Abstract
The Wnt pathway regulates the early dorsal–ventral axis in Xenopus through a complex of β-catenin and HMG box transcription factors of the Lef/Tcf family. We show that the promoter of the dorsalizing homeo box gene siamois is a direct target for the β-catenin/XTcf-3 complex, establishing a link between the Wnt pathway and the activation of genes involved in specifying the dorsal axis. By injecting siamois reporter constructs into the animal pole of Xenopus embryos, we show that a 0.8-kb fragment of the siamois promoter is strongly activated by β-catenin. The proximal 0.5 kb, which is also activated by β-catenin, contains three Lef/Tcf-binding sites. Mutations in these sites eliminate the β-catenin-mediated activation of siamois and show that siamois is regulated by the β-catenin/XTcf-3 complex, in combination with additional transcriptional activators. When expressed at the equator of the embryo, the siamois promoter is activated to much higher levels on the dorsal side than the ventral side. Ectopic ventral expression of β-catenin raises the ventral expression of the siamois promoter to the dorsal levels. Conversely, ectopic dorsal expression of dominant-negative XTcf-3 abolishes the dorsal activation of the siamois promoter. Furthermore, elimination of the Lef/Tcf sites elevates the ventral expression of siamois, revealing a repressive role for XTcf-3 in the absence of β-catenin. Finally, we find that the endogenous siamois activator, although present throughout the dorsal side of the embryo, is most potent in the dorsal vegetal region. We propose that the dorsal activation of siamois by the β-catenin/XTcf-3 complex combined with the ventral repression of siamois by XTcf-3 results in the restriction of endogenous siamois expression to the dorsal side of Xenopus embryos.
Keywords: Xenopus, homeobox, HMG box, dorsoventral patterning, Wnt, Lef/Tcf, siamois
Specification of the dorsal–ventral axis is a crucial early patterning event in vertebrate development, yet the molecular mechanisms that initiate this process in the embryo remain elusive. In Xenopus laevis, dorsal axis formation begins shortly after fertilization when the radial symmetry of the egg is broken by a rotation of the cortical cytoplasm relative to the inner cytoplasm (Gerhart et al. 1989). This cortical rotation repositions maternal factors from the vegetal pole to the future dorsal side of the embryo (Fujisue et al. 1993; Kikkawa et al. 1996; Sakai 1996; Rowning et al. 1997), establishing a dorsal signaling center that induces the Spemann organizer at the dorsal equator of the embryo. Intriguingly, prefertilization vegetal cortical cytoplasm has Wnt-like activity (Holowacz and Elinson 1995), consistent with previous demonstrations that members of the Wnt family can induce a dorsal axis in Xenopus embryos (McMahon and Moon 1989).
The identity and position of proteins within the Wnt signaling pathway was first established through genetic analysis of the homologous wingless (wg) pathway in Drosophila (Siegfried et al. 1994). Studies of the vertebrate Wnt pathway have characterized many of the biochemical mechanisms involved in transducing the signal from the cell-surface to the nucleus (Miller and Moon 1996). In Xenopus, Wnt signaling is thought to repress the activity of glycogen synthase kinase-3 (Xgsk-3) (Dominguez et al. 1995; He et al. 1995; Pierce and Kimelman 1995), which negatively regulates β-catenin stability by directly phosphorylating it and/or phosphorylating the APC protein (Rubinfeld et al. 1996; Yost et al. 1996). Artificially raising β-catenin levels on the ventral side of Xenopus embryos, either by inactivating Xgsk-3 or by ectopically expressing β-catenin, results in the formation of a second dorsal axis (Dominguez et al. 1995; Funayama et al. 1995; He et al. 1995; Pierce and Kimelman 1995). Conversely, depletion of β-catenin transcripts from Xenopus oocytes with synthetic antisense oligonucleotides inhibits dorsal axis formation (Heasman et al. 1994). A recent study analyzing the location and subcellular distribution of the endogenous protein has found elevated levels of β-catenin on the dorsal side of Xenopus embryos as early as the four-cell stage (Larabell et al. 1997). Notably, some of the β-catenin is found in the nucleus (Funayama et al. 1995; Schneider et al. 1996; Yost et al. 1996; Larabell et al. 1997), suggesting that Wnt signaling could modify gene expression by regulating the level of nuclear β-catenin.
The discovery that β-catenin interacts with high mobility group (HMG) box transcription factors of the Lef/Tcf class explains how β-catenin could affect transcription. Lef and Tcf were originally identified as factors binding to lymphocyte enhancers (Travis et al. 1991; van de Wetering et al. 1991; Waterman et al. 1991) but have recently been identified as components of the Drosophila wg pathway (Brunner et al. 1997; van de Wetering et al. 1997). In Xenopus, several Tcfs (called XTcf-3, b, c, d) are present as maternal transcripts that are not localized within the early embryo (Molenaar et al. 1996). In cell culture, both Lef-1 and XTcf-3 form a complex with β-catenin (Behrens et al. 1996; Huber et al. 1996; Molenaar et al. 1996). Dominant-negative mutants of both Lef-1 and XTcf-3 disrupt dorsal development indicating that these proteins are required for normal axial formation in Xenopus (Behrens et al. 1996; Molenaar et al. 1996). A major unresolved issue is the identity of the targets of the β-catenin/XTcf-3 complex in dorsal axis formation. The promoter of the Spemann organizer gene goosecoid (gsc) has a Wnt-responsive element (Watabe et al. 1995), however, it neither contains a Lef/Tcf consensus binding site nor does it bind Lef-1 in vitro (M. Gomperts and R.T. Moon, unpubl.). These results suggest that another transcription factor is an intermediate between β-catenin/XTcf-3 and gsc.
We have focused on the Xenopus homeo box gene siamois as a potential downstream target of β-catenin. siamois is expressed in the dorsal vegetal cells soon after the start of zygotic transcription and ectopic siamois expression induces complete axis duplication as does ectopic expression of Wnt and β-catenin (McMahon and Moon 1989; Funayama et al. 1995; Lemaire et al. 1995). Unlike gsc, siamois can be induced by Wnt and β-catenin in the absence of transforming growth factor β (TGF-β) signaling (Brannon and Kimelman 1996; Carnac et al. 1996; Yang-Snyder et al. 1996; Fagotto et al. 1997), and it can be induced by β-catenin in the absence of cell–cell contact (Brannon and Kimelman 1996). These results suggest that siamois might be directly regulated by a β-catenin/XTcf-3 complex.
We report here the isolation of the siamois promoter and show that it is a direct target for the β-catenin/XTcf-3 complex. The promoter contains a β-catenin-responsive element within 300 bp of the start of transcription and several enhancing elements contained within a 0.8-kb fragment. The β-catenin-responsive element contains three Lef/Tcf-binding sites that are required for the response to β-catenin. Activation of the siamois promoter is strongest in the dorsal vegetal region of embryos and this activation is dependent on XTcf-3. In addition, we show that the Lef/Tcf sites are involved in repressing siamois expression on the ventral side of Xenopus embryos. Our results provide the first link between the β-catenin/XTcf-3 complex and the transcriptional activation of a gene involved in dorsal axis specification.
Results
Functional domains of the siamois promoter
To understand the molecular mechanism of β-catenin-induced transcription during dorsal–ventral axis formation in Xenopus, we have studied the siamois promoter. Primer extension analysis indicated that siamois has a short 5′-untranslated region (UTR) and no introns within the first 233 bp of siamois coding sequence (data not shown). By use of a probe from the first 246 bp of the siamois cDNA, three recombinant phage were isolated from a Xenopus genomic DNA library. Nucleotide sequence analysis showed that all three clones contain a region exactly matching the published siamois cDNA sequence (Lemaire et al. 1995; data not shown).
A DNA fragment from the largest clone was inserted into a luciferase reporter vector to assay for transcriptional activity. As a preliminary analysis indicated that this fragment was β-catenin-responsive (data not shown), a 5′ nested deletion series was generated in an attempt to identify regions of the siamois promoter containing cis-acting regulatory elements that respond to β-catenin (Fig. 1A). Our previous work had shown that β-catenin, like Wnt (Carnac et al. 1996; Yang-Snyder et al. 1996; Fagotto et al. 1997), is a potent inducer of the endogenous siamois gene in animal cap assays (Brannon and Kimelman 1996). Thus, to measure the response of the siamois promoter, the deletion constructs were injected into the animal pole of Xenopus embryos at the two-cell stage, in the presence and absence of synthetic β-catenin RNA. Injected embryos were cultured beyond the mid-blastula transition (MBT; stage 8), which marks the onset of zygotic transcription, and harvested at stage 10, when endogenous siamois expression peaks (Lemaire et al. 1995). Whole embryos were used to assay the level of luciferase activity induced by β-catenin as experiments with isolated animal caps or whole embryos showed no difference in background levels of luciferase activity (data not shown).
