Abstract
Aspergillus terreus has been difficult to identify in cases of aspergillosis, and clinical identification has been restricted to the broad identification of aspergillosis lesions in affected organs or the detection of fungal carbohydrates. As a result, there is a clinical need to identify species-specific biomarkers that can be used to detect invasive A. terreus disease. Monoclonal antibodies (MAbs) were developed to a partially purified preparation of cytolytic hyphal exoantigens (HEA) derived from A. terreus culture supernatant (CSN). Twenty-three IgG1 isotype murine MAbs were developed and tested for cross-reactivity against hyphal extracts of 54 fungal species. Sixteen MAbs were shown to be specific for A. terreus. HEA were detected in conidia, hyphae, and in CSN of A. terreus. HEA were expressed in high levels in the hyphae during early stages of A. terreus growth at 37°C, whereas at room temperature the expression of HEA peaked by days 4 to 5. Expression kinetics of HEA in CSN showed a lag, with peak levels at later time points at room temperature and 37°C than in hyphal extracts. Serum spiking experiments demonstrated that human serum components do not inhibit detection of the HEA epitopes by MAb enzyme-linked immunosorbent assay (ELISA). Immunoprecipitation and proteomic analysis demonstrated that MAbs 13E11 and 12C4 immunoprecipitated a putative uncharacterized leucine aminopeptidase (Q0CAZ7), while MAb 19B2 recognized a putative dipeptidyl-peptidase V (DPP5). Studies using confocal laser scanning microscopy showed that the uncharacterized leucine aminopeptidase mostly localized to extracellular matrix structures while dipeptidyl-peptidase V was mostly confined to the cytoplasm.
INTRODUCTION
Aspergillus terreus is a filamentous fungus associated with organic detritus decay in the soil rhizosphere. Since the identification of A. terreus by Thom in 1918 (59), the species has been utilized in the biotechnology industry as a source of organic acids (11, 20), the serum cholesterol-lowering compound lovastatin (7, 62), and proteases and peptides that hydrolyze proteins (32). In contrast to its economic importance in biotechnology, A. terreus causes significant losses by spoiling food and agricultural products (2, 14, 31). Furthermore, A. terreus has been reported as a human pathogen and can cause superficial, cutaneous, and subcutaneous mycoses that affect the nail bed (28), outer ear canal (60), and skin (16). More recently, A. terreus has been associated with postoperative osteomyelitis (43), endophthalmitis (22), and peritonitis (63), and the species has been identified as an emerging opportunistic pathogen in immunocompromised populations (3, 4). Knowledge of A. terreus pathogenesis is limited; however, the species is thermotolerant and produces aleurioconidia that may provide an advantage in dissemination (48). A. terreus also produces terrelysin, a hemolytic protein that has been recently reported as a putative virulence factor (10, 44). The species also produces secondary metabolites that may have toxic effects on host cells and may help facilitate invasive disease (19, 39, 41).
Species-specific diagnosis of A. terreus opportunistic infections is clinically important due to this pathogen's resistance to the primary antifungal therapeutic amphotericin B (13, 56, 66). To date, the identification of A. terreus infections has challenged the most seasoned clinicians (29). Clinical diagnosis of A terreus infection is subjective and has been restricted to macroscopic and microscopic characterization of tissue samples (72), computed tomography imaging (15), and detection of serum galactomannan or 1,3-β-d-glucan (30, 58). These diagnostic methods are limited and prevent the identification of the specific causative agent (30). Other more specific methodologies such as PCR have been recently developed but are limited by a number of other confounding factors (6, 30). Due to increasing A. terreus infections reported in the literature, resistance to amphotericin B, and the high mortality rate associated with infection, it is critical to develop sensitive and specific diagnostic tests (33). In this study, we describe the production and characterization of species-specific monoclonal antibodies (MAbs) to a partially purified cytolytic preparation of hyphal exoantigens (HEA) from A. terreus. Using the MAbs, we characterized the cross-reactivity profiles, kinetics of antigen expression, and identity of several of the exoantigens. These data demonstrate that these MAbs may be useful for immunodiagnostic assays to detect invasive A. terreus disease.
MATERIALS AND METHODS
Preparation of A. terreus hyphal exoantigens and rabbit polyclonal antibodies.
A. terreus was purchased from the American Type Culture Collection (ATCC 1012; Manassas, VA). Conidia were inoculated in tryptic soy broth (TSB) and grown in liquid culture for 7 days. HEA were partially purified from TSB using molecular sieve and gel filtration steps to isolate the cytolytic fraction, as previously described (65).
Polyclonal antibodies to HEA were generated in rabbits and affinity purified by Bethyl Laboratories (Montgomery, TX) using HEA immobilized on activated Sepharose columns.
Production of monoclonal antibodies to HEA.
Five 10- to 14-week-old BALB/c mice (The Jackson Laboratory, Bar Harbor, ME) were housed under controlled environmental conditions in HEPA-filtered ventilated polycarbonate cages on autoclaved hardwood beta chip bedding. Mice were provided Teklad 7913 rodent chow (Harlan Laboratories, Madison, WI) and autoclaved tap water ad libitum. Sentinel mice housed in the animal quarters were free of viral pathogens, parasites, mycoplasma, and Helicobacter spp. The animal protocol was approved by the National Institute for Occupational Safety and Health (NIOSH) Animal Care and Use Committee. The NIOSH animal facility is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC).
Mice were immunized (six times) intraperitoneally at biweekly intervals with 25 μg of HEA emulsified in TiterMax adjuvant (TiterMax USA, Norcross, GA). Mice received a final boost (7th immunization) of 25 μg of HEA without adjuvant 3 days prior to hybridoma production. Pre- and postbleed mouse IgG-specific titers to HEA were tested using an indirect enzyme-linked immunosorbent assay (ELISA) method. Briefly, 96-well Nunc Immuno MaxiSorp microplates (Thermo Fisher Scientific, Rochester, NY) were coated with 1 μg/ml HEA in carbonate coating buffer (CCB; 60 mM sodium carbonate, 140 mM sodium bicarbonate, pH 9.6). Antibody binding from mouse serum was detected using biotin-SP-conjugated goat anti-mouse IgG Fcγ (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) and alkaline phosphatase (AP)-conjugated streptavidin (Jackson ImmunoResearch Laboratories, Inc.). ELISA plates were developed with 4-nitrophenyl substrate (Sigma, St. Louis, MO) and read at 405 nm after 30 min as previously described (45, 51).
Three days following the final boost, the immunized mice were euthanized by CO2 asphyxiation, the spleens were removed, and splenocytes were fused with SP2/0-Ag 14 ATCC myeloma cells (ATCC CRL-1581). Hybridomas were selected by growing the cells in Dulbecco's modified Eagle medium (DMEM) (Life Technologies, Rockville, MD) supplemented with 1 mM sodium pyruvate, 100 U/ml penicillin, 100 mg/ml streptomycin, 0.292 mg/ml l-glutamine, 100 mM sodium hypoxanthine, 16 mM thymidine, 10% fetal calf serum (FCS) (HyClone, Logan, UT), and 100 U/ml interleukin-6 (IL-6; Boehringer, Mannheim, Germany). DMEM was additionally supplemented with azaserine for selective propagation of hybridomas. After 10 to 14 days of hybridoma cell growth, the medium from individual wells was replenished with fresh medium. The supernatant fluid from individual hybridoma colonies was tested using a modification of a previously described indirect ELISA (45). Supernatant fluid from individual hybridoma clones was tested in duplicate to confirm the presence of HEA-specific MAbs. Positive hybridoma clones were further cloned twice by limiting dilution, and single positive clones were screened and selected for bulk MAb production. Hybridoma cell lines of individual clones were frozen in 10% dimethyl sulfoxide (DMSO) and 10% FCS, stored at −80°C for 2 weeks, and then transferred to a liquid nitrogen facility for long term storage.
