Abstract
Small organic molecules have proven to be invaluable tools for investigating biological systems, but there is still much to learn from their use. To discover and to use more effectively new chemical tools to understand biology, strategies are needed that allow us to systematically explore ‘biological-activity space’. Such strategies involve analysing both protein binding of, and phenotypic responses to, small organic molecules. The mapping of biological-activity space using small molecules is akin to mapping the stars — uncharted territory is explored using a system of coordinates that describes where each new feature lies.
To understand a system, you need to perturb it. This principle underlies most of the experimental sciences and explains why our depth of understanding of biological systems has been largely determined by the availability of tools that can be used to disrupt them. The development of molecular genetics in the twentieth century advanced our understanding of the molecules that control living systems. Now, molecular genetics allows investigators to eliminate specific proteins by ‘knocking out’ genes; to increase the concentrations of particular proteins by increasing the number of copies of the corresponding genes or by using a more active promoter on such genes; or to alter the function of a protein by introducing specific mutations in the corresponding gene1,2.
Although these methods have proved to be powerful in model organisms such as Saccharomyces cerevisiae and Drosophila melanogaster, mammals are more difficult to study using genetic-screening approaches because of their slower rates of reproduction, large physical sizes and large genomes. An alternative approach that has been gaining momentum in recent years is the use of small organic molecules instead of mutations. This approach is referred to as chemical genetics and is used to illuminate the molecular mechanisms underlying biological processes3–7. Because small molecules can alter the functions of proteins by binding to them and inhibiting or activating their normal functions, they can be used to perturb living systems and to reveal the molecular ‘wiring diagrams’ of these systems. There have been notable successes using this approach, although technical hurdles remain3,4.
The use of small molecules can complement gene-based methods of perturbing protein function, and in some cases, can offer advantages over such methods. For example, a protein may have several functions in a cell. In the case of a deletion mutation, all these functions are lost. However, it is possible to find small molecules that perturb only one of several functions of a protein, resulting in a level of understanding of protein function that would not be possible through gene-based perturbation8. In addition, it is easier to exert temporal control of protein function with small molecules because they can be added to induce an effect and later washed away to return a cell to its wild-type state. Finally, although most small molecules are not drugs, the occasional development of a small molecule into a drug can motivate researchers to use small-molecule tools to study biology.
To fully exploit the potential of chemical genetics, it will be necessary to create collections of small molecules that are suited to modulating the functions of many different proteins. However, each protein class generally requires a different type of small-molecule modulator. Thus, key aims should be to determine the full range of protein classes that occur in biology and to understand what type of small molecule interacts with each class. A similar argument can be made for determining the full range of phenotypes or observable properties of cells and organisms that occur in biology, given that the molecular basis of phenotypes is what we are ultimately hoping to understand. A central challenge facing the field of chemical genetics is therefore the mapping of ‘biological-activity space’, which involves analysing both protein binding of, and phenotypic responses to, small molecules. My aim here is to describe the challenges — including the design of synthetic chemicals, protein-binding and phenotypic assays, and ensuring quality control — that must be overcome to create a comprehensive map of biological-activity space using small molecules. Other systematic approaches to investigating biological systems, such as the use of RNA interference (RNAi), in which synthetic RNA fragments are designed to interfere with the expression of specific genes9, or antibodies10, are not covered here, but in many cases, could offer complementary information on systems of interest.
Assembling the ‘ideal’ chemical library
If small molecules are to be used as analogues of genetic mutations for studying mammalian systems, they must show the same generality as mutations. That is, they need to be applicable to the study of most or all proteins in an organism7. However, the specific chemical structure needed to bind to each protein is necessarily different: the requisite structure is determined by the shape of the available binding pockets on each protein. So, if we wish to create an ‘ideal’ chemical library for chemical genetics — one that contains a small-molecule ligand or binding partner for each protein — structures that bind to each protein need to be identified.
Of course, no existing chemical library contains compounds that bind selectively to every protein. Furthermore, there are many proteins for which no small-molecule ligand has yet been identified. Identifying new compounds with differing selectivities, or that bind to novel proteins, typically involves some type of screening experiment in which a library of compounds is assessed for the property of interest. Here, I focus primarily on understanding the biological effects of ‘active’ small molecules; that is, those molecules that possess a property of interest. A discussion of the screening approaches used to identify such molecules from the many that have no activity of interest is described in Box 1. The differences between high-throughput screening for modulators of a particular protein (a core activity of the pharmaceutical industry) and performing global analyses of the biological effects of a library of small molecules (a core activity in chemical genetics) are discussed in Box 2.
Box 1 Screening for new ligands.
When no ligand for a particular protein is known, screening of chemical libraries is often undertaken in the hope of identifying compounds that bind to the protein with reasonable affinity. Two distinct but complementary approaches can be applied: experimental (usually high-throughput; see Box 2) screening and structure-based virtual screening.
In one type of experimental screening, the protein is expressed and purified and used in a high-throughput screen to find small molecules that bind to it. This can be a time-consuming and expensive endeavour, and for many proteins it can fail to yield an effective ligand. Alternatively, in structure-based virtual screening, an atomic resolution structure of the protein is obtained using X-ray crystallography or nuclear magnetic resonance (NMR) spectroscopy. This protein structure is then used in a computer-based experiment to find small molecules predicted to bind to the protein. Using programs such as AutoDock, DOCK, FlexX, FRED, GOLD and Glide, millions of compounds can be examined in silico for their propensity to interact with the target protein, and the relative fit of each candidate scored66–70. This virtual screening approach has been used to generate ligands for casein kinase II using DOCK and SCORE71, and for the BCR–ABL oncoprotein using DOCK72. Although this is a useful emerging technology, current success rates are low because it is difficult to predict how small molecules will interact with a protein; there is flexibility in the torsion angles in both the protein and small molecule, causing uncertainty regarding the three-dimensional structure of both. Improvements in the predictive accuracy of such programs will affect virtual screening, and so the discovery of novel protein ligands.
Although these two approaches to ligand discovery are distinct, they can be used together to enhance the chances of finding an active compound. In particular, within the pharmaceutical industry, the use of virtual screening as a ‘filter’ to select compounds from very large virtual libraries for experimental screening has become increasingly common. This filtering process can use various types of information (for example, the crystal structures of the protein itself), with the aim of enriching the library that is experimentally screened with ‘active’ structures. Furthermore, computational filters can also be used to remove compounds that have inappropriate properties from the screening library, as discussed in Box 3. A review of this topic is given in ref. 73.
Box 2 High-throughput screens versus global analyses.
In a high-throughput screen, many different chemicals (or other test reagents) are evaluated in the same biological test for their effects on a protein or cellular process. The term ‘screen’ is used to indicate that many different chemicals are tested but only a small number of them are expected to be active. The term ‘high-throughput’ is used to indicate that many chemicals are put through this process in a short period of time. There are, however, two types of analyses that can be performed on large data sets: screens and global analyses. Both approaches involve collecting a large amount of data on the effects of specific compounds or other reagents in the same assay. However, the goals of the two approaches differ: screens seek simply to identify several active reagents that can be investigated further in subsequent experiments, whereas global analyses seek to draw meaningful conclusions regarding all the reagents that were tested in the screen. Thus, a high rate of false negatives and false positives can be tolerated in a screen because as long as a few true positives can ultimately be confirmed, the screen is successful. Unfortunately, the same is not true for global analyses, which require low false-positive and false-negative rates for the data to be meaningfully interpreted. Those setting out to perform a global analysis would be wise to consider performing numerous replicates of each compound at several concentrations. It is perhaps only a slight exaggeration to state that academic scientists frequently wish to understand fundamental property relationships between structure and activity, whereas industry scientists often seek to identify a few lead compounds that can be pursued as drug candidates. Nonetheless, the goals of individual researchers performing large numbers of chemical tests differ, and therefore the required data quality, the necessary number of tests and the properties of the tested compounds will be different for each investigator.
