Abstract
There are numerous examples of shared enhancers interacting with just a subset of target promoters. In some cases, specific enhancer–promoter interactions depend on promoter competition, whereby the activation of a preferred target promoter precludes expression of linked genes. Here, we employ a transgenic embryo assay to obtain evidence that promoter selection is influenced by the TATA element. Both the AE1 enhancer from the Drosophila Antennapedia gene complex (ANT-C) and the IAB5 enhancer from the Bithorax complex (BX-C) preferentially activate TATA-containing promoters when challenged with linked TATA-less promoters. In contrast, the rho neuroectoderm enhancer (NEE) does not discriminate between these two classes of promoters. Thus, certain upstream activators, such as Ftz, prefer TATA-containing promoters, whereas other activators, including Dorsal, work equally well on both classes of promoters. These results provide in vivo evidence that different core promoters possess distinct regulatory activities. We discuss the possibility that an invariant TFIID complex can adopt different conformations on the core promoter.
Keywords: Drosophila, TATA box, promoter–enhancer interactions, core promoter, initiator elements
Complex enhancers, or cis regulatory modules, direct stripes, bands, and tissue-specific patterns of gene expression in the early Drosophila embryo. Such enhancers are typically 300–900 bp in length and contain clustered binding sites for both transcriptional activators and repressors (for review, see Gray and Levine 1996a; Rivera-Pomar 1996). Given the importance of these enhancers in development, we have become interested in the next level of cis organization, namely, the regulation of enhancer–promoter interactions within complex genetic loci.
Recent studies suggest that there are at least two mechanisms for regulating enhancer–promoter interactions (for summary, see Fig. 1). First, an insulator DNA can specifically block the interaction of a shared enhancer with gene B and not interfere with the activation of gene A (Fig. 1A; Hagstrom et al. 1996; Zhou et al. 1996; Mihaly et al. 1997). Second, according to a promoter competition scenario, the shared enhancer can activate both genes, but prefers the promoter region associated with gene A (Choi and Engel 1988; Foley and Engel 1992). The interaction of the enhancer with gene A precludes activation of gene B (Fig. 1B). Recent studies provide evidence for both mechanisms, as discussed below.
Figure 1.
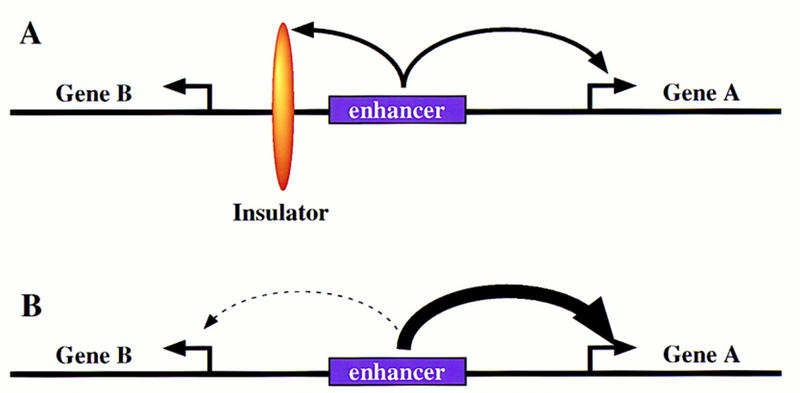
Regulation of enhancer–promoter interactions. The diagrams depict two divergently transcribed genes, A and B, with a common enhancer located in the intergenic region. (A) An insulator DNA is located between gene B and the enhancer. In principle, this blocks interactions of the enhancer with gene B, without altering the activation of gene A. (B) Promoter competition. In principle, the enhancer can activate both gene A and gene B, but prefers the promoter region of gene A. Enhancer–gene A interactions preclude activation of gene B.
The major Hox gene clusters in Drosophila, the Bithorax complex (BX-C) and Antennapedia complex (ANT-C), contain vast arrays of tissue-specific enhancers that interact with specific target genes (e.g., Celniker et al. 1990; Gindhart et al. 1995; Lewis et al. 1995). For example, the AE1 autoregulatory element in the ANT-C specifically interacts with the fushi tarazu (ftz) promoter, but does not activate the equidistant Sex combs reduced (Scr) gene (Schier and Gehring 1992; Gindhart et al. 1995; Gorman and Kaufman 1995). Removal of the ftz gene and associated promoter region permits AE1 to activate inappropriate target genes within the ANT-C, including the zen-related gene, z2 (Rushlow and Levine 1988). In the present study, we present evidence for promoter competition, whereby AE1–ftz interactions preclude the activation of the linked Scr gene.
We also investigate the regulation of the IAB5 enhancer, which is located ∼60-kb upstream of the abd-A gene and ∼60 kb downstream of the Abd-B gene within the BX-C (Busturia and Bienz 1993). IAB5 preferentially activates Abd-B, and is responsible for initiating Abd-B expression in the presumptive abdomen, particularly in parasegments (PS) 10, 12, and 14. Like AE1, IAB5 is thought to be activated by the Ftz protein. IAB5 may be inactive in PS 2, 4, 6, and 8 (Ftz stripes 1 through 4) by various gap protein repressors, including Hunchback and Krüppel (see Busturia and Bienz 1993). Although IAB5 is located at comparable distances from abd-A and Abd-B, it preferentially activates the Abd-B promoter.
In this study we investigate the role of core promoter elements, particularly TATA, in regulating interactions between the AE1 and IAB5 enhancers with specific target promoters. There is considerable information regarding the organization and composition of the core transcription complex (for review, see Roeder 1996; Verrijzer and Tjian 1996). TFIID is a critical regulatory component of the complex. It is composed of TATA-binding protein (TBP) and associated factors (TAFs). The binding of TFIID to the core promoter appears to be a pivotal rate-limiting step in transcriptional activation (for review, see Burley and Roeder 1996; Manley et al. 1996; Stargell and Struhl 1996). Sequence-specific upstream activators have been shown to make direct contact with different components of the TFIID complex, including specific TAFs as well as TBP itself (e.g., Sauer et al. 1996; for review, see Ptashne and Gann 1997). Different core promoters appear to interact with an invariant TFIID complex, so it is unclear whether they possess distinct regulatory activities. There are numerous examples of combinatorial interactions between upstream activators. For example, the Bicoid and Hunchback transcription factors function synergistically to specify head structures and initiate the segmentation cascade (e.g., Simpson-Brose et al. 1994; Arnosti et al. 1996; Sauer et al. 1996), whereas Dorsal and bHLH activators initiate the differentiation of the embryonic mesoderm and neurogenic ectoderm (for review, see Rusch and Levine 1996). It is currently unclear whether different upstream activators collaborate with specific core promoter elements to specify cell fate during embryogenesis.
