Abstract
In plants, animals, and fungi, DNA methylation is frequently associated with gene silencing, yet little is known about the role of the methylation in silencing. In Neurospora crassa, repeated sequences are silenced by repeat-induced point mutation (RIP) and genes that have suffered numerous GC → AT mutations by RIP are typically methylated at remaining cytosines. We investigated possible effects on transcription from methylation associated with RIP by taking advantage of 5-azacytidine, which prevents most methylation in Neurospora and a dim-2 mutation that abolishes all detectable methylation. Northern analyses revealed that methylation prevents the accumulation of transcripts from genes mutated by RIP. Measurements of transcription rates in vivo showed that methylation inhibits transcription severely but does not influence mRNA stability. Results of nuclear run-on experiments demonstrated that transcription initiation was not significantly inhibited by the dense methylation in the promoter sequences. In contrast, methylation blocked transcription elongation in vivo.
Keywords: DNA methylation, Neurospora, RIP, transcription, 5-azacytidine, dim
Many eukaryotes modify their DNA by the addition of a methyl group to the five position of selected cytosine residues. The correlation between DNA methylation and gene inactivity has been documented extensively, especially in animals, and has led to the idea that methylation plays a role in controlling gene expression (for review, see Bird 1992; Tate and Bird 1993; Eden and Cedar 1994). Transfection experiments with DNA methylated in vitro demonstrated that methylation can inhibit gene expression (Vardimon et al. 1982; Keshet et al. 1985; Yisraeli et al. 1988). Conversely, prevention of DNA methylation with 5-azacytidine can lead to activation of some silenced genes (Jones 1984; Ferguson et al. 1995). The importance of DNA methylation for mammals was demonstrated when it was found that disruption of the murine methyltransferase gene blocks embryogenesis (Li et al. 1992). DNA methylation has been implicated in epigenetic processes such as X-chromosome inactivation (Singer-Sam and Riggs 1993) and genomic imprinting (Bartolomei et al. 1993; Li et al. 1993; Razin and Cedar 1994), providing a possible basis for this observation.
It is generally assumed that DNA methylation inhibits gene expression at the level of transcription initiation (Bird 1992; Tate and Bird 1993; Eden and Cedar 1994). Although decisive evidence is lacking, the assumption is based on observations that methylation required for, or at least associated with, gene silencing tends to be at the 5′ end of genes (Keshet et al. 1985; Levine et al. 1992; Ngô et al. 1996). Two types of models have been proposed to explain how methylation can inhibit transcription. In the first, methylation interferes directly with the binding of trans-acting factors to their recognition sites. A number of trans-acting factors, including AP-1 (Comb and Goodman 1990), c-Myc/Myn (Prendergast et al. 1991), and NF-κB (Bednarik et al. 1991), do not bind to their recognition sites when these sites include a methylated CpG. Other transcription factors, such as Sp1 and CTF, are insensitive to methylation of their sites, however (Ben-Hattar et al. 1989). Furthermore, a number of studies have suggested that methylation does not prevent transcription directly (Levine et al. 1992; Rhodes et al. 1994). In the second class of models, methylation inhibits transcriptional activity indirectly, for example, by inducing an alteration in chromatin structure or by attracting proteins that bind to methylated DNA in a sequence-independent manner. The inhibitory effect of methylation has been demonstrated to require the packaging of a methylated template into chromatin (Buschhausen et al. 1987), and a number of proteins that bind methylated sequences have been described (Tate and Bird 1993). Two of these methyl–DNA-binding proteins, MeCP-1 (methyl–CpG-binding protein-1) and MeCP-2, have been implicated in the repression of transcription from methylated sequences (Boyes and Bird 1991; Nan et al. 1997).
The filamentous fungus Neurospora crassa offers a simple system to study the effect of DNA methylation on transcription in eukaryotes. One advantage Neurospora offers is that methylation is dispensable as revealed by the isolation of a mutant, dim-2 (defective in DNA methylation), that eliminates all detectable methylation (Foss et al. 1993). Most of the methylation in the wild-type Neurospora genome is associated with sequences that have been altered by the genetic process named RIP (repeat-induced point mutation), which operates in the premeiotic haploid nuclei of the developing fruiting body (Selker et al. 1987; Selker 1990, 1997). RIP surveys the genome for duplicated sequences, and when found, riddles both copies with GC → AT mutations (Selker and Garrett 1988; Cambareri et al. 1989). If a duplicated sequence includes a gene, the damage by RIP typically leaves both copies of the gene nonfunctional. Genes that have suffered numerous point mutations by RIP are usually methylated at most remaining cytosines (Selker and Stevens 1985; Selker et al. 1993; Singer et al. 1995). This methylation is not confined to symmetrical sequences (Selker and Stevens 1985; Selker et al. 1993) and sometimes extends for a short distance beyond the region mutated by RIP (Miao et al. 1994; Singer et al. 1995; Irelan and Selker 1997). The function of the methylation resulting from RIP is not known.
Indirect evidence suggesting that methylation can influence gene expression in Neurospora came from studies on the behavior of an allele of the am gene (NADP-specific glutamate dehydrogenase) that resulted from insertion of the Tad retrotransposon in the gene’s upstream region. Strains with this allele, am::Tad3-2, display a highly unstable Am−/+ phenotype (Kinsey and Helber 1989). Methylation associated with the 5′ region of the Tad3-2 element was found to be necessary for am expression; preventing methylation with 5-azacytidine or dim-2 eliminated reversion to Am+ (Cambareri et al. 1996). To further elucidate the function of DNA methylation in N. crassa, we explored the effect of methylation associated with genes altered by RIP. We found that the methylation can prevent accumulation of transcripts. In vivo labeling experiments demonstrated that the failure to accumulate transcripts from methylated alleles is attributable to reduced transcription rather than reduced mRNA stability. Nuclear run-on experiments demonstrated that initiation of transcription was not inhibited significantly despite extensive methylation in the proximal promoter regions but rather, showed that methylation inhibited transcription elongation.
Results
Reduced transcript levels from methylated amRIP alleles
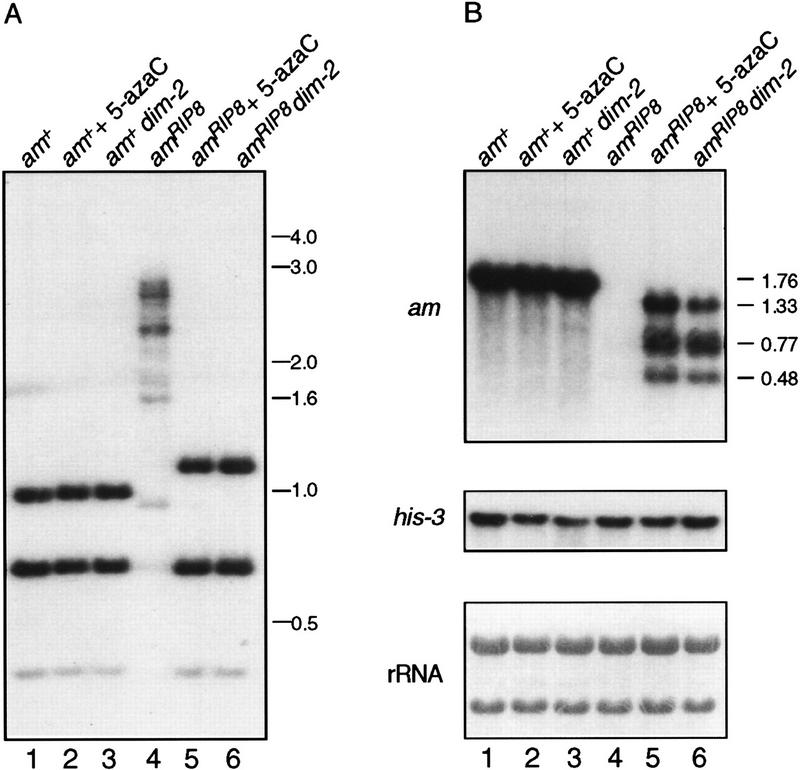
To assess the effects of methylation associated with genes altered by RIP, we first analyzed the production of transcripts from four characterized alleles of the am gene generated by RIP: amRIP2, amRIP5, amRIP6, and amRIP8 (Singer et al. 1995). These four alleles, which have 21, 84, 86, and 158 mutations created by RIP, respectively, were chosen because in each case the mutations lie downstream of the start sites of transcription. The chosen strains lacked the ectopic copy of am that had provoked RIP. Although not mutated, the promoter regions of amRIP5, amRIP6, and amRIP8 were heavily methylated (Selker et al. 1993; Singer et al. 1995). The methylation status of these alleles in strains grown 3 hr from conidia (asexual spores), assessed by digesting genomic DNA with the methylation-sensitive restriction enzyme Sau3AI, is shown in Figure 1A. The wild-type and amRIP2 alleles showed no sign of methylation at any of the Sau3AI sites (lanes 1,3). The amRIP5, amRIP6, and amRIP8 alleles showed heavy methylation at Sau3AI sites throughout the gene and extended beyond the sequences altered by RIP, as indicated by the appearance of bands representing fragments larger than 3 kb (lanes 5,7,9).
Figure 1.