Whether the goal is to find a ligand for a particular protein, or to use the global effects of a library to elucidate biological processes, the composition of the library used in the screening experiment is a key factor. Libraries can be assembled from available compounds or synthesized de novo. In practice, there are two types of chemical libraries that can be synthesized today: ‘focused libraries’ and ‘diversity-oriented libraries’11,12 (Fig. 1). Focused libraries are designed around a specific piece of a small molecule, known as a scaffold, and are used to target a specific class of proteins. Often, such scaffolds may be chemically related to endogenous ligands for particular protein classes. Recent examples of focused libraries include those targeted against G-protein coupled receptors (GPCRs)13, proteases14, phosphatases15 and kinases16. In contrast, diversity-oriented libraries are not targeted to any specific protein class and are often used in broad screens in which the target proteins are not known. Because the goal of diversity-oriented synthesis (DOS) is to create a maximally diverse collection of compounds, the synthetic planning algorithms required are distinct from those used to create single compounds or focused libraries17,18. Recent examples of DOS include the synthesis of tricyclic compounds using Ferrier and Pauson–Khand reactions with a glycal template19, and the synthesis of tetrahydroquinoline20 and hydroxyindole21 derivatives.
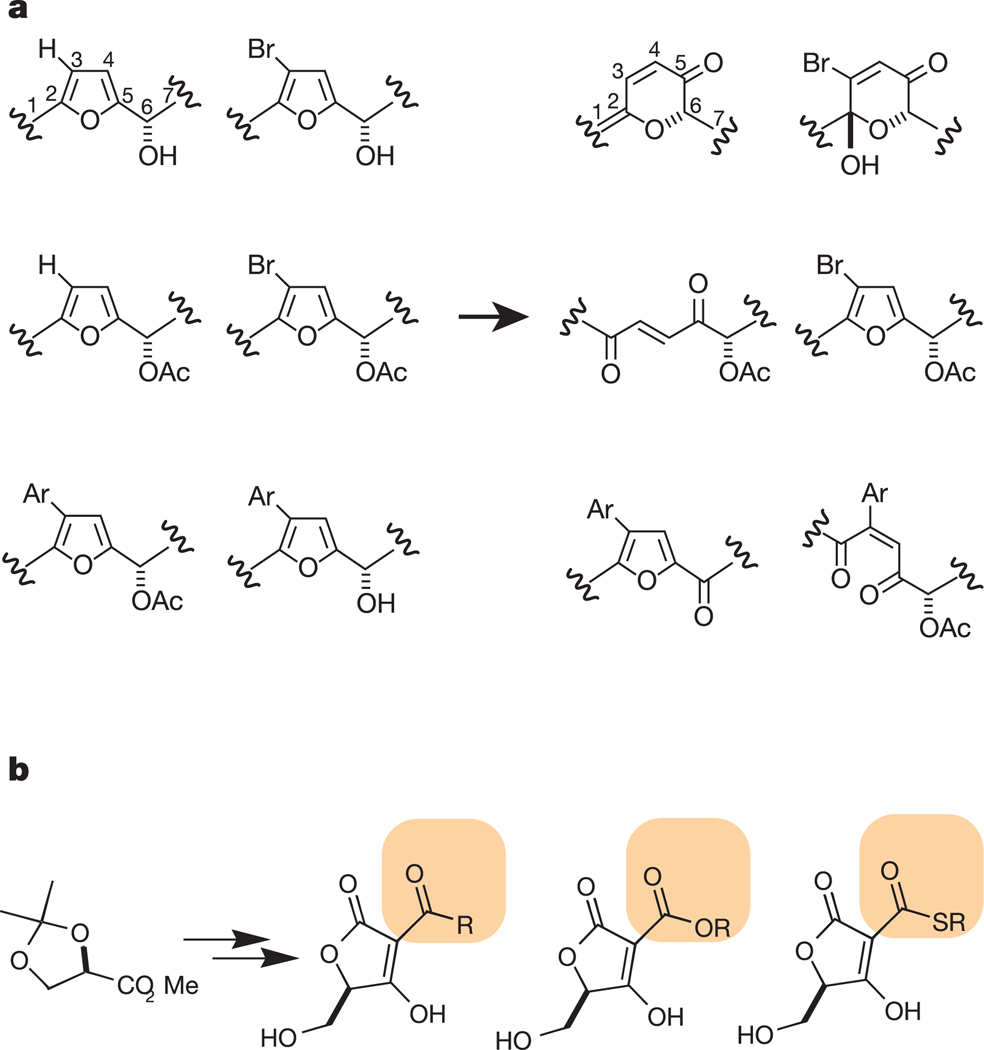
Figure 1.
Comparison of diversity-oriented synthesis (DOS) and focused library synthesis (FLS). a, The goal of DOS is to create collections of compounds that are maximally diverse, thereby increasing the probability that different proteins will be targeted by different compounds in the library. In the example shown, Burke et al65 created a library of compounds with different core structures (skeletons) starting from a common set of precursors (left). The six compounds on the right have different connectivity and are likely to interact with different proteins. b, The goal of FLS is to create analogues of the same core structure to optimize binding to a target or class of targets. If the compounds created are too diverse, they may lose their propensity to interact with the designated target protein. In this example, Sodeoka et al15 created a collection of acyltetronic acids that act as phosphate mimetics and so are likely to inhibit phosphatases. Their synthesis resulted in a library of compounds that are identical except for the portion highlighted in orange.
Each approach to chemical-library design has its advantages and disadvantages. Compounds in focused libraries are more likely than random compounds to be active, but they only target proteins in a known class. Diversity-oriented libraries, in contrast, offer the possibility of targeting entirely new classes of proteins, but any individual compound has a lower probability of activity. The pharmaceutical industry, being justifiably risk-averse, has moved towards the use of focused libraries. Practitioners argue that fewer compounds of greater quality and with a greater probability of becoming drugs are more valuable than larger libraries with compounds that are not likely to become drugs (see Box 3 for a discussion of additional factors considered by the pharmaceutical industry when assembling screening libraries, some of which could also be important for libraries for chemical genetics). Some academic groups, however, without the same constraints of industry, are pursuing higher-risk strategies centred on diversity-oriented approaches. The two approaches are ultimately complementary: a ligand to a new protein class discovered from a diversity-oriented library can serve as the basis for a future focused library that explores the structure–function relationships for compounds targeting this new class of proteins.
Box 3 Additional factors to consider in library design.
A number of other properties of small molecules are important to their use as a tool or potential drug, in addition to their ability to bind potently and specifically to particular protein targets. Such properties include their ability to cross biological membranes, to be substrates for drug efflux pumps in cells, their chemical stability, and their solubility in water and dimethyl sulphoxide (a common organic solvent). There has been much interest in the pharmaceutical industry in engineering such ‘drug-like’ properties and discarding candidate compounds that are unlikely to be effective drugs, even before they are synthesized. The most widely used of these drug-like property rules are those formulated by Lipinski et al., who compared the computed properties of marketed drugs with those of non-drugs74.
In recent years, there has been a trend towards creating libraries of compounds that are predicted to be lead-like’ rather than drug-like. This is in recognition of the fact that as a compound progresses from being a drug lead to an actual drug, its properties tend to change in a consistent way: drugs are typically larger and more hydrophobic than leads75. This reflects the practical fact that medicinal chemists tend to add chemical matter rather than remove it during lead optimization. Better predictions of drug-like and lead-like properties will have an important impact on the creation of both drug candidates and chemical tools; chemical tools also need to be soluble, stable and able to penetrate across biological membranes.
More effective chemical libraries for chemical genetics would contain compounds that affect specific proteins and phenotypes but not other closely related proteins and phenotypes. These compounds should also collectively affect a diverse range of proteins and phenotypes. The design of more effective libraries would be aided by assessments of the specificity and diversity of existing libraries, and of each new chemical library as it is designed and synthesized. This would mean that optimal libraries for a given purpose could be rationally assembled from members of other libraries.
Chemical-diversity analysis is routinely carried out today using commercial software packages that catalogue the diversity of structures present in a library (Box 4). But more relevant is the diversity of biological activities shown by a library of compounds. For example, consider a library of ten compounds that have dramatically different structures but that all bind to the protein tubulin: this is a library with significant chemical diversity, but minimal diversity of biological activity. Although there is often a correlation between chemical diversity and the diversity of biological activity, there is not a simple one-to-one correspondence.