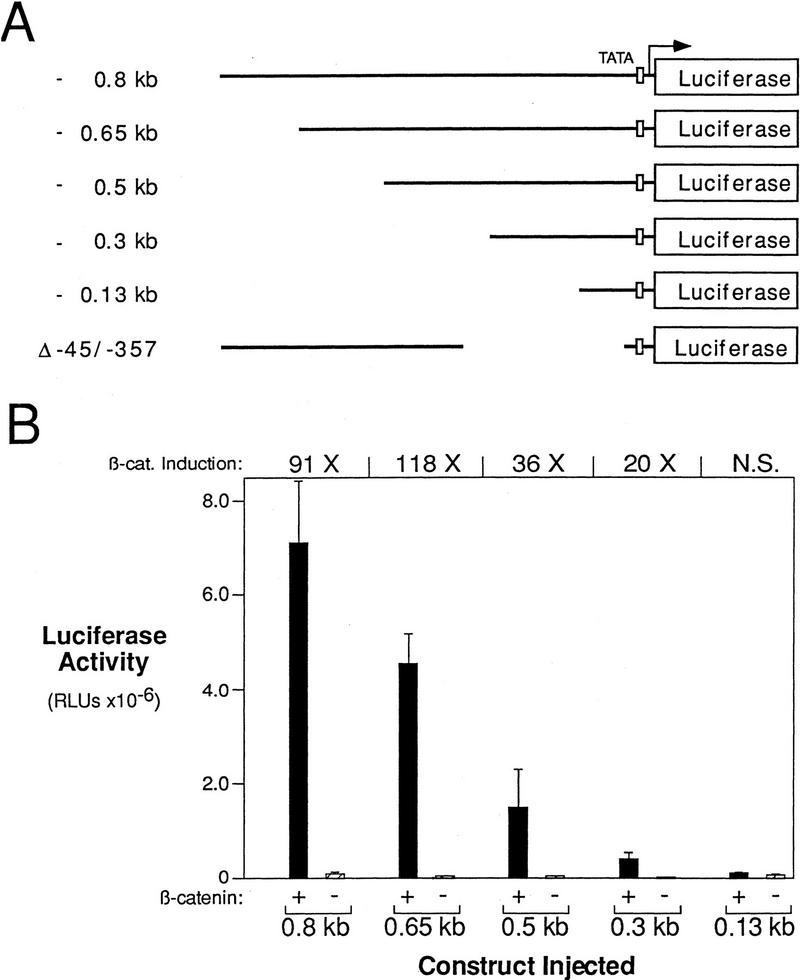
Figure 1.
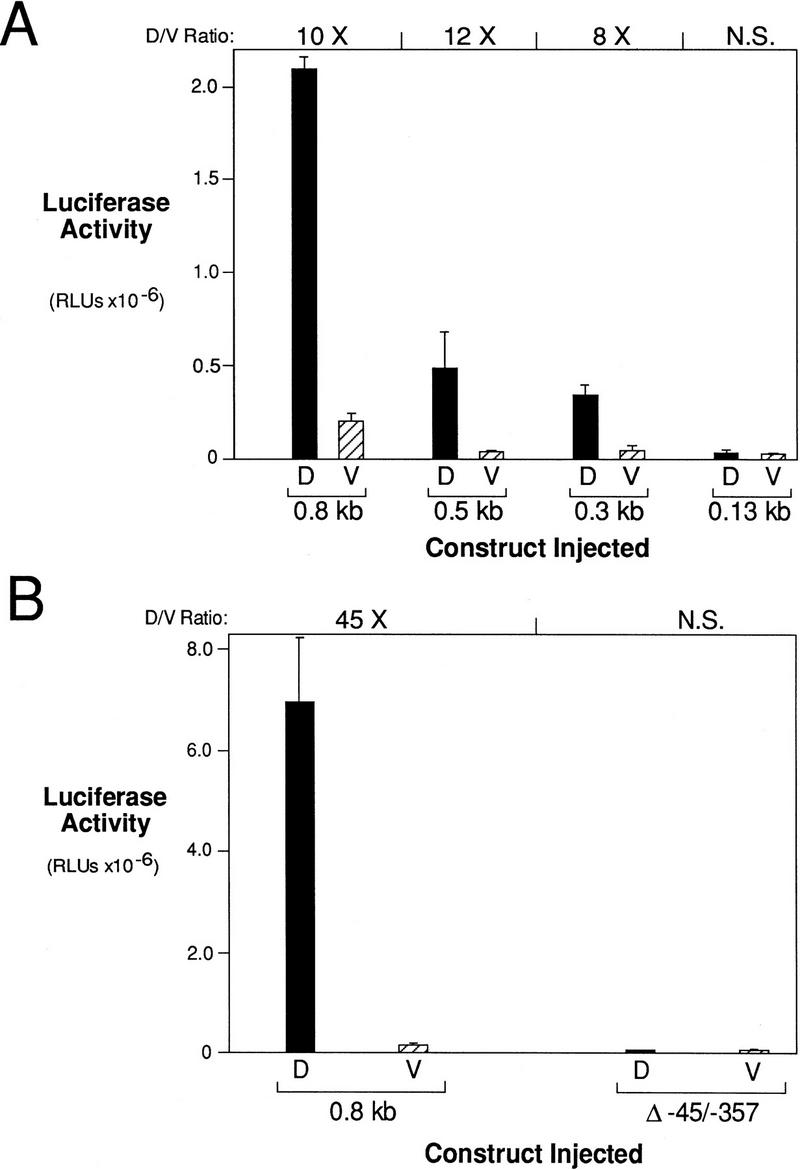
β-Catenin responsive regions of the siamois promoter. (A) Diagram of the siamois promoter deletion constructs used in this study. The length that each promoter fragment extends upstream of the siamois translation start site is indicated at left; each horizontal line represents the relative length of the promoter fragments. Δ−45/−357 represents a 312-bp deletion of the siamois promoter proximal to the TATA box, which is represented by an open box. Each siamois promoter fragment was fused to a luciferase reporter gene at the same point, as indicated. (B) β-Catenin responsive regions of the siamois promoter. The indicated constructs were injected into the animal pole of both blastomeres of two-cell embryos in the presence (+) or absence (−) of β-catenin RNA. Three pools of five stage 10 Xenopus embryos were assayed, and the mean and standard error of the resulting luciferase activities, in relative luciferase units (RLUs), are shown. The average fold induction by β-catenin (β-cat. Induction) is indicated above each data set. (N.S.) No significant β-catenin induction.
All the siamois promoter fragments, with the exception of the −0.13-kb fragment, respond to β-catenin in this assay (Fig. 1B). β-Catenin induced the expression of the −0.8- and −0.65-kb constructs to ∼100-fold over background levels. Although there was a significant drop in β-catenin inducibility when the promoter fragment is reduced to −0.3 kb, this construct was still induced 20-fold. Superimposed on this pattern of β-catenin inducibility, we found that each successive deletion decreased the absolute activity of the promoter. These data indicate that a β-catenin response element is located within −0.3 kb of the start of transcription, and that additional β-catenin response and/or enhancing elements are located between −0.3 kb and −0.8 kb.
In the Xenopus embryo, siamois expression is restricted to the dorsal equatorial and subequatorial regions (Lemaire et al. 1995). This expression pattern appears to depend on maternally deposited dorsal determinants (Brannon and Kimelman 1996). To map the sequences in the siamois promoter responsive to an endogenous dorsalizing activity, in the absence of ectopic β-catenin, we compared the levels of luciferase activity resulting from dorsal versus ventral injection of the 5′ nested deletion series. The −0.8-kb siamois promoter fragment was strongly responsive to an endogenous activity present in the dorsal, but not the ventral, marginal zone of early Xenopus embryos (10-fold; Fig. 2A). Deletion of the −0.8-kb fragment resulted in lower absolute levels of expression, as was observed with the β-catenin response (Fig. 1B). Whereas the −0.3-kb element showed a significant dorsal versus ventral response (eightfold), this difference was lost upon deletion to −0.13 kb (Fig. 2A). Our results show that the −0.3-kb fragment of the siamois promoter is activated by both β-catenin and a dorsally localized activity. Moreover, the −0.8-kb siamois promoter fragment contains multiple elements required for the normal expression of the siamois gene.
Figure 2.
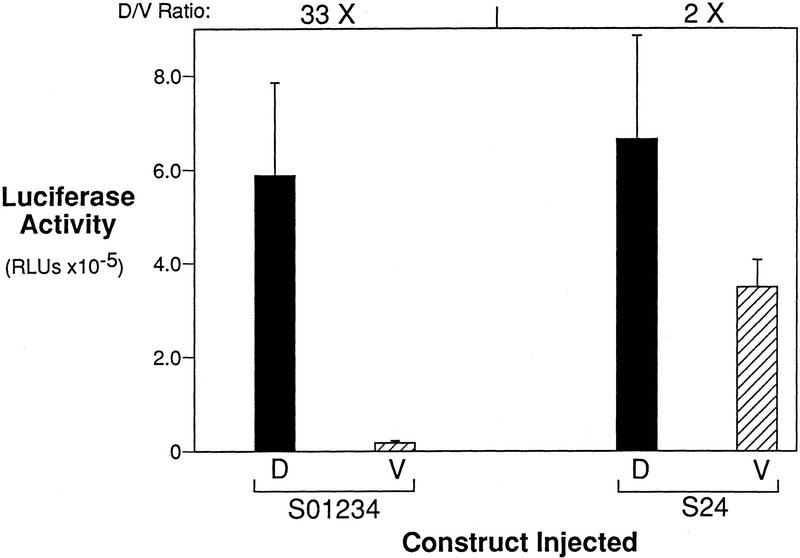
Determination of siamois promoter regions responsive to a dorsally localized endogenous activity. (A) Regions of the siamois promoter responsive to the dorsalizing activity. The indicated constructs were injected into the marginal zone of dorsal (D) or ventral (V) blastomeres at the four-cell stage. The mean and standard error, in RLUs, from three pools of five embryos each are shown. The average fold induction (D/V Ratio) for each construct is indicated above each data set. (N.S.) No significant dorsal vs. ventral difference. (B) siamois activation by the endogenous dorsalizing activity requires the proximal promoter region. The 0.8-kb and Δ−45/−357 reporter constructs were assayed as described above. The siamois promoter is no longer responsive to the endogenous dorsalizing activity when the proximal region containing the three Lef/Tcf sites is deleted.