Treatment with 5-azaC results in an accumulation of transcripts from the methylated amRIP alleles. DNA and RNA were isolated from strains grown in the presence (+) or absence (−) of 24 μm 5-azaC. (A) 5-AzaC treatment results in demethylation of the methylated amRIP alleles. Genomic DNA (750 ng) from strains N150 [am+ (lanes 1,2)], N665 [amRIP2 (lanes 3,4)], N673 [amRIP5 (lanes 5,6)], N675 [amRIP6 (lanes 7,8)], and N617 [amRIP8 (lanes 9,10)] was digested with the methylation-sensitive restriction enzyme Sau3A1, subjected to Southern blotting, and hybridized to an am probe. Size standards (kb) are indicated at left. The Southern blot was stripped and reprobed with the unmethylated mtr gene to confirm complete digestion of the DNA (data not shown). (B) Corresponding Northern analyses. The blot at top was hybridized to a wild-type am probe, whereas the blot in the middle was hybridized to a his-3 probe. (Bottom) Methylene blue staining of the rRNA of the blot from the top. (C) Map of the am gene. The am locus (horizontal line) with the exons (thick bars), transcription start sites (arrow), Sau3A1 restriction sites (ticks below the horizontal line), and scale bar are indicated. The mutations (vertical lines) for the amRIP2, amRIP5, amRIP6, and amRIP8 alleles are depicted below the diagram of the am locus. The asterisk (*) marks the Sau3A1 site that is mutated in the amRIP2 and amRIP8 alleles.
Northern analysis of total RNA from the amRIP2 strain showed that the level of mRNA from this unmethylated allele was comparable with that of the wild-type am gene (Fig. 1B, lanes 1,3). In sharp contrast, analysis of RNA from strains with the methylated alleles, amRIP5, amRIP6, and amRIP8, showed extremely reduced transcript levels (Fig. 1B, lanes 5,7,9). A control probing for his-3 transcripts and methylene blue staining of the rRNA demonstrated that the RNAs were evenly loaded (Fig. 1B).
5-Azacytidine treatment increases transcript levels from amRIP5, amRIP6, and amRIP8
To determine whether methylation was responsible for the low transcript levels from the amRIP5, amRIP6, and amRIP8 alleles, the strains were treated with 5-azacytidine (5-azaC), which efficiently prevents DNA methylation in Neurospora (Selker and Stevens 1985). DNA and RNA were extracted from conidia germinated in the presence of 24 μm 5-azaC. Treatment with 5-azaC prevented, almost completely, methylation of the amRIP5, amRIP6, and amRIP8 alleles, as shown by the disappearance of the high-molecular-weight Sau3AI fragments (Fig. 1A, lanes 6,8,10). The 5-azaC treatment had little or no effect on mRNA levels from the wild-type am allele and the unmethylated amRIP2 allele (Fig. 1B, lanes 1–4). In contrast, 5-azaC treatment of the strains containing the amRIP5, amRIP6, or amRIP8 alleles led to a significant accumulation of mRNA (Fig. 1B, lanes 6–10). The levels of amRIP5 and amRIP6 mRNAs in the 5-azaC-treated strains were lower than those from the unmethylated wild-type and amRIP2 alleles. Both of these alleles have nonsense mutations early in their coding sequence [codons 28 and 53, respectively (Singer et al. 1995)]; therefore, it seems likely the reduction in mRNAs from these alleles was attributable to a process called nonsense-mediated mRNA decay, which has been described for Saccharomyces cerevisiae and other organisms (Peltz et al. 1994; Beelman and Parker 1995). RIP might have also created one or more signals that trigger rapid decay of transcripts, equivalent to the AU-rich elements in the 3′-untranslated region of many short-lived mammalian transcripts (Zubiaga et al. 1995). Unlike amRIP5 and amRIP6, the amRIP8 allele in the 5-azaC-treated strain produced a high level of transcripts but failed to produce a full-length transcript, instead producing three smaller transcripts. The numerous C → T transition mutations on the coding strand (Singer et al. 1995) may have caused termination or led to aberrant post-transcriptional processing, yielding the three transcripts. The latter possibility seems likely because all three transcripts were polyadenylated and initiated at or very near the normal am transcription start sites as determined by RT–PCR and hybridization experiments (data not shown).
Accumulation of amRIP6 and amRIP8 mRNA in a dim-2 background
It seemed likely that the increased levels of amRIP5, amRIP6, and amRIP8 mRNAs in response to 5-azaC were attributable to inhibition of DNA methylation, but it remained possible that the increased mRNA levels resulted indirectly from an effect of 5-azaC on another cellular function (Shin et al. 1992; Crosthwaite et al. 1995). We therefore examined the effect of preventing methylation genetically, using a mutation isolated in our laboratory that prevents DNA methylation (Foss et al. 1993). Results of a Southern hybridization confirmed that the dim-2 (defective in DNA methylation) mutation crossed into amRIP6 and amRIP8 strains successfully prevented all detectable methylation of the alleles (Fig. 2A; data not shown). Northern analysis of total RNA demonstrated that dim-2 resulted in accumulation of mRNA from amRIP6 (data not shown) and amRIP8 (Fig. 2B, lane 6) at levels comparable with those observed after 5-azaC treatment (lane 5). Thus, both methods to prevent methylation resulted in increased levels of steady-state mRNA. We conclude that the methylation of genes mutated by RIP can prevent mRNA accumulation.
Figure 2.
Prevention of methylation by the dim-2 mutation increases amRIP8 transcript levels. (A) Southern analysis of genomic DNA from the strains N150 [am+; dim+ (lane 1)], N150 treated with 5-azaC (lane 2), N540 [am+; dim-2 (lane 3)], N617 [amRIP8; dim+ (lane 4)], N617 treated with 5-azaC (lane 5), and N618 [amRIP8; dim-2 (lane 6)]. Genomic DNA (750 ng) was digested with the methylation-sensitive restriction enzyme Sau3A1, subjected to Southern blotting, and hybridized to an am probe. Size standards (kb) are indicated at right. The Southern blot was stripped and reprobed with the unmethylated mtr gene to ensure complete digestion of the DNA (data not shown). (B) Northern analysis of total RNA (10 μg) from the strains depicted in the Southern blot. The blot at top was hybridized to a wild-type am probe. The size (kb) of the wild-type am transcript and of the three amRIP8 transcripts is indicated right of the top panel. The blot in the middle was hybridized to a his-3 probe. (Bottom) The methylene blue staining of the rRNA of the blot from the top.
Extent of methylation of amRIP8
As a step to understand the inhibitory effect of methylation, we extended our methylation analysis of the amRIP8 allele. In prior work, genomic sequencing showed methylation at >80% of the cytosines within six regions of amRIP8, including the promoter consisting of the putative TATA box and start sites of transcription (Selker et al. 1993). The genomic sequencing was conducted on DNA from a strain (N617) grown for 16 hr, whereas the Northern results described above were from cultures germinated for 3 hr. Because methylation levels are known to change during vegetative growth (Russell et al. 1987; Roberts and Selker 1995; M. Rountree, H. Foss, and E. Selker, unpubl.), we analyzed the level of methylation at a number of sites within and surrounding the amRIP8 allele after just 3 hr. Our results are summarized in Figure 3. Sites within and flanking the mutated region displayed a high level of methylation, including sites around the promoter, consistent with the genomic sequencing data. Sites farther upstream, however, were only lightly methylated or not methylated at all. Expression of the am gene is driven by two upstream enhancer-like elements, amα and amβ (Frederick and Kinsey 1990a), which have been demonstrated to bind nuclear factors (Frederick and Kinsey 1990b; Chen and Kinsey 1994). Restriction sites close to these enhancer-like sequences were not methylated. These results suggest that methylation could not directly prevent binding of factors to upstream activator sequences but might interfere with interactions at the proximal promoter.
Figure 3.
Methylation analysis of amRIP8. The amRIP8 allele is represented by the horizontal line. The upstream enhancer-like sequences (α and β) are represented by open boxes. The start sites of transcription are indicated by the arrow. The mutations in amRIP8 are indicated by the ticks above the horizontal line. The approximate ends of the three amRIP8 transcripts are indicated by the open diamonds, whereas the end of the wild-type am transcript is indicated by the black diamond. The recognition sites for the methylation-sensitive restriction enzymes BamHI (B), BglII (G), Bsp106-I (S), NaeI (N), NarI (A), NsiI (I), PstI (P), PvuII (V), XbaI (X), and XhoI (H) are indicated below the horizontal line. The approximate level of methylation at each restriction enzyme site is depicted in black in a pie chart below each site.
Decreased rate of transcription from the methylated amRIP8 allele
It was conceivable that the reduced steady-state transcript levels resulted from an effect of methylation on transcription, transcript stability, or both. To investigate these possibilities, we carried out in vivo labeling experiments using [3H]uridine, as described in the Materials and Methods. To obtain good uptake of the [3H]uridine, we took advantage of a mutation in the pyrimidine (pyr) biosynthesis pathway (Caroline and Davis 1969). In addition, we substituted NaNO3 for NH4NO3 in the medium because uptake of uridine has been reported to be enhanced in the presence of a poor nitrogen source (Buxton and Radford 1982). We also determined the minimal level of uridine supplementation required to give normal growth 3 hr after inoculation with conidia at 33°C (37.5 μg/ml). Northern analysis of the dim+ (N1223) and dim-2 (N1224) amRIP8; pyr-1 sibling strains grown under these conditions showed transcript levels comparable with those observed in the experiments described above (data not shown).