Box 4 Calculating chemical diversity.
Small organic molecules come in all shapes and sizes. The diversity of a library is a quantitative description of how different these compounds are from each other. Consider library A with ten compounds that all look identical except for the nature of one side-chain, compared to library B with ten compounds that have dramatically different sizes and shapes. Intuitively, most people agree that library A is in some way less diverse than library B. However, to be rigorous it is necessary to specify the attributes that are more or less diverse in these two libraries. For example, if we were to calculate the range of molecular masses in the two libraries and to find that library A has molecular masses that range from 300 to 350 daltons but that library B has molecular masses that range from 200 to 500 daltons, we could say that in terms of molecular mass, library B covers six times the range of molecular masses in library A. Similarly, we could calculate the differences in the ranges of other properties, such as charge, number of atoms, number of rotatable bonds and so on. Such properties, called descriptors, can readily be calculated using commercially available software. These descriptors allow for a quantitative description of chemical diversity. Unfortunately, an additional complication is that diversity of chemical structure does not necessarily imply diversity of biological activity. Finding descriptors for biological activity is necessary to describe the diversity of biological activities for compounds present in a library.
To assess the biological-activity diversity of a compound library, it is necessary to evaluate the range of biological activities shown by the library. This involves parameterizing ‘biological-activity space’, or creating ‘metrics’ that characterize the activity and specificity of each compound in a library. Protein-binding is a useful metric because many small molecules exert their biological effects by interacting with specific proteins in cells. Phenotypic activity is also useful to measure because ultimately we are interested in understanding how protein binding relates to phenotypic changes.
Indeed, such approaches have been implemented by several groups. Kauvar et al.22 reported a protein-affinity map of ‘chemical space’ and showed that the pattern of protein binding by small molecules can be used to cluster compounds. Greenbaum et al.23 used a similar approach, which they termed affinity fingerprinting. They used this approach to characterize the affinity of a library of peptidic epoxides for numerous proteases and thus to group these proteases by reactivity. Finally, Weinstein et al.24 used an analogous approach with a phenotypic assay. By measuring the effects of compounds on the proliferation of a panel of 60 tumour cell lines, Weinstein and colleagues24 discovered that compounds with similar structures or similar mechanisms of action had similar phenotypic profiles (that is, inhibited the growth of a similar set of tumour cell lines). In the remainder of this review, I will consider the status of the methods available for further exploring ‘biological-activity space’ and consider some of the key challenges inherent in this endeavour.
Protein-binding assays
Methods have been created to measure the ability of small molecules to bind to specific proteins25 (Table 1). In recent years, there has been a trend towards testing the specificity of a compound for binding one protein relative to related proteins of the same class (for example, kinases)26,27. Such protein-binding assays can be divided into two types: those that use labelled compounds and those that are label-free (a label is a fluorescent or radioactive group that is added to a test compound). Although labels make protein–ligand interactions easier to observe, they can also be difficult to introduce into a compound, which increases the time and expense associated with measuring protein binding. A brief description of the main assay formats of each type can be found in Table 1, together with references that contain further information on each type of assay.
Table 1.
Methods for measuring the affinity of small molecules for proteins
| Method | Label | Throughput | Phase | Protein amount | Description | References |
|---|---|---|---|---|---|---|
| Fluorescence polarization | Yes | High | Solution | High | Measures the change in tumbling rate for a compound when it is bound to a protein using loss-of-polarization of incident light. |
57 |
| Fluorescence perturbation | No | Low | Solution | High | Measures the change in fluorescence (usually of a tryptophan residue) on a protein caused by proximity of a bound compound. |
58 |
| Fluorescence correlation spectroscopy |
Yes | High | Solution | Medium | Measures the correlated changes in fluorescence properties that occur when a labelled compound is bound to a labelled protein. |
59 |
| Radioligand binding assay | Yes | High | Solid | High | Measures retention of a compound on a surface holding a protein. Detection is by means of a radioactive isotope that is incorporated into the compound. |
60 |
| Nuclear magnetic resonance |
No | Low | Solution | High | Measures changes in the nuclear spin of protons on a protein that occur when a compound is bound nearby. |
61 |
| Mass spectrometry | No | Medium | Solution | Medium | Measures the change in molecular weight observable in a mass spectrometer when a compound is bound to a protein. |
62 |
| Surface plasmon resonance |
No | Low/medium | Solid | High | Measures the change in refractivity of a metal surface when a compound binds to a protein that is immobilized on that surface. |
63 |
| Isothermal titration calorimetry |
No | Low | Solution | High | Measures the very small amount of heat produced when a compound binds to a protein. |
64 |
| Differential scanning calorimetry |
No | Low | Solution | High | An alternative method of measuring the heat released when a compound binds to a protein. |
64 |
| Small-molecule microarray | Yes | High | Solid | Medium | Compounds are immobilized on a surface. A protein is incubated with the surface and localizes to where the protein-interacting compounds are bound. |
28–30 |
| Protein microarray | Yes | High | Solid | Low/none | Proteins are immobilized on a surface and a labelled compound is incubated with the surface. The compound localizes to where the compound-binding proteins are bound. |
34 |
| Three-hybrid system | Yes | High | In cells | None | A compound is covalently linked to an anchor compound that interacts with a DNA-binding protein. If the test compound can bind to a test protein that is fused to a DNA activation domain, transcription of a reporter gene is initiated. |
41 |
Although methods are available for measuring the binding of a small molecule to a protein or to a handful of related proteins, few methods systematically measure the binding of small molecules to hundreds or thousands of proteins (Box 2). Such high-throughput protein-binding measurements are required if we are to capture the range of activities shown by small molecules. Label-free detection methods are preferred because they do not require the extra synthetic chemistry involved in introducing a label, and because introducing a label may change the properties of a molecule. However, such measurements can be more difficult to perform: without a label, a larger amount of both protein and compound must often be produced, and the instruments used for label-free measurements are slow (Table 1).
Recent attempts to create high-throughput assays for measuring protein–ligand interactions require the use of labels. One class of high-throughput assay involves immobilizing each test compound on a surface and then incubating these immobilized compounds with a soluble labelled protein28–30. Many compounds can be immobilized side by side on a surface, so this method can measure thousands of protein–small-molecule interactions (Fig. 2a). Kuruvilla et al.8 used this technique successfully to screen 3,780 compounds for those that bind to the transcriptional repressor Ure2p, and found a compound that disrupts one of the functions of Ure2p.
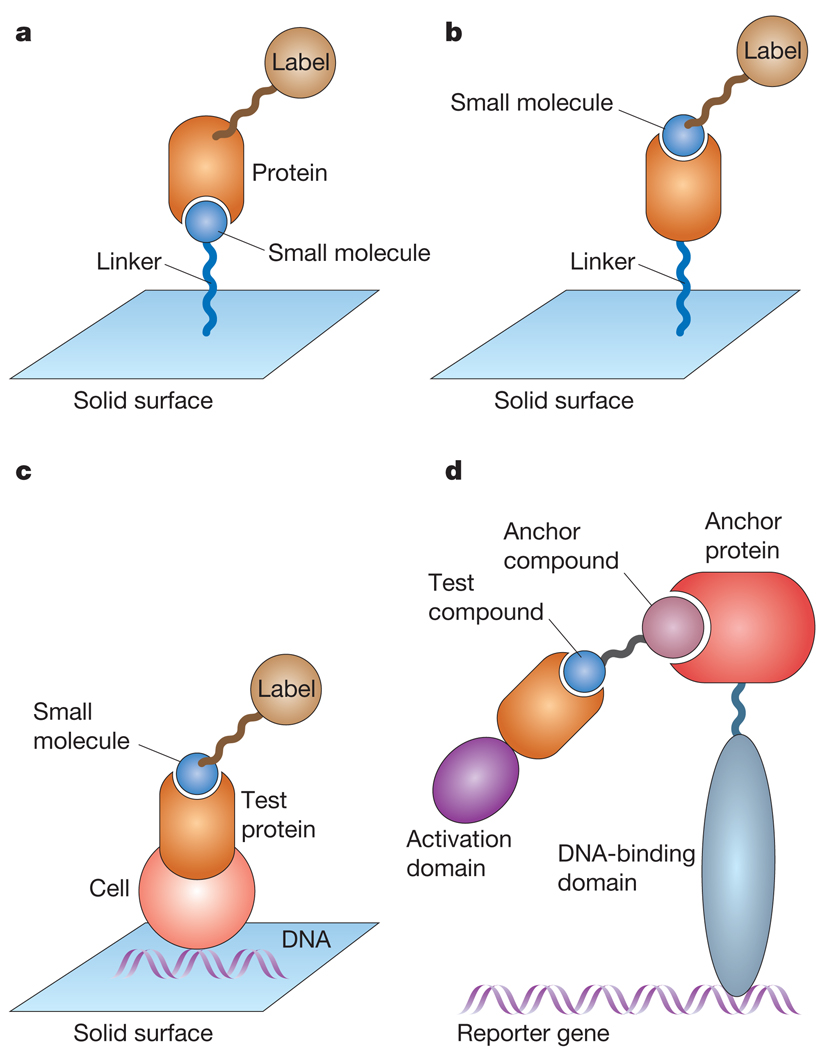
Figure 2.
