Abstract
Many or all of the sites of pseudouridine (Ψ) formation in eukaryotic rRNA are selected by site-specific base-pairing with members of the box H + ACA class of small nucleolar RNAs (snoRNAs). Database searches previously identified strong homology between the rat nucleolar protein Nap57p, its yeast homolog Cbf5p, and the Escherichia coli Ψ synthase truB/P35. We therefore tested whether Cbf5p is required for synthesis of Ψ in the yeast rRNA. After genetic depletion of Cbf5p, formation of Ψ in the pre-rRNA is dramatically inhibited, resulting in accumulation of the unmodified rRNA. Protein A-tagged Cbf5p coprecipitates all tested members of the box H + ACA snoRNAs but not box C + D snoRNAs or other RNA species. Genetic depletion of Cbf5p leads to depletion of all box H + ACA snoRNAs. These include snR30, which is required for pre-rRNA processing. Depletion of Cbf5p also results in a pre-rRNA processing defect similar to that seen on depletion of snR30. We conclude that Cbf5p is likely to be the rRNA Ψ synthase and is an integral component of the box H + ACA class of snoRNPs, which function to target the enzyme to its site of action.
Keywords: RNA modification, pre-rRNA processing, ribosome synthesis, yeast
The rRNAs of all organisms undergo extensive covalent nucleotide modification. In eukaryotes, the rRNAs are generated by post-transcriptional processing of large pre-rRNA species, and these modified nucleotides are formed in the pre-rRNAs, rather than in the mature rRNAs. The most numerous modifications are methylation of the 2′-hydroxyl residue in the ribose moieties (2′-O-methylation) and isomerization of uracil residues to pseudouridine (Θ). Formation of Θ residues is thought to occur through base rotation about the C3–C6 axis after cleavage of the glycosyl bond (Goldwasser and Heinrikson 1966; for review, see Ofengand et al. 1995). Additionally, a few positions are modified at the base level; the best described example being the universally conserved m62Am62A doublet at the 3′-end of the 18S rRNA (Lafontaine et al. 1995). Recent data have shown that, in eukaryotes, the sites of both 2′-O-methylation (Cavaillé et al. 1996; Kiss-László et al. 1996; Nicoloso et al. 1996) and Θ formation (Ganot et al. 1997a; Ni et al. 1997) in the rRNAs are selected by site-specific base-pairing of small nucleolar RNAs (snoRNAs) to the pre-rRNAs (for review, see Maden 1996; Tollervey 1996; Bachellerie and Cavaillé 1997; Smith and Steitz 1997). The snoRNAs involved can be separated into two major groups, which are designated box C + D snoRNAs and box H + ACA snoRNAs on the basis of conserved sequence elements (Balakin et al. 1996). In the case of 2′-O-methylation, base-pairing of a member of the box C + D class of snoRNAs across the site of methylation positions a conserved sequence element, box D, at a precise distance of 5 bp from the nucleotide to be modified (Cavaillé et al. 1996; Kiss-László et al. 1996; Nicoloso et al. 1996). Presumably, proteins associated with box D use this positional information to select the site of modification. In the case of Θ formation, a member of the box H + ACA class of snoRNAs base pairs to nucleotides flanking the substrate uracil, leaving the base of the nucleotide free for interaction with the modifying enzyme (Ganot et al. 1997a; Ni et al. 1997).
Both major groups of snoRNAs are associated with specific proteins in small nucleolar ribonucleoprotein (snoRNP) particles. All box C + D snoRNAs are associated with the protein fibrillarin (Nop1p in yeast) (Lischwe 1985; Schimmang et al. 1989; Ganot et al. 1997b; for review, see Maxwell and Fournier 1995), and mutations in Nop1p can globally block 2′-O-methylation of the yeast rRNAs (Tollervey et al. 1993). Similarly, all box H + ACA snoRNAs are associated with Gar1p (Girard et al. 1992; Balakin et al. 1996; Ganot et al. 1997b), and mutations in Gar1p can globally inhibit Θ formation in the rRNA (Bousquet-Antonelli et al. 1997). Nop1p and Gar1p are not required for the synthesis or stability of the snoRNAs with which they are associated. Gar1p is, however, required for the stable association of the box H + ACA snoRNAs with the pre-rRNA (Bousquet-Antonelli et al. 1997); this association presumably underlies the requirement for Gar1p in Θ formation.
Both classes of snoRNAs include species that probably do not act to select sites of rRNA modification but are required for processing of the pre-rRNA. Genetic depletion of the box C + D snoRNAs U3 (Hughes and Ares 1991) or U14 (Li et al. 1990) or of the box H + ACA snoRNA snR30 (Morrissey and Tollervey 1993), inhibits the early pre-rRNA processing reactions at sites A0, A1, and A2, preventing the synthesis of the 18S rRNA (see Fig. 1). For this reason each of these snoRNAs is essential for cell viability. In contrast, none of the snoRNAs that direct rRNA modification is essential for cell viability, although the absence of the Θ guide snoRNA, snR10, leads to some cold sensitivity (Tollervey 1987; Ni et al. 1997).
Figure 1.
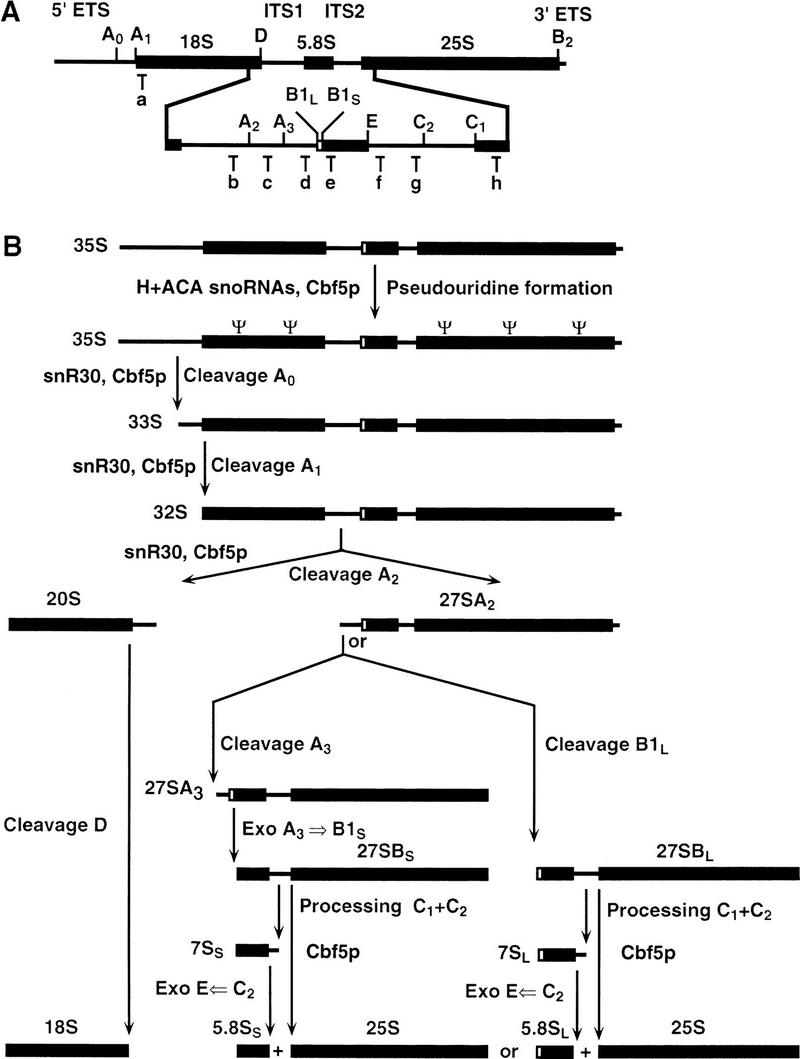
Structure of the yeast pre-rRNA and its processing pathway. (A) The 35S pre-rRNA. The sequences encoding the mature 18S, 5.8S, and 25S rRNAs (thick lines) are flanked by the 5′ and 3′ ETSs and separated by internal transcribed spacers 1 and 2 (ITS1 and ITS2). Sites of pre-rRNA processing are indicated with uppercase letters (A0 to E) and the positions of hybridization of the oligonucleotides used are indicated with lowercase letters (a–h). (B) The pre-rRNA processing pathway. Processing of the primary 35S precursor starts at site A0, yielding 33S pre-rRNA. This molecule is subsequently processed at sites A1 and A2, giving rise successively to the 32S pre-rRNA and to the 20S and 27SA2 precursors. Cleavage at A2 separates the pre-rRNAs destined for the small and large ribosomal subunit. The 20S precursor is then endonucleolytically cleaved at site D to yield the mature 18S rRNA. The 27SA2 precursor is processed by two alternative pathways, forming the mature 5.8S and 25S rRNAs. The major pathway involves cleavage at a second site in ITS1, A3, rapidly followed by exonucleolytic digestion to site B1S, generating the 27SBS precursor. Approximately 15% of the 27SA2 molecules are processed by the second pathway at site B1L, producing the 27SBL pre-rRNA. At the same time as processing at B1 is completed, the 3′ end of mature 25S rRNA is generated by processing at site B2. The subsequent processing of both 27SB species appears to follow a similar pathway. Cleavage at sites C1 and C2 releases the mature 25S rRNA and the 7S pre-rRNAs, which undergo rapid 3′ → 5′ exonuclease digestion to site E generating the mature 3′ end of 5.8S rRNA. Cbf5p is required for the early cleavages at sites A1 and A2; loss of these cleavages inhibits formation of the 20S and 27SA2 pre-rRNA preventing synthesis of 18S rRNA. Cbf5p is also required for efficient processing at site A0 and efficient processing of the 27SB and 7S pre-rRNAs in ITS2.