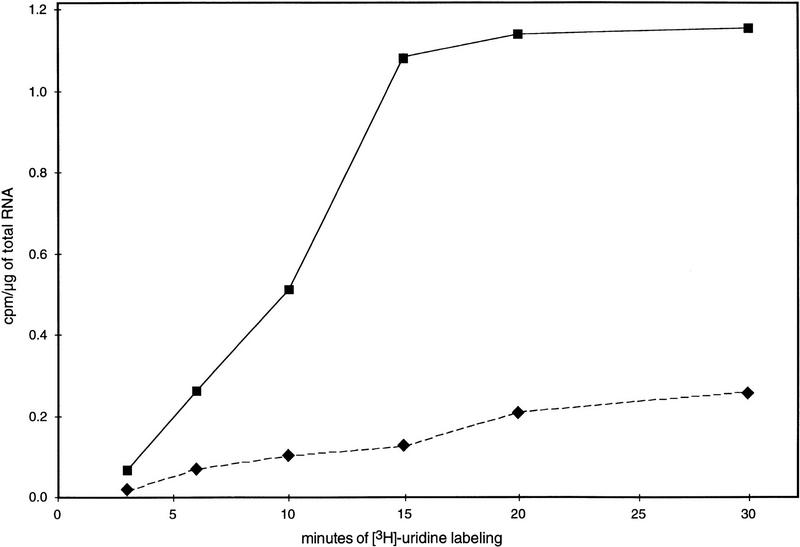
Labeling of RNA in the dim-2 and dim+; amRIP8 strains was initiated 3 hr after inoculation, and the cultures were sampled 3, 6, 10, 15, 20, and 30 min thereafter. The 3H-labeled RNA extracted from the samples was used to probe denatured plasmid DNA containing genes of interest, and the RNA bound was measured by scintillation counting as described in Materials and Methods. After 30 min of labeling, only 40% of the [3H]uridine had been consumed by the cells, suggesting that uridine was not limiting in these experiments. Transcription rates of the methylated and unmethylated amRIP8 alleles were standardized to the unmethylated histone H4 gene. The rate of transcription from the unmethylated (dim-2) amRIP8 allele (0.063 cpm/μg of total RNA/min) was more than five times greater than that of the methylated (dim+) amRIP8 allele (0.012 cpm/μg of total RNA/min; Fig. 4). Nearly identical results were obtained when the experiment was repeated (0.066 and 0.012 cpm/μg of total RNA/min for the unmethylated and methylated alleles, respectively; data not shown). The observed difference between the transcription rates from amRIP8 in the dim-2 (N1224) and dim+ (N1223) strains was comparable with the difference between the transcript levels of this allele in N617 and N618 (data not shown). Furthermore, the half-lives of the amRIP8 transcripts were similar in the methylated (dim+) and unmethylated (dim-2) backgrounds (data not shown). We conclude that the difference in the steady-state levels of the amRIP8 transcripts was attributable to a difference in transcription rate rather than a difference in mRNA stability.
Figure 4.
In vivo labeling analysis of amRIP8 transcription in dim+ and dim-2 backgrounds. Time course of [3H]uridine incorporation into amRIP8 mRNA from N617 [amRIP8; dim+ (♦)] and N618 [amRIP8; dim-2 (▪)]. The transcription rates of the amRIP8 allele in the methylated (N617) and unmethylated (N618) states was estimated from the slope of the best-fit line over the first three time points. Transcription rates of the unmethylated histone H4 gene were very similar in the sibling strains, N1223 (0.194 cpm/μg of total RNA/min) and N1224 (0.188 cpm/μg of total RNA/min).
Transcription initiation occurs from the methylated amRIP8 promoter
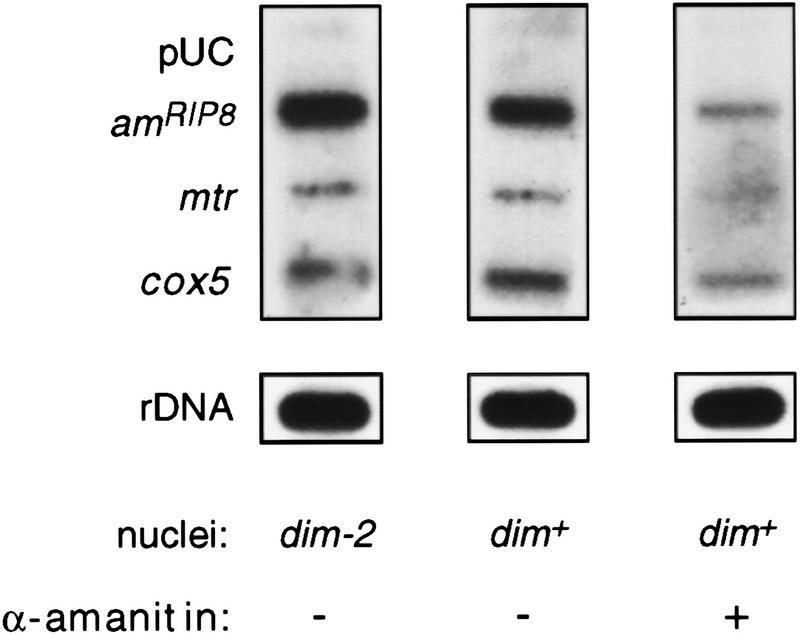
The in vivo labeling experiments demonstrated that methylation inhibited transcription of the amRIP8 allele but did not identify the level at which this effect was occurring. To investigate whether initiation of transcription was inhibited, we performed nuclear run-on assays, which measure transcription from polymerases engaged when the nuclei are isolated (Groudine et al. 1981; Schilling and Farnham 1994). The nuclear run-on assay has been used successfully to demonstrate transcriptional regulation of Neurospora genes (Paietta 1989; Sommer et al. 1989; Loros and Dunlap 1991). To validate the assay for the am gene, we took advantage of a strain containing a promoter deletion that eliminates the production of am transcripts (M. Rountree and E. Selker, unpubl.). Nuclear run-on reactions performed with nuclei from this strain showed no am transcriptional activity, whereas reactions performed with nuclei from wild-type cells showed robust signals, suggesting that the am signal results from normal initiation dependent on the am promoter (data not shown). Labeled RNA samples produced in the nuclear run-on reactions performed with the methylated (dim+) amRIP8strain (N617) or with the unmethylated (dim-2) amRIP8 strain (N618) were used to probe slotblots with denatured plasmids containing genes of interest (Fig. 5). The signals obtained for genes assayed for positive controls, mtr, cox5, and rDNA, appeared equal using RNA produced with the dim-2 or dim+ nuclei. Interestingly, the signals obtained for the methylated and unmethylated amRIP8 templates were also similar. The pUC19 plasmid, which was included as a control for nonspecific hybridization, did not give a significant signal. Four independent sets of reactions produced equivalent results. Quantitation of the results depicted in Figure 5, standardized with mtr, showed that the signal from the unmethylated amRIP8 allele was 69% of that from the methylated allele. This difference cannot account for the greater than fivefold difference in transcript levels and transcription rates measured for the unmethylated and methylated amRIP8 allele. Thus, these results suggest that transcription initiation from the amRIP8 allele is not significantly inhibited by methylation.
Figure 5.
Analysis of amRIP8 transcription in dim+ and dim-2 backgrounds by the nuclear run-on assay. Slot–blots containing 5 μg of linearized denatured plasmids or 5 μg of a denatured rDNA fragment were probed with [32P]UTP-labeled transcripts isolated from nuclear run-on reactions. The pUC19 plasmid was included as a negative control, whereas the rDNA fragment and the plasmids containing the mtr and cox5 genes served as positive controls. The first two panels show representative blots probed with labeled RNA from nuclear run-on reactions conducted with nuclei isolated from N617 (amRIP8; dim+) and N618 (amRIP8; dim-2). The right panel shows a blot probed with labeled RNA from a run-on reaction performed with N617 nuclei in the presence of 1 mg/ml of α-amanitin.
Although the control reactions with the am promoter deletions suggested that the observed signals were specific, we did one additional control experiment to eliminate the possibility that the strong signals obtained for the amRIP8 allele were somehow attributable to nonspecific hybridization of the abundant rRNA. To test this possibility, we checked whether the signals were inhibited by α-amanitin under conditions that were known to inhibit RNA polymerase II by up to 80% (but not polymerase I or III; Tyler and Giles 1985). As expected, the α-amanitin did not cause a reduction in the rRNA signals in the nuclear run-on experiments (Fig. 5). Signals from the RNA polymerase II-transcribed genes mtr, cox5, and amRIP8 were dramatically reduced, however, demonstrating that the amRIP8 signal was not attributable to cross-hybridization of the rRNA (Fig. 5; data not shown).