HEA capture ELISA.
In brief, 96-well Nunc Immuno MaxiSorp microplates (Thermo Fisher Scientific, Rochester, NY) were coated with 100 μl/well rabbit anti-HEA polyclonal antibody (1 μg/ml) in CCB and incubated overnight at room temperature (RT). Wells were washed three times by incubation with 200 μl/well phosphate-buffered saline (pH 7.4)-0.05% Tween 20 (PBST) for 10 min. The plates were then blocked for 1 h at RT with 200 μl/well of PBSTM (phosphate-buffered saline, pH 7.4, 0.05% Tween 20, 3% dry milk). Wells were then incubated for 1 h at 37°C with 100 μl/well HEA (1 μg/ml) to bind HEA. Plates were then processed for the detection of IgG antibody by the addition of hybridoma culture supernatant (CSN), followed by goat anti-mouse IgG (45). Negative-control values were obtained by replacing hybridoma CSN with complete DMEM.
Isotyping and quantification of IgG antibodies.
Isotyping of individual MAbs was determined by an indirect ELISA. Plates were coated with CSN from A. terreus diluted in CCB (1 μg/ml) and incubated overnight. Plates were then blocked with PBSTM and incubated with MAb solutions from individual hybridomas. MAbs bound to HEA were detected using biotin-SP-conjugated AffiniPure goat anti-mouse IgG1, IgG2a, IgG2b, and IgG3 secondary antibodies (Jackson ImmunoResearch Laboratories Inc.) at a dilution of 1:5,000 in PBSTM. ELISA plates were developed using methods described previously.
For quantification, MAbs were serially diluted and captured on ELISA plates coated with AffiniPure goat anti-mouse IgG Fc fragment of subclass 1, 2a, 2b, or 3 at a concentration of 1 μg/ml (Jackson ImmunoResearch Laboratories Inc.). IgG1, IgG2a, IgG2b, and IgG3 standards (Sigma) were used as a standard curve for quantification purposes. AP-conjugated goat anti-mouse secondary antibodies (1:5,000) diluted in PBSTM were used and developed using methods described previously.
Preparation of A. terreus extracts for characterization of MAbs. (i) Conidial extracts.
Conidia were collected from 10- to 14-day-old A. terreus cultures grown on malt extract agar (MEA) by rolling approximately 1 g of 0.5-mm glass beads (BioSpec Products Inc., Bartlesville, OK) over the plate. Glass beads with conidia were collected into a 2-ml screw cap microcentrifuge tube and processed in a Mini Bead Beater (BioSpec Products Inc.). Mechanical bead beating was carried out for 2 min to disrupt the outer cell wall of the conidia. Conidium proteins were extracted in 50 mM ammonium bicarbonate buffer, pH 8.0, containing 0.5 M EDTA, 0.1 M phenylmethylsulfonyl fluoride, and Complete Mini Protease Inhibitor Cocktail (Roche Diagnostics, Mannheim, Germany). The suspension was centrifuged at 4,100 × g for 10 min, and the supernatant fluid was collected and lyophilized overnight. Lyophilized conidium protein extract was resuspended in PBS, pH 7.4, and stored at −80°C.
(ii) Hyphal extracts and culture supernatants.
A. terreus conidia were inoculated in 50 ml of minimal medium consisting of 1% glucose, nitric salts, and trace elements (27). Viability of conidia was determined by a Live/Dead BacLight viability kit (Invitrogen, Corp, Carlsbad, CA) as per the manufacturer's instructions and as previously reported (17). Cultures were seeded with 2.5 × 107 viable conidia and grown at RT or 37°C with shaking (200 rpm) for various intervals of time. For MAb reactivity assays, cultures were grown for 6 days, and for the kinetic assays, cultures were grown for 12 days, with samples collected at 24-h intervals. Mycelial cultures were harvested by centrifugation at 4,100 × g for 5 min, and the hyphae (pellet) and CSN were collected and concentrated by lyophilization. The lyophilized CSN was reconstituted in 5 ml of PBS, pH 7.4, containing Complete Mini Protease Inhibitor Cocktail and stored at −80°C for further analysis. Lyophilized hyphae were macerated in a mortar containing liquid N2 and suspended in cold PBS, pH 7.4, containing Complete Mini Protease Inhibitor Cocktail, and proteins were extracted overnight at 4°C on a rocker. The mycelial extract (ME) was centrifuged at 4,100 × g for 5 min, and the supernatant fluid was collected, aliquoted, and stored at −80°C for further analysis. Protein concentrations in all fungal extract preparations were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Thermo Scientific, Wilmington, DE).
Characterization of HEA expression in fungi.
For cross-reactivity analyses, 46 fungal species (Table 1) were inoculated on MEA medium and grown for 7 to 10 days. The conidia (1.25 × 108) were collected and inoculated into 50 ml of TSB medium. Cultures were grown for 4 days at RT and centrifuged at 4,100 × g for 5 min to collect the mycelial pellet. The pellet was washed three times with cold PBS, pH 7.4, macerated, and then centrifuged at 4,100 × g for 5 min as previously described. The supernatant fluid was collected, protein concentration was determined by NanoDrop spectrophotometer, and antigen concentration was tested using the previously described indirect ELISA method. Positive reactivity was determined by an optical density (OD) of ≥0.2 (negative-control value plus 3 standard deviations). The ODs of the negative controls ranged from 0 to 0.18 for different fungal extracts.
Table 1.