One of the major unresolved questions concerning these systems of snoRNA-directed modification is the relationship between the snoRNAs and the modifying enzymes. Specifically, are the enzymes free components that recognize the structure created by the snoRNA–pre-rRNA duplexes in much the same way tRNA-modifying enzymes and bacterial rRNA-modifying enzymes recognize their RNA substrates? Or are the enzymes physically associated with the snoRNAs, which act directly to target the enzymes to their sites of action?
In contrast to Θ formation in the eukaryotic pre-rRNA, the formation of Θ in tRNAs and bacterial rRNAs is not known to involve RNA cofactors. In these cases, multiple Θ synthases exist; four tRNA Θ synthases have been characterized in Escherichia coli (truA, truB, rsuA, and rluA), each of which modifies with high specificity a single site or a number of sites with very similar structures (Kammen et al. 1988; Nurse et al. 1995; Wrzesinski et al. 1995a,b; Simos et al. 1996). A database search (Koonin 1996) revealed that each of these enzymes is a member of a distinct, evolutionarily conserved family of Θ synthases. E. coli truB/P35, which converts U55 to Θ55 in the m5UΘCG loop in most tRNAs (Nurse et al. 1995), is strongly homologous to two yeast proteins, Cbf5p and YNL480 (Koonin 1996). Recent data have shown that YNL480 (now designated Pus4p) is the yeast tRNA Θ55 synthase (Becker et al. 1997). Yeast Cbf5p was originally characterized as an essential protein that showed in vitro binding to centromeres and microtubules (Jiang et al. 1993). Subsequently, Cbf5p was found to be highly homologous (64% identity, 79% homology) to the rat nucleolar protein Nap57p and to be localized to the yeast nucleolus (Meier and Blobel 1994). This suggested that Cbf5p and Nap57p might be the rRNA Θ synthases in yeast and mammals, respectively. We have therefore investigated this possibility and report here an analysis of the role of Cbf5p in pre-rRNA processing and formation of Θ in the rRNA.
Results
Construction of a conditional CBF5 allele
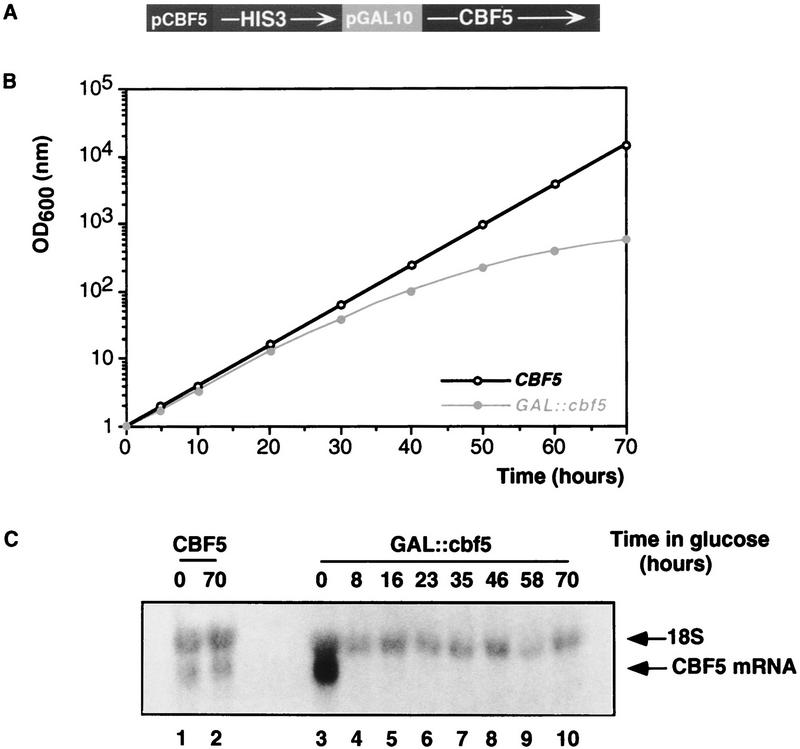
Because CBF5 is an essential gene (Jiang et al. 1993), we first constructed a conditional allele by placing its expression under the control of an inducible GAL10 promoter using the one-step PCR method described previously (Lafontaine and Tollervey 1996) (Fig. 2A). On permissive medium [2% raffinose, 2% sucrose, and 2% galactose (rsg)], the growth rate of the GAL::cbf5 strain is identical to that of the otherwise isogenic parental CBF5 strain (doubling every 3 hr). Under these conditions, the level of the CBF5 mRNA in the GAL::cbf5 strain is approximately fivefold higher than in the CBF5 control strain (Fig. 2C, lane 3). This level is expected for a gene whose transcription is driven by the strong GAL10 promoter. Following transfer of the GAL::cbf5 strain to glucose (glu) medium the CBF5 mRNA was rapidly depleted, no mRNA was detected 8 hr after transfer to glu medium (Fig. 2C), and growth slowed progressively, commencing 20 hr after transfer (Fig. 2B). This slow onset in the growth impairment is characteristic of GAL depletion of components required for ribosome synthesis.
Figure 2.
Genetic depletion of Cbf5p. (A) Schematic representation of the GAL::cbf5 allele. (B) Growth of the GAL::cbf5 (•) and CBF5 (○) strains following transfer to glucose medium. Cell density was measured at regular intervals, and the cultures were periodically diluted to be continuously kept in exponential growth. (C) Northern hybridization of the CBF5 mRNA from the CBF5 strain (lanes 1,2) and the GAL::cbf5 strain (lanes 3–10), following growth on rsg medium (0-hr lanes) and at intervals following transfer to glu medium (8 to 70-hr lanes). The positions of the CBF5 mRNA and 18S rRNA are indicated. As expected from the size of its ORF (1452 bp), the CBF5 mRNA is detected slightly under the 18S rRNA.
Cbf5p is required for pre-rRNA processing
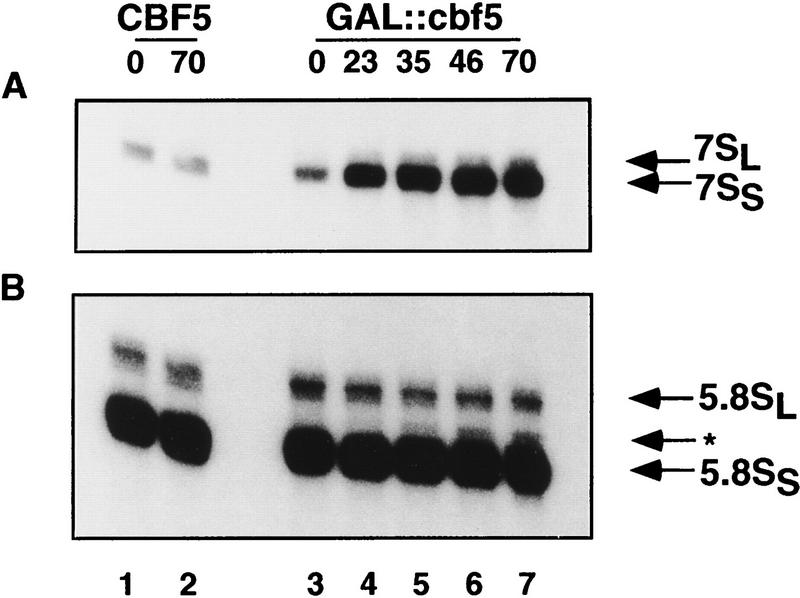
Northern hybridization (Fig. 3) shows that the levels of the mature 18S and 25S rRNAs, and all pre-rRNA species are identical in the CBF5 control strain and in the GAL::cbf5 strain grown in permissive medium (0 hr lanes in Fig. 3). Following transfer to glu medium (Fig. 3A, 23- to 46-hr lanes) the mature 18S rRNA is progressively depleted in the GAL::cbf5 strain; at later time points depletion of the 25S rRNA is also observed. Analysis of the pre-rRNAs (Fig. 3B–E) shows that the 35S primary transcript is strongly accumulated, while the 27SA2 and 20S pre-rRNAs are depleted and an aberrant RNA species, the 23S pre-rRNA, is detected. The 23S RNA extends from the 5′ end of the 35S to site A3 (Fig. 1) and is generated by direct cleavage of the 35S pre-rRNA at site A3 in the absence of prior cleavage at sites A0, A1, and A2. These effects are characteristic of mutations that inhibit the early pre-rRNA cleavages at sites A0, A1, and A2 (see Fig. 1) and were observed following depletion of several different snoRNAs, including the box H + ACA snoRNAs snR10 (Tollervey 1987) and snR30 (Morrissey and Tollervey 1993), and on depletion of Gar1p (Girard et al. 1992), which is associated with the entire class of box H + ACA snoRNAs (Balakin et al. 1996). Depletion of Cbf5p also leads to accumulation of the 27SB pre-rRNAs (Fig. 3E), consistent with the reduction in the levels of the 25S rRNA. A shorter time course following transfer to glucose medium (Fig. 3F) shows that the level of the 35S pre-rRNA is elevated and 27SA2 is reduced as early as 8 hr (<3 generations) after transfer to glu medium, indicating that these effects rapidly follow the loss of Cbf5p. Analysis of low molecular weight RNA (Fig. 4A) shows that the 7S pre-rRNA is also strongly accumulated following depletion of Cbf5p. This accumulation of the 27SB and 7S pre-rRNAs was not observed on depletion of any characterized snoRNA. In addition, a 5.8S rRNA species intermediate in length between 5.8SL and 5.8SS was observed on depletion of Cbf5p (* in Fig. 4B). This has also not been observed previously in snoRNA mutants.