Distribution of RNA polymerase II along the methylated and unmethylated amRIP templates
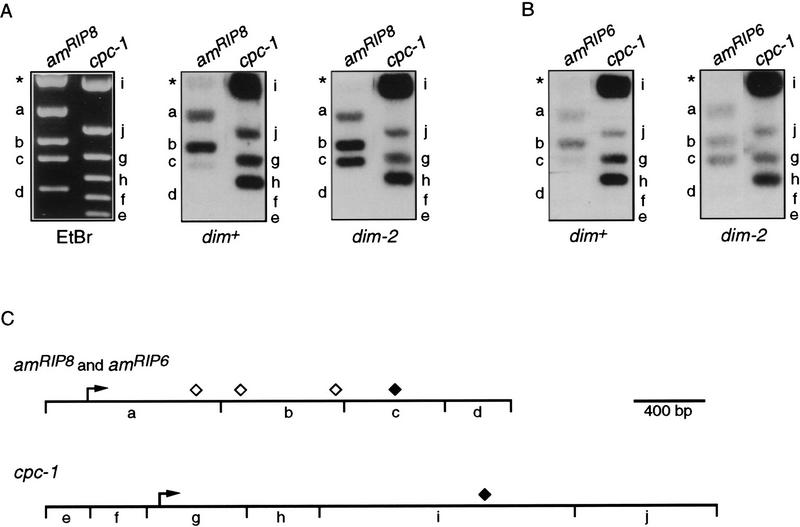
Results described above indicated that methylation inhibited transcription of amRIP8 without significantly affecting transcription initiation. To determine whether the effect of methylation was at the level of transcription elongation, we used the nuclear run-on assay to localize transcriptional activity of the amRIP8 allele, when methylated or unmethylated. Plasmids containing amRIP8 or cpc-1 sequences were digested with restriction enzymes to divide these genes into fragments. The fragments were separated by gel electrophoresis, visualized with ethidium bromide (Fig. 6A), transferred to membranes, and probed with RNA labeled in nuclear run-on reactions conducted with nuclei from the methylated amRIP8 (dim+) strain (N617) or the unmethylated amRIP8 (dim-2) strain (N618) in the presence of limiting uridine triphosphate. Labeled RNA from the dim-2 strain (N618) gave strong signals for all three of the amRIP8 fragments (Fig. 6A, a–c). In contrast, the labeled RNA from the dim+ strain (N617) showed little hybridization to the downstream fragment, c (Fig. 6A), but gave strong signals for fragments a and b. The comparable signals obtained for the upstream amRIP8 fragments (a and b) for the dim+ and dim-2 strains underscores the lack of transcriptional inhibition by methylation at the level of initiation. Little or no signal was obtained for the fragment beyond the region normally transcribed (d) or the vector (Fig. 6A,*) using RNA made from either the dim+ (N617) or dim-2 (N618) strains. Results of probing the cpc-1 gene fragments with RNA from the two strains were equivalent (Fig. 6A). No signal was seen for either of the two fragments that lie upstream of the cpc-1 transcription start site (Fig. 6A, e and f). These results were reproducible in four independent sets of reactions (data not shown). In addition, results equivalent to these with amRIP8 were obtained with amRIP6 in reactions conducted using nuclei from dim+ (N1421) or dim-2 (N1422) strains (Fig. 6B). That the signals obtained for the most upstream fragments of amRIP8, amRIP6, and cpc-1 (Fig. 6), as well as arg-2 (data not shown), were low compared with the downstream fragments suggests that the engaged RNA polymerases progressed along the template before the addition of [32P]UTP in the run-on reactions, perhaps during isolation of the nuclei. In some cases, progression of polymerases was significant enough to result in very little signal for the most 5′ fragment, confirming that little or no initiation was occurring in these reactions (data not shown). Increasing the concentration of EDTA in the nuclei isolation buffer and decreasing the time for nuclei isolation appeared to reduce this premature movement of the polymerases (data not shown). Altogether, these results demonstrate that methylation prevented elongation until after the nuclei were isolated. We conclude that DNA methylation can affect the distribution of RNA polymerases along a methylated gene.
Figure 6.
Nuclear run-on analysis of amRIP8 and amRIP6 transcription elongation. Plasmids (5 μg) containing amRIP8 (pBM7), wild-type am (pMS2), or cpc-1 (pCPC1-2) were digested with the appropriate enzymes: pBM7 and pMS2 with BamHI and XmnI and pCPC1-2 with Bsu36I and PstI. Fragments were separated on 1.5% agarose gels, transferred to nylon membranes, and probed with [32P]UTP-labeled transcripts from nuclear run-on reactions. (A) A representative ethidium bromide-stained gel is shown at left with the restriction fragments of amRIP8 and cpc-1 labeled with lowercase letters corresponding to the fragments indicated in the restriction maps (C). Blots probed with [32P]UTP-labeled transcripts from nuclear run-on reactions performed with dim+ (amRIP8) nuclei (middle) or dim-2 (amRIP8) nuclei (right). (B) Blots containing separated wild-type am and cpc-1 fragments were probed with [32P]UTP-labeled transcripts from nuclear run-on reactions performed with dim+ (amRIP6) nuclei (left) or dim-2 (amRIP6) nuclei (right). The weaker signals obtained for amRIP6, when compared with amRIP8, were most likely attributable to hybridization of the amRIP6 transcripts to wild-type am sequences. (C) Maps of amRIP8, amRIP6, and cpc-1. (⋄) The approximate ends of the amRIP8 transcripts; (♦) the ends of the amRIP6 and cpc-1 transcripts.
Same effect at a second locus
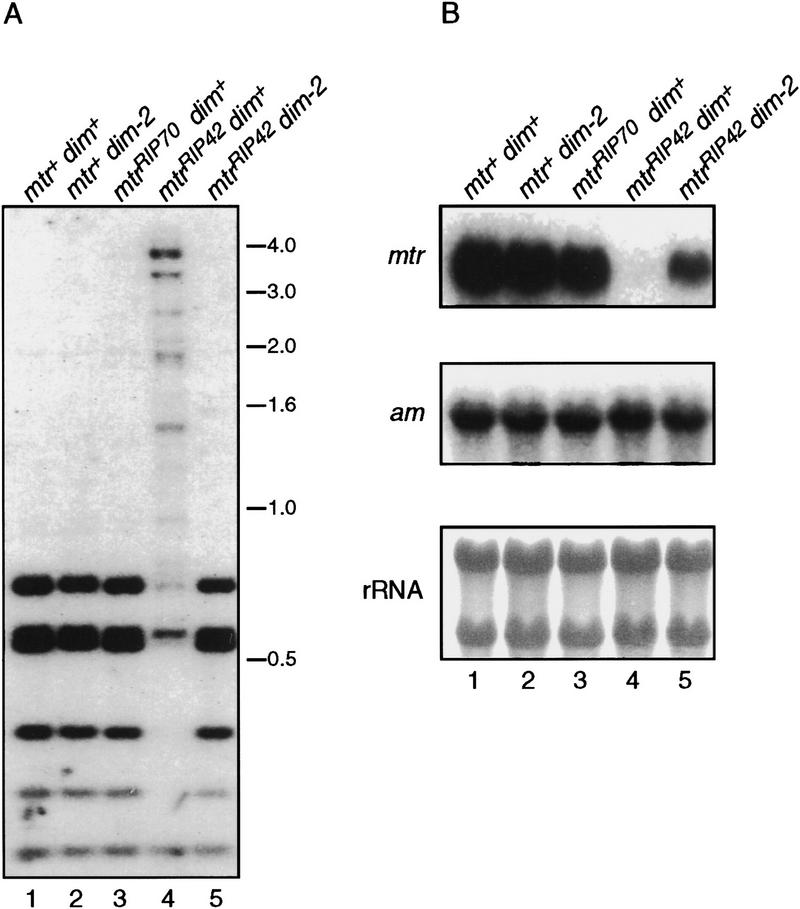
To explore the possibility that the inhibitory effect of methylation on transcription described above is a general function of methylation associated with genes altered by RIP, we analyzed a methylated allele (mtrRIP42; Fig. 7A, lane 4) from a second gene that is normally unmethylated, mtr, the structural gene for the Neurospora neutral amino acid permease (Koo and Stuart 1991). This allele and a control allele lacking detectable methylation (mtrRIP70; Fig. 7A, lanes 1–3) were obtained by first building a strain with an ectopic copy of the mtr gene, crossing the strain to unleash RIP, and then selecting progeny that had inherited a single, mutated, allele of mtr (M. Rountree, A. Hagemann, and E. Selker, unpubl.). The methylation associated with the mtrRIP42 allele was upstream of the transcription start site in the presumptive promoter region. Treatment of the mtrRIP42 strain with 5-azaC resulted in only partial reduction of methylation, perhaps because of the permease defect of this strain (data not shown). The mtrRIP42 allele was therefore crossed into a dim-2 background to test the effect of the methylation (Fig. 7A, lane 5). Northern analysis of total RNA from this strain showed the transcript level from the unmethylated mtr allele (mtrRIP70) was equivalent to that of the wild type in both dim+ and dim-2 backgrounds (Fig. 7B, lanes 1–3). No mtr transcripts were detected from the methylated (dim+) mtrRIP42 allele (Fig. 7B, lane 4); however, when methylation was prevented by the dim-2 mutation, transcripts accumulated (Fig. 7B, lane 5).
Figure 7.
The effect of methylation on mtrRIP42 transcript accumulation. (A) Southern analysis of genomic DNA from strains N1164 [mtr+; dim+ (lane 1)], N540 [mtr+; dim-2 (lane 2)], N1165 [mtrRIP70; dim+ (lane 3)], N1185 [mtrRIP42; dim+ (lane 4)], and N1183 [mtrRIP42; dim-2 (lane 5)]. Genomic DNA (750 ng) was digested with the methylation-sensitive restriction enzyme Sau3A1 and hybridized to an mtr probe. Size standards (kb) are indicated at right. The Southern blot was stripped and reprobed with the unmethylated am gene to ensure complete digestion of the DNA (data not shown). (B) Corresponding Northern analysis of total RNA (10 μg) from the strains depicted in the Southern blot. The blot at top was probed with mtr, whereas the equivalent blot in the middle was probed with am. The blot at bottom is a methylene blue staining of the rRNA from the blot at top.
Transcription from the mtrRIP42 allele was also assessed using nuclear run-on assays with nuclei isolated from methylation proficient (dim+) and methylation deficient (dim-2) strains. The signals obtained for the methylated and unmethylated templates were very similar (data not shown), as with amRIP8. Thus again, despite promoter methylation, initiation of transcription was not appreciably impaired by the DNA methylation.