The binding of the TFIID complex to a target promoter depends on at least three different core promoter elements located within a 50- to 60-bp sequence flanking the transcription start site (e.g., Burke and Kadonaga 1996; for review, see Smale 1997), the TATA box, the initiator element (Inr), and the downstream promoter element (Dpe). In general, promoters that lack a TATA sequence must possess conserved copies of the Inr and/or Dpe. Conversely, promoters containing optimal TATA sequences do not require Inr and Dpe elements for the binding of TFIID (e.g., Burke and Kadonaga 1996). The present study provides evidence that TATA versus Inr/Dpe promoters possess distinct regulatory activities in development.
We show that both the AE1 and IAB5 enhancers preferentially activate TATA-containing promoters when challenged with linked TATA-less promoters. The analysis of chimeric core promoter sequences reveals the importance of the TATA element in these selective enhancer–promoter interactions. The rhomboid (rho) neuroectoderm enhancer (NEE) (Ip et al. 1992), however, does not discriminate between TATA-containing and TATA-less promoters, thereby providing evidence that certain upstream activators, such as Ftz, prefer TATA-containing promoters, whereas other activators, including dorsal and bHLH proteins, are promiscuous and work equally well on both classes of promoters. We propose that TFIID can adopt different conformations, and thereby expose distinct basal targets for interaction with upstream activators.
Results
This study involves the analysis of two minimal enhancers, AE1 and IAB5, which are located within the ANT-C and BX-C, respectively. The 430-bp AE1 is located in the middle of the Scr–ftz interval, ∼7 kb from both promoters (Fig. 2; Pick et al. 1990). AE1 preferentially activates ftz, but not Scr, and a number of experiments were conducted to determine whether the distinct core promoter sequences associated with the two genes play a role in this regulatory specificity. In particular, the ftz promoter contains an optimal TATA sequence, but lacks both Inr and Dpe initiator elements. In contrast, Scr contains a potential Dpe element (Burke and Kadonaga 1996) but lacks TATA. The 1-kb IAB5 enhancer selectively activates Abd-B, not abd-A, although neither promoter contains an obvious TATA element (Martin et al. 1995; Lewis et al. 1995).
Figure 2.
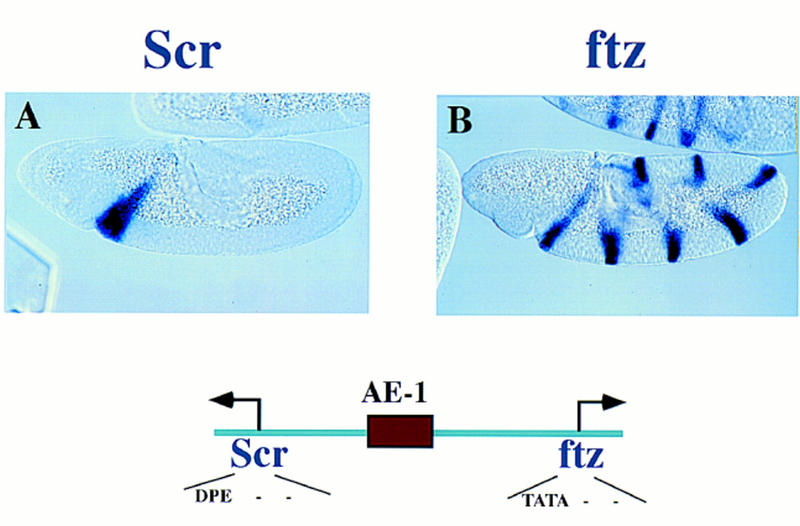
Regulatory specficity in the Scr–ftz interval of the ANT-C. Embryos were hybridized with either a dioxigenin-labeled Scr or ftz antisense RNA probe and visualized via histochemical staining. The embryos are undergoing the rapid phase of germ-band elongation (4–5 hr postfertilization) and are oriented with anterior to the left and dorsal up. Scr is expressed within the primordia of parasegment 2 (PS 2), which gives rise to regions of the labial and prothoracic segments. ftz is expressed in a series of pair–rule stripes. The diagram below the embryos shows the location of the AE1 enhancer within the Scr–ftz interval. AE1 specifically interacts with the ftz promoter to maintain the seven-stripe pattern. It does not activate the linked Scr gene.
The AE1 and IAB5 enhancers were inserted into P-transformation vectors that contain at least two different reporter genes, including lacZ and white. These reporters were placed under the control of different core promoter sequences, and gene expression was monitored in transgenic embryos via in situ hybridization with digoxigenin-labeled RNA probes.
AE1 can activate different classes of core promoters
Related core promoter sequences were initially used for the analysis of AE1. ftz and eve contain optimal TATA sequences, but lack Inr (INIT) and Dpe (DPE) elements (see Fig. 3A,B). Both the eve/CAT and ftz/lacZ fusion genes are expressed in a series of seven stripes in response to the endogenous ftz activator. Similarly, AE1 activates both white and lacZ when the two reporter genes are regulated by the white and Tp promoters (Fig. 3C,D). Both core promoters contain conserved copies of the INIT and DPE sequences, but either lacks a TATA sequence (white) or contains a suboptimal TATA (Tp). These results indicate that AE1 can simultaneously activate linked TATA-containing promoters or linked INIT/DPE-containing promoters. Additional experiments investigated the consequences of placing AE1 between different classes of promoters.
Figure 3.
