Abstract
Glanders, caused by the Gram-negative, nonmotile bacterium Burkholderia mallei, is a contagious and highly fatal disease of equines. During the last decade, the number of glanders outbreaks has increased steadily. The disease also has high zoonotic significance and B. mallei is listed biological warfare agent. The complement fixation test (CFT) is a routinely used and internationally recognized test to screen equine sera for the glanders. However, discrepant results have been observed using the CFT. The low sensitivity and specificity of the CFT and enzyme-linked immunosorbent assay (ELISA) have been linked to the use of crude test antigens. We expressed a novel recombinant Burkholderia intracellular motility A (rBimA) protein in Escherichia coli for the diagnosis of equine glanders. Purified rBimA was used in an indirect ELISA format. All of the 21 true-positive serum samples used in the study tested positive, whereas only 17 of the 1,524 potentially negative sera tested positive by indirect ELISA, thus exhibiting 100% sensitivity and 98.88% specificity. Also, rBimA protein did not react with melioidosis patient and normal healthy human serum samples, showing its high specificity. The developed assay can be used as a simple and rapid tool for diagnosis of glanders in equine serum samples. An Indian patent (1328/DEL/2010) has been filed for the reagent.
INTRODUCTION
Glanders is highly contagious disease of horses, donkeys, and mules, caused by Burkholderia mallei, a Gram-negative bacterium. The World Organization of Animal Health (OIE) has been notified of the disease. B. mallei is thought to be an obligate mammalian pathogen, with solipeds serving as the reservoir for infection (14). At the turn of the 20th century, glanders was an important cause of death among horses, and there were secondary, often fatal infections in humans (19). The disease has now been eradicated from most countries of Western hemisphere through countermeasures such as intensive blood testing, rigorous killing of positive animals, and strict trading restrictions. However, glanders is still endemic in Asia, the Middle East, and Central and South America (13). Recent outbreaks have been reported from Turkey, the United Arab Emirates, Iraq, Iran, Pakistan, China, Brazil, Bahrain, and India (1, 2, 3, 6, 10, 11).
Glanders affects upper respiratory tract and lungs of equines which develop granulomas and ulcers. Further symptoms include purulent nasal discharge, pneumonia, and poor general conditions. The cutaneous form appears on the limbs and body (14). In humans, glanders is primarily an occupational disease that affects individuals who have close contact with infected animals, such as veterinarians and farmers (12, 16). The human infection can be severe and life-threatening and is always fatal if untreated or misdiagnosed (19). No vaccines are available against glanders, and little is known about the appropriate antibiotic regimen (8). As a consequence and also because of the lethal and contagious nature of the disease, B. mallei is considered an ideal agent for biological warfare (26).
Clinical and bacteriological diagnosis of glanders in equines is difficult in the early stages of the disease or if disease is not apparent. Nearly 90% of infections exist as nonclinical or latent (12). The mallein test, a hypersensitivity test is one of the most commonly test used for the diagnosis of glanders. However, various forms of mallein test have limitations of sensitivity particularly in clinically advanced cases (13). False-positive results have been reported. The mallein test is a field test, and results are available only after 48 h. The complement fixation test (CFT) is a favored serological diagnostic test and has also been recommended by the OIE. The test has a sensitivity of 97% compared to gold standard pathology. However, false-negative and false-positive results have been reported (5, 13). The CFT reagent is not easily available commercially and test is labor-intensive. Moreover, the CFT uses a crude antigen reagent preparation that is primarily lipopolysaccharide (14). Other serological tests, e.g., agglutination, precipitation, indirect hemagglutination, immunodiffusion, counterimmunoelectrophoresis, and enzyme-linked immunosorbent assay (ELISA), have also been described (4, 9, 17, 25), but they have limitations. ELISAs are unable to differentiate serologically between B. mallei and B. pseudomallei (14). The avidin-biotin dot ELISA has been described (25), but it has not yet been widely used or validated. The Rose Bengal plate agglutination test has been described for the diagnosis of glanders in horses and other susceptible animals; this test has been validated in Russia only. Recently, polysaccharide microarray technology has offered a new promising approach to improve sensitivity in serology (15). A Western blot technique using lipopolysaccharide antigen has been shown to have higher specificity than the currently used CFT (7).