Fungal species tested for cross-reactivity
| Fungal species | Culture collection identification no.a | Reactive MAb(s) |
|---|---|---|
| Aspergillus species | ||
| A. terreus | ATCC 1012 | All |
| FGSC A1156 | All | |
| ATCC 16794 | All | |
| SRC 2174 | All | |
| A. chevalieri | NRRL 78 | None |
| A. clavatus | NIOSH 6-22-78 | None |
| A. flavus | ATCC 24689 | 67G7 |
| A. fumigatus | FGSC A1100 | None |
| A. nidulans | NIOSH 15-22-08 | None |
| A. niger | FGSC A1144 | None |
| A. parasiticus | ATCC 26691 | 67G7 |
| A. repens | NRRL 13 | 24D7, 38B6, 40C6, 52G7, 64B3, 67G7 |
| A. sydowii | ATCC 9507 | 24D7, 67G7 |
| A. ustus | NRRL 275 | None |
| A. versicolor | ATCC 44408 | None |
| Other | ||
| Acremonium strictum | ATCC 46646 | 22D9 |
| Alternaria alternata | ATCC 11612 | None |
| Alternaria brassicicola | ATCC 96836 | None |
| Botrytis cinerea | ATCC 11542 | None |
| Chaetomium globosum | ATCC 6205 | None |
| Cladosporium herbarum | ATCC 6506 | None |
| Cladosporium cladosporioides | ATCC 11288 | None |
| Epicoccum nigrum | ATCC 34929 | None |
| Exserohilum rostratum | ATCC 26856 | None |
| Fusarium moniliforme | PS M6131 | None |
| Geotrichum candidum | UAMH 7863 | None |
| Memnoniella echinata | NRRL 2373 | None |
| Memnoniella subsimplex | ATCC 32888 | None |
| Myrothecium verrucaria | NRRL 2003 | None |
| Paecilomyces variotii | ATCC 66705 | None |
| Penicillium aurantiogriseum | NRRL 971 | None |
| Penicillium expansum | NRRL 973 | None |
| Penicillium fellutanum | NRRL 746 | None |
| Penicillium purpurogenum | NRRL 1062 | None |
| Penicillium roqueforti | NRRL 844 | None |
| Rhizopus stolonifer | NIOSH 17-59-14 | None |
| Stachybotrys albipes | ATCC 18873 | None |
| Stachybotrys bisbyi | ATCC 18825 | None |
| Stachybotrys chartarum | IBT 7711 | None |
| IBT 9290 | None | |
| IBT 9460 | None | |
| IBT 9466 | None | |
| IBT 9631 | None | |
| IBT 9633 | None | |
| IBT 14915 | None | |
| IBT 14916 | None | |
| Stachybotrys chlorohalonata | ATCC 201863 | None |
| Stachybotrys cylindrospora | ATCC 16276 | None |
| Stachybotrys kampalensis | ATCC 22705 | None |
| Stachybotrys nephrospora | ATCC 18839 | None |
| Stachybotrys oenanthes | CBS 252.76 | None |
| Stachybotrys parvispora | CBS 100155 | None |
| Scopulariopsis brumptii | ATCC 16278 | None |
| Stemphylium botryosum | ATCC 26881 | None |
| Trichoderma viride | ATCC 16640 | None |
| Wallemia sebi | NIOSH 26-41-01 | None |
Sources for 46 species representing 20 different genera of fungi are as follows: ATCC, American Type Culture Collection; NRRL, Agricultural Research Service Culture Collection; NIOSH, National Institute for Occupational Safety and Health; FGSC, Fungal Genetics Stock Center; UAMH, University of Alberta Microfungus Collection and Herbarium, Canada; PS, Pennsylvania State University; IBT, Instituttet for Bioteknologi, Denmark; CBS,-Centraalbureau voor Schimmelcultures, Netherlands.
The kinetics of antigen expression during culture was also examined. Individual MAbs were tested against A. terreus spore, mycelial, and CSN extracts, used at 100 μg/ml total protein to determine the level of antigen expression during different phases of A. terreus growth. A polyclonal antibody (PAb)-based capture ELISA was used for this analysis, and all MAbs were normalized to 500 ng/ml in PBSTM. Plates were developed using secondary antibodies and reagents as previously described.
HEA detection assay in spiked serum.
Detection of HEA by the MAbs in the presence of human serum was studied using the ELISA methods described earlier. CSN was collected from a 6-day culture of A. terreus and was diluted to 100 μg/ml of total protein. Pooled human serum (Sigma) was spiked with CSN and assayed in the capture ELISA at a final concentration of 50% total volume using 100 μl of sample. Additionally, samples were also incubated in PBS and served as controls for this experiment. All samples including controls were incubated at 37°C for 1 h before analysis in capture ELISA method.
Western blot assay.
A. terreus CSN (100 μg/ml total protein) obtained from a 6-day TSB culture was separated on a 12% polyacrylamide gel using SDS-PAGE under reducing conditions. Proteins were transferred overnight at 30 V to a 0.2-μm-pore-size nitrocellulose membrane. The membrane was blocked with 3% bovine serum albumin (BSA) in PBST for 2 h. The membrane was washed in PBST and then assembled on a Mini-Protean II multiscreen apparatus (Bio-Rad Laboratories, Hercules, CA). Each lane on the membrane was incubated with an individual MAb (500 ng/ml) for 1 h with shaking. The membrane was washed with PBST and incubated with AP-conjugated goat anti-mouse IgG(H+L) (Promega, Madison, WI) diluted 1:5,000 in PBST for 1 h on a shaker. The membrane was then washed with PBST and developed using 1-Step NBT/BCIP (nitroblue tetrazolium/5-bromo-4-chloro-3-indolylphosphate) substrate (Promega). The substrate reaction was developed for 15 to 20 min and stopped by washing the membrane with distilled water.
Immunoprecipitation and proteomic analysis. (i) Immunoprecipitation.
Immunoprecipitation of antigens recognized by MAbs 13E11, 12C4, and 19B2 was performed using Dynabeads protein G (Invitrogen Dynal AS, Oslo, Norway) per the manufacturer's instructions. Briefly, protein G beads were washed twice with wash and binding (W&B) buffer (0.1 M Na-phosphate, 0.01% Tween 20, pH 8.2) and incubated with 5 μg of the individual MAbs diluted in W&B buffer for 10 min with rotation at RT. Antigen capture was performed after washing the protein G beads with PBS and incubation overnight at 4°C with A. terreus hyphal extract. The beads were then washed thoroughly in PBS, and the immunoprecipitate was eluted with Laemmli sample buffer (Bio-Rad, Hercules, CA) containing 2-mercaptoethanol (5% vol/vol). Immunoprecipitate samples were analyzed by SDS-PAGE and stained with Imperial Protein Stain (Thermo Scientific, Rockford, IL). Bands of interest, identified by Western blotting, were excised from the SDS-PAGE gels and processed for proteomic analysis.
(ii) Proteomic analysis.
All chemicals were obtained from Sigma (St. Louis, MO), unless otherwise noted, and all incubations were performed at 37°C with shaking at 300 rpm. Excised bands of interest were reduced, alkylated, and digested with porcine trypsin according to methods published previously (55, 57). Enzymatic peptides were separated on a nanoAcquity ultraperformance liquid chromatography (UPLC) system (Waters, Milford, MA). The eluent from the UPLC system was directed to the nanoelectrospray source of a Waters Synapt MS quadrupole time-of-flight (qTOF) mass spectrometer (MS). Spectra were acquired in a multiplexed tandem MS (MSe) format. Data were analyzed with ProteinLynx Global Server (PLGS), version 24 (Waters), using the default PLGS search engine to query a custom A. terreus database downloaded from UniProtKB/Swiss-Prot and UniProtKB/TrEMBL. Protein assignments were confirmed via manual inspection of tandem mass spectra.
Confocal scanning laser microscopy localization of A. terreus MAb reactive antigens.
Immunolocalization of leucine aminopeptidase and dipeptidyl-peptidase V was studied using methods previously described with slight modifications (47, 69). Briefly, A. terreus cultures were grown on alcohol-sterilized coverslips in six-well tissue culture plates containing minimal medium. Cultures were incubated at 37°C for 24 h without shaking. Coverslips were fixed with 8% formalin-buffered saline containing 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) ([PIPES] pH 6.7), 25 mM EGTA, 1% DMSO, and 5 mM MgSO4. Cells were fixed for 1 h at RT and rinsed with MTSB (50 mM PIPES, pH 6.7, 5 mM EGTA, and 5 mM MgSO4). Cell wall digestion was carried out for 1 h at RT with an enzyme solution containing 2.5% Driselase (Sigma, St. Louis, MO), 1% lysozyme from chicken egg white (Sigma), and 2 mM EGTA. Cells were rinsed with H2O and treated with 0.1% Triton X-100 in Tris-buffered saline (TBS), pH 7.4, for 10 min. Cells were quickly rinsed once in MTSB and TBS. Cells were blocked with 3% BSA in TBS overnight at 4°C with gentle shaking. Coverslips were then incubated with MAb 13E11 or 19B2 at 3 μg/ml in 3% BSA in TBS for 3 h with gentle shaking. An IgG1 isotype MAb 9B4 that was previously developed in our laboratory against a Stachybotrys chartarum conidial surface protein (51) was used as a negative control. Cells were washed thoroughly in TBS containing 0.05% Tween 20 (T-TBS) and stained with Alexa Fluor 594-conjugated goat anti-mouse IgG(H+L) (Molecular Probes, Inc., Eugene, OR) diluted 1:50 (vol/vol) in 3% BSA in TBS for 1 h at RT. Cells were washed thoroughly in T-TBS, and coverslips were placed onto clean slides containing ProLong antifade reagent with 4′,6′-diamidino-2-phenylindole ([DAPI] Molecular Probes, Inc.). Cells were observed with a Zeiss LSM-510 Meta Confocal Microscope System (Axioplan 2 Stand) (Zeiss, Thornwood, NY), and images were acquired with LSM-510 software.