High-throughput-assay formats for detecting small molecule-protein interactions. a, Small molecules can be covalently linked to a surface. Meanwhile, a test protein in solution is brought into contact with the surface. The protein binds to small molecules on the surface with high affinity. If the protein is tagged with a label, these interactions can be detected. b, Proteins can similarly be immobilized on a surface and brought into contact with a labelled small molecule in solution. High-affinity interactions between the small molecule and specific proteins can then be detected by imaging the locations to which the small molecule binds. c, DNA expression plasmids can be arrayed on a surface and cells subsequently plated on top of these expression plasmids. The cells take up the DNA and produce the proteins encoded by each plasmid. Thus, this method allows for the creation of a microarray of cells that overexpress defined proteins. When a labelled compound is brought into close proximity of the array, it localizes to where cells are overexpressing these high-affinity compound-binding proteins. d, Yeast three-hybrid system. Transcription factors that regulate gene expression can be divided into DNA-binding domains and transcription-activation domains. It is possible to fuse the complementary DNA sequence of a DNA-binding domain to the cDNA of an anchor protein that interacts with a known small molecule (anchor compound). The anchor compound is then chemically fused to a new test compound. If the cDNA of an activation domain is fused to the cDNA of a test protein, it is possible to determine whether the test protein interacts with the test compound with high affinity by determining whether transcription of a reporter gene has been activated.
A related method involves immobilizing compounds on a surface and then detecting the binding of a protein to each compound using surface plasmon resonance31 (Table 1). These surface-based methods can be useful for measuring the ability of many compounds to bind to one or several proteins. For example, Birkert et al. used such a method to measure the binding of immobilized triazines to antibodies and to screen 384 compounds for those that act as thrombin inhibitors32,33.
It is possible to invert these surface-based methods and to immobilize thousands of proteins side by side on a surface34,35. A small molecule with a label, such as a fluorescent or radioactive group, can be applied to the surface, washed away and detected by measuring the remaining label (Fig. 2b). Some applications of protein microarrays include the use of an array of most yeast proteins to assess the global pattern of protein activities found in yeast cells36, the discovery of novel protein–protein interactions in human cells37 and an analysis of interactions between human 49 leucine zipper transcription factors38.
A variation of this technology involves creating arrays of expression plasmids, which encode the information required to produce each protein of interest. Creating DNA arrays has become routine in the past decade and is preferable to creating arrays of proteins directly, primarily because DNA can be amplified and because thousands of different DNA-expression constructs will have similar chemical properties (solubility, stability, and so on). In contrast, thousands of different proteins will show idiosyncratic properties that are unique to each protein. It is possible to either place cells on this DNA array and cause proteins to be produced inside the cells39, or to use a cell lysate (produced from cells that have been broken open) to produce an array of proteins in vitro (Fig. 2c)40. In either case, the net result is a protein array without the added complication of purifying and immobilizing each protein. However, post-translational modifications and protein complexes that are physiologically relevant will not be captured in these formats. So far, only proof-of-principle experiments have been performed with these more recent technologies.
A final high-throughput method for measuring the binding of many proteins to one or more small molecules also has the advantage of not requiring protein purification. This is the three-hybrid system, which is typically carried out in yeast or bacterial cells41. In such systems, a test protein is fused to the activation domain of a transcription activator, and the test small molecule is synthetically linked to an ‘anchor’ compound that will interact with a protein containing a DNA-binding domain (Fig. 2d). So, if the test small molecule is able to interact with the test protein, the transcription activation domain will be brought into close proximity with the DNA-binding domain, and expression of the reporter gene that is controlled by the system will be activated. This method was used succesfully by Liberles et al.42 to create a mutant version of the FKBP-rapamycin binding domain (FRB), which binds to a modified, non-toxic version of rapamycin.
Although several high-throughput methods have been developed for measuring protein–ligand interactions, many desirable features are not found in these systems. First, measuring the binding of small molecules to target proteins in solution is preferable to using a surface-based method that may interfere with protein–ligand binding. Unfortunately, most high-throughput methods involve immobilizing either the ligand or the protein on a solid surface to allow parallel processing of all samples with a single solution. Second, it is better to avoid the use of labels on both small molecules and proteins because of the added time and expense needed to introduce such labels into thousands of compounds or proteins, and because the labels may change the activity of the compound or protein. Third, it is easier to use only minute quantities of protein, or better still, to manipulate only the corresponding DNA sequences and allow the system to produce the desired proteins in situ. This obviates the need to purify many proteins, each with their own solubility requirements. Fourth, it would be useful to have a system that is ‘scalable’, both in terms of the binding of small molecules and of the proteins; ideally, it should be possible to automate the detection of the binding of thousands of proteins to thousands of ligands without the need for idiosyncratic modifications to the system for each ligand or each protein. Finally, all these technologies require significant investments in capital equipment and knowledge bases, which limit their adoption by many users. Thus, although each of these problems may ultimately be solved, significant barriers will prevent the widespread adoption of these technologies in the near future.
Phenotypic outcomes
In assessing the biological activities of small molecules, it is useful to consider not only protein binding but also phenotypic effects. Cellular phenotypes that are affected by small molecules include varied phenomena, such as cell death, cell migration, cell proliferation, gene expression, vesicle sorting and axonal sprouting. Organismal phenotypes affected include body weight, tumour formation, joint inflammation and the capacity for learning and memory, among many others. In fact, although there is a finite number of proteins within a given organism, theoretically an infinite number of phenotypes may be assessed for an organism. Given the infinite number of phenotypes that can exist, phenotypic assays performed for library-assessment purposes need to be prioritized in some way. Usually, this prioritization is based on ease of measurement.
It is useful to consider how phenotypic measurements can be automated and undertaken in a high-throughput fashion to characterize the biological activity and specificity of chemical libraries. Most phenotypic measurements cannot be performed in high-throughput simply because they involve time-consuming measurements that use whole organisms, such as mice, worms, flies or zebrafish. In fact, measuring the effect of a single compound on a phenotype in mice typically involves several months of work and costs tens of thousands of dollars. For example, my laboratory recently discovered a compound, indoprofen, which has potential relevance to the pediatric genetic disease spinal muscular atrophy (SMA)43. To test this compound on mouse SMA phenotypes, we needed to evaluate potential routes of administration, achievable concentrations in the plasma, brain and in utero embryos given various doses, and toxicity and teratogenicity in pregnant mice. Although we found that this compound had a modest effect at extending the survival of embryos with an SMA genotype, such assays are expensive and time-consuming to perform. Such phenotypic measurements can be valuable for specific compounds of interest but they are not compatible with assessing the activity and specificity of large compound libraries. For this purpose, high-throughput phenotypic assays are needed (Fig. 3).