Burkholderia spp. are known to form the giant cell formation through utilization of actin polymerization to propel themselves inside the host cells. Burkholderia intracellular motility A (BimA) protein, also called the hemagglutinin domain-containing protein, is located on the pole of bacterium B. pseudomallei and helps in actin tail formation in the cell. A homolog for BimA has been reported in B. mallei and B. thailandensis. It has further been reported that the amino-terminal sequence of BimA in B. mallei, B. pseudomallei, and B. thailandensis differ markedly, whereas the C-terminal sequences are conserved (21). Unique DNA sequence at 5′ end of bimA have also been exploited for designing specific PCR assays for B. mallei (23, 24). BimA is a autosecreted protein, and the amino regions are exposed at the bacterial surface (21). Therefore, we reasoned that an antibody response against this protein may be of value for diagnosis. Further, none of the tests thus far reported for the diagnosis of glanders use a defined purified antigen. The objective of present study was to generate a specific recombinant antigen of B. mallei that can be used for detecting the anti-glanders antibodies with high sensitivity and specificity. We describe here a novel recombinant BimA protein for the diagnosis of equine glanders.
MATERIALS AND METHODS
Equine serum samples.
The study was conducted on a total of 1,545 equine samples that were either collected by or presented to National Research Centre on Equines, Hisar, India, between 2007 and 2009 for routine serological investigation of the disease. The negative or positive status of the samples was determined by combination of clinical symptoms, culture, and the CFT. Group I consisted of 21 true-positive sera. The positive samples were collected from outbreaks in the Maharashtra state (10). The CFT titer of positive samples ranged from 1:16 to 1:256. CFT reagent was purchased from Bioveta, S.C. Invanovice, Hane, Czech Republic, and testing was carried out according to the OIE-recommended procedure (14). Group II consisted of the remaining 1,524 samples and were determined to be negative by the CFT. The animals also did not have any clinical presentation of glanders.
Human serum samples.
Sera from 10 culture-positive melioidosis patients (group III) were collected at Kasturba Medical College, Manipal, India. The Burkholderia isolates were identified using Mini API ID 32GN automated system. All isolates of B. pseudomallei were sensitive to ceftazidime, amoxicillin-clavulanic acid, cotrimoxazole, doxycycline, and carbapenems and resistant to gentamicin and polymyxin B (300 U). Serum samples from apparently normal healthy individuals (n = 10) (group IV) with no history of fever in past 3 months were also collected and used as negative controls.
Bacterial strains and WCS antigen.
B. mallei strain NCTC 10230 and B. pseudomallei strain NCTC 4845 were obtained from Central Public Health Laboratory, Colindale, London, England. B. mallei and B. pseudomallei were grown in glucose dextrose broth and Ashdown medium, respectively, at 37°C for 48 h. The genomic DNA was prepared by using a QIAamp minikit (Qiagen). For the preparation of whole-cell sonicated (WCS) antigen, the growth was harvested and sonicated on ice, followed by centrifugation at 8,000 × g for 20 min. The supernatant was collected and used as a WCS antigen for ELISA. The live cultures were handled in a biosafety level 3 laboratory.
Cloning of the bimA gene.
Alignment of BimA protein sequences of B. mallei, B. pseudomallei, and B. thailandensis at amino-terminal region was carried out using CLUSTAL W tool at http://www.ebi.ac.uk/Tools/msa/clustalw2/ (Fig. 1). Part of the bimA gene (accession no. YP_105472) at 5′ end was amplified by PCR using the primers HDCPF (5′-TATGCACATATGTTGGTTCTATACATCCGTAT-3′) and HDCPR (5′-TAGCAGAAGCTTTCACGGGACTTTGTTTGGTA3′) from genomic DNA of B. mallei strain NCTC 10230. The primers had NdeI and HindIII restriction sites (underlined) at the 5′ end. The DNA amplification was accomplished in the presence of 50 ng of template DNA, a 1.0 μM concentration of each primer, 1.25 U of Taq DNA polymerase, 200 μM deoxynucleoside triphosphate, and 1.5 mM MgCl2 in 1× PCR buffer in final volume of 25 μl. The reaction mixture was denatured for 5 min at 95°C in the beginning, followed by 35 cycles each comprising of the three segments of at 95°C for 1 min, 55°C for 1 min, and 72°C for 2 min. This was followed by one cycle at 72°C for 10 min. The PCR product was digested with NdeI and HindIII and cloned in a pET28a+ expression vector digested with the same enzymes. Transformation was carried out in E. coli BL21(DE3) cells, and the transformants were selected on LB agar plates in the presence of 30 μg of kanamycin/ml.