RESULTS
Characterization of MAb reactivity to fungal extracts.
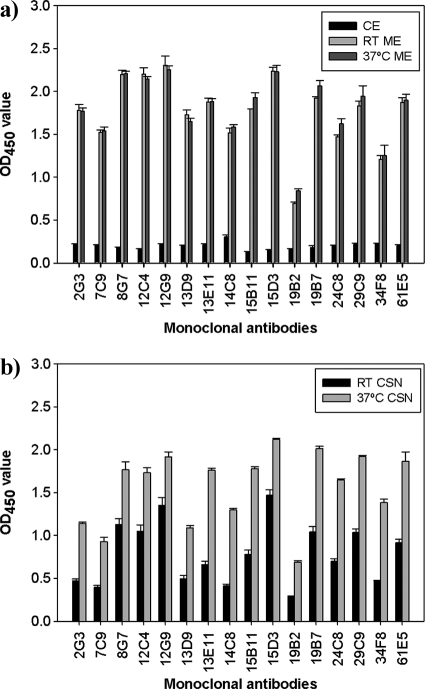
Twenty-three murine IgG1 isotype hybridomas were developed against HEA (Table 1). Since the MAbs were developed against a partially purified cytolytic extract, reactivity of the MAbs to A. terreus culture extract antigen was tested. MAb reactivity was highest to mycelial extracts; however, weak reactivity to conidial extracts was also observed (Fig. 1a). As expected, the MAbs also showed significant reactivity to 6-day culture supernatant from cultures grown at room temperature and greater reactivity to CSN grown at 37°C (Fig. 1b). Collectively these results suggest that the MAbs react with hyphal antigens that are secreted into CSN.
Fig. 1.
MAb reactivity to A. terreus extracts. (a) MAb reactivity to A. terreus conidial and hyphal extracts. Conidial and mycelial extracts (CE and ME, respectively) of A. terreus grown at RT and 37°C were tested against 16 specific MAbs. (b) MAb reactivity to A. terreus CSN collected from cultures grown at RT and 37°C was tested against 16 specific MAbs. Error bars represent the standard deviations of duplicate determinations from three independent experiments.
Cross-reactivity of MAbs toward different fungi.
Table 1 lists the 46 fungal species from 20 different genera that were tested for cross-reactivity in the capture ELISA. Positive reactivity was defined as an OD at 450 nm (OD450) of ≥0.2. All MAbs showed reactivity to ME from each A. terreus strain tested in this study. Of the 23 MAbs that react with HEA, 16 (70%) did not cross-react with any of the fungal species tested (Table 1). Seven MAbs cross-reacted with other fungi. Most of the cross-reactivity observed in these seven MAbs was limited within the genus Aspergillus, except for MAb 22D9 that also cross-reacted with ME derived from Acremonium strictum. Five MAbs (22D9, 38B6, 40C6, 52G7, and 64B3) cross-reacted with ME from at least one other fungus, while 24D7 cross-reacted with two different fungi (Table 1). The MAb 67G7 exhibited the greatest cross-reactivity, reacting to hyphal extracts derived from four different Aspergillus species. Of the fungal species tested, Aspergillus sydowii and Aspergillus repens hyphal extracts showed highest cross-reactivity.
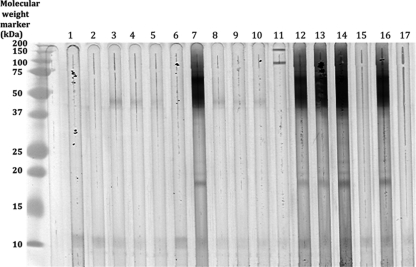
Western blot analysis of HEA.
Figure 2 shows the Western blot analysis of A. terreus CSN using the 16 species-specific MAbs and the cross-reactive MAb 67G7. The results demonstrated differences within the MAb reactivities to antigens in A. terreus CSN. The MAbs 13E11, 19B7, 24C8, 29C9, and 61E5 showed similar patterns with strong reactivity to bands at ∼18 kDa, 45 kDa, and ∼70 kDa. The MAb 19B2 identified unique high-molecular-weight bands at ∼100 kDa and ∼150 kDa. It is possible that the immune reactivity to the 10-kDa and the 45-kDa antigens is due to binding of antibodies to shared epitopes or to carbohydrate antigens.
Fig. 2.
Western blot analysis of anti-HEA MAbs. Lane 1, MAb 2G3; lane 2, MAb 7C9; lane 3, MAb 8G7; lane 4, MAb 12C4; lane 5, MAb 12G9; lane 6, MAb 13D9; lane 7, MAb 13E11; lane 8, MAb 14C8; lane 9, MAb 15B11; lane 10, MAb 15D3; lane 11, MAb 19B2; lane 12, MAb 19B7; lane 13, MAb 24C8; lane 14, MAb 29C9; lane 15, MAb 34F8; lane 16, MAb 61E5; lane 17, MAb 67G7.
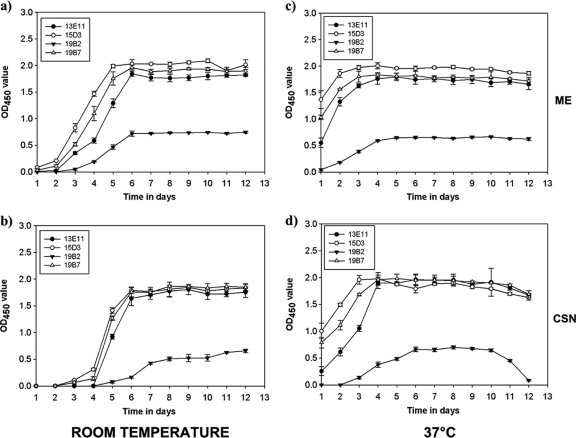
Kinetics of A. terreus HEA expression.
Four MAbs (13E11, 15D3, 19B2, and 19B7) were chosen for further study based on differences in patterns of reactivity in ELISA and Western blotting. Figure 3 illustrates the time course of HEA expression in A. terreus cultures. For MAbs 13E11, 15D3, and 19B7, reactivity to HEA preparation rapidly increased in ME during days 3 to 5 and peaked at day 6 (Fig. 3a). Reactivity to HEA in the CSN followed a similar pattern but appeared to lag behind that in ME by 24 h (Fig. 3b). At 37°C, HEA MAb reactivity was readily detectable by 24 h in both ME (Fig. 3c) and CSN (Fig. 3d) and peaked by days 3 and 4, respectively. Interestingly, HEA MAb reactivity could be detected in both CSN and ME beyond day 6 but appeared to decline in CSN after 10 days at 37°C. The MAb 19B2 antigen, while having a similar pattern as the other MAbs, appeared to lag in expression, suggesting that this MAb may recognize a different antigen than other MAbs tested in this study. Also, we observed a more rapid degradation of MAb 19B2 antigen after day 10 in CSN of A. terreus cultures grown at 37°C. This could probably be due to proteolytic degradation of the antigen.