Figure 3.
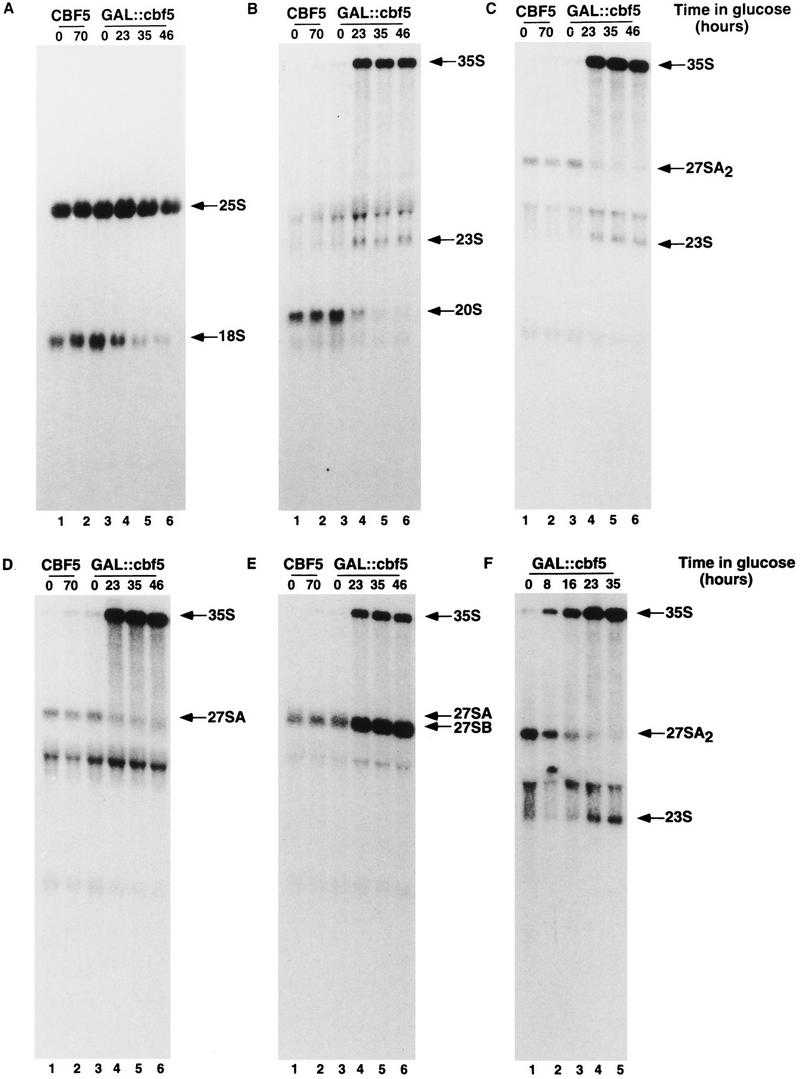
Northern analysis of rRNA and pre-rRNA synthesis in a GAL::cbf5 strain. (A) Probes against mature 25S and 18S rRNA (oligonucleotides a and h); (B) probe against the 5′ region of ITS1 (oligonucleotide b), (C); probe against ITS1 between sites A2 and A3 (oligonucleotide c); (D) probe against the 3′ region of ITS1 (oligonucleotide d); (E) probe against the 5′ region of ITS2 (probe f). (F) Shorter time course following transfer to glu medium, hybridized with oligonucleotide c. The oligonucleotides used are depicted in Fig. 1A. RNA was extracted from the CBF5 and GAL::cbf5 strains following growth on rsg medium (0-hr lanes) and at intervals following transfer to glu medium (8–70-hr lanes) and separated on a 1.2% agarose gel containing formaldehyde.
Figure 4.
Northern analysis of rRNA and pre-rRNA synthesis in a GAL::cbf5 strain. (A) Probe against ITS2 (probe f); (B) probe against mature 5.8S rRNA (probe e). The oligonucleotides used are depicted in Fig. 1A. RNA was extracted from the CBF5 and GAL::cbf5 strains following growth on rsg medium (0-hr lanes) and at intervals following transfer to glu medium (8- to 70-hr lanes) and separated on an 8% polyacrylamide gel containing 8 m urea. (*) 5.8S species with intermediate length.
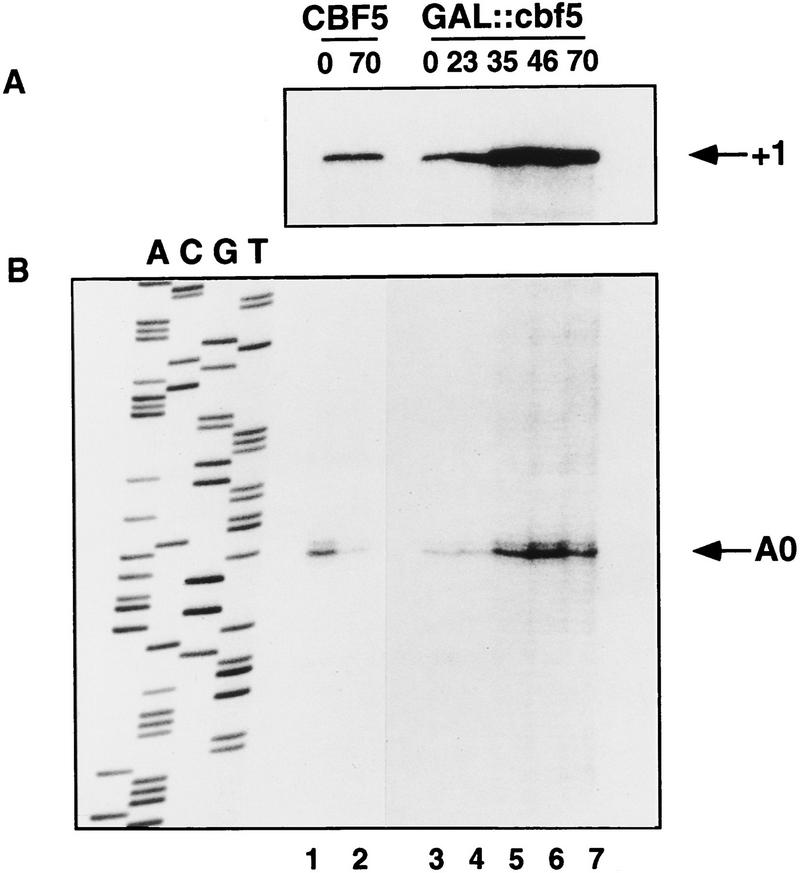
Pre-rRNA processing was further analyzed by primer extension. Analysis of the 5′ external transcribed spacer (ETS) shows an increase in the stop corresponding to the 5′ end of the 35S pre-rRNA (position +1, Fig. 5A) in the GAL::cbf5 strain following transfer to glu medium (23- to 70-hr lanes), consistent with the 35S accumulation detected by Northern hybridization (Fig. 3B–F); however, this was not accompanied by a loss of the pre-rRNA cleaved at site A0 (Fig. 5B). The primer-extension stop at site A0 is elevated on depletion of Cbf5p. The accumulation of the 35S pre-rRNA and 23S RNA indicates that cleavage at site A0 is at least delayed on depletion of Cbf5p. We interpret this observation as showing that cleavages at sites A1 and A2 are more sensitive to the depletion of Cbf5p than is cleavage at site A0. This phenomenon was observed in strains depleted of snR10, snR30, or Gar1p, but not in strains depleted of U3 (Beltrame et al. 1994).
Figure 5.
Primer-extension analysis of pre-rRNA processing in a GAL::cbf5 strain. (A) The 5′ end of the 35S primary transcript at site +1. (B) Site A0. RNA was extracted from the CBF5 and GAL::cbf5 strains following growth on rsg medium (0-hr lanes) and at intervals following transfer to glu medium (8- to 70-hr lanes) and analyzed by primer extension with oligonucleotide a (Fig. 1A). A DNA sequence made with the same primer is shown as a size marker. Site +1 lies 730 nucleotides from the primer and the sequence is not useful.
Primer extension through sites in ITS1 shows the loss of the stop corresponding to cleavage at site A2, the 5′ end of the 27SA2 pre-rRNA, and shows the increase of the stops at sites B1S and B1L, the 5′ ends of both the 27SBS and 27SBL and 7SS and 7SL pre-rRNAs, respectively (Fig. 6). These observations are in good agreement with the results of Northern hybridization in Figures 3 and 4. The accumulation of the long and short forms of 27SB and 7S, shown by the relative stops at B1S and B1L, are not obviously different (Fig. 6, cf. A with C, which shows a shorter exposure of the same gel). The level of the 27SA3 pre-rRNA, shown by the primer-extension stop at site A3, is elevated on depletion of Cbf5p (Fig. 6B). This elevation was also observed in strains depleted of snR30, but was not seen on depletion of any other snoRNP component tested (Morrissey and Tollervey 1993).