Fig. 1.
Alignment of amino-terminal BimA sequences of B. mallei ATCC 23344 (accession no. YP_105472), B. pseudomallei isolates 668 (accession no. YP_001063112), 1106a (accession no. ABN93400), and K96243 (accession no. CAH38965), and B. thailandensis isolate E264 (accession no. ABC34450) with the BimA sequence of B. mallei strain NCTC 10230. The amino acid sequences were aligned by using CLUSTAL W. Alignment scores representing the level of identity to the B. mallei NCTC 10230 BimA sequence are shown in parentheses.
Protein expression and purification.
Transformants were screened for expression in small culture, and one clone (henceforth called pETBimA), which showed maximum expression after 4 h of induction by 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), was selected for further work. The integrity of the cloned sequence was confirmed by restriction digestion and DNA sequencing. Expressed protein was obtained in insoluble fraction (inclusion bodies) and purified by immobilized metal affinity chromatography (IMAC) using a Ni+4-NTA fast-flow, chelating Sepharose column. The purified protein was dialyzed against 0.01 M phosphate-buffered saline (PBS; pH 7.2) containing decreasing concentrations of urea. The protein concentration of purified protein was determined with reference to standard bovine serum albumin (BSA) by using a BCA assay kit (Pierce), and the endotoxin content was determined by using a Limulus amebocyte lysate QCL-1000 kit (Lonza).
Western blot analysis.
In order to detect the expression of rBimA in E. coli, induced and uninduced lysates were resolved on SDS-15% PAGE and electroblotted onto nitrocellulose membrane by using the wet transfer method (22). Membrane was blocked overnight at 4°C in 5% skimmed milk and incubated with mouse anti-His tag monoclonal antibody at a dilution of 1:1,000 in blocking solution for 1 h at 37°C. Three washes were performed with PBS containing 0.05% Tween 20 (PBS-T). The membrane was then incubated with rabbit anti-mouse IgG horseradish peroxidase (HRP) conjugated at a dilution of 1:5,000 in blocking solution for 1 h at 37°C. Membrane was washed three times with PBS-T, followed by development in 20 ml of PBS containing 8.8 mM H2O2 and 10 mg of 3,3′-diaminobenzidine/ml. Development was carried out for 2 to 3 min until bands of the desired intensity appeared, and thereafter the membrane was washed under tap water and dried.
Indirect ELISA.
Indirect ELISA was used to detect the antibody against rBimA in equine sera. The optimum concentration of rBimA antigen and serum dilution to be used in indirect ELISA was determined by checkerboard titration using known glanders-positive and -negative horse sera (n = 10 each). The indirect ELISA was carried out by standard method. Briefly, an ELISA plate (Nunc-Immuno Plate MaxiSorp surface; Nunc, Denmark) was coated with rBimA (0.075 μg/well) overnight at 4°C in 0.05 M carbonate bicarbonate buffer (pH 9.6). The plates were washed twice with 0.01 M PBS (pH 7.2) containing 0.05% Tween 20 (PBS-T) and blocked with 200 μl of 1% BSA in PBS for 2 h at 37°C. Test sera diluted 1:400 in PBS containing 0.1% BSA were appended in duplicate wells (100 μl/well) for 1 h at 37°C. The wells were washed five times with PBS-T. Rabbit anti-horse (IgG) antibodies conjugated to HRP (Sigma) diluted 1:10,000 were added to the wells, followed by incubation for 1 h at 37°C. After five washes, the plate was developed in the dark for 20 min with ortho-phenylenediamine (0.4 mg/ml) and H2O2 (6%, 0.4 μl/ml) in citrate phosphate buffer (pH 4.5). The reaction was stopped by the addition of 2.5 M H2SO4 (50 μl/well), and the absorbance was measured at 492 nm in an ELISA reader.