Lef-1 and XTcf-3 bind to the siamois promoter
To identify elements conferring β-catenin responsiveness and dorsal specificity, we determined the nucleotide sequence of the −0.8-kb siamois promoter fragment (Fig. 3). Sequence analysis revealed a TATA-like basal promoter element located 34-bp upstream of the transcription start site. Whereas a Wnt-responsive element was identified in the gsc promoter (Watabe et al. 1995), no sequence of significant homology was found in the siamois promoter. Instead, we identified three consensus Lef/Tcf-binding sites, CTTTGA/TA/T [site 0 (S0), S1, and S3], within 360 bp upstream of the transcription start site (Fig. 3). Two of these sites (S1 and S3) are located within the proximal 300 bp that responds to β-catenin and exhibits dorsal–ventral differences. Two additional sequences (S2 and S4), which differ from the Lef/Tcf core consensus sequence at the 3′ most base, were also identified within the −360-bp region.
Figure 3.

Nucleotide sequence of the Xenopus siamois promoter −0.8-kb fragment. The first 23 nucleotides of the siamois coding sequence with its deduced amino acids and 817 nucleotides of 5′ siamois promoter sequence are shown. The transcription start site is indicated with an arrow and the TATA box is underlined. The possible Lef/Tcf-binding sites are boxed and identified above the sequence, as S0, S1, S2, S3 and S4. S0, S1, and S3 conform to the Lef/Tcf-binding site consensus, whereas S2 and S4 diverge at the 3′-most base. Nucleotides mutated in the Lef/Tcf-binding sites are underlined. The GenBank accession no. for the Xenopus siamois promoter is AF016226.
To determine whether XTcf-3 and Lef-1 bind to the siamois promoter, we performed electrophoretic mobility shift assays. Lef-1 and XTcf-3, transcribed and translated in vitro, were tested for binding to a radiolabeled 246-bp siamois promoter restriction fragment containing four of the possible Lef/Tcf-binding sites (S1234). One major band was observed when Lef-1 or XTcf-3 was incubated with this siamois promoter fragment (Fig. 4A,B). Lef-1 and XTcf-3 binding was completely abolished by the addition of 500-fold molar excess of a double-stranded oligonucleotide containing a consensus Lef/Tcf-binding site but was unaffected by addition of the same concentration of an oligonucleotide in which the Lef/Tcf binding site is mutated (Fig. 4A, cf. lane 6 with lane 11; data not shown). These results show that Lef-1 and XTcf-3 bind to the siamois promoter in a sequence-specific manner.
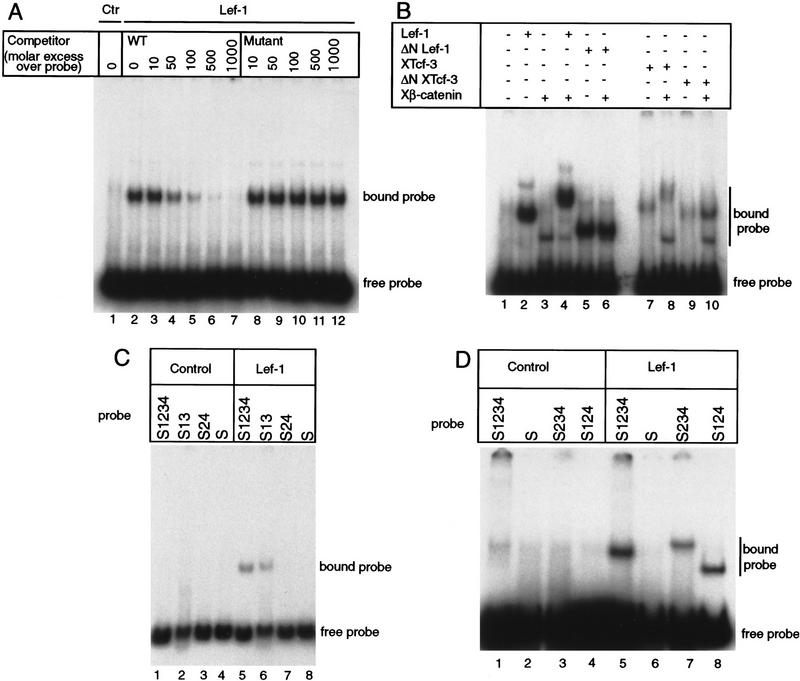
Figure 4.
Lef-1 and XTcf-3 specifically bind to the siamois promoter. (A) Lef-1 binding to the siamois promoter is competed by an oligonucleotide containing a consensus Lef/Tcf-binding site. Double-stranded competitor oligonucleotides containing either a consensus Lef/Tcf-binding site (WT) or a mutated site were incubated with Lef-1 in the presence of the siamois promoter probe S1234, which contains four potential binding sites. Binding to the promoter was analyzed by electrophoretic mobility shift assays. The concentrations of competitor oligonucleotide are indicated. [(Ctr) control; uncharged reticulocyte lysate]. (B) Lef-1 and XTcf-3 form a ternary complex with β-catenin on the siamois promoter. Control lysate or lysate containing either Lef-1, ΔNLef-1, XTcf-3, or ΔNXTcf-3 were incubated with the siamois promoter probe S1234 in the presence or absence of recombinant β-catenin. β-Catenin decreases the mobility of the S1234 probe in the presence of full-length Lef-1 or XTcf-3 but not in the presence of amino-terminally truncated proteins (cf. lanes 4 and 8 with lanes 6 and 10). A nonspecific band is observed in lanes containing recombinant β-catenin. (C) Lef/Tcf consensus S1 and S3 are required for Lef-1 to bind the siamois promoter. Control reticulocyte lysate (lanes 1–4) or lysate containing Lef-1 (lanes 5–8) was incubated with the following siamois promoter probes. Probe S1234 is wild-type for all sites, S13 contains the consensus Lef/Tcf-binding S1 and S3, S24 contains the imperfect S2 and S4, and S contains none of these sites. (D) Lef-1 binds to both consensus Lef/Tcf sites S1 and S3 and causes DNA bending. Control lysate or lysate containing Lef-1 was incubated with siamois promoter probes S1234, S234, or S124, as indicated. Because sites S2 and S4 do not bind Lef-1 (as shown in Fig. 4C), binding of Lef-1 to either S1 or S3 independently is being analyzed. Note the different migration rates of probes S234 (lane 7) and S124 (lane 8) in the presence of Lef-1, which is consistent with Lef-1 DNA bending.
Because XTcf-3 and Lef-1 both interact with β-catenin and form a ternary complex on oligonucleotides containing consensus Lef/Tcf-binding sites (Behrens et al. 1996; Huber et al. 1996; Molenaar et al. 1996), we analyzed the effect of recombinant β-catenin on Lef-1 and XTcf-3 binding to the siamois promoter. β-Catenin alone did not bind the S1234 promoter fragment at any concentration tested, but it did reduce the mobility of both Lef-1 and XTcf-3 promoter complexes (Fig. 4B, cf. lane 4 with 2 and lane 8 with 7). Previous studies have shown that truncation of the amino-terminal region of Lef-1 and XTcf-3 eliminates the interaction of these proteins with β-catenin but does not affect their ability to bind DNA (Behrens et al. 1996; Huber et al. 1996; Molenaar et al. 1996). Even at the highest β-catenin concentration tested, we observed no change in the mobility of amino-terminally deleted Lef-1 or XTcf-3 (Fig. 4B, cf. lane 6 with 5 and lane 10 with 9). This result indicates that the β-catenin-induced supershift is caused by an interaction with the amino-terminal domains of XTcf-3 and Lef-1 and is not the result of a nonspecific interaction.
Site-directed point mutations were used to determine whether consensus Lef/Tcf-binding sites S1 and S3 are required for Lef-1 and XTcf-3 to bind the siamois promoter. Probe S1234 is wild-type, probe S13 retains the consensus sites S1 and S3, probe S24 retains the imperfect sites S2 and S4, and probe S is mutated at all four sites (diagrammed in Fig. 6A, below). Whereas probes S1234 and S13 produced a single-shifted band when incubated with Lef-1 (Fig. 4C, lanes 5,6), probes S24 and S did not bind Lef-1 (Fig. 4C, lanes 7,8). Because we observed a single major band only when using probes known to contain two consensus Lef/Tcf-binding sites, probes S124 and S234 were generated to measure Lef-1 binding to S1 and S3 independently. Both of these probes were found to bind Lef-1 (Fig. 4D, lanes 7,8) with similar affinity (data not shown). The greater mobility of the Lef-1/S124 probe complex (lane 8) compared with the Lef-1/S234 probe complex (lane 7) is consistent with Lef-1 induced DNA bending (Behrens et al. 1996) at a site near the end of the probe (S1), compared with bending at a centrally located site (S3). In separate gel shift assays the consensus S0 also specifically bound Lef-1 (data not shown).