Figure 6.
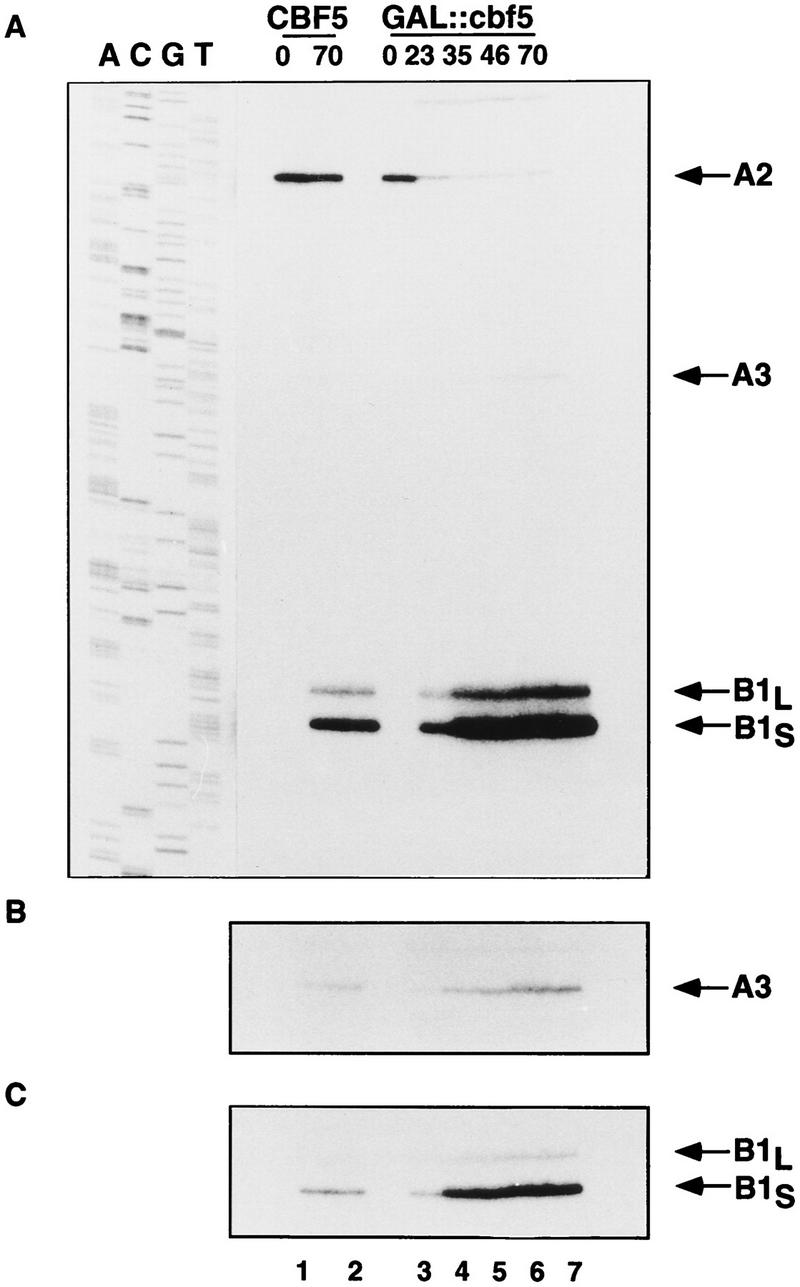
Primer-extension analysis of pre-rRNA processing in a GAL::cbf5 strain. (A) Primer extension through sites B1S, B1L, A3, and A2. (B) Longer exposure of A showing the level of stop at site A3. (C) Shorter exposure of A showing the level of stop at site B1. RNA was extracted from the CBF5 and GAL::cbf5 strains following growth on rsg medium (0-hr lanes) and at intervals following transfer to glu medium (8- to 70-hr lanes) and analyzed by primer extension using oligonucleotide g (Fig. 1A). A DNA sequence made with the same primer is shown as a size marker.
No defects in pre-tRNA processing or accumulation of mature tRNAs were detected by Northern hybridization with probes specific for the intron-containing precursor of tRNATrpCCA, tRNAProUGG, tRNATyrGΘA, and tRNAPheGAA and probes specific for mature tRNATrpCCA and tRNAProUGG (data not shown).
Cbf5p is required for formation of Θ in the pre-rRNA
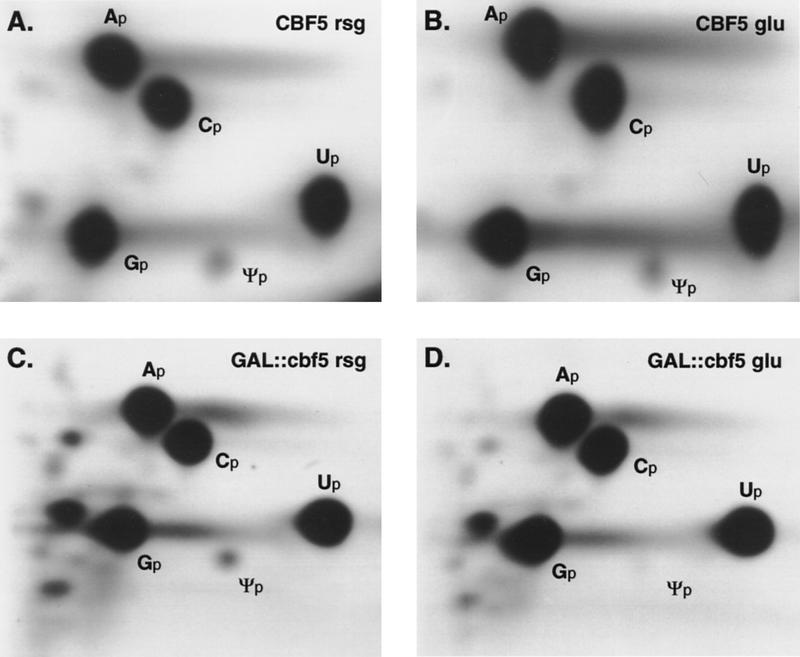
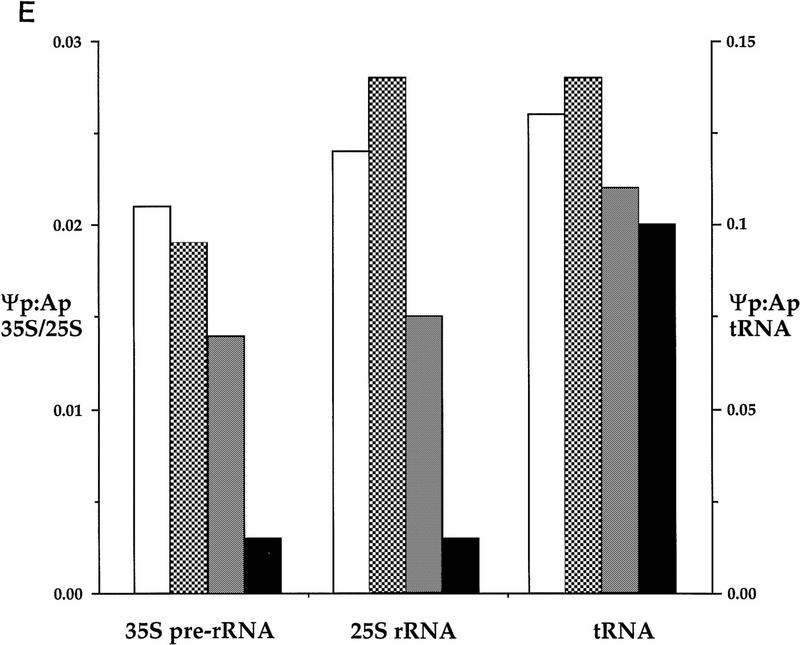
The effects of depletion of Cbf5p on Θ formation were assessed by pulse labeling of the newly synthesized pre-rRNA and rRNA with 32P. Cells were labeled following growth in permissive rsg medium or following transfer to glu medium for 24 hr. Labeled RNAs, 35S pre-rRNA, mature 25S rRNA, and bulk tRNA, were gel purified, and the nucleotide composition was analyzed by two-dimensional thin layer chromatography (TLC). Θp content was determined by comparison with other nucleotides present in the same RNA samples. Figure 7E shows the data expressed as the ratio of incorporation into Θp compared to Ap; comparison of Θp to Gp or Cp gave similar results (data not shown). The results presented are from one set of experiments; an independent analysis gave similar data (data not shown). The Θp:Ap ratio in the 25S rRNA from the Cbf5p-depleted strain is ∼11% of the value obtained for the wild-type 25S rRNA. This would correspond to a residual level of ∼3 Θ residues per 25S rRNA molecule (the wild-type 25S has 30 Θ residues) (Ofengand et al. 1995). The Θp:Ap ratio in the 35S rRNA from the Cbf5p-depleted strain is 16% of the wild-type value, corresponding to ∼7 Θ residues (the wild-type 35S has 43 Θ residues) (Ofengand et al. 1995). It is not clear whether this corresponds to low residual modification of all sites or whether some sites are preferentially modified. Because the 18S rRNA is not synthesized in the Cbf5p-depleted strain this species cannot be analyzed directly. The reduced modification of the 35S pre-rRNA, however, strongly indicates that formation of Θ in the region of the pre-rRNA corresponding to the mature 18S rRNA is also inhibited by depletion of Cbf5p.