Discussion
We investigated possible effects of DNA methylation on transcription of genes altered by RIP in Neurospora crassa. Availability of the dim-2 mutation, which abolishes methylation in Neurospora (Foss et al. 1993), allowed us to distinguish between effects of mutations and effects of DNA methylation. None of the methylated amRIP alleles or the methylated mtrRIP42 allele showed appreciable levels of steady-state transcripts. When methylation was prevented with 5-azaC or dim-2, however, steady-state transcript levels from these alleles increased dramatically. It is formally possible that loss of methylation activated a regulator of the am and mtr genes, but this is unlikely for several reasons. Despite extensive genetic research on these genes, no such regulator has been identified. Also, no methylated normal (i.e., non-RIPed) protein coding genes have been found in Neurospora. Furthermore, our observation that loss of methylation throughout the genome, induced either by 5-azaC treatment or by using the dim-2 mutation, had no effect on the steady state transcript levels from the unmethylated wild-type am and mtr genes (see Fig. 2 and Fig. 7), argues against the existence of a methylation-sensitive regulator of these genes. Thus, almost certainly, methylation of the amRIP alleles and the mtrRIP42 allele was responsible for reduced transcript levels. It was conceivable that the reduction of transcripts caused by methylation resulted from reduced transcription or reduced transcript stability. By labeling RNA in vivo, we determined that the unmethylated amRIP8 allele was transcribed over five times faster than the methylated version, whereas the half-lives of their transcripts were equivalent. Thus, methylation inhibited transcription per se. To determine the level at which methylation was inhibiting transcription, we performed nuclear run-on experiments. Our results corroborated reports that nuclear run-on assays detect transcription from transcription complexes established before isolation of the nuclei (Groudine et al. 1981; Schilling and Farnham 1994). Elongation, but not initiation, of transcription occurred in vitro. We verified that run-on signals for the am gene required the am promoter and were sensitive to α-amanitin. Unexpectedly, our results demonstrated that dense promoter methylation did not appreciably interfere with initiation of transcription. Additional assays revealed a marked difference in the distribution of active RNA polymerase complexes along the unmethylated and methylated amRIP8 and amRIP6 templates. Active complexes were detected along the entire length of the unmethylated templates. The methylated templates lacked transcriptional activity in the downstream portions of the gene, however, implying that methylation affects elongation of RNA polymerase complexes.
Polymerases paused or arrested in the 5′ portion of the methylated amRIP8 and amRIP6 templates were activated apparently under the conditions of the nuclear run-on assays. A similar override of transcriptional regulation in nuclear run-on assays has been seen for the uninduced Drosophila hsp70 and hsp26 genes (Rougvie and Lis 1988; Vazquez et al. 1993), the murine dhfr gene (Schilling and Farnham 1994), and the human proto-oncogene c-myc (Eick et al. 1994). In each case the signals obtained in the run-on assays for the nonactivated genes was shown to be attributable to artificial activation of RNA polymerase molecules paused near the start of transcription. Movement of the RNA polymerase molecules before and during the nuclear run-on assays made it impossible to determine where polymerase molecules had stalled on the methylated template in vivo.
The results of our first nuclear run-on experiments are reminiscent of some results with transgenic plants that showed gene silencing by introduced homologous sequences (de Carvalho et al. 1992; Dehio and Schell 1994; Ingelbrecht et al. 1994; Smith et al. 1994; Mueller et al. 1995). In many cases, the silenced genes were methylated, and like the methylated alleles that resulted from RIP (e.g., amRIP5, amRIP6, amRIP8, and mtrRIP42), they produced abnormally low levels of stable mRNA. In the plant systems, it is not yet known whether methylation is necessary for the reduction in transcript levels. Some silenced plant genes appear to be transcribed as assayed by the nuclear run-on procedure, and this led investigators to conclude that the genes were silenced post-transcriptionally. In light of our findings, it may be prudent to reexamine this conclusion. It is generally assumed that inhibition of transcription by DNA methylation occurs primarily, if not exclusively, at the level of initiation. There are previous findings, however, that have suggested that methylation can inhibit elongation. Results of transfection experiments have shown that methylation of just the promoter–distal portion of a gene can, in some cases, interfere with expression (Keshet et al. 1985). Additional evidence comes from a study of methylation induced premeiotically (MIP) in the fungus Ascobolus immersus. MIP is a gene silencing process similar to RIP that results in extensive methylation without mutations (Rhounim et al. 1992; Rossignol and Faugeron 1994). Barry and colleagues (1993) showed that methylation of a portion of the coding region of a gene can block transcription elongation. The effect of methylation by MIP on transcription initiation was not tested. The results of our nuclear run-on assays indicate that functional transcription complexes are able to assemble at methylated promoters. It is noteworthy in this context that methylation of all CpG sites within the mouse metallothionein 1 promoter does not prevent binding of the transcription factors TFIID and TFIIA to the TATA region in vitro (Levine et al. 1992).
We do not yet know how methylation effects transcription elongation. In principle, methylation could directly or indirectly interfere with transcription elongation by blocking progression of the polymerase or by preventing activation of the transcription complex bound at the promoter. Some transcriptional activators, such as those responsible for activating the c-myc gene, have been shown to enhance transcription by increasing the processivity of transcription complexes (Yankulov et al. 1994). Although the possibility that methylation could prevent the activation of the transcription complex is intriguing, there are hints that this was probably not the case in our system. First, the methylation associated with amRIP8 does not extend appreciably beyond the proximal promoter into the region of the upstream enhancer-like elements. Second, preliminary run-on experiments using a strain with a deletion of the am upstream enhancer-like elements revealed that this region is required for efficient transcription initiation (M. Rountree and E. Selker, unpubl.). Thus, it seems unlikely that methylation was interfering with transcription elongation from the amRIP alleles by preventing the action of the enhancer-like elements. It remains possible, however, that methylation prevented the binding of a transcription factor required for elongation, but not for initiation, such as TFIIH (Yankulov et al. 1996). It is also possible that methylation interfered with elongation, per se. If so, the fact that heavily methylated templates were efficiently transcribed in our nuclear run-on reactions suggests that the inhibitory effect was indirect.
Although methylation can directly interfere with binding of some transcription factors (Kovesdi et al. 1987; Watt and Molloy 1988) and may directly inhibit transcription initiation in some cases, there is precedent for indirect effects of methylation on transcription. For example, Graessmann and Graessmann (1993) showed that methylated herpes simplex virus thymidine kinase gene introduced into nuclei of rat cells remains active for many hours. Bird and his colleagues showed that inhibition of transcription by methylation in vitro is an indirect effect and presented evidence that MeCPs are responsible for inhibition of transcription by methylation (Boyes and Bird 1991, 1992; Nan et al. 1997). Although it is generally assumed that the effect of methyl-DNA-binding proteins is on promoter function, they might also inhibit elongation. The effects of methyl-DNA-binding proteins may depend in each case on several factors including the distribution and density of methylated sites in the promoter and coding regions as well as the characteristics of the promoter (Bird 1992). The inhibition of transcription elongation that we observed in Neurospora involved templates with high levels of methylation within the promoter and coding region; they were not particularly CG poor, and most cytosines were methylated (Selker et al. 1993). Of course it is also possible that methyl-DNA-binding proteins work differently in different organisms.
Our finding that the DNA methylation frequently associated with sequences mutated by RIP blocks transcription raises both mechanistic and evolutionary questions. Could inhibition of transcription from a thoroughly mutated gene be advantageous? One can imagine scenarios in which transcripts from defective genes, such as those resulting from RIP, would be detrimental to an organism. For example, a transcript from a mutated retroelement might still be copied by reverse transcriptase and then inserted into the genome. In addition, the possibility that a mutant transcript could be translated into a poisonous product cannot be dismissed. It is also conceivable that an aberrant transcript might itself have negative effects. It has been suggested, for example, that gene silencing in vegetative cells of Neurospora by homologous transgenes (“quelling”) may be caused by aberrant transcripts from the introduced sequences (Cogoni et al. 1996). Thus, methylation of sequences altered by RIP, like RIP itself, may limit damage to the structure and function of the genome.
Materials and methods
Strains, media, and genetic procedures
Neurospora strains used in this study are listed in Table 1. Standard Neurospora culture conditions and genetic techniques were used (Davis and DeSerres 1970), except as indicated.
Table 1.
Neurospora crassa strains
| Strain
|
Genotype
|
Source or reference
|
|---|---|---|
| N150 | wild type | FGSCa 2489 |
| N193 | pyr-1; mtr; cot-1; pan-2; nic-3 | R. Metzenberg (Stanford University, CA) |
| N665 | amRIP2; lys-1; inl | Singer et al. (1995) |
| N673 | amRIP5; lys-1 | Singer et al. (1995) |
| N675 | amRIP6; lys-1; inl | Singer et al. (1995) |
| N1421 | amRIP6 | this study |
| N1422 | amRIP6; dim-2 | this study |
| N617 | amRIP8; lys-1 | Singer et al. (1995) |
| N619 | am132; dim-2 | laboratory collection |
| N618 | amRIP8; dim-2 | this study |
| N1223 | amRIP8; pyr-1; mtr | this study |
| N1224 | amRIP8; dim-2; pyr-1; mtr | this study |
| N540 | dim-2 | laboratory collection |
| N1164 | trp-2 | this study |
| N1165 | mtrRIP70; trp-2 | this study |
| N1185 | mtrRIP42; trp-2 | this study |
| N1183 | mtrRIP42; dim-2; trp-2 | this study |
Fungal Genetics Stock Center.