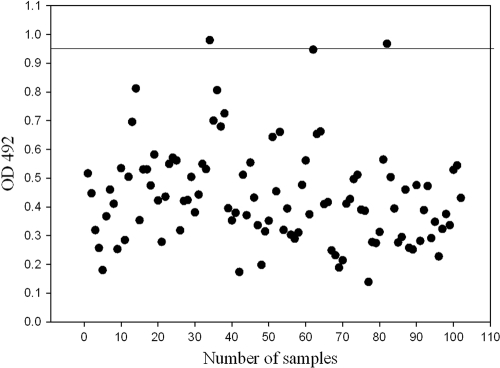
Determination of cutoff for indirect ELISA.
To determine the cutoff, 102 randomly picked glanders-negative serum samples (group II) were tested by indirect ELISA essentially by the above-described method. The cutoff optical density at 492 nm (OD492) was determined by the addition of three standard deviations (SD) to the mean OD492 of these samples, and test samples with an OD492 greater than the cutoff were treated as positive.
Cross-reactivity of rBimA.
The melioidosis serum samples (group III) were screened for any cross-reactivity with rBimA by indirect ELISA. Normal healthy control sera (group IV) were tested as negative controls. The procedure for indirect ELISA was same as described above except that rBimA was coated in the ELISA wells at a higher concentration (10 μg/ml). The WCS antigen of B. mallei or B. pseudomallei (10 μg/ml) was also used for coating as control. Anti-human IgG-HRP conjugate (Dako) was used at 1:6,000.
Stability of the purified recombinant protein.
The rBimA was kept in aliquot of 1 ml each at 4°C, room temperature (RT), and 40°C and analyzed by SDS-15% PAGE at weekly (first month), fortnightly (second month), and monthly intervals for 9 months to monitor the integrity of the protein.
RESULTS
Alignment of BimA protein sequences.
A total of 90 amino acids of BimA protein at the amino-terminal end of the B. mallei strain NCTC 10230 were aligned with homologues sequence of different strains of B. mallei, B. pseudomallei, and B. thailandensis using CLUSTAL W, and the sequence of B. mallei was found to be unique. The BimA sequence of strain NCTC 10230 had only 14% identity with corresponding amino acid sequence of B. pseudomallei strain K96243 and even less identity (6%) with strain 1106a. The sequence identity with B. thailandensis strain E264 was found to be 41%. However, the selected sequence shared 97% identity with B. pseudomallei strain 668 (Fig. 1).
Overexpression of truncated rBimA.
A band of 297 bp was observed on amplification of truncated bimA gene from genomic DNA of B. mallei strain NCTC 10230. No PCR product was obtained when genomic DNA of B. pseudomallei strain NCTC 4845 was used as a template DNA (data not shown). The purified PCR product was cloned in the pET28a+ vector, and the recombinant plasmid pETBimA was transformed into E. coli host strain BL21(DE3). Protein expression screening of small-scale cultures of transformants revealed many positive clones capable of expressing the predicted ∼12.2-kDa recombinant protein. The expression was obtained at 0.5 mM IPTG. No overexpression at the calculated rBimA size was seen in uninduced cells (Fig. 2A). These results suggested that rBimA was successfully overexpressed. Western blot analysis with anti-His tag antibody detected a unique ∼12.2-kDa band in the induced bacterial lysate, representing rBimA, but the same band was not seen in the uninduced bacterial lysate (Fig. 2B).
Fig. 2.
