Abstract
Planctomycetes represent a remarkable clade in the domain Bacteria because they play crucial roles in global carbon and nitrogen cycles and display cellular structures that closely parallel those of eukaryotic cells. Studies on Planctomycetes have been hampered by the lack of genetic tools, which we developed for Planctomyces limnophilus.
TEXT
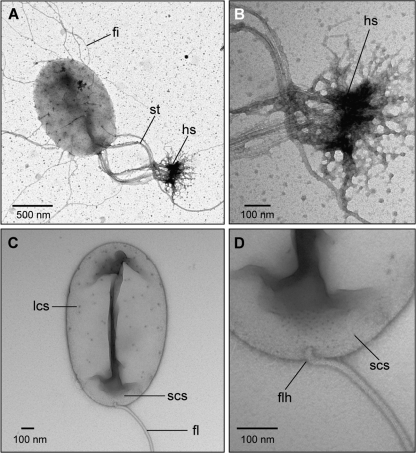
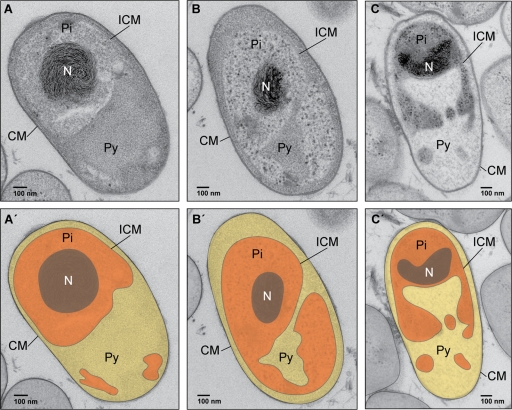
The Planctomycetes represent a remarkable clade within the domain Bacteria. They display cellular features once thought to be the sole domain of members of Eukarya. Some species have elaborate membrane-enclosed compartments, at times resembling the eukaryotic nucleus, and the capability to carry out endocytosis using membrane coat-like proteins (3, 6, 7, 9). In addition, Planctomycetes are major players in the global nitrogen and carbon cycles and perform reactions such as the anaerobic oxidation of ammonium with the aid of subcellular organelles known as anammoxosomes (6, 11, 13). Despite recent progress in revealing the complex subcellular structures of several Planctomycetes, in-depth analyses of these unusual features have been hampered by the lack of genetic tools. Since not all Planctomycetes are available as pure cultures and some species have doubling times of up to 30 days (12), selecting an appropriate model organism in which to develop a genetic toolbox is essential. After analyzing several species (see Table S1 in the supplemental material), we chose Planctomyces limnophilus to develop methods for gene transfer and mutagenesis. The key criteria for selecting this species were (i) it displays one of the fastest growth rates among the cultured Planctomycetes (6 to 14 days to detect a colony), (ii) plasmids and bacteriophage have been described (5, 16, 15), and (iii) the genome sequence is available. However, the unusual cell biology is a key hallmark of Planctomycetes, and little was known about P. limnophilus in this regard (4, 6). Thus, we first set out to perform cell biological analyses of this species using high-pressure cryofixation, freeze substitution, and thin sectioning in conjunction with transmission electron microscopy (TEM) (6). Cryofixation of entire colonies scraped from agar plates was performed with a Leica EM PACT-2 followed by freeze-substitution in acetone with 2% osmium tetroxide at −80°C for 50 h. Afterward, the temperature was increased over a span of 14 h to −20°C and finally brought to +20°C over 22 h. Samples were washed twice for 30 min in fresh acetone and then embedded in TAAB epon. Sections were cut at 50 nm and poststained with uranyl acetate and lead citrate, and subsequent analysis was done with a Tecnai G2 Spirit Bio TWIN microscope. The life cycle of P. limnophilus involves the production of two distinct cell types, somewhat reminiscent of the phylogenetically very distant Caulobacter crescentus. There are sessile cells with a holdfast (hs) and a noncellular stalk that allows surface anchoring (Fig. 1A and B). The second cell type contains a single polar flagellum (Fig. 1C and D). Although these findings had been preliminarily described before (4), negative staining of whole cells points toward two types of crateriform structures (large [LCS] and small [SCS]) on the cell surface of P. limnophilus. Such structures are common among Planctomycetes and had been used for taxonomic purposes in the past (10). Interestingly, our micrographs (Fig. 1C and D) indicate