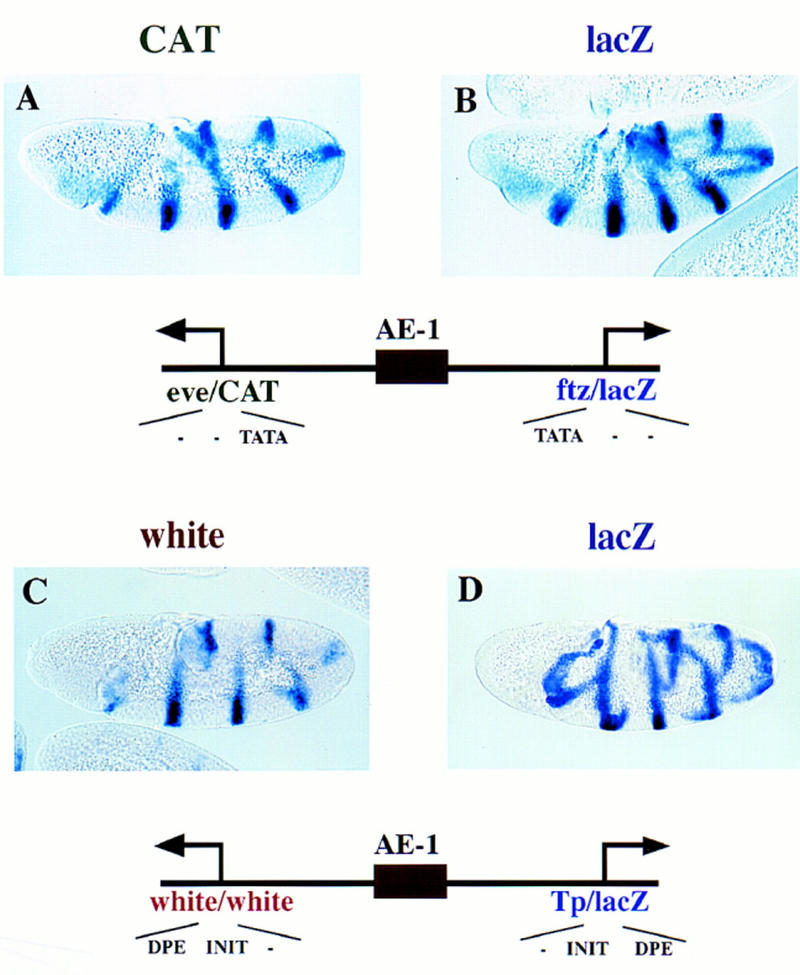
AE1 can coactivate linked reporter genes. Transgenic embryos carry fusion genes containing the 430-bp AE1 enhancer placed between divergently transcribed reporter genes that can be independently assayed. Embryos are undergoing the rapid phase of germ-band elongation. (A,B) Transgenic embryos carry a fusion gene with linked CAT and lacZ reporter genes. The arrows indicate the location and orientation of the transcription start sites. The leftward CAT gene was linked to the eve promoter, whereas the rightward lacZ reporter gene was attached to the ftz promoter. A was hybridized with a CAT antisense RNA probe; B was hybridized with a lacZ probe. Both reporter genes are expressed in a series of seven stripes, indicating that AE1 activates both the ftz and eve promoters. The diagrams indicate that the promoters contain TATA sequences, but lack optimal Inr (INIT) and Dpe (DPE) sequences. (C,D) Transgenic embryos carry a fusion gene with linked white and lacZ reporter genes. The white gene contains a mini-white promoter sequence, whereas lacZ was placed under the control of the core promoter sequence from the transposase gene (Tp) located within the P-element vector. C was hybridized with a white antisense RNA probe; D was hybridized with a lacZ probe. Both reporter genes are activated by AE1 and expressed in a series of stripes. The diagrams indicate that the promoters lack TATA sequences, but contain INIT and DPE elements.
Promoter competition
There is a substantial reduction in the white staining pattern when the Tp promoter (Fig. 3D) is replaced with the core eve promoter sequence (Fig. 4A,B). The eve/lacZ reporter gene is expressed in a series of seven stripes in response to the AE1 enhancer (Fig. 4B). This AE1–eve interaction appears to block the expression of the linked white gene (Fig. 4A). In the absence of eve, white is fully active (Fig. 3C). These observations are compatible with a promoter-competition mechanism whereby AE1–eve interactions inhibit white (see Discussion).
Figure 4.
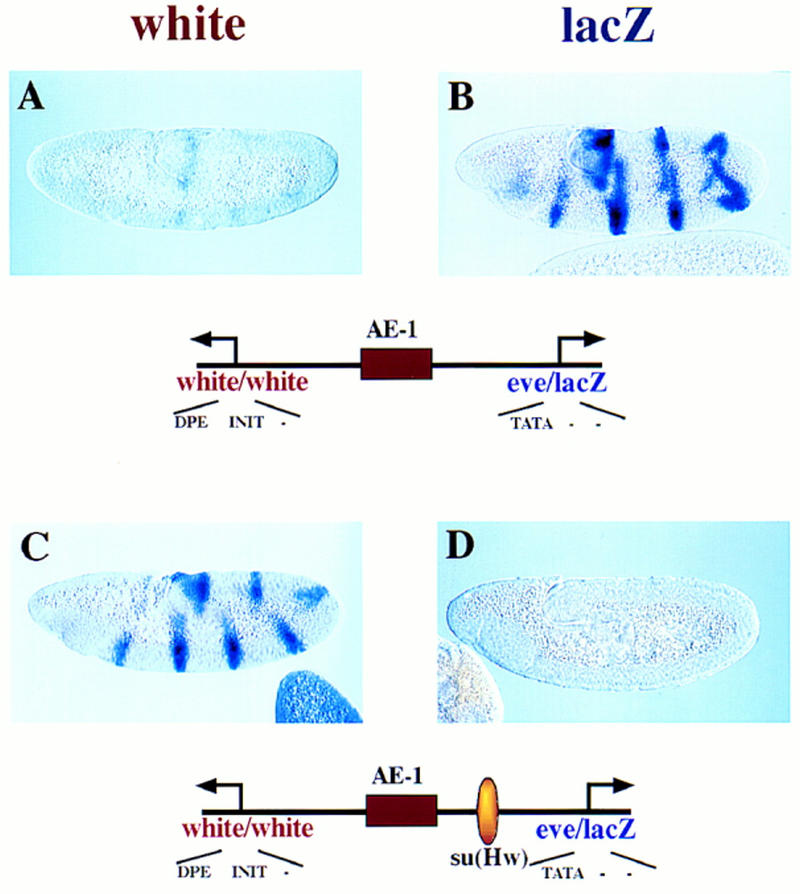
Promoter competition influences AE1 activity. Transgenic embryos contain a P transposon with divergently transcribed white and lacZ reporter genes that are under the control of different core promoter sequences. The AE1 enhancer was placed between the linked genes, as summarized in the diagrams below the stained embryos. The embryos were hybridized with digoxigenin-labeled white or lacZ antisense RNA probes. (A,B) The white and lacZ reporter genes are driven by minimal white and eve promoter sequences, respectively. The eve/lacZ gene is expressed in a series of seven stripes, but the white gene exhibits just residual staining. It would appear that AE1-eve interactions preclude activation of the linked white gene, because AE1 can activate white in the absence of eve (e.g., see Fig. 3C). (C,D) Same as A and B except that the 340-bp su(Hw) insulator DNA from the gypsy retrotransposon was placed between the AE1 enhancer and eve/lacZ fusion gene. This silences lacZ staining and results in the activation of white.
Previous studies have shown that the gypsy retrotransposon contains an insulator DNA that blocks the interactions of distal, not proximal, enhancers with a target promoter (Geyer and Corces 1992; Dorsett 1993; Cai and Levine 1995; Scott and Geyer 1995). The lacZ reporter gene is blocked when the gypsy insulator is placed between AE1 and the eve promoter (Fig. 4D). The insulator redirects AE1 to the less preferred white gene, which is now expressed in a series of stripes (Fig. 4C). These results raise the possibility that a combination of promoter competition and insulator DNAs regulates enhancer–promoter interactions within complex loci.