Fig. 3.
Kinetic assay of HEA in A. terreus hyphae and culture supernatant. (a) Mycelial extract collected from A. terreus cultures grown at room temperature. (b) CSN collected from A. terreus cultures grown at room temperature. (c) Mycelial extract collected from A. terreus cultures grown at 37°C. (d) CSN collected from A. terreus cultures grown at 37°C. All samples were collected at 24-h interval for 12 days and tested in capture ELISA. Data represent the mean and standard deviations following analysis of duplicate ELISA determinations from three independent experiments.
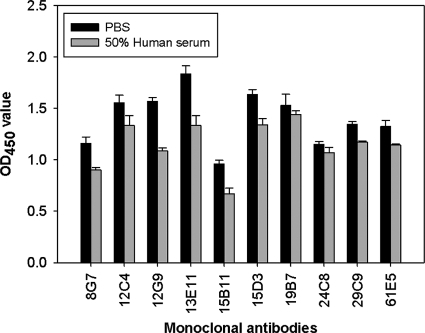
Human serum detection assay.
To determine the ability of these MAbs to be used in an immunodiagnostic assays, we tested 10 MAbs for reactivity to A. terreus antigens spiked in pooled human serum (Fig. 4). CSN grown at 37°C was mixed with human serum and assayed using a capture ELISA. Overall, there was a slight reduction in the detection of HEA when HEA was spiked into human serum compared to PBS for all MAbs tested; however, there did not seem to be any significant binding to serum components for any of the antibodies.
Fig. 4.
Detection of HEA by MAbs in the presence of serum. A. terreus CSN was spiked in 50% human serum and detected using capture ELISA. PBS served as a control in the experiment. Error bars represent the standard deviations of duplicate determinations from three independent experiments.
Immunoprecipitation and proteomic analysis.
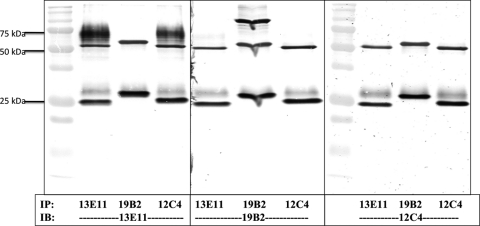
Three MAbs (13E11, 19B2, and 12C4) were selected for further analysis based on differential patterns of reactivity to A. terreus extract (Fig. 2). The MAbs were incubated with A. terreus hyphal extract, and protein-antibody complexes bound to the protein G beads were eluted and separated using SDS-PAGE. Using MAb 13E11 to stain the immunoblot showed immunoprecipitation of an ∼65-kDa band, slightly higher than the antibody heavy chain band. Interestingly, MAb 13E11 also showed this same band in the immunoprecipitate of MAb 12C4 but not that of MAb 19B2 (Fig. 5). This suggests that MAb 13E11 and MAb 12C4 recognized similar antigens. In contrast, MAb 19B2 demonstrated immunoreactivity to an antigen localized at ∼100 kDa (middle panel). While MAb 12C4 precipitated a 66-kDa band that was recognized by 13E11 (left panel), this MAb does not recognize this band when it is used to stain the Western blot (right panel), indicating that it may recognize a conformational epitope.
Fig. 5.
Immunoprecipitation of HEA. MAbs 13E11, 19B2, and 12C4 were used for immunoprecipitation of their respective antigens from A. terreus CSN. Immunoprecipitates were analyzed using individual MAbs to identify cross-identification of antigens between MAbs. IP, immunoprecipitate; IB, immunoblot. MAbs are indicated below the panels.
Specific bands were excised from a parallel SDS-PAGE gel on which immunoprecipitated samples from each MAb were separated. Samples were subjected to UPLC tandem MS (MS/MS) analysis to determine the identity of the proteins. By comparing peptide masses of recovered peptides in silico to the generated database for A. terreus, we identified peptides for a putative uncharacterized protein (Q0CAZ7) in immunoprecipitates from both MAbs 13E11 and 12C4. The protein Q0CAZ7 has >60% sequence homology to a leucine aminopeptidase found in other Aspergillus species such as Aspergillus fumigatus, Aspergillus nidulans, Aspergillus oryzae, and Aspergillus flavus. Peptides for a probable dipeptidyl peptidase V (Q0C8V9) were also identified following the UPLC MS/MS analysis of MAb 19B2 immunoprecipitates (Table 2). Both of these proteolytic enzymes contain putative N-glycosylation sites and are secreted after processing of a signal peptide.
Table 2.
Proteomic analysis of HEA antigens
| MAb(s) | HEAa | Sequence coverage (%) | N-glycosylation sitesb | Predicted signal peptide site(s) (method)b,c |
|---|---|---|---|---|
| 13E11, 12C4 | Putative uncharacterized leucine aminopeptidase (Q0CAZ7) | 10 | 232, 349, 432, 435, 466 | Between 17 and 18 (NN), between 21 and 22 (HMM) |
| 19B2 | Probable dipeptidyl-peptidase V (Q0C8V9) | 52 | 37, 79, 97, 154, 255, 339, 381, 451, 509, 608 | Between 19 and 20 (NN and HMM) |
Protein identification numbers are given in parentheses.
Numbers represent amino acid sites on preprotein.
NN, neural networks; HMM, hidden Markov model.
Immunolocalization of HEA.
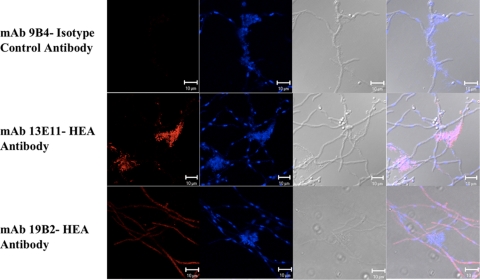
The immunolocalizations of A. terreus leucine aminopeptidase (MAb 13E11) and dipeptidyl peptidase V (MAb 19B2) in the hyphae were determined using confocal laser scanning microscopy. Interestingly, the leucine aminopeptidase (13E11) was localized in extracellular matrix (ECM) structures (Fig. 6). Immunostaining was also observed within the A. terreus hyphae. MAb 19B2 immunostaining for a probable dipeptidyl peptidase V was diffuse but uniform over the entire hypha, suggesting that this protein might be present in the cytoplasm of A. terreus hyphae. No staining was observed in the extracellular matrix for MAb 19B2. The MAb 9B4 served as an isotype control for these studies and did not stain any structures.
Fig. 6.
Immunolocalization of HEA. Immunolocalization of HEA was determined by Alexa Fluor 594-labeled goat anti-mouse secondary antibodies (red), and nuclear staining was identified by DAPI (blue). MAb 9B4 acted as an IgG1 isotype control antibody in this study.
DISCUSSION
Invasive aspergillosis develops in the lungs of immunocompromised subjects following the inhalation of viable Aspergillus conidia from the environment (33, 70, 73). Upon germination, the conidia differentiate into a vegetative hyphal form that can cause damage to the host tissue and may allow the fungus to disseminate to other parts of the body (52). To date, the diagnostic methods have been limited to identifying macroscopic and microscopic characteristics in lung biopsy specimens (30, 53). Serological diagnostics have also been developed but are limited to the detection of galactomannan and β-d-glucan (30, 53). Although this methodology may confirm a fungal infection, it does not identify the specific pathogenic species involved. Previous studies have detected Aspergillus antigens in the sera and urine of patients diagnosed with invasive aspergillosis (18, 25, 26, 36). These results suggest that during infection, Aspergillus species secrete proteins that could be used to serologically detect the organism. Based on the limitations of available detection methodologies, it is critical to identify biomarkers that could be used to serologically identify individual pathogenic Aspergillus species.