that the putative LCS are distributed across the entire cell, while putative SCS are limited to the pole where the flagellum (FL) is located. The subcellular analysis in Fig. 2A shows what appears to be close to a typical Blastopirellula-type of cell organization (2), with a cytoplasmic membrane (CM) confining the paryphoplasm (Py) and an intracytoplasmic membrane (ICM) dividing it from the pirellulosome (Pi). The ICM and CM both are about 6-nm-wide lipid bilayers (see Fig. S3 in the supplemental material). Interestingly, different cells have different shapes of Py and Pi. Serial sections of individual cells (see Fig. S1 in the supplemental material) revealed that the shapes of Py and Pi change along the z axis, and thus P. limnophilus subcellular structures are not rotationally symmetrical. In summary, the finding that P. limnophilus displays the characteristic subcellular compartmentalization of the Planctomycetes provided us with the necessary impetus to develop genetic tools for its manipulation. Such tools would make P. limnophilus relevant as a model for investigating the molecular basis of planctomycete compartmentalization in general.
Fig. 1.
TEM analysis of negative-stained P. limnophilus cells in either sessile (A and B) or swarmer (C and D) state of the life cycle, which is similar to yet distinct from that previously reported (12a). (A) The holdfast substance (hs) is positioned at the end of the multifiber stalk (st) and attaches cells to a surface. Attachment is further supported by fibrous structures (fi). (B) High-resolution micrographs of the stalk attachment at the cell pole and of the holdfast structure. (C) Swarmer cell with flagellum (fl) and so-called large crateriform structures (lcs) dispersed all over the cell surface. Further magnification (D) reveals in addition small crateriform structures (scs) at the pole of the cell, where the flagellar hook (flh) is attached.
Fig. 2.
TEM analysis of thin sections from P. limnophilus cells revealed an intracytoplasmic membrane (ICM) that divides the cytoplasm into a paryphoplasm (Py) and a pirellulosome (Pi). Within the ICM of the Pi, the nucleoid is not covered by a further membrane but is always condensed, while the size and organization of Py and Pi differ between individual cells (A to C). (A′ to C′) Colors of cell compartments are false (for illustration only).
Development of a gene transfer system and transposon mutagenesis.
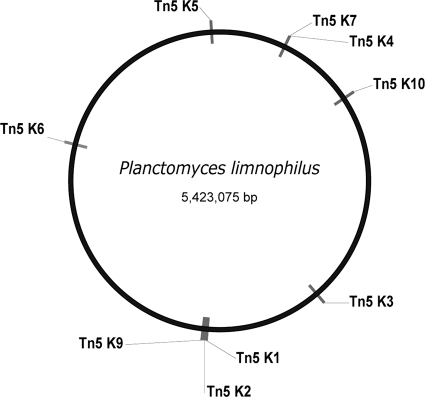
We choose the Epicentre EZ-Tn5 R6Kγori/Kan-2 Tnp transposome kit to develop our gene transfer protocol. Unlike plasmids, which have host ranges that are often limited, EZ-Tn5 insertion does not require any form of replication. In addition, a comparable approach was recently employed to demonstrate gene transfer into Verrucomicrobium spinosum, which belongs to the same superphylum as P. limnophilus (1, 14). After determining that P. limnophilus was indeed sensitive to kanamycin (see Fig. S2 in the supplemental material), fresh electrocompetent cells were prepared by washing cells from 400 ml of a broth culture at an optical density at 600 nm (OD600) of 0.4 (PYGV medium; http://www.dsmz.de/microorganisms/media_list.php), containing about 4 × 108 cells, twice with equal volumes of ice-cold 10% glycerol. Cells were resuspended in 400 μl of washing buffer, and aliquots of 100 μl were dispensed into 0.2-mm gapped electroporation cuvettes along with 1 μl of EZ-Tn5 solution and 1 μl of TypeOne restriction inhibitor (Epicentre). Electroporation was performed with a Bio-Rad Gene Pulser Xcell (capacity [C], 25 μF; resistance [PC], 200 Ω; voltage [V], 1.0 kV). Cells were immediately diluted in 1 ml PYGV medium and plated onto PYGV agar supplemented with 30 μg/ml kanamycin. After 14 days of incubation at 30°C in the dark, 3.75 × 107 CFU/ml (±1.8 × 107) could be detected on control plates without antibiotic, indicating that about 40% of cells survived electroporation. Colonies on kanamycin-containing plates arose only when the mixtures had contained EZ-Tn5; not a single colony was observed in the mixtures without DNA. In general, up to 1.5 × 103 transformants were obtained per microgram of DNA using 108 cells. Thus, transformation efficiency is by far lower than average Escherichia coli performance (107 CFU/μg DNA and better). From three independent transformations, we picked 10 random colonies and used those cells to inoculate fresh liquid medium. To verify Tn5 integration, DNA was isolated using the Qiagen DNeasy blood and tissue kit, and Tn5 insertion sites within the genome were analyzed by modified arbitrary PCR (8). We used primer ARB1 (8) and CJ318 (5′-CAGACCGTTCCGTGGCAAAGCAAA-3′) for the first PCR (95°C for 5 min; 6 rounds of 95°C for 30 s, 30°C for 30 s, and 72°C for 1 min and 30 s; and 30 rounds of 95°C for 30 s, 45°C for 30 s, 72°C for 2 min, and 72°C for 4 min), while oligonucleotides ARB2 (8) and CJ319 (5′-ACCTACAACAAAGCTCTCATCAACC-3′) were used for the second PCR (95°C for 1 min and 30 rounds of 95°C for 30 s, 50°C for 30 s, 72°C for 2 min, and a final elongation at 72°C for 4 min). The DNA sequence of purified PCR products (Qiagen MiniElut gel extraction kit) revealed nine insertion sites within the genome, as shown in Fig. 3 and Table 1. Thus, mutagenesis was effective as evidenced by disruption of the genes listed in Table 1. While chromosomal insertions showed some regional selectivity, no significant regional bias among the nine different disrupted genes was found (Fig. 3, Table 1). Interestingly neither the ICM nor the condensed nucleoid as described in the present study formed a barrier for gene transfer and chromosomal integration. This is the first report of gene transfer and mutagenesis of a Planctomycetes thus far, and it opens the door to in-depth genetic analyses of this bacterium, such as generation of defined deletion mutants or expression of green fluorescent protein (GFP) fusion proteins.
Fig. 3.
P. limnophilus genome map with highlighted positions of Tn5 transposon insertions as revealed by modified arbitrary PCR. The distribution appears random, although the sample number of nine does not allow statistical analysis.
Table 1.
Predicted functions of genes in the P. limnophilus genome that have been disrupted by Tn5 transposon insertions of 10 representative analyzed mutantsa
| Mutant no. | Disrupted gene | Predicted function of disrupted gene |
|---|---|---|
| 1 | Plim_2159 | SppA (signal peptide peptidase) |
| 2 | Plim_2165 | Hypothetical protein |
| 3 | Plim_1621 | Putative thioredoxin-like protein |
| 4 | Plim_3595 | Hypothetical protein |
| 5 | Plim_4202 | Tetratricopeptide (TPR) |
| 6 | Plim_3308 | Heme-binding protein |
| 7 | Plim_0300 | Hypothetical proteinb |
| 8 | ND | ND |
| 9 | Plim_2177 | Acriflavine resistance protein A |
| 10 | Plim_0649 | Phosphoglycerate mutase |
Gene functions were annotated by five iterations of Ψ-BLAST against the NCBI database. ND, not determined.
A Planctomycetes-specific protein.