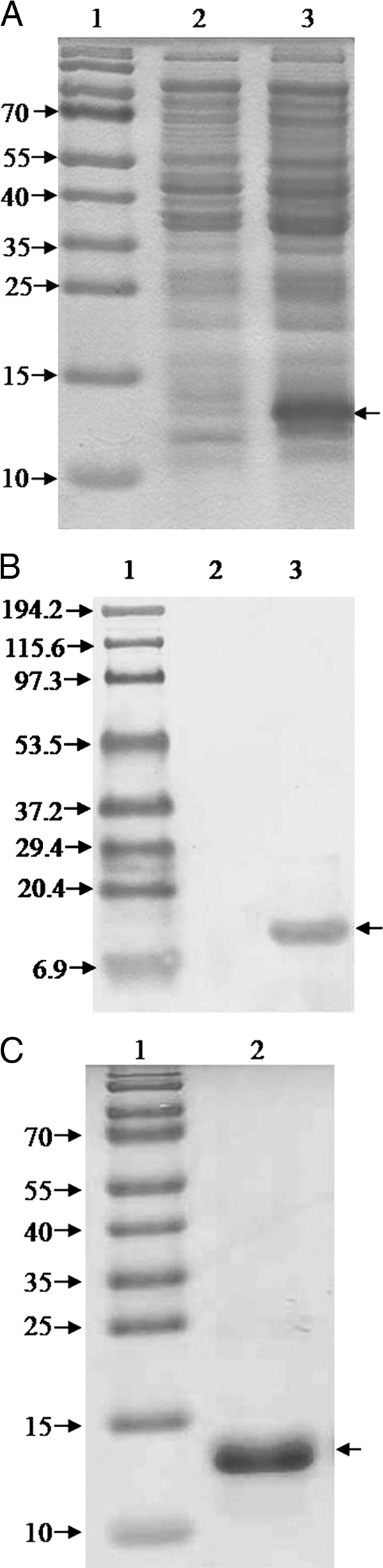
SDS-PAGE analysis of BimA protein expression (A) and its reactivity with anti-His antibody (B). Lane 1, protein molecular mass marker (in kilodaltons); lane 2, uninduced cells; lane 3, IPTG-induced cells. The arrow at the right of each panel indicates the position of the rBimA protein. (C) SDS-PAGE (15%) profile of the purified rBimA protein. Lane 1, protein molecular mass marker (in kilodaltons); lane 2, purified rBimA protein.
Purification of rBimA.
Lysis under native conditions revealed association of recombinant protein with the pellet fraction, demonstrating that the rBimA is insoluble. Pellet was solubilized in buffer containing 8 M urea, and rBimA was purified by IMAC under denaturing conditions. The expressed protein (Fig. 2C) was eluted using a pH 4.0 elution buffer, and SDS-PAGE analysis showed it to be almost pure. During dialysis the purified protein was found to precipitate in 0.01 M PBS (pH 7.2) without urea; therefore, dialysis was terminated at a 1 M urea concentration. The rBimA protein was stored in PBS with 1 M urea. The protein yield of purified rBimA was found to be 20 mg/liter of bacterial culture, and the endotoxin content was <0.25 EU/μg. rBimA protein was found to be stable for 9 months at 4°C and 6 months at RT. The protein degraded at 40°C after 2 months.
Determination of the cutoff for indirect ELISA.
Randomly picked negative serum samples (n = 102) were tested by indirect ELISA at a 1:400 dilution. The OD492s of the samples ranged from 0.139 to 0.980, with a mean OD492 (± the SD) of 0.437 ± 0.169. The cutoff OD492 (mean OD492 + 3 SD) for indirect ELISA was 0.94. A total of 75% of these samples had OD492 values less than 0.530 (Fig. 3).
Fig. 3.
Determination of the cutoff for the rBimA protein ELISA. The horizontal line indicates the cutoff for ELISA, which is the mean + 3 SD.
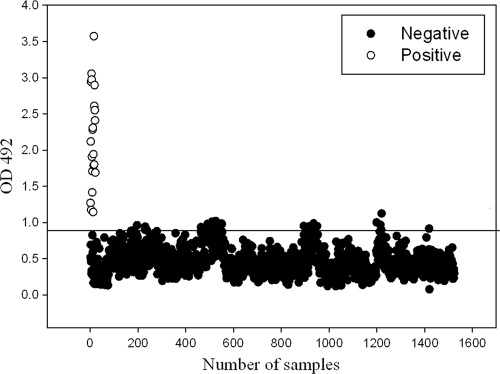
Evaluation of equine sera by indirect ELISA.