Nucleic acid isolation
Conidia (∼1 × 107 conidia/ml) were germinated in 50 ml of Vogel’s liquid media with the appropriate supplements at 33°C, shaking at 200 rpm for 3 hr. The culture was then split into two 30-ml Corex tubes: one for RNA isolation containing ∼10 ml of crushed ice and one for DNA isolation. Total RNA was extracted from the germinated conidia using a procedure adapted from McKinney et al. (1993). The germinated conidia were pelleted by centrifugation at 5000 rpm in a Sorvall SS34 rotor for 5 min at 4°C, resuspended in 1 ml of cold 10 mm Tris (pH 7.6), 1 mm EDTA (TE), and transferred to a microcentrifuge tube. The germinated conidia were pelleted by centrifugation for 1 min at 4°C. Approximately 300 μl of acid-washed 240- to 300- μm glass beads in sterile water, 350 μl of phenol/chloroform/isoamyl alcohol (25:24:1), and 350 μl of NETS (300 mm NaCl, 1 mm EDTA, 10 mm Tris-HCl, 0.2% SDS) were added to the pelleted germlings and vortexed at top speed in a multitube vortexer for 10 min. The aqueous phase was separated from the organic phase of each sample by centrifugation for 5 min at 4°C. The aqueous phase was removed and split between two microcentrifuge tubes containing 700 μl of ice-cold 95% ethanol to precipitate the RNA. After incubation on ice for ∼15 min, the RNA was collected by centrifugation, washed with 70% ethanol, and resuspended in sterile water treated with diethylpyrocarbonate (DEPC). Genomic DNA was isolated as described previously (Oakley et al. 1987) except that the germinated conidia were disrupted by vortexing for 10 min in the salt extraction solution with ∼300 μl of glass beads.
Northern analysis
Total RNA (10 μg) was denatured and fractionated through 1.2% agarose gels containing MOPS buffer as described (McKinney et al. 1993), except formaldehyde was omitted from the agarose gels (Liu and Chou 1990) and the RNA was transferred onto a nylon membrane (Zetabind; Cuno, Inc.) in 5× SSC. The RNA was affixed to the membrane by UV cross-linking (FisherBiotech UV Crosslinker). The rRNA was visualized by soaking the blots in methylene blue stain (0.03% methylene blue, 0.3 m NaOAc at pH 5.2) with gentle agitation for 2–3 min, followed by four to five rounds of rinsing in deionized water (Wilkinson et al. 1990). Membranes were prehybridized for 1–2 hr at 60°C in 250 mm sodium phosphate (pH 7.4), 7% SDS, 1 mm EDTA, and 5% dextran sulfate. Hybridization probes were prepared by the random hexamer method (Feinberg and Vogelstein 1983) and added to the prehybridization solution. Hybridizations were performed at 60°C for 16 hr. Membranes were washed three or four times for 20 min each in 0.1× SSC, 0.5% SDS at 60°C and exposed to film with intensifying screens at −70°C.
Southern analysis
Genomic DNA (750 ng) was digested with at least a fivefold excess of restriction enzyme and analyzed by standard Southern hybridization methods (Sambrook et al. 1989). Before hybridization, the DNA was UV cross-linked to the Zetabind membranes. The membranes were then baked at 60°C for ∼1 hr and then washed for 1 hr at 65°C in 0.1× SSC, 0.5% SDS. The “1-kb ladder” set of standards was purchased from GIBCO BRL. Hybridization probes were made by the random hexamer method (Feinberg and Vogelstein 1983). After hybridization, the blots were washed three to four times in Zeta-wash (50 mm NaCl, 20 mm NaHPO4 at pH 6.8, 1 mm EDTA, 0.1% SDS) at 60°C and exposed to film. Southern blots were stripped and reprobed with an unmethylated sequence to verify that the restriction digests were complete.
Methylation analysis
To estimate the level of methylation at a single methylation-sensitive restriction site, genomic DNA was digested to completion with a given methylation-sensitive restriction enzyme (e.g., BamHI) in conjunction with a methylation-insensitive restriction enzyme (EcoRV or MboI) that has sites flanking the assay site. The methylation level was estimated using an Ambis Optical Imaging System (Ambis, Inc.). The lack of methylation at some of the sites flanking amRIP8 was indicated by complete digestion in single enzyme digests.
Nuclear run-on assays
Crude nuclei were isolated as described by Loros and Dunlap (1991), except that germinated conidia were used in place of mycelium. Conidia (∼1 × 108/ml) were germinated for 3 hr and pelleted in a 30-ml Corex tube. All steps in the isolation of nuclei were performed at 4°C. Cold acid-washed glass beads (750 μl) and 4 ml of solution A (1 m sorbitol, 7% Ficoll, 20% glycerol, 5 mm MgCl2, 10 mm CaCl2, 1% Triton X-100, 5 mm EDTA at pH 7.5) were added to the pellet. The germinated conidia were disrupted by vortexing for 15 sec four times with 30 sec on ice between the pulses. The solution was then clarified by low-speed centrifugation (1500g) for 10 min at 4°C and aliquoted to microcentrifuge tubes. The nuclei were pelleted by centrifugation (15,000g) for 10 min at 4°C, the supernatant was removed, and the nuclei were frozen on dry ice then stored at −70°C. Run-on reactions were performed as described (Hollick and Gordon 1993), with some modifications. Nuclei were thawed on ice and pelleted by a brief spin in a microcentrifuge and resuspended in 100 μl of nuclei resuspension buffer (50 mm Tris-HCl at pH 7.5, 5 mm MgCl2, 50% glycerol, 10 mm 2-mercaptoethanol). Reactions were initiated by the addition of 100 μl of the reaction mix [80 mm (NH4)2SO4, 4 mm MgCl2, 0.5 mm ATP, GTP, and CTP, 5.0 μm UTP, 0.3 μm phosphocreatine, 25 μg/ml of creatine phosphokinase, 200 μCi of [α-32P]UTP (3000 μCi/mmole; NEN), and 100 units of RNase inhibitor (U.S. Biochemical)] to the resuspended nuclei (∼107). In the reactions shown in Figure 6, the nuclei were isolated in solution A containing 25 mm EDTA and unlabeled UTP was not included. Reactions were incubated at 33°C for 10 min. The RNA polymerase II inhibitor α-amanitin (1 mg/ml) was added to some of the reactions to test the sensitivity of the production of the RNA polymerase II transcripts (Loros and Dunlap 1991). The nuclei were pelleted at 6500 rpm in a microcentrifuge for 30 sec. They were then resuspended in 500 μl of NMC [75 mm (NH4)2SO4, 4 mm MgCl2, plus 50 units of RNase inhibitor] and incubated in the presence of 20 μg/ml of RNase-free DNase (Pharmacia) at 37°C for 30 min. The nuclei were pelleted as before and resuspended in 200 μl of TES (10 mm Tris-HCl at pH 7.6, 5 mm EDTA, 1% SDS) containing 100 μg/ml of proteinase K and incubated at 42°C for 30 min. The solution was phenol/chloroform/IAA (25:24:1) extracted, and the organic phase was back-extracted with 200 μl of TES. The two aqueous fractions were pooled and chloroform-extracted. The RNA was then precipitated in ethanol in the presence of 2 m NH4OAc. A second DNase treatment was performed in 200 μl of NMC, followed by a phenol/chloroform/IAA extraction/back-extraction and ethanol precipitation as before. Plasmids (5 μg) were either linearized by digestion with a restriction enzyme, denatured with 0.4 n NaOH, and transferred through a slot–blot apparatus onto a nylon membrane (Zetabind) or restriction fragments were separated on a 1.5% agarose gel before transfer to nylon membranes. Blots were cross-linked and prehybridized in 0.5 m sodium phosphate (pH 7.0), 7% SDS, 5 mm EDTA at 64°C for 2–3 hr in 2-ml screw-top microcentrifuge tubes before the labeled RNA (typically 5 × 106 cpm/ml) was added. Hybridization was performed at 64°C for 10 min, and then the temperature was lowered to 55°C for 36 hr. Blots were washed twice for 15 min in 1× SSC, 1% SDS at room temperature and twice for 15 min in 0.1× SSC, 1% SDS at 55°C. The blots were exposed to film with an intensifying screen at −70°C.