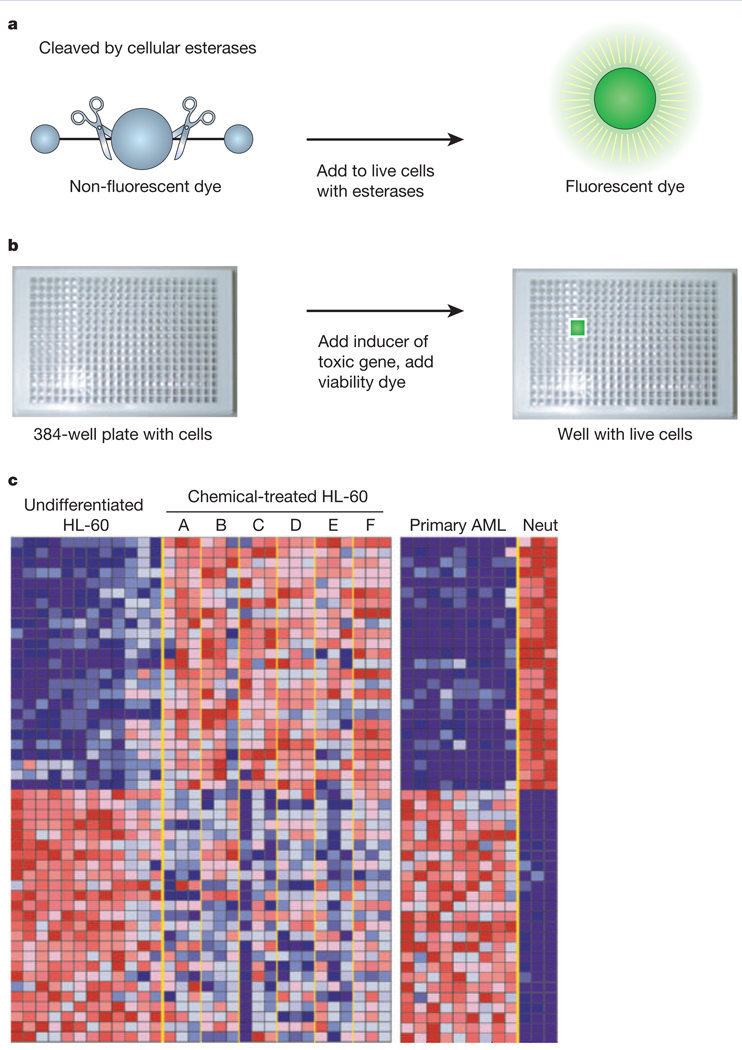
Figure 3.
Examples of high-throughput phenotypic screens. These are measurements of properties of cells that can be performed in a parallel fashion and so allow for the testing of many different chemicals at once. a, Fluorescence-based viability can be used to measure the number of living cells in a miniaturized test tube. The non-fluorescent dye calcein acetoxymethyl ester, shown schematically in blue, can be cleaved by intracellular esterases to create a fluorescent compound (shown in green). b, Such a dye can be used to measure the number of live cells in 384-well plates, which hold 384 individual miniature chambers for growing cells. For example, if a toxic gene is introduced, cells will die unless they are treated with a chemical that is able to prevent this cell death. In this example, the wells holding cells treated with such a chemical are bright green because the viability dye becomes fluorescent on being cleaved by esterases from live cells. c, A pattern of gene expression can be used as a signature of the state of a cell. In this example by Stegmeier et al47, gene-expression signatures were obtained for: (1) human neutrophil precursors (HL-60 tumour cells, left) that have failed to differentiate and have become tumour cells; (2) primary acute myelogenous leukaemia (AML) cells from patients (right); and (3) differentiated human neutrophils (Neut, far right). A screen was performed to identify compounds that convert the signature of the HL-60 tumour cell line into the signature of differentiated neutrophils, with the goal of rendering the HL-60 tumour cells non-tumorigenic. Six compounds (of approximately 2,000 tested) were found to induce this switch in gene signatures (labelled ‘Chemical-treated HL-60, A to F’). Each row in this table shows the expression level of a different gene under these different conditions (the columns). The colour indicates whether expression in the sample is high (red) or low (blue). The six compounds shown revert the gene-expression pattern of HL-60 tumour cells to that of differentiated neutrophils.
A number of high-throughput phenotypic assays have been developed, including assays that measure cell viability or proliferation3–5. Such assays measure the presence of intact cell membranes, the abundance of cellular energy (ATP concentration), or the presence of cellular reductases or esterases, which are found in nearly all cells. Such viability assays have been extended to the analysis of synthetic lethal effects: a compound is tested for its ability to kill cells in the presence, but not in the absence, of a defined element, such as another compound or a gene of interest44. Identifying compounds that have genotype-selective activity is of interest both because such compounds can be developed into safer drugs with fewer side effects and because they can reveal the molecular consequences of oncogenic mutations in tumour cells. Moreover, viability assays can be used to search for chemical suppressors; a compound is tested for its ability to prevent the lethality of another compound or a toxic gene product. For example, Wang and Dreyfuss45 screened for compounds that prevent the cell death that occurs when the survival motor neuron (SMN)-gene protein is eliminated from mammalian cells. Similarly, Aiken et al.46 screened for compounds that prevent apoptotic cell death caused by the mutant huntingtin protein in PC12 cells.
Recently, gene-expression signatures have been developed into high-throughput, phenotypic assays47. In this approach, a gene-expression profile is measured using DNA microarrays for two cell states of interest, such as undifferentiated neutrophil (a type of granular white blood cell) precursors and differentiated neutrophils. Then the profiles are compared and a gene signature is created which determines whether the cell is in one state or the other. By measuring the effects of small molecules on the appearance of this gene signature, it is possible to determine whether each compound changes the cell state (for example, induces differentiation of neutrophil precursors into neutrophils).
Another emerging trend in high-throughput phenotypic assays involves imaging cells using an automated microscope48. Such an approach allows for the detection of phenotypes that can be measured using microscopy. For example, Yarrow et al.49 recently used an imaging-based screen to identify compounds that affect cell migration during wound healing; Kau et al.50 used this technique to screen for compounds that prevent nuclear export of FOXO transcription factors. Image-analysis algorithms then allow for the automated processing of these images so that conclusions regarding the effects of compounds on these phenotypes can be extracted. Imaging-based phenotypes could allow for the digitization and clustering of otherwise unrelated phenotypes. Because any image consists of a series of pixels with distinct values, the relationship between any two images can be quantified mathematically.
Finally, the concentration of a particular messenger RNA or protein, such as the SMN protein, can represent a phenotype of interest. For example, patients with the disease SMA have a low SMN protein phenotype. Finding mechanisms and compounds that convert these cells to producing abundant SMN protein is of interest. This concept of molecular phenotypes can be extended to include the measurements of thousands of proteins or mRNAs simultaneously. The global pattern of these proteins or mRNAs represents a quantifiable state of a cell. Thus, measuring the abundance of thousands of proteins, mRNAs or metabolites can be used to create cell signatures or phenotype measurements. Unfortunately, it is not yet feasible to perform such global measurements of protein, mRNA or metabolite abundance in high-throughput. Moreover, some phenotypes do not involve significant transcriptional changes, whereas others do not involve significant changes in protein or metabolite concentrations. New methods for automating and rapidly performing such measurements would be of value.
Creation and use of biological-activity matrices
After collecting a large amount of data on the ability of the members of a chemical library to bind to a set of proteins and affect a set of phenotypes, the data can be analysed to determine the relationship between chemical structure and biological activity. Each compound can be assigned a vector that describes the quantitative level of binding to each protein, and the quantitative effect this has on each phenotype. Comparing these parameters for different libraries could reveal how specific scaffolds and functional elements influence specificity and diversity. Figure 4 shows an example of how a compound might be evaluated for its ability to bind to nine different kinases. Although this evaluation has not been performed, it should be straightforward to do so.
