/0092-8674(83)90388-4. [DOI] [PubMed] [Google Scholar]
- Shpakovski GV, Acker J, Wintzerith M, Lacroix JF, Thuriaux P, Vigneron M. Four subunits that are shared by three classes of RNA polymerase are functionally interchangeable between Homo sapiens and Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:4702–4710. doi: 10.1128/mcb.15.9.4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinn E, Wang Z, Kovelman R, Roeder RG. Cloning and characterization of a TFIIIC2 subunit (TFIIICb) whose presence correlates with activation of RNA polymerase III-mediated transcription by adenovirus E1A expression and serum factors. Genes & Dev. 1995;9:675–685. doi: 10.1101/gad.9.6.675. [DOI] [PubMed] [Google Scholar]
- Söderlund H, Pettersson U, Vennström B, Philipson L. A new species of virus-coded low molecular weight RNA from cells infected with adenovirus type 2. Cell. 1976;7:585–593. doi: 10.1016/0092-8674(76)90209-9. [DOI] [PubMed] [Google Scholar]
- Struhl K. Duality of TBP, the universal transcription factor. Science. 1994;263:1103–1104. doi: 10.1126/science.8108728. [DOI] [PubMed] [Google Scholar]
- Thompson CM, Young RA. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J, Sollner-Webb B. Polymerase III transcription factor B activity is reduced in extracts of growth-restricted cells. Mol Cell Biol. 1988;8:1001–1005. doi: 10.1128/mcb.8.2.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Roeder RG. Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2–related domains. Proc Natl Acad Sci. 1995;92:7026–7030. doi: 10.1073/pnas.92.15.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— TFIIIC1 acts through a downstream region to stabilize TFIIIC2 binding to RNA polymerase III promoters. Mol Cell Biol. 1996;16:6841–6850. doi: 10.1128/mcb.16.12.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Three human RNA polymerase III-specific subunits form a subcomplex and participate in specific transcription initiation. Genes & Dev. 1997;11:1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- Weinmann R. Low molecular weight viral RNAs transcribed by RNA polymerase III during adenovirus 2 infection. Cell. 1976;7:557–566. doi: 10.1016/0092-8674(76)90206-3. [DOI] [PubMed] [Google Scholar]
- Werner M, Chaussivert N, Willis IM, Sentenac A. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J Biol Chem. 1993;268:20721–20724. [PubMed] [Google Scholar]
- White RJ. RNA polymerase III transcription. Austin, TX: R.G. Landes Company; 1994. [Google Scholar]
- White RJ, Scott D, Rigby PWJ. Regulation of RNA polymerase III transcription in response to F9 embryonal carcinoma stem cell differentiation. Cell. 1989;59:1081–1093. doi: 10.1016/0092-8674(89)90764-2. [DOI] [PubMed] [Google Scholar]
- White RJ, Khoo BCE, Inostroza JA, Reinberg D, Jackson SP. Different regulation of RNA polymerase I, II, and III by the TBP-binding repressor Dr1. Science. 1994;266:448–450. doi: 10.1126/science.7939686. [DOI] [PubMed] [Google Scholar]
- Wingender E, Jahn D, Seifart KH. Association of RNA polymerase III with transcription factors in the absence of DNA. J Biol Chem. 1986;261:1409–1413. [PubMed] [Google Scholar]
- Woychik NA, Liao SM, Kolodziej PA, Young RA. Subunits shared by eukaryotic nuclear RNA polymerases. Genes & Dev. 1990;4:313–323. doi: 10.1101/gad.4.3.313. [DOI] [PubMed] [Google Scholar]
- Yoon J, Murphy S, Bai L, Wang Z, Roeder RG. Proximal sequence element-binding transcription factor (PTF) is a multisubunit complex required for transcription of both RNA polymerase II- and RNA polymerase III-dependent small nuclear RNA genes. Mol Cell Biol. 1995;15:2019–2027. doi: 10.1128/mcb.15.4.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga ST, Boulanger PA, Berk AJ. Resolution of human transcription factor TFIIIC into two functional components. Proc Natl Acad Sci. 1987;84:3585–3589. doi: 10.1073/pnas.84.11.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Feigenblum D, Schneider RJ. A late adenovirus factor induces eIF-4E dephosphorylation and inhibition of cell protein synthesis. J Virol. 1994;68:7040–7050. doi: 10.1128/jvi.68.11.7040-7050.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]