Labeling RNA in vivo
Conidia (5 × 107 conidia/ml) were germinated at 33°C with shaking (200 rpm) in a modified Vogel’s liquid medium, in which the NH4NO3 was replaced by 1.5 times (moles) as much NaNO3 (Buxton and Radford 1982), and containing alanine (1 mg/ml) and uridine (37.5 μg/ml). After 3 hr, prewarmed [5,6-3H]uridine (35–50 Ci/mmole; NEN) was added to 35 μCi/ml. At various times, 10-ml aliquots were removed and incorporation was arrested by the addition of two volumes of ice-cold ethanol plus cycloheximide (100 μg/ml, final concentration) in a −70°C dry-ice ethanol bath (Crabeel et al. 1990). For the “chase” portion of the experiment, unlabeled uridine (0.5 mg/ml) was added to the cultures after 30 min of labeling. RNA was extracted from the samples as described above and treated with DNase I as described for the nuclear run-on experiments. Labeled total RNA (typically 200 μg) was hybridized to blots as described in the nuclear run-on reactions at 60°C for 36 hr. Blots were washed for 15 min in 0.1× SSC, 1% SDS at room temperature, 30 min in 1× SSC at 60°C, 30 min in 1× SSC containing 20 μg/ml of RNase A at room temperature, and 30 min in 1× SSC at room temperature. The blots were dried at room temperature, cut into individual strips, and placed into scintillation vials. The [3H]RNA hybridized to each slot was measured as described (Schilling and Farnham 1994). The RNA was hydrolyzed by incubation in 500 μl of 0.1 n NaOH at 60°C for 1 hr and neutralized with 50 μl of 1 n HCl, before scintillation cocktail (Ecolume) was added, and samples were counted three times in a scintillation counter (Beckman LS6800). The signal obtained for the pUC19 vector on each blot was subtracted from the signals for gene-containing strips. The corrected amRIP8 (pBM7) signal was normalized to the signal obtained for the histone H4 gene (pPG21) at each time point. The rate of transcription was determined from the slope of a best-fit line generated for the first 10 min of labeling (Microsoft Excel). The approximate half-life of a transcript was determined by the approach to steady-state labeling method and checked by the classical pulse–chase method (Greenberg 1972; Crabeel et al. 1990; Herrick et al. 1990).
Plasmids
The following plasmids were used in these studies: pUC19 (Yanisch et al. 1985), pMS2 (containing the wild-type am gene; Miao et al. 1994), pBM7 (containing the amRIP8 allele; Singer et al. 1995), pCVN2.9 (containing the mtr gene; Koo and Stuart 1991), pSRcox5 (containing the cox5 gene; gift from M. Sachs, Oregon Graduate Institute, Portland), pCPC1-2 (containing the cpc-1 gene; Paluh et al. 1988), and pKH1 (containing the entire 9.2-kb rDNA repeat unit; K. Haack and E. Selker, unpubl.).
Acknowledgments
We thank Karen Sprague, Stephen Baylin, Michael Freitag, Jeremy Graff, and Diane Hawley for comments on the manuscript. We are grateful to Jay Hollick, Joe Horecka, Brian Margolin, Ann Hagemann, and Charles Foulds for technical assistance and useful discussions. This work was supported by U.S. Public Health Service grant GM-35690 from the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL selker@oregon.uoregon.edu; FAX (541) 346-5011.
References
- Barry C, Faugeron G, Rossignol J-L. Methylation induced premeiotically in Ascobolus: Coextension with DNA repeat lengths and effect on transcript elongation. Proc Natl Acad Sci. 1993;90:4557–4561. doi: 10.1073/pnas.90.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS, Webber AL, Brunkow ME, Tilghman SM. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes & Dev. 1993;7:1663–1673. doi: 10.1101/gad.7.9.1663. [DOI] [PubMed] [Google Scholar]
- Bednarik DP, Duckett C, Kim SU, Perez VL, Griffis K, Guenther PC, Folks TM. DNA CpG methylation inhibits binding of NF-kB proteins to the HIV-1 long terminal repeat cognate DNA motifs. New Biol. 1991;3:969–976. [PubMed] [Google Scholar]
- Beelman CA, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- Ben-Hattar J, Beard P, Jiricny J. Methylation in CTF and Sp1 recognition sites of an HSV tk promoter: Effects on transcription in vivo and on factor binding in vitro. Nucleic Acids Res. 1989;17:10179–10190. doi: 10.1093/nar/17.24.10179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- ————— Repression of genes by DNA methylation depends on CpG density and promoter strength: Evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11:327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhausen G, Wittig B, Graessmann M, Graessmann A. Chromatin structure is required to block transcription of the methylated herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci. 1987;84:1177–1181. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton FP, Radford A. Nitrogen metabolite repression of fluoropyrimidine resistance and pyrimidine uptake in Neurospora crassa. Mol & Gen Genet. 1982;186:259–262. doi: 10.1007/BF00331859. [DOI] [PubMed] [Google Scholar]
- Cambareri EB, Jensen BC, Schabtach E, Selker EU. Repeat-induced G-C to A-T mutations in Neurospora. Science. 1989;244:1571–1575. doi: 10.1126/science.2544994. [DOI] [PubMed] [Google Scholar]
- Cambareri EB, Foss HM, Rountree MR, Selker EU, Kinsey JA. Epigenetic regulation of a transposon-inactivated gene in Neurospora is dependent on DNA methylation. Genetics. 1996;143:137–146. doi: 10.1093/genetics/143.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caroline DF, Davis RH. Pyrimidine synthesis in Neurospora crassa: Regulation of enzyme activities. J Bacteriol. 1969;100:1378–1384. doi: 10.1128/jb.100.3.1378-1384.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Kinsey JA. Sequential gel mobility shift scanning of 5′ upstream sequences of the Neurospora crassa am (GDH) gene. Mol & Gen Genet. 1994;242:399–403. doi: 10.1007/BF00281789. [DOI] [PubMed] [Google Scholar]
- Cogoni C, Ireland JT, Schumacher M, Schmidhauser TJ, Selker EU, Macino G. Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA:DNA interactions or DNA methylation. EMBO J. 1996;15:3153–3163. [PMC free article] [PubMed] [Google Scholar]
- Comb M, Goodman HW. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18:3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabeel M, Lavalle R, Glansdorff N. Arginine-specific repression in Saccharomyces cerevisiae: Kinetic data on ARG1 and ARG3 mRNA transcription and stability support a transcriptional control mechanism. Mol Cell Biol. 1990;10:1226–1233. doi: 10.1128/mcb.10.3.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosthwaite SK, Loros JJ, Dunlap JC. Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell. 1995;81:1003–1012. doi: 10.1016/s0092-8674(05)80005-4. [DOI] [PubMed] [Google Scholar]
- Davis RH, DeSerres FJ. Genetic and microbial research techniques for Neurospora crassa. Methods Enzymol. 1970;17A:47–143. [Google Scholar]
- de Carvalho F, Gheysen G, Kushnir S, Montagu MV, Inze D, Castresana C. Suppression of β-1,3-glucanase transgene expression in homozygous plants. EMBO J. 1992;11:2595–2602. doi: 10.1002/j.1460-2075.1992.tb05324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehio C, Schell J. Identification of plant genetic loci involved in a posttranscriptional mechanism for meiotically reversible transgene silencing. Proc Natl Acad Sci. 1994;91:5538–5542. doi: 10.1073/pnas.91.12.5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden S, Cedar H. Role of DNA methylation in the regulation of transcription. Curr Opin Genet Dev. 1994;4:255–259. doi: 10.1016/s0959-437x(05)80052-8. [DOI] [PubMed] [Google Scholar]
- Eick D, Kohlhuber F, Wolf DA, Strobl LJ. Activation of pausing RNA polymerases by nuclear run-on experiments. Anal Biochem. 1994;218:347–351. doi: 10.1006/abio.1994.1190. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Ferguson AT, Lapidus RG, Baylin SB, Davidson NE. Demethylation of the estrogen receptor gene in estrogn receptor-negative breast cancer cells can reactivate estrogen receptor gene expression. Cancer Res. 1995;55:2279–2283. [PubMed] [Google Scholar]
- Foss HM, Roberts CJ, Claeys KM, Selker EU. Abnormal chromosome behavior in Neurospora mutants defective in DNA methylation. Science. 1993;262:1737–1741. doi: 10.1126/science.7505062. ; correction 267: 316. [DOI] [PubMed] [Google Scholar]
- Frederick GD, Kinsey JA. Distant upstream regulatory sequences control the level of expression of the am (GDH) locus of Neurospora crassa. Curr Genet. 1990a;18:53–58. doi: 10.1007/BF00321115. [DOI] [PubMed] [Google Scholar]
- ————— Nucleotide sequence and nuclear protein binding of the two regulatory sequences upstream of the am (GDH) gene in Neurospora. Mol & Gen Genet. 1990b;221:148–154. doi: 10.1007/BF00261714. [DOI] [PubMed] [Google Scholar]
- Graessmann M, Graessmann A. DNA methylation, chromatin structure and the regulation of gene expression. In: Jost JP, Saluz HP, editors. DNA methylation: Molecular biology and biological significance. Boston, MA: Birkhäuser Verlag; 1993. pp. 404–424. [DOI] [PubMed] [Google Scholar]
- Greenberg JR. High stability of messenger RNA in growing cultured cells. Nature. 1972;240:102–104. doi: 10.1038/240102a0. [DOI] [PubMed] [Google Scholar]
- Groudine M, Peretz M, Weintraub H. Transcriptional regulation of hemoglobin switching in chicken embryos. Mol Cell Biol. 1981;1:281–288. doi: 10.1128/mcb.1.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick D, Parker R, Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick JB, Gordon MP. A poplar tree proteinase inhibitor-like gene promoter is responsive to wounding in transgenic tobacco. Plant Mol Biol. 1993;22:561–572. doi: 10.1007/BF00047398. [DOI] [PubMed] [Google Scholar]