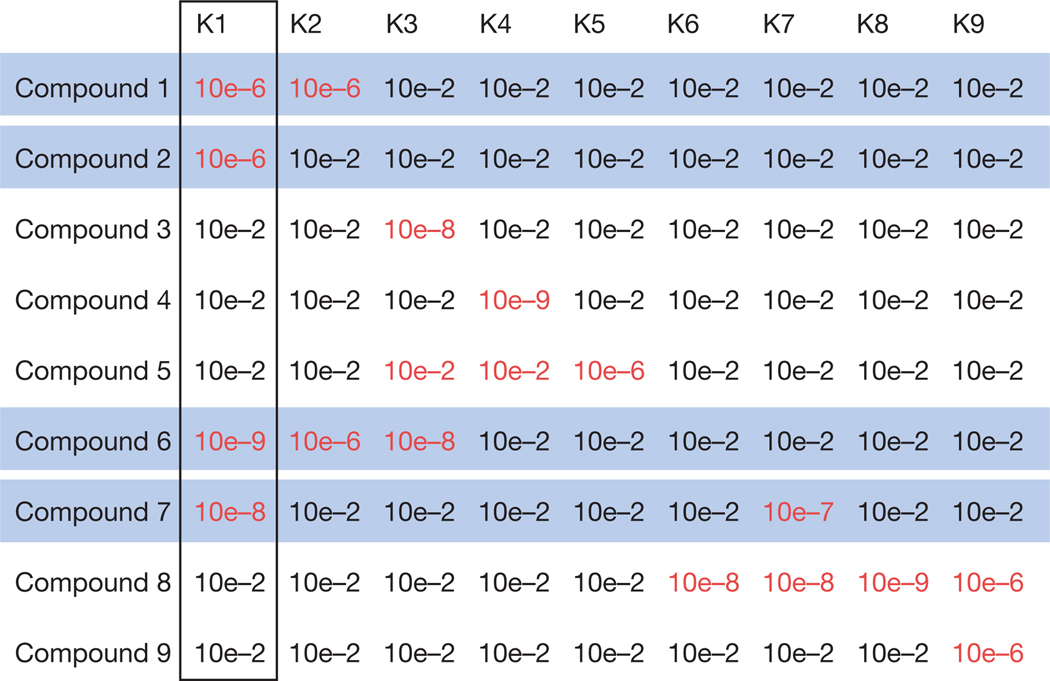
Figure 4.
Using biological-activity matrices to determine the proteins that regulate phenotypes. A hypothetical activity matrix for a library of nine kinase inhibitors. Each row lists the affinity (that is, the equilibrium dissociation constant, written in scientific notation, where 10e – 6 represents 0.000001 M) of one compound for each of nine different kinase proteins. Smaller numbers indicate higher affinity. The affinities less than or equal to 10e – 6 are highlighted in red because these correspond to high-affinity compounds for these targets. The kinase proteins are labelled K1 to K9. The same affinity matrix can be used to determine which kinases are involved in specific biological processes. In this hypothetical example, if the four compounds highlighted in blue are all capable of inhibiting the growth of a tumour cell line, the K1 kinase is probably responsible for the ability of these compounds to inhibit the growth of this cell line: this is the only kinase to be targeted by all four compounds.
Such data sets can be used to generate hypotheses regarding the molecular mechanisms underlying biological phenotypes51. For example, if each compound in a library has been annotated with a pattern of protein-binding activity, then it is possible to determine whether binding to any specific protein is correlated with the ability to induce a phenotypic change. In validating such an approach, Root et al.51 rediscovered that small molecules that bind to tubulin are highly likely to inhibit tumour-cell proliferation. This approach can be extended to targets other than proteins: Root et al.51 also found that compounds that bind to small ions, such as potassium, are able to selectively inhibit the proliferation of lung tumour cells relative to other cells. By annotating compound libraries with high-quality target binding and phenotypic profiles, it is possible to extract information regarding the molecules that regulate these phenotypes.
Further challenges
Specificity of small molecules
One limitation of small molecules is their frequent lack of specificity for a single target protein. This can be problematic when using small molecules both as therapeutic agents and as chemical probes: a lack of specificity can lead to unexpected toxicity, preventing the development of an otherwise promising compound into a drug, and can also confound interpretation of the effects of a compound. This problem of non-specificity is often dose-dependent: at higher concentrations, compounds interact with additional proteins. In addition, specific functional groups and scaffolds have been found to be promiscuous, in the sense that they allow binding to a wide range of proteins or non-specific killing of a wide-range of cell types52. Such chemical functions need to be identified and removed from future library designs.
There are several strategies for overcoming the problem of specificity. First, it is preferable to identify and use potent compounds (that is, compounds that are likely to modulate a target protein at low nanomolar or picomolar concentrations) because at such low concentrations they are less likely to affect other proteins. Second, measuring the binding specificity of compounds in the type of large-scale protein-binding assays described above should identify some of the alternative protein targets of compounds. Third, it is always critical to confirm the putative mechanism of action of a compound using either additional compounds or other reagents, such as small interfering RNAs (siRNAs)53,54. Although the phenotypic consequences of an RNAi reagent and a small molecule targeting the corresponding mRNA are not always the same, their effects are often sufficiently similar to make this comparison useful. RNAi itself can lack specificity, and it is necessary to test numerous RNAi reagents designed against a target mRNA sequence55. Finally, a large collection of RNAi reagents can be a useful tool for high-throughput screens9. By using such collections, it should eventually be possible to measure the phenotypic consequences of turning off expression of each gene in an organism.
Building redundancy into a set of probe molecules is an effective way of dealing with the problem of specificity. That is, it is desirable to have not just one compound that inhibits each protein, but rather dozens of compounds that inhibit each protein. If inhibition of protein X causes phenotype Y, we would expect — in an ideal world — all the small molecules in our collection that inhibit protein X to cause phenotype Y. In the real world, not every protein-X inhibitor will be effective, because some will bind protein X in slightly different ways or be metabolized differently in different cell types. Nonetheless, our confidence that the modulation of protein X causes phenotype Y should be proportional to the percentage of our protein-X inhibitors that cause phenotype Y. Thus, the problem of specificity can be overcome by assembling a sufficiently redundant set of probe compounds: even if no single compound is specific for one target protein, the collection as a whole contains the requisite information on the effects of modulating each target protein.
Finally, given that compounds have different specificities at different concentrations, it would be preferable to collect information on the effects of each compound at multiple concentrations; a full dose–response curve for each compound would be ideal. Unfortunately, the added time and expense associated with collecting this additional information usually makes it impractical. Therefore, new technologies that allow an increase in the number of tests performed per unit time would be valuable. Alternatively, a smaller number of compounds may be tested with more replicates and a full dose–response curve. This trade off between the number of compounds tested and the quality and completeness of the data set collected for each compound needs to be optimized in each project.
Quality control
When collecting large-scale data sets, attention to quality control is crucial. However, there is an inherent trade off between the level of throughput and data quality in large-scale data collection. A minimum level of quality is necessary to ensure that reliable conclusions are extracted from such data sets. However, attention to data quality has not been a priority for many researchers engaged in high-throughput chemical screens, simply because the data quality required for a screen is much lower than the data quality required for a global analysis56 (Box 2).
In addition, it is important to eliminate artefacts through the use of counter screens for properties that could interfere with the assay readout, such as intrinsic compound fluorescence or compound aggregation. In general, a counter screen is performed on the compounds that emerge from an initial screen, and compounds that are active in the counter screen are not taken further. For example, in a screen that uses the fluorescent dye calcein as a detection method (Fig. 3), any compound that shows the same colour of fluorescence as calcein will appear to be a positive compound from the screen; a counter screen would involve testing each compound for its intrinsic fluorescence to eliminate those compounds that were falsely active because of this property.
Finally, it is important to assess the solubility and stability of each tested compound or protein, and to confirm that the chemical being tested is the desired one. Solubility can be measured using nephelometry, which detects insoluble particles in solution, and compound identities can be confirmed using liquid chromatography and mass spectrometry. All these methods of improving data quality increase the time and expense associated with large-scale data collection but are crucial if meaningful conclusions are to be drawn.
Outlook
Designing better tools with which to perturb biological systems requires a systematic evaluation of the properties of existing tools. Although large-scale measurements of the effects of small molecules on proteins and phenotypes can be challenging, the resulting data sets can be useful in probing biological-activity diversity. New ways to increase the complexity and sophistication of the phenotypic assays and protein-binding measurements that can be performed on vast arrays of molecules will prove valuable. Moreover, more comprehensive and effective compound libraries will allow us to perturb an increasing percentage of the macromolecules that make up living systems. In so doing, we may move closer to understanding the roles of the diverse molecules that are responsible for life, death and disease.