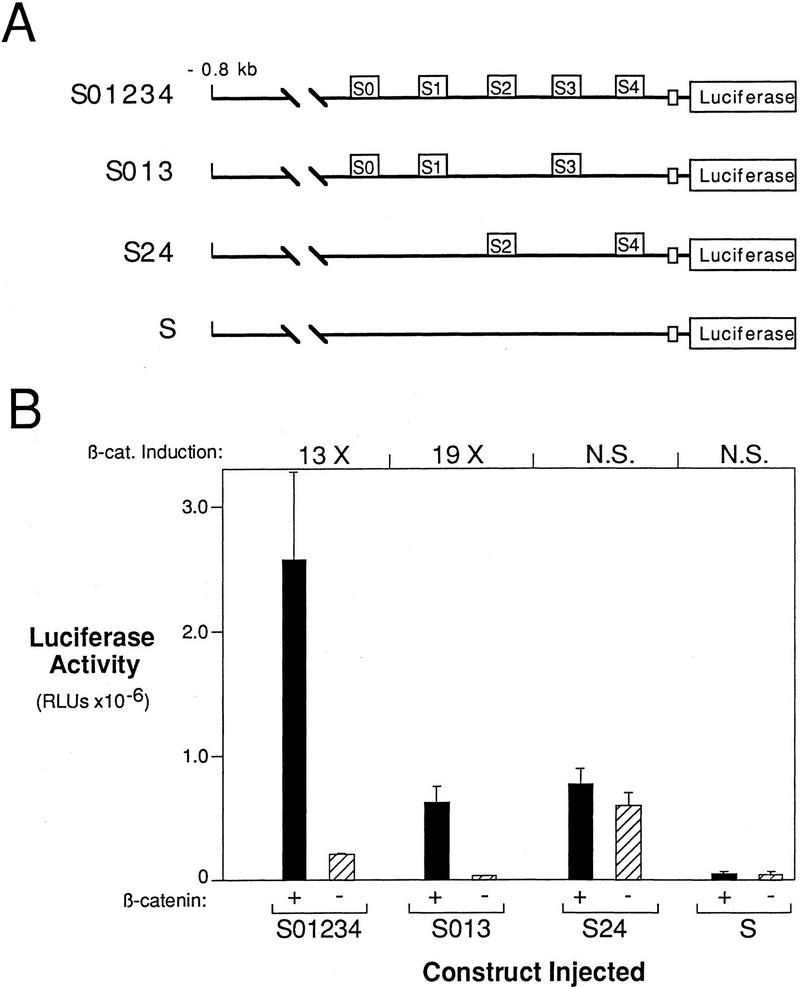
Figure 6.
Lef/Tcf site mutations eliminate the β-catenin response of the siamois promoter. (A) Schematic representation of the constructs that result from introducing site-directed mutations into the −0.8-kb siamois promoter. S01234 contains no mutations, S013 is mutant at S2 and S4, S24 is mutant at S0, S1, and S3, and S is mutant at all sites. (B) Lef/Tcf consensus sites S0, S1, and S3 are β-catenin response elements, whereas S2 and S4 have a general activating function. The indicated promoter constructs were injected into the animal pole of both blastomeres of two-cell-stage embryos in the presence (+) or absence (−) of β-catenin. The mean luciferase activities and standard errors, in RLUs, from three pools of five embryos each are shown. Average fold inductions by β-catenin (β-cat. Induction) are indicated above each data set. Note that basal luciferase levels resulting from injection of S24 are greater than those for S01234. (N.S.) No significant β-catenin induction.
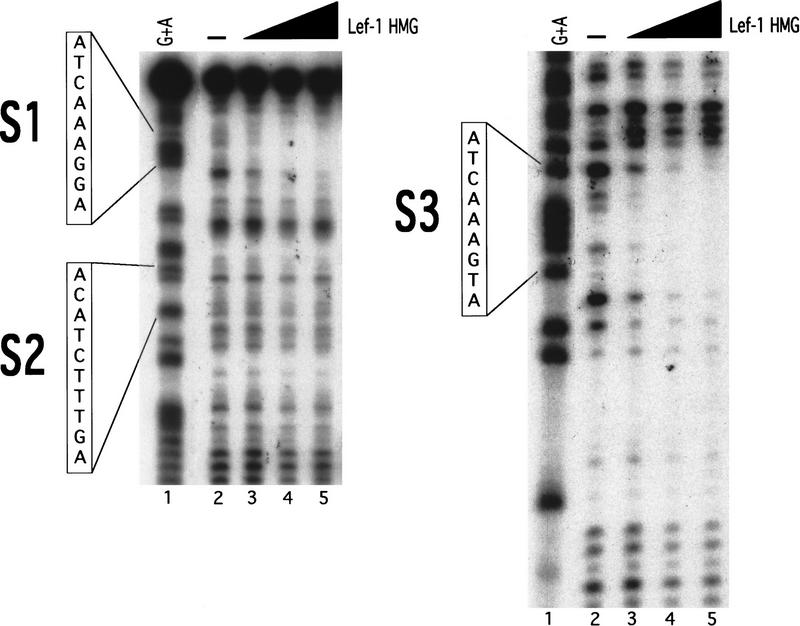
To further show the specificity of Lef-1 binding to the siamois promoter we performed DNase I footprinting assays by use of Lef-1 HMG domain and the S1234 restriction fragment used in the band shift assays described above. Under conditions of excess protein to probe, we observed DNase I-protected regions centering on S1 and S3, but not S2 or S4 (Fig. 5, cf. lane 2 with lane 5; data not shown). The gel shift and DNase I footprinting data indicate that S0, S1, and S3 are specific binding sites for Lef-1 and XTcf-3 class transcription factors.
Figure 5.
DNase I footprint analysis of the S1234 siamois promoter fragment. S1234 was 3′-end labeled and incubated in the presence of 0.1 μg BSA (−, lane 2), or 0.025 μg (lane 3), 0.05 μg (lane 4), or 0.1 μg (lane 5) of Lef-1 HMG domain protein. (Lane 1) Maxam-Gilbert G + A sequence reaction of the same DNA fragment. The positions and sequences of S1, S2, and S3 are indicated. DNase I-protected regions can be observed at S1 and S3 in the presence of 0.1 μg Lef-1 HMG protein (cf. lanes 2 and 5) but not S2 (shown) or S4 (not shown).
Contribution of the Lef/Tcf sites to the activation of siamois by β-catenin
Although no additional Lef/Tcf consensus sites are located upstream of the 5′-most site, S0, our deletion series shows that other sites in this region must contribute to the overall activation of siamois. By use of construct Δ−45/−357 (Fig. 1A), however, in which the region containing S0–S4 is deleted, we determined that the upstream region of the promoter is unable to activate siamois expression on its own (Fig. 2B). Therefore, we focused our analysis on the region of the siamois promoter containing the Lef/Tcf-binding sites. To evaluate the contribution of the Lef/Tcf sites to the activation of siamois expression by β-catenin, we analyzed the site-directed mutations described for the gel shift assays within the context of the −0.8 kb siamois reporter construct. When all potential Lef/Tcf sites were eliminated, β-catenin responsiveness was abolished, and the basal level of promoter activity was reduced (Fig. 6B, S). Elimination of S2 and S4 reduced the absolute level of the promoter activity in the presence and absence of β-catenin (Fig. 6B, S013), indicating that these sites have a general enhancing role.
Elimination of any one of the consensus Lef/Tcf sites (S0, S1, and S3) reduced the response to β-catenin only weakly, whereas elimination of any pair of sites reduced most of the β-catenin response (data not shown). Elimination of all three Lef/Tcf-binding sites abolished the β-catenin responsiveness of the siamois promoter (Fig. 6B, S24). Intriguingly, we always observed a significant increase in the basal level of the promoter, in the absence of β-catenin, when all three Lef/Tcf sites are mutated. These results (and see below) suggest that XTcf-3 binding to the Lef/Tcf consensus sites is important for repressing the siamois promoter in the absence of β-catenin.
Lef/Tcf site contribution to siamois activation by the endogenous dorsalizing activity
In addition to β-catenin activation in the animal pole, the siamois promoter responds to an activity present in the dorsal marginal region of early Xenopus embryos (Fig. 2). Because endogenous β-catenin is elevated on the dorsal side of the embryo (Schneider et al. 1996; Larabell et al. 1997) and required in early Xenopus embryos for siamois expression (Fagotto et al. 1997) and dorsal axis formation (Heasman et al. 1994), we postulated that the relatively lower ventral level of β-catenin accounts for the absence of ventral siamois expression. To test this hypothesis, the wild-type siamois reporter construct S01234 was injected with β-catenin RNA into the marginal zone of both ventral blastomeres at the four-cell stage and compared with S01234 injected both dorsally and ventrally in the absence of β-catenin. The dose of β-catenin RNA used in this experiment (250 pg) consistently caused complete axis duplication (data not shown). The ventral expression of S01234 was 30-fold less than the dorsal expression (Fig. 7A). Coinjection of β-catenin RNA induced the ventral expression of S01234 103-fold, a level exceeding that observed for S01234 activated by the endogenous dorsal activity (Fig. 7A). This result shows the potent ability of β-catenin to fully activate the siamois promoter in the marginal zone and implies that the differential localization of β-catenin in early Xenopus embryos restricts activation of siamois gene expression to the dorsal side of the embryo.