IAB5 prefers the eve promoter
The 1-kb IAB5 enhancer also exhibits a preference for TATA-containing promoters. IAB5 was placed downstream of an eve/lacZ fusion gene; the linked CAT reporter gene was placed under the control of the mini-white promoter (Fig. 5). There is strong expression of the lacZ reporter gene in the presumptive abdomen (Fig. 5B), whereas CAT is not expressed above background levels (Fig. 5A). This result suggests that IAB5 prefers the eve promoter over white. As shown below, IAB5 continues to select the eve promoter even when it is attached to the distal CAT reporter gene (see Fig. 6).
Figure 5.
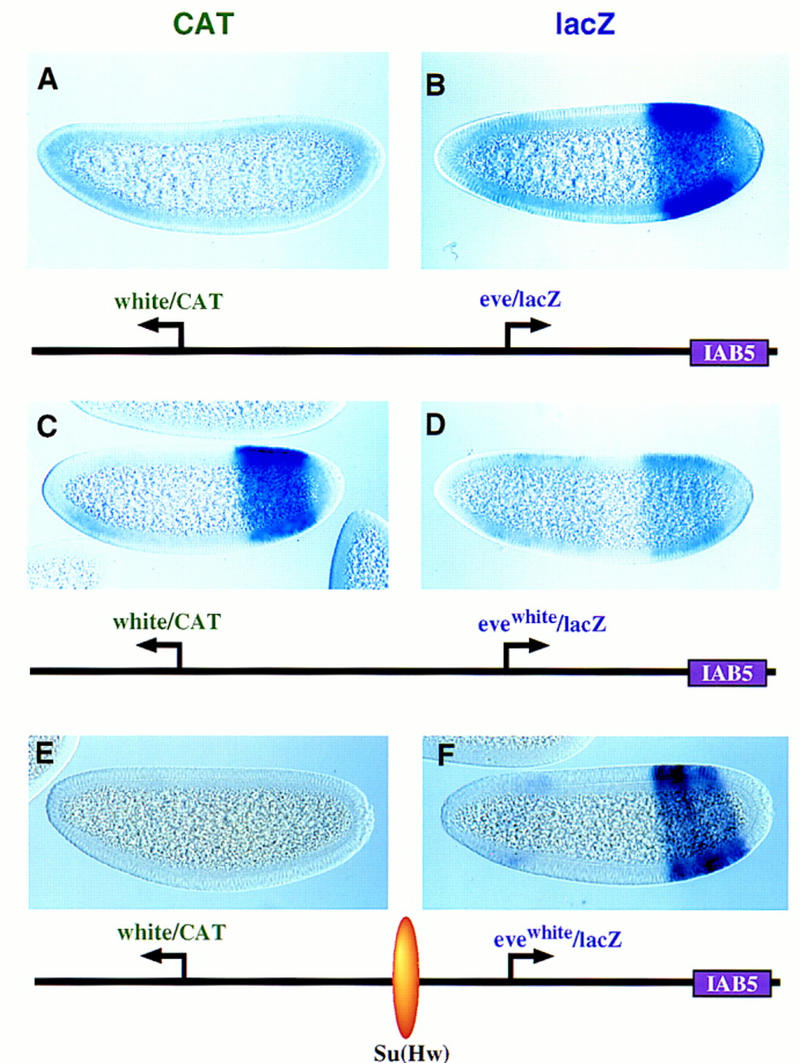
The IAB5 enhancer prefers TATA-containing promoters. Transgenic embryos were stained and oriented as described in the previous figure legends, except that these are younger embryos (between cellularization and the onset of gastrulation). The IAB5 enhancer was placed downstream of the rightward lacZ reporter gene. The distal CAT gene is under the control of the white promoter. The proximal lacZ gene is driven by eve (B) or an evewhite chimeric promoter (D,F) whereby the eve TATA region was replaced with the corresponding sequences in white. (A,B) CAT (A) and lacZ (B) staining patterns obtained with linked white/CAT and eve/lacZ genes. The IAB5 enhancer selects eve over white, so that the eve/lacZ reporter gene exhibits strong expression whereas white/CAT is silent. (C,D) Same as A and B except that the proximal lacZ gene is under the control of the evewhite chimeric promoter (D). There is only weak expression of the lacZ reporter in the presumptive abdomen (D). IAB5 now mediates strong expression of the distal white/CAT fusion gene (C). The residual lacZ staining observed in anterior regions (D) may be a position effect resulting from the site of P insertion. (E,F) Same as C and D except that the 340-bp gypsy insulator DNA [su(Hw)] was placed between the leftward CAT gene and rightward lacZ reporter. The insulator blocks IAB5–white interactions, so that CAT is not expressed above backround levels. Instead, IAB5 directs strong expression of the evewhite/lacZ in the presumptive abdomen, indicating that the chimeric promoter is not defective. The weak staining seen in head regions is caused by sequences contained within the P-transformation vector (Small et al. 1992).
Figure 6.
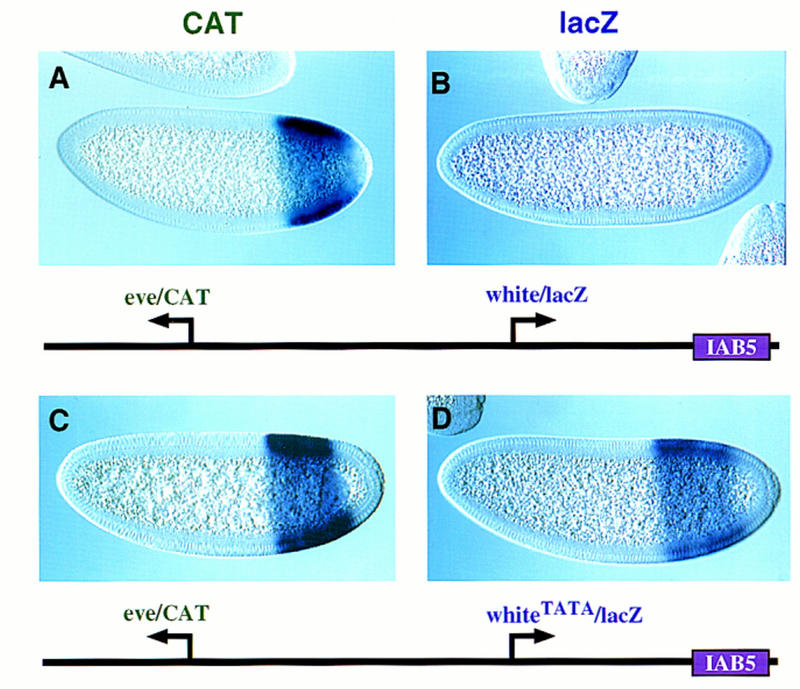
TATA is an important determinant of IAB5–eve interactions. Transgenic embryos carry the indicated P transposons and are oriented as described in the legend to Fig. 5. (A,B) Nuclear cleavage 14 embryos that carry a P transposon with the distal CAT gene driven by the core eve promoter and the rightward lacZ gene driven by mini-white. The IAB5 enhancer selectively interacts with the eve promoter, and directs intense expression of the CAT reporter in the presumptive abdomen (A). In contrast, the white/lacZ reporter gene is not expressed above background levels (B). (C,D) Cellularizing embryos carrying the same P transposon as A and B except that a synthetic TATA sequence was inserted into the mini-white promoter. IAB5 activates the lacZ reporter gene in the presumptive abdomen (D). The eve/CAT fusion gene is also activated by IAB5 (C). These results suggest that the whiteTATA promoter is almost as active as eve.