Although A. fumigatus is the most widely known etiological agent of invasive aspergillosis, A. terreus has emerged as an opportunistic pathogen that has been attributed to a variety of infections including fatal disseminated aspergillosis (4). To our knowledge, no immunodiagnostics have been developed for the specific detection of A. terreus in clinical samples. In this study, 23 IgG1 MAbs were produced using a partially purified cytolytic HEA preparation that was isolated using a methodology previously used to purify stachylysin from Stachybotrys chartarum (65). The MAbs developed in the present study specifically detect antigens localized in A. terreus conidia and hyphae, but, more importantly, these hemolytic antigens were detected in the CSN fluid. These findings demonstrate that these antigens may be actively secreted during hyphal differentiation and growth and may be candidate biomarkers for immunodiagnostic assays. Detection of hemolytic antigen in higher concentrations in hyphae is consistent with our previous studies of stachylysin (50, 61, 65, 71). Sixteen MAbs were found to be species specific while seven cross-reacted with other species. The species-specific MAbs did not cross-react with mycelial extracts from other Aspergillus pathogenic species including A. fumigatus, A. flavus, Aspergillus niger, and A. nidulans. No cross-reactivity was observed with other fungal species belonging to the genera Penicillium and Fusarium. Most cross-reactivity was minimal with OD450 values of ≤0.5.
Previously, differences were reported in metabolic activities, growth rates, and virulence capabilities of different A. terreus strains depending on their environmental source (49). We were curious to see if any of these differences were reflected in altered expression of HEA. All MAbs reacted with the mycelial extracts from the four A. terreus strains; however, comprehensive testing with additional clinical strains of A. terreus will be critical prior to the development of diagnostic screening methods for use in the clinical setting. Moreover, other species that are closely related to A. terreus such as Aspergillus carneus, Aspergillus niveus, and the newly identified Aspergillus alabamensis, as well as other unrelated species including Scedosporium and Rhizopus stolonifer (5, 38), should be tested for cross-reactivity.
A. terreus growth is accompanied by conidial germination during favorable nutrient and environmental conditions. This process involves the swelling of conidia, initiation of primary metabolism, and hyphal extension and aggregation. In this study, we observed that the antigens were detected earlier in hyphal extracts than in CSN, suggesting an active secretion of these proteins. The concentration of these antigens appeared to correlate with the total biomass of the culture and protein concentration (data not shown) during HEA kinetic experiments. MAb reactivity to HEA was also observed to increase proportionally with increases in the mycelial pellet size. Most importantly, HEA were continuously detected in CSN at 37°C, emphasizing the relative stability of these antigens to proteolytic degradation for longer period of time.
Detection of the antigens in CSN may not fully reflect antigen production during invasive disease. Furthermore, secreted antigens may bind to serum proteins or other factors that alter the confirmation of the protein and subsequently reduce the availability of the epitope for MAb detection. Certain fungal proteins are known to bind serum components in vitro (21). In tests with pooled human serum, there was only a slight reduction in the detection of epitopes using our MAbs. This suggests that the HEA epitopes do not interact with serum components, and this may have potential use for serodetection of invasive A. terreus disease.
Leucine aminopeptidase and dipeptidyl-peptidase V are both predicted to possess putative N-glycosylation sites as determined by N-Glycosite (74). These proteins are secreted with putative signal peptides as determined by SignalP, version 3.0, in silico analysis (9, 23, 46). This has been confirmed experimentally by us in this study and previously by others for A. terreus and other fungal species (24, 64). Homologues of dipeptidyl-peptidase V in other fungal species have been reported as a potential virulence factor or allergen and as important for tissue invasion and modulation of host immune responses (8, 34, 35, 54, 64, 67, 68).
Immunolocalization studies demonstrated that the putative leucine aminopeptidase identified by MAb 13E11 was localized to extracellular structures containing DNA. Similar structures containing extracellular DNA have been reported in vitro and in vivo and have been identified as the extracellular matrix of fungi (1, 37, 42). ECMs have only been recently identified, and there is little information on their role in the pathogenesis of fungal infections. MAb 13E11 may be a useful tool in studying ECMs, and, more importantly, the putative leucine aminopeptidase may function as a biomarker of invasive A. terreus disease. In contrast, MAb 19B2 recognized a probable dipeptidyl peptidase V, and immunostaining was primarily localized within the cytoplasm. These MAbs also have the potential to be used for the immunofluorescent detection of A. terreus in bronchoalveolar lavage samples.
In conclusion, we observed that HEA were released from vegetative hyphae and into CSN in a time-dependent manner. The MAbs developed in this study recognized these antigens, and their binding was not inhibited by any serum components in spiking experiments. Collectively, the data suggest that the MAbs developed to HEA may have potential diagnostic value for cases of A. terreus invasive aspergillosis. Serological detection of A. terreus-specific antigens would obviate the need to obtain clinical specimens by invasive methods to identify the causal agent. Previously, dipeptidyl-peptidase V has been reported as one of two major antigens with the greatest serodiagnostic potential for detecting aspergillosis due to A. fumigatus infection (8, 12, 34, 40). These methodological developments may aid in the development of standardized immunoassays for rapid identification of pathogenic species in clinical samples.
ACKNOWLEDGMENTS
The findings and the conclusions in this report are those of the authors and do not necessarily represent the views of the National Institute for Occupational Safety and Health.
This work was supported in part by an interagency agreement with the National Institute of Environmental Health Sciences (Y1-ES0001-06).
Footnotes
Published ahead of print on 6 July 2011.