The glanders-positive and -negative sera were evaluated by indirect ELISA for anti-rBimA antibody, and the OD492s were was determined. All of the 21 positive serum samples (group I) tested positive by rBimA-based indirect ELISA. The ELISA ODs ranged from 1.14 to 3.57, with a median OD492 of 2.11. A total of 1,507 serum samples from group II had ELISA OD492s less than cutoff value, and only 17 samples were found to have an OD492 value greater than the cutoff. Group II serum sample ELISA OD492s ranged from 0.07 to 1.1, with a median OD492 of 0.559. The results are shown in Fig. 4. The calculated sensitivity and specificity of the test were 100 and 98.88%, respectively.
Fig. 4.
Analysis of equine serum samples by ELISA. Hollow spheres represent the true-positive group I serum samples. Solid spheres represent the group II equine serum samples. Samples with OD values below the cutoff line are negative.
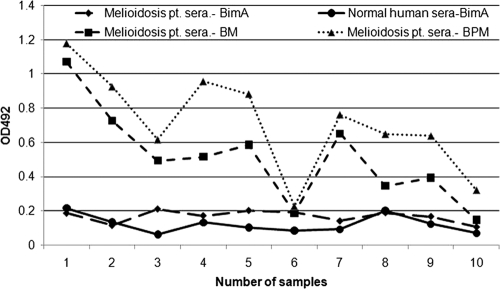
Cross-reactivity of rBimA with melioidosis patient sera.
The reactivity of rBimA with melioidosis patient sera was observed by indirect ELISA. The OD492s of the melioidosis patient (group III) sera were found to be <0.2. Sera from normal healthy individuals (group IV) also had OD492 values of <0.2, suggesting that none of the group III or group IV sera reacted with the rBimA protein (Fig. 5). This shows the high specificity of the rBimA protein for equine glanders. All except one melioidosis patient sera reacted with WCS antigens prepared from either B. pseudomallei or B. mallei, further indicating that the crude antigens of the two species are cross-reactive in nature.
Fig. 5.
ELISA showing the reactivity of rBimA protein with melioidosis patient sera or normal healthy human control sera. The reactivity of the melioidosis patient sera is also shown with the WCS antigen of B. pseudomallei or B. mallei as a control.
DISCUSSION
The limitations of currently available serological tests for laboratory diagnosis of glanders have necessitated the development and evaluation of highly specific and sensitive tests. A crude antigen, which is largely lipopolysaccharide, is used in the CFT, and no serological test is available that uses purified recombinant antigen. In the present study, part of the bimA gene was amplified by PCR, cloned in pET vector, and expressed using E. coli. A purified protein of 12.2 kDa was obtained in an insoluble fraction which reacted specifically with anti-His monoclonal antibody.
rBimA was used in an indirect ELISA format to screen the sera. A panel of more than 100 sera from group II sera were randomly taken to determine the cutoff for ELISA. The OD492 of the majority (80%) of these samples was <0.6. The optimized ELISA was used to evaluate the sera of different groups. Group I, which represented true-positive equine glanders serum samples, were all determined to be positive by rBimA-based indirect ELISA. Therefore, the ELISA had a sensitivity of 100%. The mean OD492 of the group I sera (2.1) was significantly higher than that of group II sera (0.45). All but 17 serum samples from group II were determined to be negative by rBimA-based indirect ELISA. The negative status of group II sera was based on clinical symptoms and the CFT. Although we assume that all of these samples represented true-negative samples, CFT false-negatives cannot be ruled out. The CFT is an internationally recognized serological test and has been used for glanders diagnosis for many years (14). The accuracy of the CFT has been reported to be 90 to 95%, with serum samples being positive within 1 week of infection and remaining positive in the case of exacerbation of the chronic process (20). Recently, however, the specificity of the CFT has been questioned (13). Considering group II as true-negative samples, the specificity of rBimA-based indirect ELISA is 98.88%.