Figure 7.
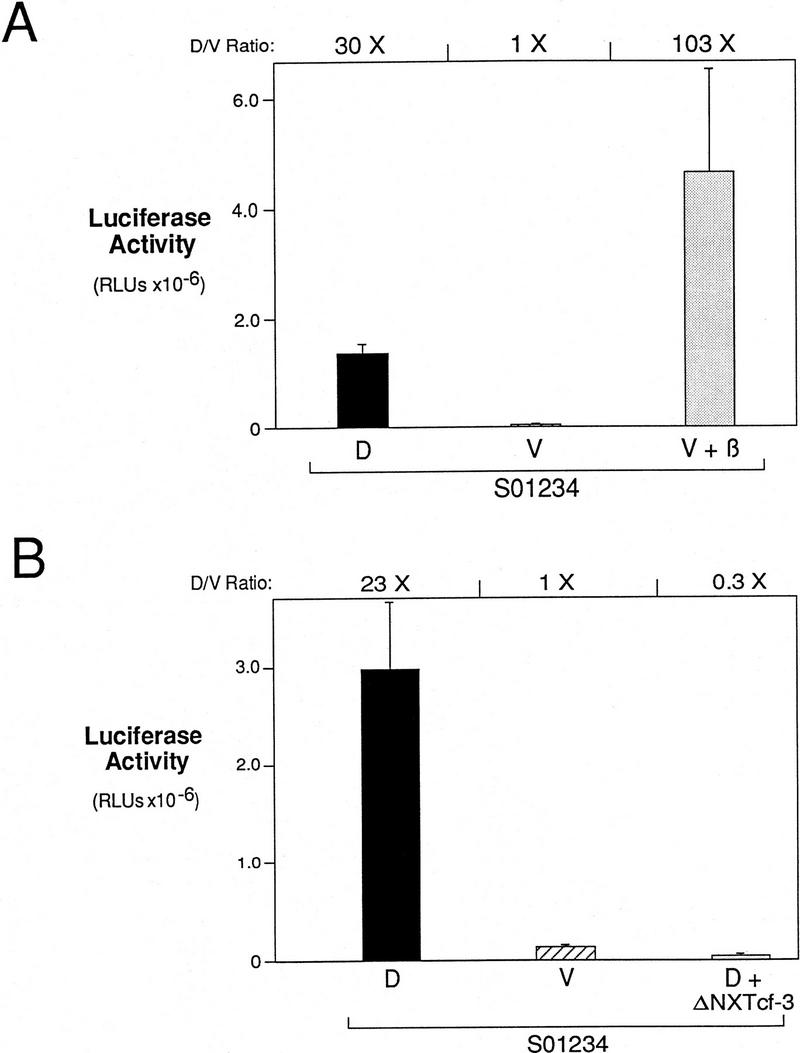
siamois promoter activation is dependent on the β-catenin/XTcf-3 complex. (A) β-Catenin activates the siamois promoter in the ventral marginal zone. Construct S01234 was injected dorsally (D), ventrally (V), or ventrally in the presence of β-catenin RNA (V + β). The mean and standard errors of luciferase activities are shown for each sample, as described previously. Average fold induction (D/V Ratio) for D vs. V and V + β vs. V is shown above the graph. (B) ΔNXTcf-3 blocks the dorsal activation of the siamois promoter. Construct S01234 was injected dorsally (D), ventrally (V), or dorsally in the presence of ΔNXTcf-3 RNA (D + ΔNXTcf-3). The mean and standard errors of luciferase activities are shown for each sample. Average fold inductions for D vs. V and D + ΔNXTcf-3 vs. V are shown above the graph.
Next, we tested the hypothesis that the endogenous activation of siamois requires the β-catenin/XTcf-3 complex. Amino-terminally truncated XTcf-3 (ΔNXTcf-3) does not bind to β-catenin and acts as a dominant–negative protein, suppressing axis formation when dorsally expressed (Molenaar et al. 1996). To determine whether ΔNXTcf-3 will block the normal activation of the siamois promoter, we injected the S01234 siamois reporter gene into the dorsal blastomeres of four-cell stage Xenopus embryos, with and without 1.6 ng of ΔNXTcf-3 RNA. This amount of ΔNXTcf-3 has a strong ventralizing effect, resulting in embryos with an average dorsoanterior index of 1.0 (Molenaar et al. 1996). Consistent with the ability of ΔNXTcf-3 to inhibit axis formation, the dorsal activation of S01234 was reduced 77-fold when coinjected with ΔNXTcf-3 compared with S01234 injected alone (Fig. 7B). This result shows that siamois promoter activation is dependent on the β-catenin/XTcf-3 complex.
Because the Lef/Tcf sites are required for β-catenin responsiveness in the animal pole assay, we predicted that the Lef/Tcf sites are also necessary for the activation of siamois gene expression in the dorsal marginal zone. To test this, the S24 reporter gene, which was mutated to eliminate the three Lef/Tcf-binding sites in the siamois promoter, was injected into the marginal zone of either dorsal or ventral blastomeres at the four-cell stage. Surprisingly, we found that the S24 reporter construct was as active on the dorsal side of the embryo as the S01234 construct (Fig. 8, D). Moreover, the S24 promoter was 20-fold more active on the ventral side of the embryo than the S01234 construct (Fig. 8, V). These results are consistent with the derepression observed for S24 in the animal pole when all three Lef/Tcf-binding sites are mutated (Fig. 6B). Thus, whereas S01234 was expressed 33-fold more strongly on the dorsal side of the embryo, S24 showed only a 2-fold dorsal–ventral difference. These results suggest that the high dorsal levels observed for S24 are the result of a lack of Lef/Tcf-mediated repression, which allows sites S2 and S4 to execute a general activating function.
Figure 8.
The Lef/Tcf-binding sites repress ventral siamois activation. Constructs S01234 and S24, which lacks the three Lef/Tcf-binding S0, S1, and S3, were injected into the marginal zone of dorsal (D) or ventral (V) blastomeres at the four-cell stage. The mean and standard errors of the resulting luciferase activities from three pools of five embryos each are shown. Average fold induction (D/V Ratio) for D vs. V is shown above both data sets. Note that construct S24 has 20-fold higher ventral activation of the siamois promoter than S01234, indicating that the siamois promoter lacking the Lef/Tcf-binding sites is no longer repressed on the ventral side of the embryo.
Localization of the endogenous siamois activator
Because the siamois promoter specifically responds to the endogenous dorsal signal, we attempted to map the location of this activity more precisely in the whole embryo. This approach was first used with the gsc promoter to show that an activin/BVg1-like activity is present throughout the vegetal hemisphere of Xenopus embryos (Watabe et al. 1995). For our experiments, the siamois promoter construct S01234 was injected into each of the four tiers of blastomeres on either the dorsal or ventral side of 32-cell stage embryos (Fig. 9A). As with the four-cell stage injections, S01234 was activated on the dorsal, but not the ventral side of embryos (Fig. 9B). The highest activity level was obtained in C and D tier blastomeres, although significant activity was present in B tier blastomeres and a low level in A tier blastomeres. These results agree well with previous studies showing that the highest dorsal axis-inducing activity is present in tiers C and D (Kageura 1990).
Figure 9.
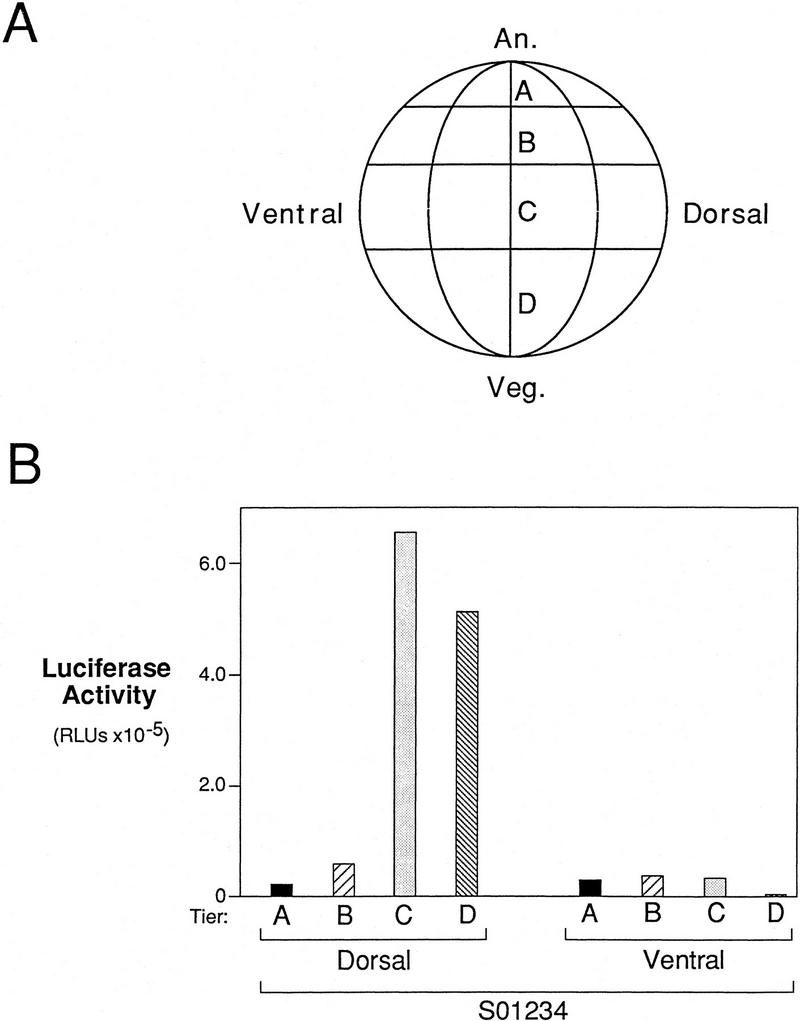
Localization of the endogenous dorsalizing activity in the embryo. (A) Diagram of a 32-cell stage Xenopus embryo showing the four tiers of blastomeres. (An) Animal pole; (Veg) vegetal pole. (B) The endogenous dorsalizing activity is highest in dorsal vegetal cells. A single dorsal or ventral blastomere at the 32-cell stage was injected with S01234. The luciferase activity of a pool of five embryos injected into the same blastomere indicates that the endogenous dorsalizing activity is present mainly in the dorsal C and D tiers (dorsal vegetal cells) of early embryos. This region of the embryo is most abundant in β-catenin (Larabell et al. 1997).