REFERENCES
- 1. Al-Fattani M. A., Douglas L. J. 2006. Biofilm matrix of Candida albicans and Candida tropicalis: chemical composition and role in drug resistance. J. Med. Microbiol. 55:999–1008 [DOI] [PubMed] [Google Scholar]
- 2. Atalla M. M., Hassanein N. M., El-Beih A. A., Youssef Y. A. 2003. Mycotoxin production in wheat grains by different aspergilli in relation to different relative humidities and storage periods. Nahrung 47:6–10 [DOI] [PubMed] [Google Scholar]
- 3. Baddley J. W., Pappas P. G., Smith A. C., Moser S. A. 2003. Epidemiology of Aspergillus terreus at a university hospital. J. Clin. Microbiol. 41:5525–5529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Balajee S. A. 2009. Aspergillus terreus complex. Med. Mycol. 47(Suppl. 1):S42–S46 [DOI] [PubMed] [Google Scholar]
- 5. Balajee S. A., et al. 2009. Aspergillus alabamensis, a new clinically relevant species in the section Terrei. Eukaryot. Cell 8:713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balajee S. A., et al. 2009. Molecular identification of Aspergillus species collected for the Transplant-Associated Infection Surveillance Network (TRANSNET). J. Clin. Microbiol. 47:3138–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barrios-Gonzalez J., Miranda R. U. 2010. Biotechnological production and applications of statins. Appl. Microbiol. Biotechnol. 85:869–883 [DOI] [PubMed] [Google Scholar]
- 8. Beauvais A., et al. 1997. Biochemical and antigenic characterization of a new dipeptidyl-peptidase isolated from Aspergillus fumigatus. J. Biol. Chem. 272:6238–6244 [DOI] [PubMed] [Google Scholar]
- 9. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 10. Berne S., Lah L., Sepcic K. 2009. Aegerolysins: structure, function, and putative biological role. Protein Sci. 18:694–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Berry D., Chmiel A., Al Obaidi Z. (ed.). 1977. Citric acid production by Aspergillus niger. Academic Press, London, United Kingdom [Google Scholar]
- 12. Biguet J., Tran van Ky P., Andrieu S. 1967. Identification d'une activité chymotrypsique au niveau de fractions remarquables d'Aspergillus fumigatus. Répercussions sur le diagnostic immunologique de l'aspergillose. Rev. Immunol. Ther. Antimicrob. 31:317–328 [PubMed] [Google Scholar]
- 13. Blum G., et al. 2008. Potential basis for amphotericin B resistance in Aspergillus terreus. Antimicrob. Agents Chemother. 52:1553–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bueno D. J., Silva J. O., Oliver G. 2001. Mycoflora in commercial pet foods. J. Food Prot. 64:741–743 [DOI] [PubMed] [Google Scholar]
- 15. Caillot D., et al. 2001. Increasing volume and changing characteristics of invasive pulmonary aspergillosis on sequential thoracic computed tomography scans in patients with neutropenia. J. Clin. Oncol. 19:253–259 [DOI] [PubMed] [Google Scholar]
- 16. Cheetham H. D. 1964. Subcutaneous infection due to Aspergillus terreus. J. Clin. Pathol. 17:251–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen C. Y., Seguin-Swartz G. 2002. A rapid method for assessing the viability of fungal spores. Can. J. Plant Pathol. 24:230–232 [Google Scholar]
- 18. Chumpitazi B. F. F., Pinel C., Lebeau B., Ambroise-Thomas P., Grillot R. 2000. Aspergillus fumigatus antigen detection in sera from patients at risk for invasive aspergillosis. J. Clin. Microbiol. 38:438–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chung C. H., Ling K. H., Tung S. S., Tung T. C. 1971. Study on fungi of the stored unhulled rice of Taiwan. II. Aflatoxin B1-like compounds from the culture of Aspergillus genus. J. Formosan Med. Assoc. 70:258–266 [PubMed] [Google Scholar]
- 20. Dowdells C., et al. 2010. Gluconic acid production by Aspergillus terreus. Lett. Appl. Microbiol. 51:252–257 [DOI] [PubMed] [Google Scholar]
- 21. Fukuchi Y. 2001. Interactions between Asp-hemolysin from Aspergillus fumigatus and blood plasma components. Yakugaku Zasshi. 121:423–432 (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 22. Garg P., Mahesh S., Bansal A. K., Gopinathan U., Rao G. N. 2003. Fungal infection of sutureless self-sealing incision for cataract surgery. Ophthalmology 110:2173–2177 [DOI] [PubMed] [Google Scholar]
- 23. Gasteiger E., et al. 2005. Protein identification and analysis tools on the ExPASy server, p. 571–607 In Walker J. M. (ed.), The proteomics protocols handbook. Humana Press, New York, NY [Google Scholar]
- 24. Han M. J., Kim N. J., Lee S. Y., Chang H. N. 2010. Extracellular proteome of Aspergillus terreus grown on different carbon sources. Curr. Genet. 56:369–382 [DOI] [PubMed] [Google Scholar]
- 25. Haynes K. A., Latge J. P., Rogers T. R. 1990. Detection of Aspergillus antigens associated with invasive aspergillosis. J. Clin. Microbiol. 28:2040–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haynes K. A., et al. 1996. Purification and characterization of a 93 kDa Aspergillus fumigatus antigen with diagnostic potential. J. Med. Vet. Mycol. 34:421–426 [PubMed] [Google Scholar]
- 27. Hill T. W., Kafer E. 2001. Improved protocols for Aspergillus minimal medium: trace element and minimal medium stock solutions. Fungal Genet. Newsl. 48:20–21 [Google Scholar]
- 28. Hilmioglu-Polat S., et al. 2005. Non-dermatophytic molds as agents of onychomycosis in Izmir, Turkey—a prospective study. Mycopathologia 160:125–128 [DOI] [PubMed] [Google Scholar]
- 29. Hope W. W., Denning D. W. 2004. Invasive aspergillosis: current and future challenges in diagnosis and therapy. Clin. Microbiol. Infect. 10:2–4 [DOI] [PubMed] [Google Scholar]
- 30. Hope W. W., Walsh T. J., Denning D. W. 2005. Laboratory diagnosis of invasive aspergillosis. Lancet Infect. Dis. 5:609–622 [DOI] [PubMed] [Google Scholar]
- 31. Horn B. W. 2006. Relationship between soil densities of Aspergillus species and colonization of wounded peanut seeds. Can. J. Microbiol. 52:951–960 [DOI] [PubMed] [Google Scholar]
- 32. Hughes R. L. 1968. Microbiological degradation of paper, p. 281–290 In Walters A. H., Elphick J. J. (ed.), Biodeterioration of materials. Elsevier Publishing Co. Ltd., New York, NY [Google Scholar]
- 33. Iwen P. C., Rupp M. E., Langnas A. N., Reed E. C., Hinrichs S. H. 1998. Invasive pulmonary aspergillosis due to Aspergillus terreus: 12-year experience and review of the literature. Clin. Infect. Dis. 26:1092–1097 [DOI] [PubMed] [Google Scholar]
- 34. Kobayashi H., Debeaupuis J. P., Bouchara J. P., Latge J. P. 1993. An 88-kilodalton antigen secreted by Aspergillus fumigatus. Infect. Immun. 61:4767–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Latge J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Latge J. P., et al. 1991. The 18-kilodalton antigen secreted by Aspergillus fumigatus. Infect. Immun. 59:2586–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Loussert C., et al. 2010. In vivo biofilm composition of Aspergillus fumigatus. Cell Microbiol. 12:405–410 [DOI] [PubMed] [Google Scholar]
- 38. Marr K. A., Carter R. A., Crippa F., Wald A., Corey L. 2002. Epidemiology and outcome of mould infections in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 34:909–917 [DOI] [PubMed] [Google Scholar]
- 39. Miller P. A., Trown P. W., Fulmor W., Morton G. O., Karliner J. 1968. An epidithiapiperazinedione antiviral agent from Aspergillus terreus. Biochem. Biophys. Res. Commun. 33:219–221 [DOI] [PubMed] [Google Scholar]