One of the major drawbacks of serological tests for glanders has been that they were unable to differentiate between B. mallei and B. pseudomallei infections (14). B. mallei and B. pseudomallei are very close at the genome level; therefore, it has always been difficult to develop a specific immunodiagnostic test for glanders. The amino-terminal region of BimA protein of B. mallei has been reported to be unique, with no homology to the corresponding B. pseudomallei or B. thailandensis proteins (21). Studies related to the development of PCR assays for the specific detection of B. mallei used 29 B. mallei and 34 B. pseudomallei isolates from various clinical sources collected over a 70-year period found this region to be conserved in B. mallei (23, 24). In present study, also, no bimA amplicon was obtained with B. pseudomallei genomic DNA. However, a 2008 study suggested that certain strains of B. pseudomallei that originated in the Northern Territory of Australia contained a B. mallei-like BimA protein (18). B. pseudomallei strain 668 was isolated from this geographical region and had 99% homology with our recombinant protein (Fig. 1). It can be assumed that our protein may give false-positive results in animals and/or humans infected with such B. pseudomallei strains. Since B. pseudomallei strains that contain B. mallei-like BimA protein are restricted to the Northern Territory of Australia, the recombinant protein described in the present study may still be useful for the diagnosis of equine glanders in regions where this disease is endemic.
The cross-reactivity of rBimA was evaluated with sera from melioidosis patients. The ELISA ODs obtained with melioidosis patient and normal healthy human sera were similar and close to 0.2, indicating that no antibody against BimA is present in these sera. This further demonstrates the high specificity of rBimA protein. However, with the results of present study, it is not possible to assure that BimA is expressed during human infection.
For the development of any protein reagent as diagnostic reagent, it is important to ascertain its stability at various temperatures. In stability studies, we found that the recombinant protein was stable for 9 months at 4°C and for 6 months at RT.
To conclude, the rBimA protein-based indirect ELISA reported here is a simple, sensitive, and specific assay. The recombinant protein also does not cross-react with the sera of the melioidosis patients, further showing its value for the specific immunodiagnosis of glanders. The stable rBimA protein may easily be used as test reagent for serodiagnosis of glanders. An Indian patent (1328/DEL/2010) has been filed for the recombinant protein described in the present study.
ACKNOWLEDGMENT
We thank the Director of Defense Research and Development Establishment, Gwalior, India, for providing necessary facilities and encouragement.
Footnotes
Published ahead of print on 13 July 2011.
REFERENCES
- 1. Al-Ani F. K., Al-Rawashdeh O. F., Ali A. H., Hassan F. K. 1998. Glanders in horses: clinical, biochemical and serological studies in Iraq. Vet. Arch. 68:155–162 [Google Scholar]
- 2. Arun S., et al. 1999. Equine glanders in Turkey. Vet. Rec. 144:255–258 [DOI] [PubMed] [Google Scholar]
- 3. Bazargani T. T., Tadjbakhs H., Badii A., Zahraei T. 1996. The outbreak of glanders in some racehorses in three states of Iran. J. Equine Vet. Sci. 16:232–236 [Google Scholar]
- 4. Boyden S. V. 1950. Absorption by erythrocytes of antigens of Pf. mallei and Pf. whitmori. Proc. Sac. Exp. Biol. Med. 73:289 [Google Scholar]
- 5. Cravitz L., Miller W. R. 1950. Immunologic studies with Malleomyces mallei and Malleomyces pseudomallei I: serological relationships between M. mallei and M. pseudomallei. J. Infect. Dis. 86:46–51 [DOI] [PubMed] [Google Scholar]
- 6. Elschner M. C., et al. 2009. Burkholderia mallei infection in a horse imported from Brazil. Equine Vet. Educ. 21:147–150 [Google Scholar]