Figure 7.
Θ formation in a GAL::cbf5 strain. Two-dimensional TLC analysis of 32P-labeled 25S rRNA digested with RNAse T2. (A) RNA extracted from the CBF5 strain following growth in rsg medium. (B) RNA extracted from the CBF5 strain 24 hr after transfer to glu medium. (C) RNA extracted from the GAL::cbf5 strain following growth in rsg medium. (D) RNA extracted from the GAL::cbf5 strain 24 hr after transfer to glu medium. Spots corresponding to Ap, Cp, Gp, Up, and Θp are indicated. (E) Nucleotides separated by two-dimensional TLC were quantitated by PhosphorImager scanning. The ratio between incorporation into Θp and Ap in 35S pre-rRNA, 25S rRNA, and bulk tRNA is shown following growth in rsg medium, and 24 hr after transfer to glu medium. The Θp:Ap ratio in tRNA (right) is shown on a different scale from the 35S and 25S RNA samples (left) because of the greater representation of Θp in tRNA compared to rRNA. (Open bars) CBF5+/rsg; (light gray bars) CBF5+glu; (dark gray bars) GAL::cbf5 rsg; (solid bars) GAL::cbf5 glu.
As an example of the data, Figure 7(A–D) presents the analysis of the 25S rRNA from the CBF5 and GAL::cbf5 strains. The spot corresponding to Θp can be seen in the CBF5 samples (Fig. 7A,B) and in the sample from the GAL::cbf5 strain grown under permissive conditions (Fig. 7C), but is not readily visible in the sample obtained following depletion of Cbf5p (Fig. 7D).
Surprisingly, the level of Θp in the GAL::cbf5 strain grown in rsg medium was consistently below the expected level. The Θp:Ap ratios in the 25S and 35S RNA samples was 62% and 67%, respectively, of the values in the corresponding wild-type samples. Northern hybridization indicates that the level of CBF5 mRNA under permissive conditions is elevated compared to the wild type (Fig. 2C). We speculate that this elevation leads to some excess in Cbf5p synthesis, which leads in turn to a form of squelching, in which excess free protein blocks the interaction of other components with the complex, interfering with its function.
Θ levels in the tRNA fraction are also mildly reduced by depletion of Cbf5p; the Θp:Ap ratio for the GAL::cbf5 strain was 85% of the wild-type ratio in rsg medium and 71% of the wild-type ratio in glu medium. E. coli truB synthesizes Θ55 in most tRNAs contributing approximately to ∼40%–50% of total Θ in bulk tRNA (H. Grosjean, pers. comm.), but in yeast, this activity is attributable to Pus4p, another homolog of truB (Koonin 1996; Becker et al. 1997). It seems probable that the reduced Θp:Ap ratio is an indirect consequence of the impaired growth of the Cbf5p-depleted strain. It is, however, notable that a number of the box H + ACA snoRNAs can be drawn in the consensus structure to act as Θ guides but do not appear to have target sites in the rRNA (Ganot et al. 1997a). The tRNA fraction represents bulk tRNA and we cannot exclude the possibility that some tRNA(s) or other small RNAs present in this fraction are specifically undermodified on depletion of Cbf5p.
We conclude that, in contrast to E. coli truB, Cbf5p is required for Θ formation in the pre-rRNA but does not synthesize Θ55 in the tRNAs.
Cbf5p is a component of the box H + ACA class of snoRNPs
To test for a physical association between Cbf5p and the box H + ACA snoRNAs, a Cbf5p–protein A carboxyl fusion was constructed and integrated at the CBF5 locus under the control of its own promoter (see Materials and Methods). In the resulting strain, Cbf5p–protein A is the only source of Cbf5p activity in the cell. This strain had a wild-type growth rate (data not shown), showing the Cbf5p–protein A fusion to be fully functional.
Immunoprecipitation of Cbf5p–protein A with IgG–agarose beads resulted in the coprecipitation of all tested box H + ACA snoRNAs (snR3, snR10, snR11, snR30, snR31, snR33, snR37, and snR42) but did not detectably coprecipitate box C + D snoRNAs (U3, U14, and snR190), RNAse MRP (Fig. 8) or the U5 snRNA (data not shown). The immunoprecipitations were performed on two independent Cbf5p–protein A strains (YDL524-18 and YDL524-19) at 150 mm and 500 mm Kacetate. Coprecipitation of the box H + ACA snoRNAs with a protein-A fusion to the snoRNP protein Nop1p (fibrillarin) is observed at 150 mm salt but is lost at the higher salt concentration (Ganot et al. 1997b). In contrast, coprecipitation of the H + ACA snoRNAs with Cbf5p was observed at both salt concentrations. No precipitation of any RNA was seen with an otherwise isogenic CBF5 strain expressing only nontagged Cbf5p (Fig. 8, lanes 1–3). The efficiency of coprecipitation of the box H + ACA snoRNAs with Cbf5p–protein A ranges from 30% to 70%, similar to the efficiency with which the box C + D snoRNAs are coprecipitated with a Nop1p–protein A fusion (data not shown).
Figure 8.
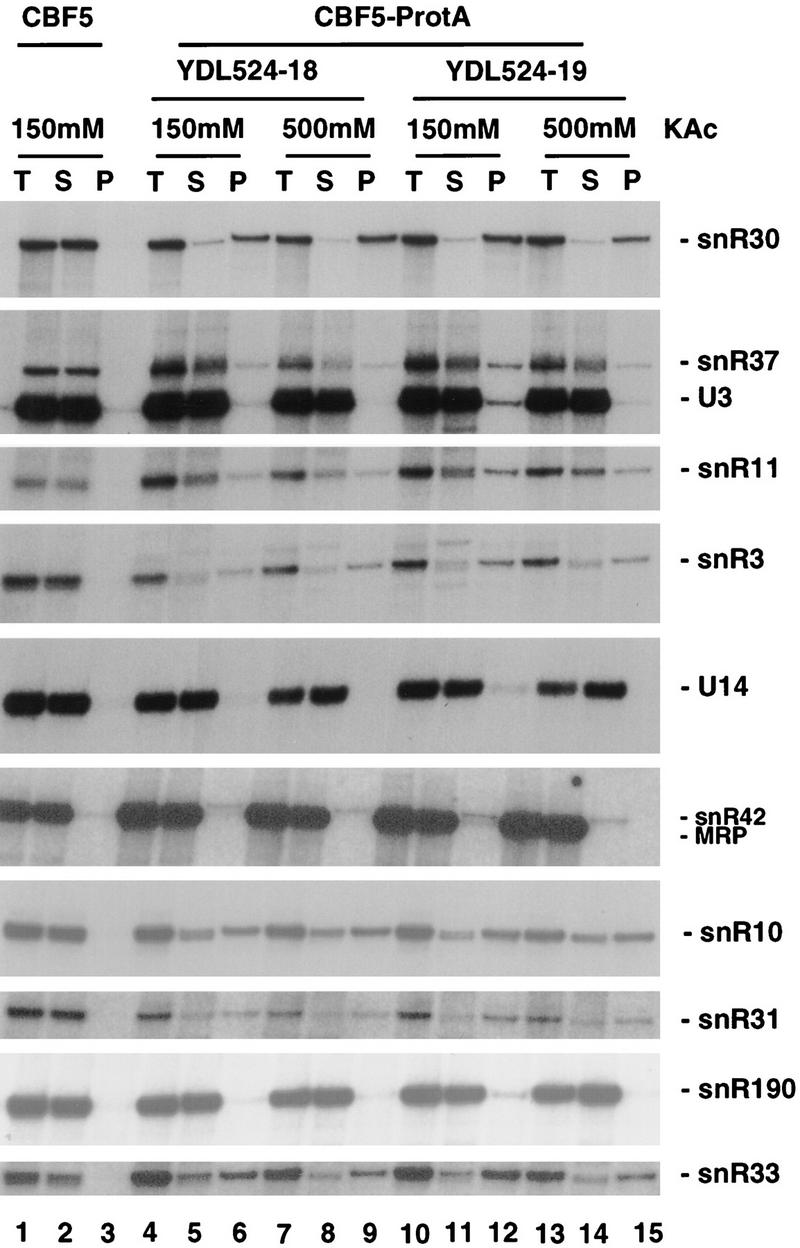
The box H + ACA snoRNAs are associated with Cbf5p–protein A (CBFS-Prot.A). Immunoprecipitations were performed at two salts concentration [150 and 500 mm Kacetate (KAc)] on two CBF5–Prot.A strains (YDL524-18 and YDL524-19) and at 150 mm Kacetate on the wild-type isogenic control (CBF5). RNA was extracted from equivalent amounts of total (T), supernatant (S), and pellet (P) fractions and separated on a 8% polyacrylamide gel containing 8 m urea. Probes used for the hybridizations are described in Materials and Methods.