An eve–white chimeric promoter was analyzed in an effort to assess the importance of the core elements, particularly the TATA sequence. An ∼20-bp region of the eve sequence (the TATA region) was replaced with the corresponding region of white. This modified eve promoter (evewhite) is attenuated and mediates only weak expression of lacZ in the presumptive abdomen (Fig. 5D). In contrast, the linked white promoter directs strong expression of CAT (Fig. 5C). These results suggest that the removal of the eve TATA releases the IAB5 enhancer so that it can now interact with the white promoter.
The selection of the white promoter over evewhite might reflect differences in the remaining core elements. The white promoter contains both Inr and Dpe sequences, whereas evewhite contains an Inr element that possesses just a 4 of 6 match with the consensus sequence (Smale 1997).
Additional experiments were done to determine whether the evewhite chimeric promoter possesses normal activity. The gypsy insulator was inserted between the distal white/CAT gene and proximal evewhite/lacZ gene (see diagram below Fig. 5E,F). The lacZ reporter gene is strongly activated in the abdomen (Fig. 5F). This level of expression is comparable to that obtained with the normal eve promoter, suggesting that the evewhite promoter is fully functional.
TATA is a prime determinant of IAB5 specificity
The preceding results suggest that the eve TATA region is important for selective interactions with the IAB5 enhancer. Additional evidence was obtained by analysis of the activities of a synthetic white promoter (whiteTATA), which contains just 9 nucleotide substitutions between −29 and −21 bp upstream of the transcription start site. These changes create an optimal TATA box (GTATAAAAG) that is identical in sequence to the eve TATA (see Materials and Methods). The whiteTATA promoter was attached to the proximal lacZ reporter gene, whereas the distal CAT gene was placed under the control of the normal eve promoter sequence (Fig. 6). As seen previously, the 3′ IAB5 enhancer selects the distal eve/CAT fusion gene, and ignores the more proximal white/lacZ reporter (Fig. 6A,B). The whiteTATA promoter, however, mediates strong induction of lacZ expression (Fig. 6, cf. D and B), which is nearly comparable to that obtained with the normal eve promoter (Fig. 6C).
Promoter specificity
Several different embryonic enhancers were challenged with linked TATA and TATA-less promoters, and the 300-bp rho NEE was found to be equally effective in activating the two classes of promoters (data not shown; see below). Additional experiments were done to determine whether the targeting of IAB5 to eve influences the activities of the nonspecific rho NEE. The latter enhancer is activated by the maternal dorsal gradient in lateral stripes within the neurogenic ectoderm (Ip et al. 1992).
A synthetic gene complex was prepared that contains both the NEE and IAB5 enhancers. white and CAT reporter genes were attached to the mini-white promoter, whereas lacZ is driven by eve (Fig. 7). The rho NEE activates all three reporter genes, so that white, CAT, and lacZ are all expressed in lateral stripes. In contrast, IAB5 primarily activates the eve promoter, so that only lacZ exhibits strong expression within the presumptive abdomen (Fig. 7C). The white reporter gene is not expressed in the abdomen (Fig. 7A), whereas CAT exhibits only residual staining in this region (Fig. 7B). These results suggest that IAB5–eve interactions do not influence the nonspecific activities of the rho NEE.
Figure 7.
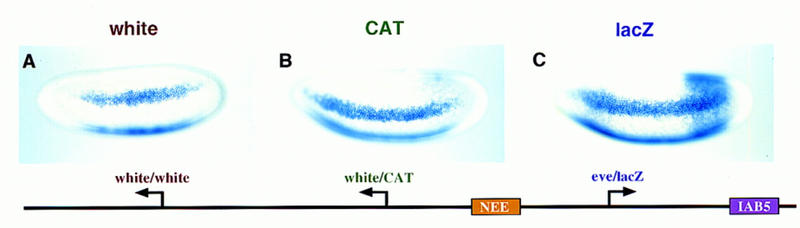
Independent activities of the IAB5 and NEE enhancers. Transgenic embryos carry the P-transformation vector shown in the diagram and are oriented as described in the previous legends to the figures. This synthetic gene complex contains three different reporter genes, white, CAT, and lacZ. The white and CAT genes are driven by the mini-white promoter, whereas lacZ contains the eve promoter. All three reporter genes exhibit robust expression in the lateral neurogenic ectoderm, indicating that the NEE enhancer interacts equally well with the mini-white and eve promoters. In contrast, lacZ is strongly activated in the presumptive abdomen (C), whereas white and CAT exhibit little or no expression in this region (A,B).
The NEE was used as an internal control to show that the selection of eve over white by IAB5 is not caused by differences in promoter strength. The NEE was placed between an eve/CAT fusion gene and lacZ reporter, whereas IAB5 was placed 3′ of lacZ (see diagrams in Fig. 8). The NEE activates eve and the chimeric evewhite promoter equally well, so that both the CAT and lacZ reporter genes exhibit lateral stripes of gene expression (Fig. 8A,B). In contrast, IAB5 mediates strong expression of the distal eve/CAT fusion gene in the presumptive abdomen (Fig. 8A), but only weakly activates the proximal evewhite promoter (Fig. 8B), which lacks both the TATA and Dpe sequences (see Fig. 5). The introduction of a synthetic TATA sequence in the white promoter (whiteTATA) results in strong IAB5–lacZ interactions (Fig. 8D), so that the eve/CAT and whiteTATA/lacZ fusion genes are expressed at comparable levels in the presumptive abdomen (Fig. 8C,D). The NEE continues to activate both reporter genes and is not influenced by changes in the lacZ promoter that control IAB5 interactions.
Figure 8.