Discussion
The Wnt pathway, mediated by β-catenin, regulates the formation of the dorsal–ventral axis in Xenopus (Heasman et al. 1994; Miller and Moon 1996). Evidence from a number of recent studies indicates that β-catenin exerts its effect by modulating transcription. β-Catenin accumulates in the nuclei of dorsal blastomeres by the 16- to 32-cell stage (Larabell et al. 1997), hours before the onset of zygotic transcription. Taken with evidence that β-catenin binds to transcription factors of the Lef/Tcf family (Behrens et al. 1996; Huber et al. 1996; Molenaar et al. 1996), one would predict that complexes of β-catenin and Lef/Tcf might regulate transcription. Consistent with this possibility, β-catenin/Lef-1 heterodimers bind to the murine E-cadherin (Huber et al. 1996) and Drosophila Ultrabithorax (Ubx) (Riese et al. 1997) promoters. Finally, a transactivation domain has been mapped to the carboxyl terminus of armadillo, the Drosophila β-catenin homolog (van de Wetering et al. 1997). Here we present evidence that β-catenin regulates siamois expression by showing that the siamois promoter is a target for the β-catenin/XTcf-3 complex. Our results provide a direct link between the accumulation of β-catenin in dorsal blastomeres and the activation of a gene involved in the specification of the Xenopus dorsal–ventral axis.
Analysis of siamois promoter activity in the animal pole, a region that does not express the endogenous siamois gene unless β-catenin is ectopically expressed (Brannon and Kimelman 1996), showed that a −0.8-kb genomic fragment is strongly induced by β-catenin. Progressive deletion from the 5′ end of this fragment showed that the −0.3-kb proximal region was still inducible by β-catenin. The −0.3-kb fragment contains four possible Lef/Tcf-binding sites, but only the two sites that conform to the Lef/Tcf consensus sequence (S1 and S3) (Grosschedl et al. 1994) bind Lef-1 and XTcf-3 and form a ternary complex with β-catenin in vitro. Introducing mutations at these sites eliminates binding in vitro and renders the siamois promoter insensitive to β-catenin in animal pole injections. These results indicate that the β-catenin induced expression of siamois in the animal pole occurs through the direct binding of the β-catenin/XTcf-3 complex to the siamois promoter.
An extensive mutational analysis was performed within the context of the −0.8-kb fragment, which contains the three consensus Lef/Tcf-binding sites (S0, S1, and S3), to determine the functional significance of these sites. Mutation of any single site reduced β-catenin inducibility of the siamois promoter two- to threefold, showing that each site contributes to the optimal expression of siamois. Mutations in any combination of two sites removed most of the β-catenin responsiveness, showing that at least two sites must be present to achieve high-level expression of siamois. When all three sites were mutated the β-catenin inducibility was eliminated, indicating that each site can activate siamois at a low level.
Similar results were obtained when the siamois promoter fragments were injected into the marginal zone. The −0.8-kb fragment showed a large difference in activity between the dorsal and ventral marginal zones. The −0.3-kb fragment also showed a dorsal–ventral difference, but as with the β-catenin inducibility in the animal pole, it was not as strongly activated as the −0.8-kb fragment. The ventral levels of expression were raised to the dorsal levels by ectopic expression of β-catenin RNA, supporting the hypothesis that the dorsal enrichment of β-catenin observed in early Xenopus embryos (Schneider et al. 1996; Larabell et al. 1997) is responsible for activating endogenous siamois on the dorsal side of the embryo. Dorsal activation of siamois is also dependent on the β-catenin/XTcf-3 complex, because dominant–negative XTcf-3 completely abolished dorsal activation of a siamois reporter construct. In summary, our results from injection of the siamois promoter into the animal pole and marginal regions of Xenopus embryos indicate that β-catenin activates siamois expression in a manner dependent on the formation of a β-catenin/XTcf-3 complex.
Injection of the siamois promoter into dorsal cells of the 32-cell stage embryo indicates that the β-catenin/XTcf-3 complex is not the only component necessary for the correct localization of siamois. Siamois is expressed in the dorsal vegetal cells of the gastrula stage embryo (Lemaire et al. 1995), which accords well with our result that maximal siamois promoter activity is observed on injection into dorsal tiers C and D, because these cells populate the gastrula stage dorsal vegetal region (Bauer and Moody 1994). The dorsal tier B cells of the 32-cell-stage embryo, however, have similar staining of cytoplasmic and nuclear β-catenin relative to the dorsal tier C cells (Larabell et al. 1997), suggesting that another factor may be important for maximizing the expression of siamois in the vegetal cells. Previously, we have suggested that a member of the TGF-β family such as Vg1 could be important for enhancing the vegetal expression of siamois (Brannon and Kimelman 1996). These data fit well with our prior hypothesis that an overlap of broadly localized factors, activated during the blastula stage, determines the specific patterns of gene expression observed during the early gastrula stage (Kimelman et al. 1992).
Regulation of the siamois promoter is complex
Whereas the major regulation of siamois expression is caused by activation by the β-catenin/XTcf-3 complex, our studies have uncovered additional regulatory mechanisms. Perhaps the most intriguing is a XTcf-3-mediated repression of siamois expression. XTcf-3 appears to be ubiquitously expressed (Molenaar et al. 1996) and binds to the three siamois promoter Lef/Tcf sites in the presence or absence of β-catenin. Elimination of these sites resulted consistently in elevated basal levels of expression in both the animal pole and marginal zone, indicating that in the absence of β-catenin, XTcf-3 binds to the siamois promoter and represses its activity. Consistent with this finding, a repressive role for Lef-1 has been shown on a synthetic promoter containing four tandem Lef-binding sites (Ho and Leiden 1990), and a recent study showed that HBP1, an HMG box transcription factor related to Lef-1, is a transcriptional repressor (Tevosian et al. 1997).
Because Lef-1 and XTcf-3 have never been shown to repress a bona fide promoter, the mechanism of repression has not been elucidated. Lef/Tcf-type factors have an architectural function that involves the induction of sharp DNA bends (Grosschedl et al. 1994) and it is possible that in the absence of bending, the siamois promoter becomes active, whereas XTcf-3-induced bending may cause repression. β-Catenin has been shown to decrease the ability of Lef-1 to bend DNA (Behrens et al. 1996) and suggests a mechanism in which β-catenin contributes to siamois activation by relieving the XTcf-3-mediated repression. Alternatively, XTcf-3 may contain a domain that represses transcription in the absence of β-catenin. It is clear that simply derepressing the siamois promoter is not sufficient for full activation, because elimination of the Lef/Tcf sites does not activate the promoter to β-catenin induced levels in the animal pole assay. Our results suggest that β-catenin has a positive role in activating transcription, which is supported by the recent finding that the β-catenin homolog armadillo contains a transactivation domain (van de Wetering et al. 1997). We are currently investigating why the siamois promoter lacking the Lef/Tcf-binding sites is more strongly derepressed in the marginal zone than in the animal pole.
The remaining regulatory regions we have identified appear to be involved in enhancing the β-catenin/XTcf-3-mediated activation of the siamois promoter. First, using the deletion series, we found that the −0.8-kb fragment showed the same degree of response to β-catenin as the −0.65-kb fragment, but both the basal and induced levels of expression of the −0.8-kb fragment were higher. This suggests the presence of a general transcriptional enhancer between −0.8 and −0.65 kb. Second, we observed an additional enhancing element between −0.8 and −0.5 kb that affected the β-catenin inducibility of the promoter. The factor binding in this region appears to function in conjunction with β-catenin. Importantly, the region between −0.8 and −0.5 kb cannot activate the siamois promoter when the proximal region containing the consensus Lef/Tcf sites is deleted, showing that the proximal region is absolutely required for siamois expression. Finally, we showed by site-directed mutagenesis that sites S2 and S4 also have a general activating function for the siamois promoter. Because they do not bind Lef-1 or XTcf-3 in vitro, we expect that these sites function by binding a different transcriptional activator. Our results show that transcriptional activation by the β-catenin/XTcf-3 complex alone is not sufficient for optimal siamois expression. Instead, this complex appears to function as part of an assembly of transcriptional activators that are brought together on the siamois promoter.