Acknowledgements
B.R.S. is supported in part by a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Footnotes
Competing interests statement The author declares that he has no competing financial interests.
References
- 1.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartwell LH. Twenty-five years of cell cycle genetics. Genetics. 1991;4:975–980. doi: 10.1093/genetics/129.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stockwell BR. Chemical genetics: ligand-based discovery of gene function. Nature Rev. Genet. 2000;1:116–125. doi: 10.1038/35038557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stockwell BR. Frontiers in chemical genetics. Trends Biotechnol. 2000;18:449–455. doi: 10.1016/s0167-7799(00)01499-2. [DOI] [PubMed] [Google Scholar]
- 5.Stockwell BR. Chemical genetic screening approaches to neurobiology. Neuron. 2002;36:559–562. doi: 10.1016/s0896-6273(02)01056-5. [DOI] [PubMed] [Google Scholar]
- 6.Schreiber SL. The small-molecule approach to biology: chemical genetics and diversity-oriented organic synthesis make possible the systematic exploration of biology. Chem. Eng. News. 2003;81:51–61. [Google Scholar]
- 7.Schreiber SL. Chemical genetics resulting from a passion for synthetic organic chemistry. Bioorg. Med. Chem. 1998;6:1127–1152. doi: 10.1016/s0968-0896(98)00126-6. [DOI] [PubMed] [Google Scholar]
- 8.Kuruvilla FG, Shamji AF, Sternson SM, Hergenrother PJ, Schreiber SL. Dissecting glucose signalling with diversity-oriented synthesis and small-molecule microarrays. Nature. 2002;416:653–657. doi: 10.1038/416653a. [DOI] [PubMed] [Google Scholar]
- 9.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 10.Moore P, Clayton J. To affinity and beyond. Nature. 2003;426:725–731. doi: 10.1038/426725a. [DOI] [PubMed] [Google Scholar]
- 11.Schreiber SL. Target-oriented and diversity-oriented organic synthesis in drug discovery. Science. 2000;287:1964–1969. doi: 10.1126/science.287.5460.1964. [DOI] [PubMed] [Google Scholar]
- 12.Young SS, Ge N. Design of diversity and focused combinatorial libraries in drug discovery. Curr. Opin. Drug Discov. Dev. 2004;7:318–324. [PubMed] [Google Scholar]
- 13.Jimonet P, Jager R. Strategies for designing GPCR-focused libraries and screening sets. Curr. Opin. Drug Discov. Dev. 2004;7:325–333. [PubMed] [Google Scholar]
- 14.Reid RC, et al. Countering cooperative effects in protease inhibitors using constrained beta-strand-mimicking templates in focused combinatorial libraries. J. Med. Chem. 2004;47:1641–1651. doi: 10.1021/jm030337m. [DOI] [PubMed] [Google Scholar]
- 15.Sodeoka M, et al. Synthesis of a tetronic acid library focused on inhibitors of tyrosine and dual-specificity protein phosphatases and its evaluation regarding VHR and cdc25B inhibition. J. Med. Chem. 2001;44:3216–3222. doi: 10.1021/jm0100741. [DOI] [PubMed] [Google Scholar]
- 16.Stahura FL, Xue L, Godden JW, Bajorath J. Molecular scaffold-based design and comparison of combinatorial libraries focused on the ATP-binding site of protein kinases. J. Mol. Graph Model. 1999;17:1–9. doi: 10.1016/s1093-3263(99)00015-7. 51–2. [DOI] [PubMed] [Google Scholar]
- 17.Burke MD, Schreiber SL. A planning strategy for diversity-oriented synthesis. Angew. Chem. Int. Edn Engl. 2004;43:46–58. doi: 10.1002/anie.200300626. [DOI] [PubMed] [Google Scholar]
- 18.Spring DR. Diversity-oriented synthesis; a challenge for synthetic chemists. Org. Biomol. Chem. 2003;1:3867–3870. doi: 10.1039/b310752n. [DOI] [PubMed] [Google Scholar]
- 19.Kubota H, Lim J, Depew KM, Schreiber SL. Pathway development and pilot library realization in diversity-oriented synthesis: exploring Ferrier and Pauson-Khand reactions on a glycal template. Chem. Biol. 2002;9:265–276. doi: 10.1016/s1074-5521(02)00099-6. [DOI] [PubMed] [Google Scholar]
- 20.Couve-Bonnaire S, Chou DT, Gan Z, Arya PA. solid-phase, library synthesis of natural-product-like derivatives from an enantiomerically pure tetrahydroquinoline scaffold. J. Comb. Chem. 2004;6:73–77. doi: 10.1021/cc030026x. [DOI] [PubMed] [Google Scholar]
- 21.Arya P, Wei CQ, Barnes ML, Daroszewska M. A solid phase library synthesis of hydroxyindoline-derived tricyclic derivatives by Mitsunobu approach. J. Comb. Chem. 2004;6:65–72. doi: 10.1021/cc0340067. [DOI] [PubMed] [Google Scholar]
- 22.Kauvar LM, Villar HO, Sportsman JR, Higgins DL, Schmidt DEJ. Protein affinity map of chemical space. J. Chromatog. B. 1998;715:93–102. doi: 10.1016/s0378-4347(98)00045-0. [DOI] [PubMed] [Google Scholar]
- 23.Greenbaum DC, et al. Small molecule affinity fingerprinting. A tool for enzyme family subclassification, target identification, and inhibitor design. Chem. Biol. 2002;9:1085–1094. doi: 10.1016/s1074-5521(02)00238-7. [DOI] [PubMed] [Google Scholar]
- 24.Weinstein JN, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- 25.Lakey JH, Raggett EM. Measuring protein-protein interactions. Curr. Opin. Struct. Biol. 1998;8:119–123. doi: 10.1016/s0959-440x(98)80019-5. [DOI] [PubMed] [Google Scholar]
- 26.Gray NS, et al. Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science. 1998;281:533–538. doi: 10.1126/science.281.5376.533. [DOI] [PubMed] [Google Scholar]
- 27.Salemme FR. Chemical genomics as an emerging paradigm for postgenomic drug discovery. Pharmacogenomics. 2003;4:257–267. doi: 10.1517/phgs.4.3.257.22692. [DOI] [PubMed] [Google Scholar]
- 28.MacBeath G, Koehler AN, Schreiber SL. Printing small molecules as microarrays and detecting protein-ligand interactions en masse. J. Am. Chem. Soc. 1999;121:7967–7968. [Google Scholar]
- 29.Winssinger N, Ficarro S, Schultz PG, Harris JL. Profiling protein function with small molecule microarrays. Proc. Natl Acad. Sci. USA. 2002;99:11139–11144. doi: 10.1073/pnas.172286899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falsey JR, Renil M, Park S, Li S, Lam KS. Peptide and small molecule microarray for high throughput cell adhesion and functional assays. Bioconjug. Chem. 2001;12:346–353. doi: 10.1021/bc000141q. [DOI] [PubMed] [Google Scholar]
- 31.Vetter D. Chemical microarrays, fragment diversity, label-free imaging by plasmon resonance—a chemical genomics approach. J. Cell Biochem. 2002;39 suppl.:79–84. doi: 10.1002/jcb.10408. [DOI] [PubMed] [Google Scholar]
- 32.Birkert O, Tunnemann R, Jung G, Gauglitz G. Label-free parallel screening of combinatorial triazine libraries using reflectometric interference spectroscopy. Anal. Chem. 2002;74:834–840. doi: 10.1021/ac0106952. [DOI] [PubMed] [Google Scholar]
- 33.Birkert O, Gauglitz G. Development of an assay for label-free high-throughput screening of thrombin inhibitors by use of reflectometric interference spectroscopy. Anal. Bioanal. Chem. 2002;372:141–147. doi: 10.1007/s00216-001-1196-4. [DOI] [PubMed] [Google Scholar]
- 34.Jona G, Snyder M. Recent developments in analytical and functional protein microarrays. Curr. Opin. Mol. Ther. 2003;5:271–277. [PubMed] [Google Scholar]
- 35.MacBeath G. Protein microarrays and proteomics. Nature Genet. 2002;32 suppl.:526–532. doi: 10.1038/ng1037. [DOI] [PubMed] [Google Scholar]