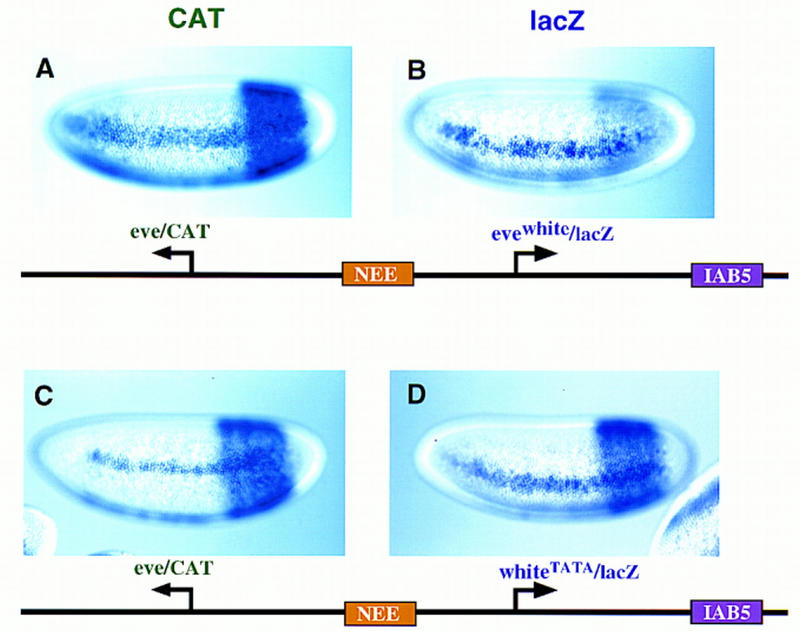
Independent activities of NEE and IAB5 on chimeric promoters. Transgenic embryos express the indicated P-transformation vectors, and are oriented as described previously. The rho NEE was placed in the intergenic region between the reporters, whereas IAB5 is located 3′ of the lacZ gene. The leftward CAT gene contains the eve promoter, whereas lacZ is driven by different chimeric promoters. (A,B) CAT and lacZ staining patterns obtained with the evewhite promoter, which contains an Inr element but lacks TATA and Dpe sequences. CAT transcripts are detected in lateral stripes and the presumptive abdomen (A), indicating activation of the eve/CAT gene by both the NEE and IAB5 enhancers. In contrast, lacZ is expressed primarily in lateral stripes; there is only residual staining in the abdomen (B). This staining pattern indicates that the evewhite promoter is strongly activated by the NEE enhancer, but only weakly interacts with IAB5. (C,D) CAT and lacZ staining patterns obtained with a synthetic white promoter (whiteTATA) that contains a TATA sequence. Both reporter genes exhibit robust expression in lateral stripes and the abdomen, indicating that the NEE and IAB5 enhancers work equally well on the eve and whiteTATA promoters.
Discussion
This study provides evidence for promoter competition within the ANT-C. Preferential interactions between AE1 and the ftz promoter may preclude activation of the linked Scr gene. Previous studies on the chicken globin cluster showed that promoter competition is an important regulatory strategy for gene switching during hematopoiesis (Choi and Engel 1988; Foley and Engel 1992; Foley et al. 1994). The analysis of AE1 and IAB5 suggest that promoter competition depends, at least in part, on core promoter elements, particularly TATA. Both AE1 and IAB5 prefer the eve promoter, which contains an optimal TATA sequence, and fail to activate a linked mini-white promoter that lacks TATA but contains both Inr and Dpe sequences. In contrast, the NEE enhancer indiscriminately activates core promoters that contain either TATA or Inr/Dpe elements. These studies suggest that TATA-containing and TATA-less core promoters possess distinct regulatory activities.
Diverse core promoters
Previous studies have identified instances of specific enhancer–promoter interactions in the Drosophila embryo (Li and Noll 1994; Merli et al. 1996). In particular, shared enhancers located between the divergently transcribed gooseberry genes interact with just one of the promoters (Li and Noll 1994), whereas 3′ decapentaplegic (dpp) enhancers fail to activate neighboring genes, such as out at first (oaf; Merli et al. 1996). These studies, however, failed to determine whether promoter competition precluded inappropriate enhancer–promoter interactions and also failed to distinguish between promoter–proximal elements or core promoter elements in the regulatory specificity. The present study provides evidence that core promoter elements influence specific enhancer–promoter interactions and suggests that there are at least two classes of core promoters (for summary, see Fig. 9).
Figure 9.
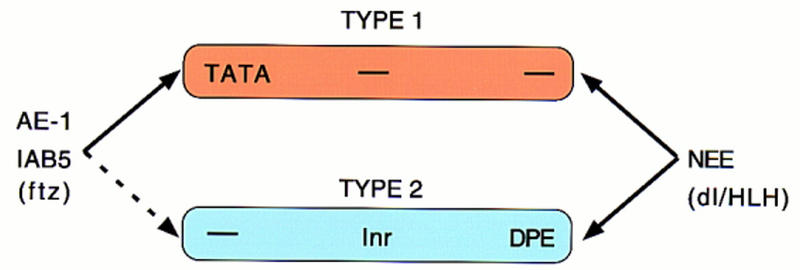
Different core promoters possess distinct regulatory activities. The IAB5 and AE1 enhancers preferentially activate TATA-containing promoters (type I) when given a choice between linked TATA and Inr/Dpe (type II) promoters. In contrast, the NEE activates both classes of promoters. These results suggest that the IAB5 and AE1 activators, particularly ftz, prefer type I promoters. NEE activators, including Dorsal (dl) and bHLH proteins, appear to be promiscuous and work equally well on both classes of core promoters. We propose that the TFIID complex adopts different conformations on type I and type II promoters. Basal targets for the Ftz activator may be displayed in a more accessible conformation when TFIID binds TATA. In contrast, basal targets for the Dorsal and bHLH activators may be equally accessible whether TFIID binds TATA or Inr/Dpe elements.
Type I promoters contain TATA, whereas type II promoters contain Inr and Dpe sequences. AE1 and IAB5 activators, presumbably including Ftz, preferentially activate type I promoters, whereas NEE activators, such as Dorsal, do not discriminate between the two classes of promoters. Future studies will determine whether there are upstream activators that selectively activate type II promoters. Possible candidates include activators that bind the T1 and VM enhancers in the ANT-C (Ginhart et al. 1995; Gorman and Kaufman 1995), which preferentially activate the TATA-less Scr promoter, but do not influence the expression of the neighboring TATA-containing ftz gene. Previous tissue culture and in vitro assays suggest that Sp1 may be more effective at activating Inr versus TATA promoters (Colgan and Manley 1995; Das et al. 1995; Emami et al. 1995).
It is unclear whether there are additional classes of core promoters. In this study we have investigated many, but not all possible combinations of the three TFIID core promoter elements. For example, the whiteTATA promoter contains TATA, Inr, and Dpe, but behaves like the eve promoter, which contains TATA but lacks Inr/Dpe sequences (see Fig. 6C,D). Perhaps TATA is dominant to Inr/Dpe in promoters that contain both types of core elements, although recent in vitro studies suggest that TATA and Inr elements work synergistically to activate transcription (Emami et al. 1997). It is also conceivable that these elements are antagonistic in transgenic embryos. For example, the proximal whiteTATA/lacZ fusion gene does not preclude the activation of a distal eve/CAT reporter (Fig. 6). In contrast, a proximal eve promoter completely blocks activation of the distal eve/CAT reporter gene (S. Ohtsuki, unpubl.). TATA may be more effective at trapping the IAB5 enhancer in the absence of Inr/Dpe elements.