- 7. Elschner M. C., et al. 2011. Use of a Western blot technique for the serodiagnosis of glanders. BMC Vet. Res. 7:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heine H. S., England M. J., Wagg D. M., Byrne W. R. 2001. In vitro antibiotic susceptibilities of Burkholderia mallei (causative agent of glanders) determined by broth microdilution and E-test. Antimicrob. Agents Chemother. 45:2119–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jana A. M., Gupta A. K., Pandya G., Verma R. D., Rao K. M. 1982. Rapid diagnosis of glanders in equines by counter-immuno-electrophoresis. Indian Vet. J. 59:5 [Google Scholar]
- 10. Malik P., Khurana S. K., Dwivedi S. K. 2010. Re-emergence of glanders in India: report of Maharashtra state. Indian J. Microbiol. 50:345–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Naureen A., et al. 2010. Antimicrobial susceptibility of 41 Burkholderia mallei isolates from spontaneous outbreaks of equine glanders in Punjab, Pakistan. J. Equine Vet. Sci. 30:134–141 [Google Scholar]
- 12. Neubauer H., Meyer H., Finke E. J. 1997. Human glanders. Rev. Int. Serv. Sante Forces Armees 70:258–265 [Google Scholar]
- 13. Neubauer H., et al. 2005. Serodiagnosis of Burkholderia mallei infections in horses: state-of-the-art and perspectives. J. Vet. Med. B 52:201–205 [DOI] [PubMed] [Google Scholar]
- 14. OIE 2010. Glanders, chapter 2.5.11. Manual of diagnostic tests and vaccines for terrestrial animals 2010. OIE, Paris, France [Google Scholar]
- 15. Parthasarathy N., Deshazer D., England M., Waag D. M. 2006. Polysaccharide microarray technology for the detection of Burkholderia pseudomallei and Burkholderia mallei antibodies. Diagn. Microbiol. Infect. Dis. 56:329–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sanford J. P. 1995. Pseudomonas species (including melioidosis and glanders), p. 2003–2009 In Mandell G. L., Bennett J. E., Dolin R. (ed.), Principles and practice of infectious diseases, 4th ed Churchill Livingstone, New York, NY [Google Scholar]
- 17. Sen G. P., Singh G., Joshi T. P. 1968. Comparative efficacy of serological tests in the diagnosis of glanders. Indian Vet. J. 45:286. [PubMed] [Google Scholar]
- 18. Sitthidet C., et al. 2008. Prevalence and sequence diversity of a factor required for actin-based motility in natural populations of Burkholderia species. J. Clin. Microbiol. 46:2418–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Srinivasan A., et al. 2001. Glanders in a military research microbiologist. N. Engl. J. Med. 345:256–258 [DOI] [PubMed] [Google Scholar]
- 20. Steele J. H. 1980. Bacterial, rickettsial and mycotic diseases, p. 339–351 In Steele J. H. (ed.), CRC handbook series in zoonoses, section A, vol. I CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 21. Stevens J. M., et al. 2005. Actin-binding proteins from Burkholderia mallei and Burkholderia thailandensis can functionally compensate for the actin-based motility defect of a Burkholderia pseudomallei bimA mutant. J. Bacteriol. 187:7857–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Towbin H., Staehelin T., Gordin J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ulrich M. P., Norwood D. A., Christensen D. R., Ulrich R. L. 2006. Using real-time PCR to specifically detect Burkholderia mallei. J. Med. Microbiol. 55:551–559 [DOI] [PubMed] [Google Scholar]
- 24. Ulrich R. L., Ulrich M. P., Schell M. A., Kim S., DeShzer D. 2006. Development of a PCR assay for the specific identification of Burkholderia mallei and differentiation from Burkholderia pseudomallei and other closely related Burkholderiaceae. Diagn. Microbiol. Infect. Dis. 55:47–55 [DOI] [PubMed] [Google Scholar]
- 25. Verma R. D., Sharma J. K., Venkateswaran K. S., Batra H. V. 1990. Development of an avidin-biotin dot enzyme-linked immunosorbent assay and its comparison with other serological tests for diagnosis of glanders in equines. Vet. Microbiol. 25:77–85 [DOI] [PubMed] [Google Scholar]
- 26. Wheelis M. 1998. First shots fired in biological warfare. Nature 395:213. [DOI] [PubMed] [Google Scholar]