Supplementary Material
Acknowledgments
We thank John Fuerst, Evgeny Sagulenko, and Rudolf Amann for scientific discussion and Elizabeth Benecci from the HMS EM Facility for technical assistance.
The project was funded by the Deutsche Forschungsgemeinschaft (JO 893/1-1) and the Marie Curie IOF Program (FP7) of the European Union (CoGniSePlanctomyces).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 1 July 2011.
REFERENCES
- 1. Domman D. B., Steven B. T., Ward N. L. 2011. Random transposon mutagenesis of Verrucomicrobium spinosum DSM 4136(T). Arch. Microbiol. 193:307–312 [DOI] [PubMed] [Google Scholar]
- 2. Fuerst J. A. 2005. Intracellular compartmentation in Planctomycetes. Annu. Rev. Microbiol. 59:299–328 [DOI] [PubMed] [Google Scholar]
- 3. Fuerst J. A., Webb R. I. 1991. Membrane-bounded nucleoid in the eubacterium Gemmata obscuriglobus. Proc. Natl. Acad. Sci. U. S. A. 88:8184–8188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hirsch P., Müller M. 1985. Planctomyces limnophilus sp. nov., a stalked and budding bacterium from freshwater. Syst. Appl. Microbiol. 6:276–280 [Google Scholar]
- 5. Labutti K., et al. 2010. Complete genome sequence of Planctomyces limnophilus type strain (Mu 290). Stand. Genomic Sci. 3:47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lindsay M. R., et al. 2001. Cell compartmentalisation in Planctomycetes: novel types of structural organisation for the bacterial cell. Arch. Microbiol. 175:413–429 [DOI] [PubMed] [Google Scholar]
- 7. Lonhienne T. G., et al. 2010. Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc. Natl. Acad. Sci. U. S. A. 107:12883–12888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. O'Toole G. A., Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461 [DOI] [PubMed] [Google Scholar]
- 9. Santarella-Mellwig R., et al. 2010. The compartmentalized bacteria of the Planctomycetes-Verrucomicrobia-Chlamydiae superphylum have membrane coat-like proteins. PLoS Biol. 8:e1000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmidt J. M., Starr M. P. 1978. Morphological diversity of freshwater bacteria belonging to the Blastocaulis-Planctomyces group as observed in natural populations and enrichments. Curr. Microbiol. 1:325–330 [Google Scholar]
- 11. Strous M., et al. 1999. Missing lithotroph identified as new planctomycete. Nature 400:446–449 [DOI] [PubMed] [Google Scholar]
- 12. Strous M., Heijnen J. J., Kuenen J. G., Jetten M. S. M. 1998. The sequencing batch reactor as a powerful tool for the study of slowly growing anaerobic ammonium-oxidizing microorganisms. Appl. Microbiol. Biotechnol. 50:589–596 [Google Scholar]
- 12a. Tekniepe B. L., Schmidt J. M., Starr M. P. 1981. Life cycle of a budding and appendaged belonging to morphotype IV of the Blastocaulis-Planctomyces group. Curr. Microbiol. 50:1–6 [Google Scholar]
- 13. van Niftrik L. A., et al. 2004. The anammoxosome: an intracytoplasmic compartment in anammox bacteria. FEMS Microbiol. Lett. 233:7–13 [DOI] [PubMed] [Google Scholar]
- 14. Wagner M., Horn M. 2006. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 17:241–249 [DOI] [PubMed] [Google Scholar]
- 15. Ward N., et al. 2006. The order Planctomycetales, including the genera Planctomyces, Pirellula, Gemmata and Isosphaera and the Candidatus genera Brocadia, Kuenenia and Scalindua, p. 757–793 In Dworkin M. (ed.), The prokaryotes, vol. 7 Springer, New York, NY [Google Scholar]
- 16. Ward-Rainey N., Rainey F. A., Wellington E. M., Stackebrandt E. 1996. Physical map of the genome of Planctomyces limnophilus, a representative of the phylogenetically distinct planctomycete lineage. J. Bacteriol. 178:1908–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.