It has been suggested that TATA-containing promoters are intrinsically stronger than TATA-less promoters, possibly because of higher affinity interactions with the TFIID complex (e.g., Zenzie-Gregory et al. 1993). The divergent activities of the IAB5 and NEE enhancers, however, are most easily interpreted on the basis of qualitative, not quantitative, differences in type I and type II core promoter sequences. For example, the insertion of a TATA sequence in the white promoter allows it to compete with a linked eve promoter, whereas the removal of TATA from eve permits activation of white. These alterations in the white and eve promoters, the insertion and removal of TATA, dramatically alter the activities of IAB5, but have virtually no effect on the NEE enhancer (Fig. 8). NEE is equally effective in activating the eve, white, evewhite, and whiteTATA promoters, and thereby serves as an internal control for normal promoter function.
Mechanisms of core specificity
We propose that different core promoter sequences induce distinct conformations of the basal transcription complex. The TFIID complex is thought to bind the core promoter through direct interactions between TBP and TATA (e.g., Burley and Roeder 1996). In the absence of TATA, TFIID appears to make alternate contacts with the promoter, at least in part, through interactions between TAF150/TAF60 and Inr/Dpe sequences (Burke and Kadonaga 1996, 1997; Kaufmann et al. 1996). Previous studies have shown that the binding of TFIID induces substantial changes in DNA structure (Oelgeschlager et al. 1996). Perhaps TFIID adopts different conformations when bound to TATA versus Inr/Dpe sequences. Ftz activators bound to AE1 and IAB5 might prefer the conformation of the TFIID complex on TATA. In contrast, basal targets for NEE activators (e.g., Dorsal) might be equally accessible when TFIID is bound to either type I (TATA) or type II (Dpe/Inr) promoters.
An alternative model is that the TFIID complex is not invariant. Instead, there may be different forms of TFIID that interact with distinct core promoters. This possibility is suggested by the recent demonstration of a tissue-specific form of human TAFII130 (Dikstein et al. 1996) and by the identification of a variant TATA-binding protein, TRF, that is expressed in just a subset of tissues (Hansen et al. 1997). Perhaps distinct, but related, TFIID complexes interact with type I and type II promoters, and the Ftz activator preferentially interacts with the type I complex.
Maintaining the integrity of Hox complexes
It is conceivable that the evolutionary conservation of Hox gene clusters stems, at least in part, from the promoter-competition mechanisms that appear to be employed for the orderly trafficking of cis regulatory elements in the ANT-C. Deletions and translocations within Hox complexes may be only rarely tolerated because of inappropriate enhancer–promoter interactions. For example, a deletion in the ftz promoter region might unlock AE1–ftz interactions, so that AE1 is now able to activate the linked Scr gene in ectopic tissues, thereby causing homeotic transformations and dominant lethality. In general, proper enhancer–promoter interactions in Hox gene complexes might depend on a combinatorial code that links specific upstream activators with particular core promoters.
Materials and methods
P-transformation assays
yw67 flies were used for all P-transformation assays. Fusion genes were introduced into the Drosophila germ line as described in Small et al. (1992). Multiple transformants were generated for each construct, and at least three independent lines were examined. Embryos were collected, fixed, and hybridized with digoxigenin-labeled white, CAT, and lacZ antisense RNA probes exactly as described by Tautz and Pfeifle (1989) and Jiang et al. (1991).
Preparation of enhancers and promoters
The AE1 enhancer is located between −2574 and −2145 bp upstream of the ftz transcription start site (Schier and Gehring 1992). This 430-bp DNA fragment was synthesized from the genomic DNA of the yw67 strain by use of conventional PCR methods. This genomic DNA was also used for the PCR amplification of the ftz promoter region. The ftz DNA fragment extends from −100 bp upstream of the transcription start site to +91 bp and includes the untranslated leader sequence and first seven codons of the protein coding region (Laughon and Scott 1984). The mini-white promoter region used in this study extends from −316 bp upstream of the start site and extends to +174 bp. The Tp promoter sequence extends from −48 bp to +502 bp and includes coding sequences in both reading frames (for review, see Kaufman and Rio 1991).
The eve promoter sequence used in these studies is ∼200 bp in length and includes just 34 bp of 5′-flanking sequence. The core eve promoter contains an optimal TATA sequence (TATAAAA) but lacks both Inr and Dpe sequences (Macdonald et al. 1986; Frasch et al. 1987). In contrast, white lacks a TATA sequence, but contains conserved copies of the Inr and Dpe sequences (Pirrotta et al. 1985). The Inr encompasses a 6-bp consensus sequence: TCAG/TTT/C (for review, see Smale 1997). The central A corresponds to the transcription start site (+1). The Dpe is located downstream of the transcription start site, between +1 and +35 (for review, see Burke and Kadonaga 1996). It includes a 7-bp consensus sequence: A/GGA/TCGTG; the central GA/TCG motif is particularly well conserved among TATA-less promoters. The core white promoter contains a 6 of 6 match to the Inr consensus and a 4 of 4 match to the central Dpe sequence.
The chimeric evewhite promoter is 202 bp in length and includes white promoter sequences from −36 bp to +2 bp and eve sequences from +3 bp to +166 bp. It lacks TATA and Dpe sequences and contains a chimeric Inr (TCAGCA) that shares only 4 of 6 matches with the consensus sequence (Smale 1997). The whiteTATA promoter is identical to the wild-type sequence (−315 bp to +174 bp) except that in vitro mutagenesis was done to create the optimal eve TATA sequence (GTATAAAAG) from −29 to −21 bp.
Construction of P-element transposons
The eve/CAT–AE1–ftz/lacZ fusion gene shown in Figure 3 (A,B) was prepared by insertion of the eve promoter region into a pBluescript plasmid containing the CAT coding region (Barolo and Levine 1997). The eve/CAT recombinant was cloned into the NotI–AscI sites of the pCasPer P transposon (Small et al. 1992). The AE1 DNA fragment was cloned into the AscI site of this P element vector, and the ftz promoter region was cloned into the AscI–XbaI sites.
The white–AE1–Tp fusion gene shown in Figure 3 (C,D) was made by cloning the 430-bp AE1 genomic DNA fragment into the unique EcoRI site of the C4PLZ P-transformation vector (Gray and Levine 1996b; Zhou et al. 1996).