- 36.Zhu H, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 37.Espejo A, Cote J, Bednarek A, Richard S, Bedford MT. A protein-domain microarray identifies novel protein-protein interactions. Biochem. J. 2002;367:697–702. doi: 10.1042/BJ20020860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- 39.Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001;411:107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 40.Ramachandran N, et al. Self-assembling protein microarrays. Science. 2004;305:86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- 41.Lefurgy S, Cornish V. Finding Cinderella after the ball: a three-hybrid approach to drug target identification. Chem. Biol. 2004;11:151–153. doi: 10.1016/j.chembiol.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 42.Liberles SD, Diver ST, Austin DJ, Schreiber SL. Inducible gene expression and protein translocation using nontoxic ligands identified by a mammalian three-hybrid screen. Proc. Natl Acad. Sci. USA. 1997;94:7825–7830. doi: 10.1073/pnas.94.15.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lunn MR, et al. Indoprofen upregulates the survival motor neuron protein through a cyclooxygenase-independent mechanism. Chem. Biol. 2004;11:1495–1503. doi: 10.1016/j.chembiol.2004.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–296. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 45.Wang J, Dreyfuss G. A cell system with targeted disruption of the SMN gene: functional conservation of the SMN protein and dependence of Gemin2 on SMN. J. Biol. Chem. 2001;276:9599–9605. doi: 10.1074/jbc.M009162200. [DOI] [PubMed] [Google Scholar]
- 46.Aiken CT, Tobin AJ, Schweitzer ES. A cell-based screen for drugs to treat Huntington’s disease. Neurobiol. Dis. 2004;16:546–555. doi: 10.1016/j.nbd.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 47.Stegmaier K, et al. Gene expression-based high-throughput screening(GE-HTS) and application to leukaemia differentiation. Nature Genet. 2004;36:257–263. doi: 10.1038/ng1305. [DOI] [PubMed] [Google Scholar]
- 48.Kapur R. Fluorescence imaging and engineered biosensors: functional and activity-based sensing using high content screening. Ann. NY Acad. Sci. 2002;961:196–197. doi: 10.1111/j.1749-6632.2002.tb03081.x. [DOI] [PubMed] [Google Scholar]
- 49.Yarrow JC, Perlman ZE, Westwood NJ, Mitchison TJ. A high-throughput cell migration assay using scratch wound healing, a comparison of image-based readout methods. BMC Biotechnol. 2004;4:21. doi: 10.1186/1472-6750-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kau TR, et al. A chemical genetic screen identifies inhibitors of regulated nuclear export of a Forkhead transcription factor in PTEN-deficient tumor cells. Cancer Cell. 2003;4:463–476. doi: 10.1016/s1535-6108(03)00303-9. [DOI] [PubMed] [Google Scholar]
- 51.Root DE, Flaherty SP, Kelley BP, Stockwell BR. Biological mechanism profiling using an annotated compound library. Chem. Biol. 2003;10:881–892. doi: 10.1016/j.chembiol.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 52.Seidler J, McGovern SL, Doman TN, Shoichet BK. Identification and prediction of promiscuous aggregating inhibitors among known drugs. J. Med. Chem. 2003;46:4477–4486. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- 53.Tuschl T. Expanding small RNA interference. Nature Biotechnol. 2002;20:446–448. doi: 10.1038/nbt0502-446. [DOI] [PubMed] [Google Scholar]
- 54.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 55.Lassus P, Rodriguez J, Lazebnik Y. Confirming specificity of RNAi in mammalian cells. Sci. STKE. 2002;147:PL13. doi: 10.1126/stke.2002.147.pl13. [DOI] [PubMed] [Google Scholar]
- 56.Root DE, Kelley BP, Stockwell BR. Global analysis of large-scale chemical and biological experiments. Curr. Opin. Drug Discov. Dev. 2002;5:355–360. [PMC free article] [PubMed] [Google Scholar]
- 57.Burke TJ, Loniello KR, Beebe JA, Ervin KM. Development and application of fluorescence polarization assays in drug discovery. Comb. Chem. High Throughput Screen. 2003;6:183–194. doi: 10.2174/138620703106298365. [DOI] [PubMed] [Google Scholar]
- 58.Timasheff SN, Andreu JM, Na GC. Physical and spectroscopic methods for the evaluation of the interactions of antimitotic agents with tubulin. Pharmacol. Ther. 1991;52:191–210. doi: 10.1016/0163-7258(91)90008-a. [DOI] [PubMed] [Google Scholar]
- 59.Bulseco DA, Wolf DE. Fluorescence correlation spectroscopy: molecular complexing in solution and in living cells. Methods Cell Biol. 2003;72:465–498. doi: 10.1016/s0091-679x(03)72022-6. [DOI] [PubMed] [Google Scholar]
- 60.Misra R. Modern drug development from traditional medicinal plants using radioligand receptor-binding assays. Med. Res. Rev. 1998;18:383–402. doi: 10.1002/(sici)1098-1128(199811)18:6<383::aid-med3>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 61.Hicks RP. Recent advances in NMR: expanding its role in rational drug design. Curr. Med. Chem. 2001;8:627–650. doi: 10.2174/0929867013373237. [DOI] [PubMed] [Google Scholar]
- 62.Siegel MM. Early discovery drug screening using mass spectrometry. Curr. Top. Med. Chem. 2002;2:13–33. doi: 10.2174/1568026023394551. [DOI] [PubMed] [Google Scholar]
- 63.Homola J. Present and future of surface plasmon resonance biosensors. Anal. Bioanal. Chem. 2003;377:528–539. doi: 10.1007/s00216-003-2101-0. [DOI] [PubMed] [Google Scholar]
- 64.Jelesarov I, Bosshard HR. Isothermal titration calorimetry and differential scanning calorimetry as complementary tools to investigate the energetics of biomolecular recognition. J. Mol. Recogn. 1999;12:3–18. doi: 10.1002/(SICI)1099-1352(199901/02)12:1<3::AID-JMR441>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 65.Burke MD, Berger EM, Schreiber SL. Generating diverse skeletons of small molecules combinatorially. Science. 2003;302:613–618. doi: 10.1126/science.1089946. [DOI] [PubMed] [Google Scholar]
- 66.Oprea TI, Matter H. Integrating virtual screening in lead discovery. Curr. Opin. Chem. Biol. 2004;8:349–358. doi: 10.1016/j.cbpa.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 67.Ewing TJ, Makino S, Skillman AG, Kuntz ID. DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J. Comput. Aided Mol. Des. 2001;15:411–428. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- 68.Osterberg F, Morris GM, Sanner MF, Olson AJ, Goodsell DS. Automated docking to multiple target structures: incorporation of protein mobility and structural water heterogeneity in AutoDock. Proteins. 2002;46:34–40. doi: 10.1002/prot.10028. [DOI] [PubMed] [Google Scholar]
- 69.Kramer B, Rarey M, Lengauer T. Evaluation of the FLEXX incremental construction algorithm for protein-ligand docking. Proteins. 1999;37:228–241. doi: 10.1002/(sici)1097-0134(19991101)37:2<228::aid-prot8>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 70.Halgren TA, et al. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 71.Vangrevelinghe E, et al. Discovery of a potent and selective protein kinase CK2 inhibitor by high-throughput docking. J. Med. Chem. 2003;46:2656–2662. doi: 10.1021/jm030827e. [DOI] [PubMed] [Google Scholar]
- 72.Peng H, et al. Identification of novel inhibitors of BCR-ABL tyrosine kinase via virtual screening. Bioorg. Med. Chem. Lett. 2003;13:3693–3699. doi: 10.1016/j.bmcl.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 73.Bajorath J. Integration of virtual and high-throughput screening. Nature Rev. Drug Discov. 2002;1:882–894. doi: 10.1038/nrd941. [DOI] [PubMed] [Google Scholar]
- 74.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 75.Hann MM, Oprea TI. Pursuing the leadlikeness concept in pharmaceutical research. Curr. Opin. Chem. Biol. 2004;8:255–263. doi: 10.1016/j.cbpa.2004.04.003. [DOI] [PubMed] [Google Scholar]