A model for siamois regulation
On the basis of our data, we propose a model to explain how the endogenous siamois expression pattern is obtained. The rotation of the cortex induced by sperm entry moves an activator of the Wnt pathway (Holowacz and Elinson 1995) from the vegetal pole to the future dorsal side of the embryo (Fujisue et al. 1993; Kikkawa et al. 1996; Sakai 1996), resulting in the stabilization of β-catenin in the dorsal region by the two-cell stage (Larabell et al. 1997). Presently it is not known whether a Wnt signal is required or whether the pathway is activated cytoplasmically (Moon et al. 1997).The stabilized β-catenin associates with XTcf-3 (Molenaar et al. 1996), and binds to the siamois promoter during the cleavage stages, before embryonic transcription begins. With the onset of transcription at MBT, the β-catenin/XTcf-3 complex activates the expression of siamois in concert with other transcription factors. In contrast, on the ventral side of the embryo, XTcf-3 bound to the siamois promoter in the absence of β-catenin represses siamois expression. This provides a molecular model linking the accumulation of β-catenin in dorsal marginal blastomeres to the potent activation of siamois, a gene involved in specifying the Spemann organizer (Carnac et al. 1996).
Materials and methods
Isolation of genomic clones containing the siamois promoter
A siamois cDNA PCR clone in pGEMT (Promega) was a gift from Jan Christian (Oregon Health Sciences University, Portland). Oligonucleotide Xsiam/GSP2 (5′-ACTCCGAGGACACCTTAAGGG-3′) was designed and used in the PCR to generate a probe containing the first 246 bp of the siamois 5′-coding sequence. This probe was used to screen ∼1 × 106 plaques from a Xenopus genomic DNA library (made from strain HD-1; a gift from Dr. Thomas D. Sargent, National Institutes of Health, Bethesda, MD) and resulted in the isolation of 3 hybridizing recombinant phage. Hybridizations were performed by standard procedures. Genomic DNA was isolated from each positive clone, digested with EcoRI or BglII and tested for hybridization to the same 246-bp probe by Southern blot analysis. Three hybridizing fragments, corresponding to −3.0, −0.8, and −0.3 kb of the siamois promoter, were gel purified, subcloned into pBluescript (Stratagene), and sequenced.
Construction of luciferase reporter plasmids
To fuse the isolated siamois promoter fragments to a luciferase reporter gene, oligonucleotide siam/luc-2 (5′-CGCAGATCTCTGTCTCCCAAAATGTTGG-3′) was designed. This oligonucleotide introduces a BglII site (underlined) and eliminates the siamois translation start site. By use of siam/luc-2 and the pBluescript reverse primer, a −3.0-kb siamois promoter fragment was amplified by PCR with Vent polymerase (New England Biolabs). The amplification product was directionally cloned into the KpnI and BglII sites of the luciferase reporter vector pGL3B (Promega) upstream of the luciferase ATG. The −0.3-kb siamois promoter fragment was similarly cloned into pGL3B with SacI and BglII sites following amplification with siam/luc-2 and the pBluescript −20 primer. The −0.8-kb promoter construct was generated by digesting the −3.0-kb/pGL3B construct with KpnI and at an internal EcoRI site to remove 2.2 kb of intervening sequence, filling in with DNA polymerase I large fragment and religating.
A series of 5′ nested deletions was generated from the −3.0-kb siamois reporter construct by use of the exonuclease III–mung bean nuclease method (Sambrook et al. 1989). Deletions ranged from the internal EcoRI site at the 5′ end of the −0.8-kb construct down to a −0.13-kb fragment. Three reporter constructs containing −0.65, −0.5, and −0.13-kb of the siamois promoter were selected.
Oligonucleotide-mediated, site-directed mutagenesis (Kunkel et al. 1987) was used to eliminate the possible Lef/Tcf sites in the −0.8-kb siamois reporter construct. The mismatched oligonucleotides used to eliminate sites S0 through S4 are as follows: (S0) 5′-GTTGGCAAGACTTGGAATTCCCTTACTTACA-3′; (S1) 5′-CAAAAGGGAGGTAAT[lTTCATGATTCTGATGAC-3′; (S2) 5′-GAATTGGCAAGGTGATATCTGTGATTTGG GGAC-3′; (S3) 5′-TGAAAAAAATATAAATAGAATTCATGTACTGTTGC-3′; (S4) and 5′-ATATCAACAGGGGAGCTCTTGTGTGTCCAGG-3′. Each mutagenic oligonucleotide introduces or eliminates a restriction site (underlined) that was used to assay for the incorporation of the mutation. These oligonucleotides were used successively or in combination to eliminate multiple sites. All constructs were sequenced to confirm that only the intended point mutations were introduced.
Preparation of synthetic RNA
β-Catenin RNA was prepared from a CS + MT vector linearized with NotI as described previously (Yost et al. 1996). ΔNXTcf-3 RNA was prepared from pT7Ts ΔNXTcf-3 linearized with XbaI as described (Molenaar et al. 1996).
Xenopus embryo manipulation and microinjection
Xenopus embryos were obtained by artificial fertilization of eggs as described previously (Pierce and Kimelman 1995). Embryos were cultured at 14°C to 23°C and staged as described previously (Nieuwkoop and Faber 1967).
Embryos were microinjected at the 2-cell stage into the animal pole of both blastomeres, at the 4-cell stage into the equator of either dorsal or ventral blastomeres, or into one of the four tiers of blastomeres on either the dorsal or ventral side at the 32-cell stage. The volume of each microinjection was 10 nl per blastomere, except for 32-cell stage injections, which were 5 nl per blastomere. For each embryo, 265 pg (13.25 μg/ml) of siamois reporter plasmid DNA was microinjected in all experiments except the dorsal versus ventral injections of the 5′ nested deletion series, which used a total of 300 pg (15 μg/ml). When coinjected with the siamois reporter constructs, synthetic β-catenin RNA was used at a final amount of 250 pg (12.5 μg/ml) per embryo. Synthetic ΔNXTcf-3 RNA was coinjected at a final amount of 1.6 ng (80 μg/ml). Each microinjection experiment was performed with embryos obtained from a single female as absolute luciferase activity levels varied from batch to batch.
Luciferase assays
All embryos were cultured to stage 10 (early gastrula) and then each set of microinjected embryos was separated into three pools of five embryos each for assay in triplicate. Excess medium was removed, embryos were homogenized in 50 μl of 1× Rapid Lysis Buffer (Promega) and cleared by a 30-sec microcentrifugation. Ten microliters of the resulting supernatant was used for luciferase activity assays that were performed according to the manufacturers protocol (Promega) with a Berthold luminometer. Experiments were repeated not less than two times. Figures 1, 2, 6, 7, 8, 9 show a single representative experiment.
Electrophoretic mobility shift and DNase I footprinting assays
XTcf-3 (Molenaar et al. 1996), hLef-1 (Waterman et al. 1991), and their derivatives were transcribed and translated in vitro with the TNT T7 coupled reticulocyte lysate system (Promega) according to the manufacturers instructions. Recombinant His-tagged β-catenin was prepared as described (Yost et al. 1996). A 246-bp BsaAI–BglII fragment containing potential Lef/Tcf-binding sites S1–S4 or a 264-bp StyI–BsaAI fragment containing Lef/Tcf S0 were end-labeled by standard procedures. Probe [10 × 103 cpm (12.5 fmoles)] was incubated with 0.5-μl equivalents of reticulocyte lysate in the presence of 1 μg of poly[d(I-C)] in 1× binding buffer (20 mm HEPES, 50 mm EDTA, 5.0 mm MgCl2, 8% glycerol, 1.0 mm DTT, at pH 8) in a total volume of 10 μl. Samples were incubated for 10 min on ice, followed by a further 20-min incubation with radiolabeled probe. Competition analyses were performed with a double-stranded oligonucleotide, 5′-GATCTAGGGCACCCTTTGAAGCTCT-3′, which contains a consensus Lef/Tcf-binding site (underlined) and an oligonucleotide containing three point mutations (in bold) identical to the mutated S1 described above, 5′-GATCTAGGGCACAATTTCAAGCTCT-3′. Electrophoresis was performed through 3.5% native polyacrylamide gels in 0.5× TBE at room temperature.
DNase I footprinting assays were performed with the Promega SureTrack footprinting kit according to the manufacturers instructions.
Acknowledgments
We are grateful to Marian Waterman for providing full-length and ΔNhLef-1 plasmids and Lef-1 HMG domain protein, and for generously providing advice and assistance throughout the course of this work. We thank Olivier Destrée for providing XTcf-3 and ΔNXTcf-3, Jan Christian for providing a siamois cDNA, and Tom Sargent for providing his genomic library. We thank Marian Waterman and Cynthia Yost for critical comments on the manuscript. This work was supported by National Institutes of Health (NIH) Predoctoral Training Grant 5T32HDO7183-17 (to M.B.), Human Frontier Science Program Long Term Fellowship (to M.G.), NIH/National Research Services Award grant HD08016 (to L.S.), and NIH grants HD27262 (to D.K.) and HD29360 (to R.T.M.). R.T.M. is an investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kimelman@u.washington.edu; FAX (206) 685-1792.