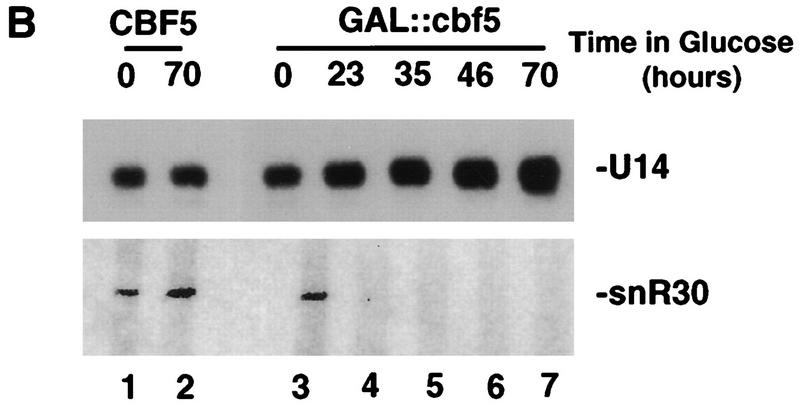
The levels of the snoRNAs were assessed during depletion of Cbf5p (Fig. 9A,B). Following growth of the GAL::cbf5 strain on rsg medium, the levels of all tested snoRNAs were the same as in the CFB5 control strain. In contrast, all tested box H + ACA snoRNAs, snR3, snR10, snR11, snR31, snR33, snR37, snR42 (Fig. 9A), and snR30 (Fig. 9B) were strongly depleted following transfer of the GAL::cbf5 strain to glu medium. The levels of the box C + D snoRNAs, U3, snR190 (Fig. 9A), and U14 (Fig. 9B) were unaffected, as were the levels of the RNAse MRP RNA (Fig. 9A) and the U5 snRNA (data not shown). Analysis of earlier time points during depletion of Cbf5p shows that the major drop in the level of snR30 occurs between 8 and 16 hr of depletion of Cbf5p (data not shown), in agreement with the onset of the inhibition of processing.
Figure 9.
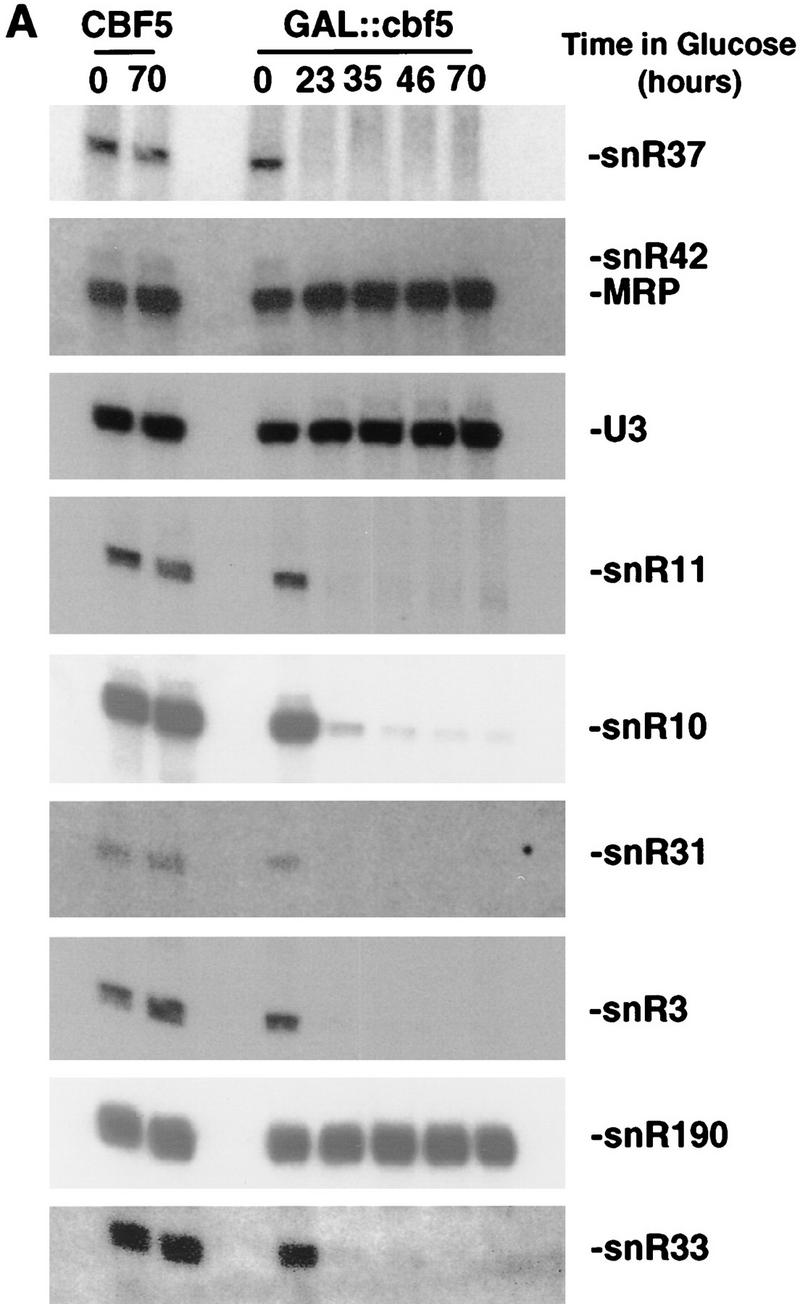
Box H + ACA snoRNP components are codepleted in a GAL::cbf5 strain. H + ACA snoRNAs (A,B) and Gar1p (C) are codepleted with Cbf5p. Probes used for the hybridizations are described in Materials and Methods. RNA was extracted from the CBF5 and GAL::cbf5 strains following growth on rsg medium (0-hr lanes) and at intervals following transfer to glu medium (8- to 70-hr lanes) and separated on an 8% polyacrylamide gel containing 8 m urea. The anti-Gar1p antibody used was described by Girard et al. (1992) and cross-reacts weakly with Nop1p.
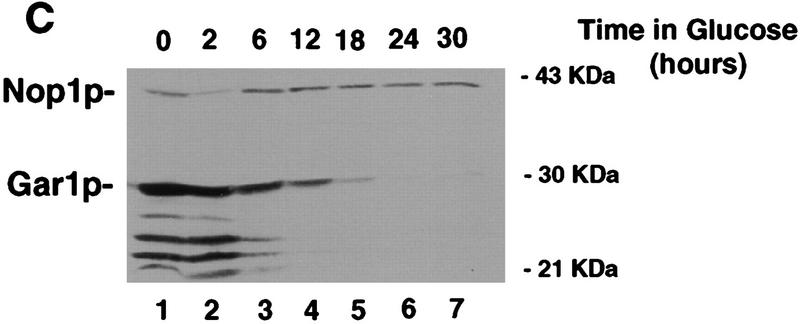
Gar1p, like Cbf5p, is associated with all known members of the family of box H + ACA snoRNAs (Girard et al. 1992). Moreover, a yeast two-hybrid screen has shown that Gar1p interacts physically with Cbf5p (Y. Henry, M. Fromont, P. Legrain, and M. Caizergues-Ferrer, unpubl.). After transfer of the GAL::cbf5 strain to glu medium, the level of Gar1p also falls dramatically (Fig. 9C).
We conclude that Cbf5p is a core component of the box H + ACA snoRNPs that is required for the stability of both the RNA and protein components of the snoRNPs.
Discussion
We report here a detailed functional analysis of Cbf5p, an essential nucleolar protein (Jiang et al. 1993) and putative Θ synthase (Koonin 1996). We found that genetic depletion of Cbf5p inhibits both pre-rRNA processing and formation of Θ in the pre-RNA. Expression of a Cbf5p–protein A fusion protein allowed the coprecipitation of all tested members of the large class of box H + ACA snoRNAs, most of which function as guides for the site-specific formation of Θ residues in the pre-rRNA (Ganot et al. 1997a; Ni et al. 1997). Members of the other major class of snoRNAs, the box C + D snoRNAs, were not detectably coprecipitated with Cbf5p–protein A nor were other small RNA species tested (U5 and MRP RNA). Moreover, the depletion of Cbf5p resulted in the codepletion of all tested box H + ACA snoRNAs but did not affect the levels of the box C + D snoRNAs or other small RNA species. We conclude that Cbf5p is an integral component of the box H + ACA class of snoRNPs.
Protein components of other small RNPs are required for the stability of the RNA components of the particles; these include the spliceosomal snRNAs (see Cooper et al. 1995 and references therein) and the RNA components of signal recognition particle (SRP; Brown et al. 1994), RNAse P and RNAse MRP (Lygerou et al. 1994; Chu et al. 1997). This observation presumably reflects a requirement for the intact structure of the RNP particles to prevent degradation of the RNAs. Unusually, we find that Cbf5p is not only required for the stability of the box H + ACA snoRNAs, but is also required for the stability of another protein component of the snoRNPs, Gar1p. The instability of Gar1p is presumably also attributable to a requirement for the intact RNP structure. In marked contrast to Cbf5p, depletion of Gar1p does not affect the stability of the box H + ACA snoRNAs (Girard et al. 1992). Gar1p depletion, however, does lead to the release of the box H + ACA snoRNA snR36 from the pre-rRNA (Bousquet-Antonelli et al. 1997), indicating that it normally plays a role in establishing the snoRNA–pre-rRNA interaction or in stabilizing the base pairing.