- 40. Monod M., Jousson O., Reichard U. 2009. Aspergillus fumigatus secreted proteases, p. 87–106 In Latge J. P., Steinbach W. J. (ed.), Aspergillus fumigatus and aspergillosis. ASM Press, Washington, DC [Google Scholar]
- 41. Moss M. O. (ed.). 1977. Aspergillus mycotoxins. Academic Press, London, United Kingdom [Google Scholar]
- 42. Muller F. M. C., Seidler M., Beauvais A. 2011. Aspergillus fumigatus biofilms in the clinical setting. Med. Mycol. 49:S96–S100 [DOI] [PubMed] [Google Scholar]
- 43. Natesan S., Abraham G., Mathew M., Lalitha M. K., Srinivasan C. N. 2007. Secondary sternal Aspergillus osteomyelitis in a diabetic hemodialysis patient with previous allograft rejection. Hemodial Int. 11:403–405 [DOI] [PubMed] [Google Scholar]
- 44. Nayak A. P., et al. 2011a. Characterization of recombinant terrelysin, a hemolysin of Aspergillus terreus. Mycopathologia 171:23–34 [DOI] [PubMed] [Google Scholar]
- 45. Nayak A. P., et al. 2011b. Production and characterization of IgM monoclonal antibodies against hyphal antigens of Stachybotrys species. Hybridoma 30:29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nielsen H., Krogh A. 1998. Prediction of signal peptides and signal anchors by a hidden Markov model, p. 122–130 In Glasgow J., et al. (ed.), Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology AAAI Press, Menlo Park, CA: [PubMed] [Google Scholar]
- 47. Osmani S. A., Engle D. B., Doonan J. H., Morris N. R. 1988. Spindle formation and chromatin condensation in cells blocked at interphase by mutation of a negative cell cycle control gene. Cell 52:241–251 [DOI] [PubMed] [Google Scholar]
- 48. Pore R. S., Larsh H. W. 1967. Aleuriospore formation in four related Aspergillus species. Mycologia 59:318–325 [Google Scholar]
- 49. Rippon J. W., Anderson D. N., Soo Hoo M. 1974. Aspergillosis, comparative virulence, metabolic rate, growth rate and ubiquinone content of soil and human isolates of Aspergillus terreus. Sabouraudia 12:157–161 [DOI] [PubMed] [Google Scholar]
- 50. Sakaguchi O., Shimada H., Yokota K. 1975. Proceedings: purification and characteristics of hemolytic toxin from Aspergillus fumigatus. Jpn. J. Med. Sci. Biol. 28:328–331 [PubMed] [Google Scholar]
- 51. Schmechel D., Simpson J. P., Beezhold D., Lewis D. M. 2006. The development of species-specific immunodiagnostics for Stachybotrys chartarum: the role of cross-reactivity. J. Immunol. Methods 309:150–159 [DOI] [PubMed] [Google Scholar]
- 52. Segal B. H. 2009. Aspergillosis. N. Engl. J. Med. 360:1870–1884 [DOI] [PubMed] [Google Scholar]
- 53. Segal B. H., Walsh T. J. 2006. Current approaches to diagnosis and treatment of invasive aspergillosis. Am. J. Respir. Crit. Care Med. 173:707–717 [DOI] [PubMed] [Google Scholar]
- 54. Slunt J. B., Taketomi E. A., Woodfolk J. A., Hayden M. L., Platts-Mills T. A. 1996. The immune response to Trichophyton tonsurans: distinct T cell cytokine profiles to a single protein among subjects with immediate and delayed hypersensitivity. J. Immunol. 157:5192–5197 [PubMed] [Google Scholar]
- 55. Speicher K. D., Kolbas O., Harper S., Speicher D. W. 2000. Systematic analysis of peptide recoveries from in-gel digestions for protein identifications in proteome studies. J. Biomol. Tech. 11:74–86 [PMC free article] [PubMed] [Google Scholar]
- 56. Steinbach W. J., et al. 2004. Infections due to Aspergillus terreus: a multicenter retrospective analysis of 83 cases. Clin. Infect. Dis. 39:192–198 [DOI] [PubMed] [Google Scholar]
- 57. Stone K. L., Williams K. R. 2002. Enzymatic digestion of proteins in solution and in SDS-polyacrylamide gels, p. 511–521 In Walker J. M. (ed.), The protein protocols handbook, 2nd ed. Humana Press, Totowa, NJ [Google Scholar]
- 58. Stynen D., Goris A., Sarfati J., Latge J. P. 1995. A new sensitive sandwich enzyme-linked immunosorbent assay to detect galactofuran in patients with invasive aspergillosis. J. Clin. Microbiol. 33:497–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Thom C., Church M. B. 1918. A. fumigatus, A. nidulans, A. terreus n. sp., and their allies. Am. J. Botany. 5:84–104 [Google Scholar]
- 60. Tiwari S., Singh S. M., Jain S. 1995. Chronic bilateral suppurative otitis media caused by Aspergillus terreus. Mycoses 38:297–300 [DOI] [PubMed] [Google Scholar]
- 61. Van Emon J. M., Reed A. W., Yike I., Vesper S. J. 2003. ELISA measurement of stachylysin in serum to quantify human exposures to the indoor mold Stachybotrys chartarum. J. Occup. Environ. Med. 45:582–591 [DOI] [PubMed] [Google Scholar]
- 62. Varga J., et al. 2005. Evolutionary relationships among Aspergillus terreus isolates and their relatives. Antonie Van Leeuwenhoek 88:141–150 [DOI] [PubMed] [Google Scholar]
- 63. Verghese S., Palani R., Thirunavakarasu N., Chellamma T., Pathipata P. 2008. Peritonitis due to Aspergillus terreus in a patient undergoing continuous ambulatory peritoneal dialysis. Mycoses 51:174–176 [DOI] [PubMed] [Google Scholar]
- 64. Vermout S., Baldo A., Tabart J., Losson B., Mignon B. 2008. Secreted dipeptidyl peptidases as potential virulence factors for Microsporum canis. FEMS Immunol. Med. Microbiol. 54:299–308 [DOI] [PubMed] [Google Scholar]
- 65. Vesper S. J., Magnuson M. L., Dearborn D. G., Yike I., Haugland R. A. 2001. Initial characterization of the hemolysin stachylysin from Stachybotrys chartarum. Infect. Immun. 69:912–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Walsh T. J., et al. 2003. Experimental pulmonary aspergillosis due to Aspergillus terreus: pathogenesis and treatment of an emerging fungal pathogen resistant to amphotericin B. J. Infect. Dis. 188:305–319 [DOI] [PubMed] [Google Scholar]
- 67. Woodfolk J. A., Slunt J. B., Deuell B., Hayden M. L., Platts-Mills T. A. 1996. Definition of a Trichophyton protein associated with delayed hypersensitivity in humans. Evidence for immediate (IgE and IgG4) and delayed hypersensitivity to a single protein. J. Immunol. 156:1695–1701 [PubMed] [Google Scholar]
- 68. Woodfolk J. A., Wheatley L. M., Piyasena R. V., Benjamin D. C., Platts-Mills T. A. 1998. Trichophyton antigens associated with IgE antibodies and delayed type hypersensitivity. Sequence homology to two families of serine proteinases. J. Biol. Chem. 273:29489–29496 [DOI] [PubMed] [Google Scholar]
- 69. Xiang X., Roghi C., Morris N. R. 1995. Characterization and localization of the cytoplasmic dynein heavy chain in Aspergillus nidulans. Proc. Natl. Acad. Sci. U. S. A. 92:9890–9894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yano S., Shishido S., Kobayashi K., Nakano H., Kawasaki Y. 1999. Bronchocentric granulomatosis due to Aspergillus terreus in an immunocompetent and non-asthmatic woman. Respir. Med. 93:672–674 [DOI] [PubMed] [Google Scholar]
- 71. Yokota K., Shimada H., Kamaguchi A., Sakaguchi O. 1977. Studies on the toxin of Aspergillus fumigatus VII. Purification and some properties of hemolytic toxin (asp-hemolysin) from culture filtrates and mycelia. Microbiol. Immunol. 21:11–22 [DOI] [PubMed] [Google Scholar]
- 72. Yuanjie Z., Jingxia D., Hai W., Jianghan C., Julin G. 2009. Primary cutaneous aspergillosis in a patient with cutaneous T-cell lymphoma. Mycoses 52:462–464 [DOI] [PubMed] [Google Scholar]
- 73. Zaas A. K., Alexander B. D. 2009. Invasive pulmonary aspergillosis, p. 293–300 In Latge J. P., Steinbach W. J. (ed.), Aspergillus fumigatus and aspergillosis. ASM Press, Washington, DC [Google Scholar]
- 74. Zhang M., et al. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229–1246 [DOI] [PubMed] [Google Scholar]