References
- Bauer DV, Moody SA. The cleavage stage origin of Spemann’s organizer: Analysis of the movements of blastomere clones before and during gastrulation in Xenopus. Development. 1994;120:1179–1189. doi: 10.1242/dev.120.5.1179. [DOI] [PubMed] [Google Scholar]
- Behrens J, von Kries JP, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- Brannon M, Kimelman D. Activation of Siamois by the Wnt pathway. Dev Biol. 1996;180:344–347. doi: 10.1006/dbio.1996.0306. [DOI] [PubMed] [Google Scholar]
- Brunner E, Peter O, Schweizer L, Basler K. pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature. 1997;385:829–833. doi: 10.1038/385829a0. [DOI] [PubMed] [Google Scholar]
- Carnac G, Kodjabachian L, Gurdon JB, Lemaire P. The homeobox gene Siamois is a target of the Wnt dorsalization pathway and triggers organiser activity in the absence of mesoderm. Development. 1996;122:3055–3065. doi: 10.1242/dev.122.10.3055. [DOI] [PubMed] [Google Scholar]
- Dominguez I, Itoh K, Sokol SY. Role of glycogen synthase kinase 3β as a negative regulator of dorsoventral axis formation in Xenopus embryos. Proc Natl Acad Sci. 1995;92:8498–8502. doi: 10.1073/pnas.92.18.8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F, Guger K, Gumbiner BM. Induction of the primary dorsalizing center in Xenopus by the Wnt/GSK/β-catenin signaling pathway, but not Vg1, Activin or Noggin. Development. 1997;124:453–460. doi: 10.1242/dev.124.2.453. [DOI] [PubMed] [Google Scholar]
- Fujisue M, Kobayakawa Y, Yamana K. Occurence of dorsal axis-inducing activity around the vegetal pole of an uncleaved Xenopus egg and displacement to the equatorial region by cortical rotation. Development. 1993;118:163–170. doi: 10.1242/dev.118.1.163. [DOI] [PubMed] [Google Scholar]
- Funayama N, Fagotto F, McCrea P, Gumbiner BM. Embryonic axis induction by the Armadillo repeat domain of β-catenin: Evidence for intracellular signaling. J Cell Biol. 1995;128:959–968. doi: 10.1083/jcb.128.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart, J.C., M. Danilchick, T. Doniach, S. Roberts, B. Rowning, and R. Stewart. 1989. Cortical rotation of the Xenopus egg: Consequences for the anteroposterior pattern of embryonic dorsal development. Development (Suppl.) 107: 37–51. [DOI] [PubMed]
- Grosschedl R, Giese K, Pagel J. HMG domain proteins: Architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- He X, Saint-Jeannet J-P, Woodgett JR, Varmus HE, Dawid I. Glycogen synthase kinase-3 and dorsoventral patterning in Xenopus embryos. Nature. 1995;374:617–622. doi: 10.1038/374617a0. [DOI] [PubMed] [Google Scholar]
- Heasman J, Crawford A, Goldstone K, Garner-Hamrick P, Gumbiner B, McCrea P, Kintner C, Noro CY, Wylie C. Overexpression of cadherins and underexpression of β-catenin inhibit dorsal mesoderm induction in early Xenopus embryos. Cell. 1994;79:791–803. doi: 10.1016/0092-8674(94)90069-8. [DOI] [PubMed] [Google Scholar]
- Ho I-C, Leiden JM. The Tα2 nuclear protein binding site from the human T cell receptor α enhancer functions as both a T cell-specific transcriptional activator and repressor. J Exp Med. 1990;172:1443–1449. doi: 10.1084/jem.172.5.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holowacz T, Elinson RP. Properties of the dorsal activity found in the vegetal cortical cytoplasm of Xenopus eggs. Development. 1995;121:2789–2798. doi: 10.1242/dev.121.9.2789. [DOI] [PubMed] [Google Scholar]
- Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann BG, Kemler R. Nuclear localization of β-catenin by interaction with transcription factor LEF-1. Mech Dev. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- Kageura H. Spatial distribution of the capacity to initiate a secondary embryo in the 32-cell embryo of Xenopus laevis. Dev Biol. 1990;142:432–438. doi: 10.1016/0012-1606(90)90365-p. [DOI] [PubMed] [Google Scholar]
- Kikkawa M, Takano K, Shinagawa A. Location and behavior of dorsal determinants during first cell cycle in Xenopus eggs. Development. 1996;122:3687–3696. doi: 10.1242/dev.122.12.3687. [DOI] [PubMed] [Google Scholar]
- Kimelman D, Christian JL, Moon RT. Synergistic principles of development: Overlapping patterning systems in Xenopus mesoderm induction. Development. 1992;116:1–9. doi: 10.1242/dev.116.Supplement.1. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Larabell CA, Torres M, Rowning BA, Yost C, Miller JR, Wu M, Kimelman D, Moon RT. Establishment of the dorso-ventral axis in Xenopus embryos is presaged by early asymmetries in β-catenin that are modulated by the Wnt signaling pathway. J Cell Biol. 1997;136:1123–1136. doi: 10.1083/jcb.136.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire P, Garrett N, Gurdon JB. Expression cloning of Siamois, a Xenopus homeobox gene expressed in dorsal-vegetal cells of blastulae and able to induce a complete secondary axis. Cell. 1995;81:85–94. doi: 10.1016/0092-8674(95)90373-9. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Moon RT. Ectopic expression of the proto-oncogene int-1 in Xenopus embryos leads to duplication of the embryonic axis. Cell. 1989;58:1075–1084. doi: 10.1016/0092-8674(89)90506-0. [DOI] [PubMed] [Google Scholar]
- Miller JR, Moon RT. Signal transduction through β-catenin and specification of cell fate during embryogenesis. Genes & Dev. 1996;10:2527–2539. doi: 10.1101/gad.10.20.2527. [DOI] [PubMed] [Google Scholar]
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destrée O, Clevers H. XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- Moon RT, Brown JD, Torres M. WNTs modulate cell fate and behavior during vertebrate development. Trends Genet. 1997;13:157–162. doi: 10.1016/s0168-9525(97)01093-7. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis. Amsterdam, The Netherlands: North-Holland Publishing Company; 1967. [Google Scholar]
- Pierce SB, Kimelman D. Regulation of Spemann organizer formation by the intracellular kinase Xgsk-3. Development. 1995;121:755–765. doi: 10.1242/dev.121.3.755. [DOI] [PubMed] [Google Scholar]
- Riese J, Yu X, Munnerlyn A, Eresh S, Hsu S-C, Grosschedl R, Bienz M. LEF-1, a nuclear factor coordinating signalling inputs from wingless and decapentaplegic. Cell. 1997;88:777–787. doi: 10.1016/s0092-8674(00)81924-8. [DOI] [PubMed] [Google Scholar]
- Rowning BA, Wells J, Wu M, Gerhart JC, Moon RT, Larabell CA. Microtubule-mediated transport of organelles and localization of β-catenin to the future dorsal side of Xenopus eggs. Proc Natl Acad Sci. 1997;94:1224–1229. doi: 10.1073/pnas.94.4.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Binding of GSK3β to the APC-β-catenin complex and regulation of complex assembly. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- Sakai M. The vegetal determinants required for the Spemann organizer move equatorially during the first cell cycle. Development. 1996;122:2207–2214. doi: 10.1242/dev.122.7.2207. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schneider S, Steinbeisser H, Warga RM, Hausen P. β-Catenin translocation into nuclei demarcates the dorsalizing centers in frog and fish embryos. Mech Dev. 1996;57:191–198. doi: 10.1016/0925-4773(96)00546-1. [DOI] [PubMed] [Google Scholar]
- Siegfried E, Wilder EL, Perrimon N. Components of wingless signaling in Drosophila. Nature. 1994;367:76–80. doi: 10.1038/367076a0. [DOI] [PubMed] [Google Scholar]
- Tevosian SG, Shih HH, Mendelson KG, Sheppard K-A, Paulson KE, Yee AS. HBP1: A HMG box transcriptional repressor that is targetted by the retinoblastoma family. Genes & Dev. 1997;11:383–396. doi: 10.1101/gad.11.3.383. [DOI] [PubMed] [Google Scholar]
- Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor α enhancer function. Genes & Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence specific HMG box. EMBO J. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Armadillo co-activates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- Watabe T, Kim S, Candia A, Rothbacher U, Hashimoto C, Inoue K, Cho KWY. Molecular mechanisms of Spemann’s organizer formation: Conserved growth factor synergy between Xenopus and mouse. Genes & Dev. 1995;9:3038–3050. doi: 10.1101/gad.9.24.3038. [DOI] [PubMed] [Google Scholar]
- Waterman ML, Fischer WH, Jones KA. A thymus-specific member of the HMG protein family regulates the human T cell receptor α enhancer. Genes & Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- Yang-Snyder J, Miller JR, Brown JD, Lai C, Moon RT. A frizzled homolog functions in a vertebrate Wnt signaling pathway. Curr Biol. 1996;6:302–306. doi: 10.1016/s0960-9822(02)70716-1. [DOI] [PubMed] [Google Scholar]
- Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT. The axis-inducing activity, stability, and subcellular distribution of β-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes & Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]