After depletion of Cbf5p, the residual level of Θ in the 25S rRNA corresponds to ∼3 Θ residues per 25S rRNA molecule, whereas the residual level of Θ in the 35S pre-rRNA is estimated at ∼7 residues per molecule. It is not clear whether the residual Θ residues are synthesized by a low residual level of Cbf5p that remains after depletion, or whether these represent a small number of sites at which Θ is synthesized by a mechanism that is independent of Cbf5p and the box H + ACA snoRNAs. Surprisingly, the level of Θ in the GAL::cbf5 strain grown under permissive conditions was also consistently below the wild-type values. This result is unlikely to be attributable to a reduced level of Cbf5p; the levels of the box H + ACA snoRNAs is not reduced and pre-rRNA processing is unaffected. The CBF5 mRNA is elevated compared to the wild type under permissive conditions, and the protein is likely to be overexpressed. We speculate that this overexpression leads to a form of squelching, a phenomenon well known from transcription studies (e.g., see Gill and Ptashne 1988; Tasset et al. 1990; Flanagan et al. 1991). Squelching often results from the overexpression of a free form of a component that normally functions in a complex with other proteins. The excess free protein blocks the interaction of other components with the complex, interfering with its function.
In contrast to the Θ guide snoRNAs, all of which are nonessential, another box H + ACA snoRNA, snR30, is essential and required for pre-rRNA processing (Bally 1988; Morrissey and Tollervey 1993). Genetic depletion of Cbf5p leads to the codepletion of snR30. The major drop in the level of snR30 occurs between 8 and 16 hr of depletion of Cbf5p, in agreement with the onset of the inhibition of processing. Presumably, as a consequence of this drop, the pre-rRNA processing reactions that require snR30, cleavage at sites A1 and A2 and to a lesser extent A0, are inhibited by Cbf5p depletion, leading to the loss of 18S rRNA synthesis. This observation explains why Cbf5p is essential for viability. In addition, depletion of Cbf5p leads to a strong delay in the processing of the 27SB and 7S pre-rRNAs, to the accumulation of a 5.8S species of intermediate length, and to a reduced level of mature 25S rRNA. This phenotype has not been observed in any snoRNA mutant and may be a consequence of the absence of Θ, possibly because of alterations in the folding of the pre-rRNA or changes in the structure of the preribosomal particle.
Other enzymes involved in RNA metabolism might also be brought to their substrates by carrier RNAs. The box C + D snoRNAs select sites of 2′-O-methylation by placing a putative protein-binding site at a precise distance from the nucleotide to be modified (Cavaillé et al. 1996; Kiss-László et al. 1996; Nicoloso et al. 1996). As the rRNA methylase has not yet been identified, it is not clear whether it associates with the box C + D snoRNAs prior to their association with the pre-rRNA. In vivo cleavage of the pre-rRNA at site A0 in the 5′ ETS is absolutely dependent on binding of the U3 snoRNA to an upstream site in the 5′ ETS (Beltrame and Tollervey 1995). A0, however, can be cleaved in vitro by Rnt1p (an RNAse III-like endonuclease) in the absence of cofactors (Abou Elela et al. 1996). It has not yet been established whether U3 carries the enzyme to its site of action in vivo. It seems possible that the role of the box H + ACA snoRNAs in targeting Cbf5p may be a model for the roles of other small RNA species.
Although the data presented here do not formally prove that Cbf5p functions as a Θ synthase within the snoRNPs, the homology to truB and the strong predictive value of the model make this possibility very likely. The E. coli homolog truB is the tRNA Θ55 synthase, indicating that the ancestral eukaryotic protein had this function and later acquired the ability to recognize sites made up of a trans-acting RNA based-paired to the target site for Θ formation. The level of Θ in the tRNA population is not strongly affected by depletion of Cbf5p, making it unlikely that Cbf5p is the eukaryotic tRNA Θ55 synthase. In fact, Pus4p, another yeast homolog of truB (Koonin 1996), is the tRNA Θ55 synthase (Becker et al. 1997). We propose that Pus4p and Cbf5p arose in early eukaryotes by gene duplication followed by divergence of function.
Materials and methods
Construction of the GAL::cbf5 and CBF5::Prot.A strains
Standard Saccharomyces cerevisiae growth and handling techniques were employed. The transformation procedure was described by Gietz et al. (1992). The wild-type strains used in this study were FY1679-28C and YDL401 (Lafontaine and Tollervey 1996). The GAL::cbf5 strain was created in the YDL401 background by use of a one-step PCR strategy (Lafontaine and Tollervey 1996). The oligonucleotides used for the amplification with plasmid pTL26 were oligonucleotide 1, 5′-TTTCAAATGATGAGATGTTTAGCTTAGGAAAAATATAATGTACTCTTGGCCTCCTCTAGT-3′ and oligonucleotide 2, 5′-CTTAATAACGAAATCCTCCTTTGACATTGTGTATATCGGTCCTCTCGAATTCCTTGAATTTTCAAA-3′. Transformants were screened for glucose sensitivity by PCR on yeast colonies and by Southern blot analysis. All the RNA analysis experiments were done in duplicate on two independently isolated GAL::cbf5 strains (YDL521-1 and YDL521-3). The analysis of the Θ content of rRNAs was made on strain YDL521-1. The CBF5::Prot.A strains expressing Cbf5–protein A were constructed in strain FY1679-28C by use of the same strategy. The oligonucleotides used for the amplification with pTL54 were oligonucleotide 3, 5′-GAAGACGGTGATTCTGAGGAAAAGAAATCTAAGAAATCTAAGAAAGGCGTGGACAACAAATTC-3′ and oligonucleotide 4, 5′-TACAAGTCGTTGATAAAGAAATTTTACGTTATTAATATACACACTCTGGGTAGAAGATCGGTC-3′. Transformants were screened by PCR on yeast colonies and by Western blot analysis (using PAP antibody, Sigma). Two independently isolated CBF5::Prot.A strains (YDL524-18 and YDL524-19) were used for the immunoprecipitation experiment presented in Figure 8.
GAL::cbf5 time course, RNA extraction, Northern hybridization, and primer extension
For depletion of Cbf5p, cells growing exponentially in permissive conditions (rsg) at 30°C were harvested by centrifugation, washed, and resuspended in 2% glucose minimal medium. During growth, cells were diluted with prewarmed medium and constantly maintained in exponential phase. RNA extraction, Northern hybridization, and primer extension were as described by Lafontaine et al. (1995). Standard 1.2% agarose/formaldehyde and 8% acrylamide gels were used to analyze the processing of the high and low molecular weight rRNAs species, respectively (Tollervey 1987). Ten percent acrylamide gels were used to analyze the tRNA processing. Nine micrograms of total RNA was used for the Northern and primer-extension experiments presented in Figures 2, 3, 5 and 6, whereas 4.5 μg was used for the Northern analysis presented in Figures 4 and 9. Oligonucleotides used for pre-rRNA hybridization were oligonucleotides a, b, c, d, g, h described previously by Lafontaine et al. (1995) as oligonucleotides d, g, h, i, k, l, respectively, and oligonucleotide f described previously as oligonucleotide b by Mitchell et al. (1996). Oligonucleotides anti-U3, U14, MRP, snR10, and snR190 were as described previously (Girard et al. 1992; Dichtl and Tollervey 1997). Oligonucleotides antimature tRNATrpCCA and tRNAProUGG and anti-intronic tRNATrpCCA, tRNAProUGG, tRNATyrGΘA and tRNAPheGAA were described by Sharma et al. (1996). Oligonucleotides anti-snR37, snR11, snR3, snR42, snR31, snR33, and U5 were 5′-GATAGTATTAACCACTACTG-3′, 5′-GACGAATCGTGACTCTG-3′, 5′-TCGATCTTCGTACTGTCT-3′, 5′-CTCCCTAAAGCATCACAA-3′, 5′-GTAGAACGAATCATGACC-3′, 5′-GATTGTCCACACACTTCT3′ and 5′-AAGTTCCAAAAAATATGGCAA-3′, respectively. Vector pT3/T7α18–snR30 (Morrissey and Tollervey 1993) was used to synthesize an snR30 riboprobe. To detect the CBF5 mRNA, a fragment spanning the whole ORF of CBF5 was generated by PCR and labeled by use of the Prime-a-Gene Labeling kit (Promega).
Analysis of Ψ levels
To determine the Θ content in rRNAs and tRNAs, strain YDL521-1 was pregrown at 30°C in rich medium containing 2% raffinose, 2% sucrose, and 2% galactose (YP–RSG). The culture was split in two, washed, and resuspended in either rich glu medium (YPD) or YP–RSG at an OD600 of 0.03. Both cultures were incubated for 6 hr at 30°C before being washed and transferred to similar medium without PO4 [following the recipe described by Warner (1991)]. After a further incubation of 18 hr, 50 ml from each culture was labeled for 15 min with 9 mCi of [32P] orthophosphate (9000 Ci/mmole). Total RNA was extracted and analyzed by electrophoresis on a 1.2% agarose–formaldehyde gel. 35S pre-rRNA, 25S rRNA, and total tRNAs were purified by electroelution, digested with RNAse T2 (in 50 mm Na acetate at pH 5.5 at 37°C), and analyzed by two-dimensional cellulose TLC as described by Filipowicz and Shatkin (1983). The procedure used was essentially as described by Bousquet-Antonelli et al. (1997).