- Ingelbrecht I, Houdt HV, Montagu MV, Depicker A. Posttranscriptional silencing of reporter transgenes in tobacco correlates with DNA methylation. Proc Natl Acad Sci. 1994;91:10502–10506. doi: 10.1073/pnas.91.22.10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irelan JT, Selker EU. Cytosine methylation associated with repeat-induced point mutation (RIP) causes epigenetic gene silencing in Neurospora crassa. Genetics. 1997;146:509–523. doi: 10.1093/genetics/146.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. Gene activation by 5-azacytidine. In: Razin A, Cedar H, Riggs AD, editors. DNA methylation: Biochemistry and biological significance. New York, NY: Springer-Verlag; 1984. pp. 165–187. [Google Scholar]
- Keshet I, Yisraeli J, Cedar H. Effect of regional DNA methylation on gene expression. Proc Natl Acad Sci. 1985;82:2560–2564. doi: 10.1073/pnas.82.9.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey JA, Helber J. Isolation of a transposable element from Neurospora crassa. Proc Natl Acad Sci. 1989;86:1929–1933. doi: 10.1073/pnas.86.6.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo K, Stuart WD. Sequence and structure of mtr, an amino acid transport gene of Neurospora crassa. Genome. 1991;34:644–651. doi: 10.1139/g91-098. [DOI] [PubMed] [Google Scholar]
- Kovesdi I, Reichel R, Nevins JR. Role of an adenovirus E2 promoter binding factor in E1A-mediated coordinate gene control. Proc Natl Acad Sci. 1987;84:2180–2184. doi: 10.1073/pnas.84.8.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A, Cantoni GL, Razin A. Methylation in the preinitiation domain suppresses gene transcription by an indirect mechanism. Proc Natl Acad Sci. 1992;89:10119–10123. doi: 10.1073/pnas.89.21.10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Liu YC, Chou YC. Formaldehyde in formaldehyde/agarose gel may be eliminated without affecting the electrophoretic separation of RNA molecules. Biotechniques. 1990;9:558. , 560. [PubMed] [Google Scholar]
- Loros JJ, Dunlap JC. Neurospora crassa clock-controlled genes are regulated at the level of transcription. Mol Cell Biol. 1991;11:558–563. doi: 10.1128/mcb.11.1.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney JD, Chang F, Heintz N, Cross FR. Negative regulation of FAR1 at the start of the yeast cell cycle. Genes & Dev. 1993;7:833–843. doi: 10.1101/gad.7.5.833. [DOI] [PubMed] [Google Scholar]
- Miao VPW, Singer MJ, Rountree MR, Selker EU. A targeted-replacement system for identification of signals for de novo methylation in Neurospora crassa. Mol Cell Biol. 1994;14:7059–7067. doi: 10.1128/mcb.14.11.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller E, Gilbert J, Davenport G, Brigneti G, Baulcombe DC. Homology-dependent resistance: Transgene virus resistance in plants related to homology-dependent gene silencing. Plant J. 1995;7:1001–1013. [Google Scholar]
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Ngô V, Gourdji D, Laverriére J-N. Site-specific methylation of the rat prolactin and growth hormone promoters correlates with gene expression. Mol Cell Biol. 1996;16:3245–3254. doi: 10.1128/mcb.16.7.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley CE, Weil C, Kretz P, Oakley B. Cloning of the riboB locus of Aspergillus nidulans. Gene. 1987;53:293–298. doi: 10.1016/0378-1119(87)90019-9. [DOI] [PubMed] [Google Scholar]
- Paietta JV. Molecular cloning and regulatory analysis of the arylsulfatase structural gene of Neurospora crassa. Mol Cell Biol. 1989;9:3630–3637. doi: 10.1128/mcb.9.9.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluh JL, Orbach MJ, Legerton TL, Yanofsky C. The cross-pathway control gene of Neurospora crassa, cpc-1, encodes a protein similar to GCN4 of yeast and the DNA-binding domain of the oncogene v-jun-encoded protein. Proc Natl Acad Sci. 1988;85:3728–3732. doi: 10.1073/pnas.85.11.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz SW, He F, Welch E, Jacobson A. Progress in nucleic acid research and molecular biology. New York, NY: Academic Press, Inc.; 1994. Nonsense-mediated mRNA decay in yeast; pp. 271–297. [DOI] [PubMed] [Google Scholar]
- Prendergast GC, Lawe D, Ziff EB. Association of Myn, the murine homolog of max, with c-myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Razin A, Cedar H. DNA methylation and genomic imprinting. Cell. 1994;77:473–476. doi: 10.1016/0092-8674(94)90208-9. [DOI] [PubMed] [Google Scholar]
- Rhodes K, Rippe RA, Umezawa A, Nehls M, Brenner D, Breindl M. DNA methylation represses the murine a1 (I) collagen promoter by an indirect mechanism. Mol Cell Biol. 1994;14:5950–5960. doi: 10.1128/mcb.14.9.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhounim L, Rossignol J-L, Faugeron G. Epimutation of repeated genes in Ascobolus immersus. EMBO J. 1992;11:4451–4457. doi: 10.1002/j.1460-2075.1992.tb05546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Selker EU. Mutations affecting the biosynthesis of S-adenosylmethionine cause reduction of DNA methylation in Neurospora crassa. Nucleic Acids Res. 1995;23:4818–4826. doi: 10.1093/nar/23.23.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol J-L, Faugeron G. Gene inactivation triggered by recognition between DNA repeats. Experientia. 1994;50:307–317. doi: 10.1007/BF01924014. [DOI] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Russell PJ, Rodland KD, Rachlin EM, McCloskey JA. Differential DNA methylation during the vegetative life cycle of Neurospora crassa. J Bacteriol. 1987;169:2902–2905. doi: 10.1128/jb.169.6.2902-2905.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schilling LJ, Farnham PJ. Inappropriate transcription from the 5′ end of the murine dihydrofolate reductase gene masks transcriptional regulation. Nucleic Acids Res. 1994;22:3061–3068. doi: 10.1093/nar/22.15.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU. Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- ————— Epigenetic phenomena in filamentous fungi: Useful paradigms or repeat-induced confusion? Trends Genet. 1997;13:296–301. doi: 10.1016/s0168-9525(97)01201-8. [DOI] [PubMed] [Google Scholar]
- Selker EU, Stevens JN. DNA methylation at asymmetric sites is associated with numerous transition mutations. Proc Natl Acad Sci. 1985;82:8114–8118. doi: 10.1073/pnas.82.23.8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU, Garrett PW. DNA sequence duplications trigger gene inactivation in Neuropora crassa. Proc Natl Acad Sci. 1988;85:6870–6874. doi: 10.1073/pnas.85.18.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU, Cambareri EB, Jensen BC, Haack KR. Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell. 1987;51:741–752. doi: 10.1016/0092-8674(87)90097-3. [DOI] [PubMed] [Google Scholar]
- Selker EU, Fritz DY, Singer MJ. Dense nonsymmetrical DNA methylation resulting from repeat-induced point mutation in Neurospora. Science. 1993;262:1724–1728. doi: 10.1126/science.8259516. [DOI] [PubMed] [Google Scholar]
- Shin TH, Paterson AJ, Grant III JH, Meluch AA, Kudlow JE. 5-azacytidine treatment of HA-A melanoma cells induces Sp1 activity and concomitant transforming growth factor α expression. Mol Cell Biol. 1992;12:3998–4006. doi: 10.1128/mcb.12.9.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MJ, Marcotte BA, Selker EU. DNA methylation associated with repeat-induced point mutation in Neurospora crassa. Mol Cell Biol. 1995;15:5586–5597. doi: 10.1128/mcb.15.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Sam J, Riggs AD. X chromosome inactivation and DNA methylation. In: Jost JP, Saluz HP, editors. DNA methylation: Molecular biology and biological significance. Boston, MA: Birkhauser Verlag; 1993. pp. 358–384. [DOI] [PubMed] [Google Scholar]
- Smith HA, Swaney SL, Parks D, Wernsman EA, Dougherty WG. Transgenic plant virus resistance mediated by unstranslatable sense RNAs: Expression, regulation, and fate of nonessential RNAs. Plant J. 1994;6:1441–1453. doi: 10.1105/tpc.6.10.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer T, Chambers JAA, Eberle J, Lauter FR, Russo VEA. Fast light-regulated genes of Neurospora crassa. Nucleic Acids Res. 1989;17:5713–5723. doi: 10.1093/nar/17.14.5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Dev. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- Tyler BM, Giles NH. Accurate transcription of cloned Neurospora RNA polymerase II-dependent genes in vitro by homologous soluble extracts. Proc Natl Acad Sci. 1985;82:5450–5454. doi: 10.1073/pnas.82.16.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardimon L, Kressmann A, Cedar H, Maechler M, Doerfler W. Expression of a cloned adenovirus gene is inhibited by in vitro methylation. Proc Natl Acad Sci. 1982;79:1073–1077. doi: 10.1073/pnas.79.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez J, Pauli D, Tissiêres A. Transcriptional regulation in Drosophila during heat shock: A nuclear run-on analysis. Chromosoma. 1993;102:233–248. doi: 10.1007/BF00352397. [DOI] [PubMed] [Google Scholar]
- Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes & Dev. 1988;2:1136–1143. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- Wilkinson M, Doskow J, Lindsey S. RNA blots: Staining procedures and optimization of conditions. Nucleic Acids Res. 1990;19:679. doi: 10.1093/nar/19.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch PC, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yankulov K, Blau J, Purton T, Roberts S, Bentley DL. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Yankulov KY, Pandes M, McCracken S, Bouchard D, Bentley DL. TFIIH functions in regulating transcriptional elongation by RNA polymerase II in Xenopus oocytes. Mol Cell Biol. 1996;16:3291–3299. doi: 10.1128/mcb.16.7.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yisraeli J, Frank D, Razin A, Cedar H. Effect of in vitro DNA methylation on β-globin gene expression. Proc Natl Acad Sci. 1988;85:4638–4642. doi: 10.1073/pnas.85.13.4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubiaga AM, Belasco JC, Greenberg ME. Nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]