Immunoprecipitation of Cbf5p–protein A
Yeast whole-cell extracts were prepared according to Séraphin and Rosbash (1989). Lysates were made in buffer A (20 mm Tris HCl at pH 8.0, 5 mm MgCl2, 1 mm DTT, 0.2% Triton X-100, 0.5 mm PMSF, and 150 or 500 mm Kacetate), and supernatants were cleared by centrifugation (56,000 rpm at 4°C for 20 min). Immunoprecipitation experiments were performed as described previously (Ganot et al. 1997b). Lysates equivalent to 37.5 OD600 of cells were incubated on a rotating wheel for 2 hr at 4°C with 100 μl of IgG–agarose beads (Sigma A2909), and pre-washed in buffer A in a total volume of 400 μl. Pellets were washed four times for 20 min in 1 ml of buffer A. Each gel lane (T, S, and P) was loaded with RNA from a fraction of the preparation equivalent to 10 OD600 of cells.
Western blotting
Total protein extracts corresponding to 106 cells were loaded in each lane. Affinity-purified anti-Gar1p antibodies were used at 1:200 dilution as described by Girard et al. (1992). The anti-Gar1p antibodies weakly cross-react with Nop1p.
Acknowledgments
We would like to thank Henri Grosjean for helpful advice and Tom Meier and Henri Grosjean for communicating results prior to publication, Ian Dix for providing the U5 oligonucleotide, and Tamás Kiss for his advice on the TLC technique. D.L.J.L. was the recipient of a fellowship from the European Commission and C.B.-A. was the recipient of a fellowship from l’Association pour la Recherche sur le Cancer (ARC). This work was supported by grants from the Wellcome Trust to D.T. and by grants from the Région Midi-Pyrénées, the ARC and La Ligue Nationale Contre le Cancer to M.C.-F.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL d.tollervey@ed.ac.uk; FAX 44 131 650 7040.
References
- Abou Elela S, Igel H, Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- Bachellerie JP, Cavaillé J. Guiding ribose methylation of rRNA. Trends Biochem Sci. 1997;22:257–261. doi: 10.1016/s0968-0004(97)01057-8. [DOI] [PubMed] [Google Scholar]
- Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell. 1996;85:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- Bally M, Hughes J, Cesareni G. snR30: A new essential small nuclear RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1988;16:5291–5303. doi: 10.1093/nar/16.12.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker HF, Motorin Y, Planta RJ, Grosjean H. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of Psi55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res. 1997;25:4493–4499. doi: 10.1093/nar/25.22.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M, Henry Y, Tollervey D. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 1994;22:5139–5147. doi: 10.1093/nar/22.23.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M, Tollervey D. Base-pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Henry Y, Gèlugne J-P, Caizergues-Ferrer M, Kiss T. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J. 1997;16:4770–4776. doi: 10.1093/emboj/16.15.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, Hann BC, Medzihradszky KF, Niwa M, Burlingame AL, Walter P. Subunits of the Saccharomyces cerevisiae signal recognition particle required for its functional expression. EMBO J. 1994;13:4390–4400. doi: 10.1002/j.1460-2075.1994.tb06759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaillé J, Nicoloso M, Bachellerie J-P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- Chu S, Zengel JM, Lindahl L. A novel protein shared by RNase MRP and RNase P. RNA. 1997;3:382–391. [PMC free article] [PubMed] [Google Scholar]
- Cooper M, Johnston LH, Beggs JD. Identification and characterization of Uss1p (Sdb23p): A novel U6 snRNA- associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 1995;14:2066–2075. doi: 10.1002/j.1460-2075.1995.tb07198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B, Tollervey D. Pop3p is essential for the activity of the RNase MRP and RNase P ribonucleoproteins in vivo. EMBO J. 1997;16:417–429. doi: 10.1093/emboj/16.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Shatkin AJ. Origin of splice junction phosphate in tRNAs processed by HeLa cell extract. Cell. 1983;32:547–557. doi: 10.1016/0092-8674(83)90474-9. [DOI] [PubMed] [Google Scholar]
- Flanagan PM, Kelleher RH, Sayre MH, Tschochner H, Kornberg RD. A mediator required for activation of RNA polymerase II transcription in vitro. Nature. 1991;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- Ganot P, Bortolin M-L, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997a;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily defined secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes & Dev. 1997b;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- Geitz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- Girard JP, Lehtonen H, Caizergues-Ferrer M, Amalric F, Tollervey D, Lapeyre B. Gar1p is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992;11:673–682. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldwasser E, Heinrikson RL. The biochemistry of pseudouridine. In: Davidson NJ, Cohn WE, editors. Progress in nucleic acid research and mol. biol. New York, NY: Academic Press; 1966. pp. 399–416. [DOI] [PubMed] [Google Scholar]
- Hughes JMX, Ares MJ. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Middleton K, Yoon H-J, Fouquet C, Carbon J. An essential yeast protein, Cbf5p, binds in vitro to centromeres and microtubules. Mol Cell Biol. 1993;13:4884–4893. doi: 10.1128/mcb.13.8.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammen HO, Marvel CC, Hardy L, Penhoet EE. Purification, structure and properties of Esherichia coli tRNA pseudouridine synthase I. J Biol Chem. 1988;263:2255–2263. [PubMed] [Google Scholar]
- Kiss-László Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of pre ribosomal RNA: A novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Pseudouridine synthases: Four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res. 1996;24:2411–2415. doi: 10.1093/nar/24.12.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D, Tollervey D. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 1996;24:3469–3472. doi: 10.1093/nar/24.17.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D, Vandenhaute J, Tollervey D. The Dim1p methylase is required for pre-rRNA processing in yeast. Genes & Dev. 1995;9:2470–2481. doi: 10.1101/gad.9.20.2470. [DOI] [PubMed] [Google Scholar]
- Li HV, Zagorski J, Fournier MJ. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischwe MA, Ochs RL, Reddy R, Cook RG, Yeoman LC, Tan EM, Reichlin M, Busch H. Purification and partial characterization of a nucleolar scleroderma antigen (Mr = 34,000; pI, 8.5) rich in NG, NG-dimethylarginine.J. Biol Chem. 1985;260:14304–14310. [PubMed] [Google Scholar]
- Lygerou Z, Mitchell P, Petfalski E, Séraphin B, Tollervey D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes & Dev. 1994;8:1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- Maden T. Click here for methylation. Nature. 1996;383:675–676. doi: 10.1038/383675a0. [DOI] [PubMed] [Google Scholar]
- Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- Meier UT, Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria.J. Cell Biol. 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Tollervey D. The 3′-end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes & Dev. 1996;10:502–513. doi: 10.1101/gad.10.4.502. [DOI] [PubMed] [Google Scholar]
- Morrissey JP, Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- Nicoloso M, Qu L-H, Michot B, Bachellerie J-P. Intron-encoded, antisense small nucleolar RNAs: The characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J Mol Biol. 1996;260:178–195. doi: 10.1006/jmbi.1996.0391. [DOI] [PubMed] [Google Scholar]
- Nurse K, Wrzensinski J, Bakin A, Lane BG, Ofengand J. Purification, cloning, and properties of the tRNA Θ55 synthase from Escherichia coli. RNA. 1995;1:102–112. [PMC free article] [PubMed] [Google Scholar]
- Ofengand J, Bakin A, Wrzesinski J, Nurse K, Lane BG. The pseudouridine residues of ribosomal RNA. Biochem Cell Biol. 1995;73:915–924. doi: 10.1139/o95-099. [DOI] [PubMed] [Google Scholar]
- Schimmang T, Tollervey D, Kern H, Frank R, Hurt EC. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989;8:4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B, Rosbash M. Identification of functional U1 snRNA–pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- Sharma K, Fabre E, Tekotte H, Hurt EC, Tollervey D. Yeast nucleoporin mutants are defective in pre-tRNA splicing. Mol Cell Biol. 1996;16:294–301. doi: 10.1128/mcb.16.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos G, Tekotte H, Grosjean H, Segref A, Sharma K, Tollervey D, Hurt EC. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO J. 1996;15:2270–2284. [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Steitz JA. Sno storm in the Nucleolus: New roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Tasset D, Tora L, Fromental C, Scheer E, Chambon P. Distinct classes of transcriptional activating domains function by different mechanisms. Cell. 1990;62:1177–1187. doi: 10.1016/0092-8674(90)90394-t. [DOI] [PubMed] [Google Scholar]
- Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Small nucleolar RNAs guide ribosomal RNA methylation. Science. 1996;273:1056–1057. doi: 10.1126/science.273.5278.1056. [DOI] [PubMed] [Google Scholar]
- Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt EC. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- Warner JR. Labeling of RNA and phosphoproteins in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:423–428. doi: 10.1016/0076-6879(91)94033-9. [DOI] [PubMed] [Google Scholar]
- Wrzesinski J, Bakin A, Nurse K, Lane BG, Ofengand J. Purification, cloning, and properties of the 16S RNA ϑ516 synthase from Escherichia coli. Biochemistry. 1995a;34:8904–8913. doi: 10.1021/bi00027a043. [DOI] [PubMed] [Google Scholar]
- Wrzesinski J, Nurse K, Bakin A, Lane BG, Ofengand J. A dual specificity pseudouridine synthase: Purification and cloning of a synthase from Escherichia coli which is specific for both Θ746 in 23S RNA and for Θ32 in tRNAPhe. RNA. 1995b;1:437–448. [PMC free article] [PubMed] [Google Scholar]