The white–AE1–eve fusion shown shown in Figure 4 was made by cloning of AE1 into the unique EcoRI site of the pEb vector (Cai and Levine 1997). A derivative of this P transposon was prepared by insertion of the 340-bp su(Hw) insulator DNA (Cai and Levine 1995) into the unique NotI site of the pEb vector described above. The white/CAT/lacZ P-transformation vector that was used for all of the experiments presented in this study is a modification of pCasPer, which contains divergently transcribed white and lacZ reporter genes (Small et al. 1992). It was modified by insertion of a CAT reporter between white and lacZ (Barolo and Levine 1997). The white reporter gene includes the 500-bp mini-white promoter sequence. Various eve, white, and chimeric promoter sequences were placed upstream of the CAT and lacZ reporters. This was done by insertion of promoter sequences into the unique AscI and BamHI sites of pBluescript plasmids containing either the CAT or lacZ coding region. CAT gene fusions were then excised from the plasmid by the combination of NotI and AscI. The resulting DNA fragment was then inserted into the unique NotI and AscI sites in the pCasPer vector. Similarly, promoter sequences were placed upstream of the lacZ coding region by use of the unique AscI and BamHI sites of a pBluescript plasmid. The lacZ fusion genes were excised from the plasmid with XbaI and AscI, and inserted into these unique sites within the pCasPer vector.
A 1-kb genomic DNA fragment containing the IAB5 enhancer (Zhou et al. 1996) was modified to include PstI restriction sites and inserted into the unique PstI site of the P-transformation vector, which is located 3′ of the lacZ coding region. Various DNA fragments were placed between the CAT and lacZ reporter genes. The P-transformation vectors used in Figure 5A–D contain a 1.6-kb spacer DNA from λ (Zhou et al. 1996). It was inserted into the unique AscI site located between CAT and lacZ. The vector used in Figure 5 (E and F) contains the 340-bp gypsy insulator DNA at this AscI site, in place of the λ spacer (Cai and Levine 1995). Finally, the vectors used in Figures 7 and 8 contain the 300-bp rho NEE enhancer (Ip et al. 1992) at the unique AscI site instead of the λ spacer DNA or gypsy insulator.
Acknowledgments
We thank James Sharpe and Robb Krumlauf for sharing unpublished results, and Jim Kadonaga and Jim Manley for helpful discussions. We also thank Anna Di Gregorio, Jumin Zhou, and Hailan Zhang for comments on the manuscript. This work was funded by a grant from the National Institutes of Health (GM34431). S.O. is a fellow of the Human Frontiers Science Program.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mlevine@uclink4.berkeley.edu; FAX (510) 642-6062.
References
- Arnosti DN, Barolo S, Levine M, Small S. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 1996;122:205–214. doi: 10.1242/dev.122.1.205. [DOI] [PubMed] [Google Scholar]
- Barolo S, Levine M. Hairy mediates dominant repression in the Drosophila embryo. EMBO J. 1997;16:2883–2891. doi: 10.1093/emboj/16.10.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TW, Kadonaga JT. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes & Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- ————— The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes & Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- Busturia A, Bienz M. Silencers in abdominal-B, a homeotic Drosophila gene. EMBO J. 1993;12:1415–1425. doi: 10.1002/j.1460-2075.1993.tb05785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Levine M. Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature. 1995;376:533–536. doi: 10.1038/376533a0. [DOI] [PubMed] [Google Scholar]
- ————— The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo. EMBO J. 1997;16:1732–1741. doi: 10.1093/emboj/16.7.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Sharma S, Keelan DJ, Lewis EB. The molecular genetics of the bithorax complex of Drosophila: Cis-regulation in the Abdominal-B domain. EMBO J. 1990;9:4277–4286. doi: 10.1002/j.1460-2075.1990.tb07876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi OR, Engel JD. Developmental regulation of β-globin gene switching. Cell. 1988;55:17–26. doi: 10.1016/0092-8674(88)90005-0. [DOI] [PubMed] [Google Scholar]
- Colgan J, Manley JL. Cooperation between core promoter elements influences transcriptional activity in vivo. Proc Natl Acad Sci. 1995;92:1955–1999. doi: 10.1073/pnas.92.6.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das G, Hinkley CS, Herr W. Basal promoter elements as a selective determinant of transcriptional activator function. Nature. 1995;374:657–660. doi: 10.1038/374657a0. [DOI] [PubMed] [Google Scholar]
- Dikstein R, Zhou S, Tjian R. Human TAFII105 is a cell type-specific TFIID subunit related to hTAFII130. Cell. 1996;87:137–146. doi: 10.1016/s0092-8674(00)81330-6. [DOI] [PubMed] [Google Scholar]
- Dorsett D. Distance-independent inactivation of an enhancer by the suppressor of Hairy-wing DNA-binding protein of Drosophila. Genetics. 1993;134:1135–1144. doi: 10.1093/genetics/134.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami KH, Navarre WW, Smale ST. Core promoter specificities of the Sp1 and VP16 transcriptional activation domains. Mol Cell Biol. 1995;15:5906–5916. doi: 10.1128/mcb.15.11.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami KH, Jain A, Smale ST. Mechanism of synergy between TATA and initiator: Synergistic binding of TFIID following a putative TFIIA-induced isomerization. Genes & Dev. 1997;11:3007–3019. doi: 10.1101/gad.11.22.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley KP, Engel JD. Individual stage selector element mutations lead to reciprocal changes in β- vs. ε-globin gene transcription: Genetic confirmation of promoter competition during globin gene switching. Genes & Dev. 1992;6:730–744. doi: 10.1101/gad.6.5.730. [DOI] [PubMed] [Google Scholar]
- Foley KP, Pruzina S, Winick JD, Engel JD, Grosveld F, Fraser P. The chicken β/ε-globin enhancer directs autonomously regulated, high-level expression of the chicken ε-globin gene in transgenic mice. Proc Natl Acad Sci. 1994;91:7252–7256. doi: 10.1073/pnas.91.15.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M, Hoey T, Rushlow C, Doyle H, Levine M. Characterization and localization of the even-skipped protein of Drosophila. EMBO J. 1987;6:749–759. doi: 10.1002/j.1460-2075.1987.tb04817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK. The role of insulator elements in defining domains of gene expression. Curr Opin Genet Dev. 1997;7:242–248. doi: 10.1016/s0959-437x(97)80134-7. [DOI] [PubMed] [Google Scholar]
- Geyer PK, Corces VG. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes & Dev. 1992;6:1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- Gindhart JG, Jr, King AN, Kaufman TC. Characterization of the cis-regulatory region of the Drosophila homeotic gene Sex combs reduced. Genetics. 1995;139:781–795. doi: 10.1093/genetics/139.2.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman MJ, Kaufman TC. Genetic analysis of embryonic cis-acting regulatory elements of the Drosophila homeotic gene sex combs reduced. Genetics. 1995;140:557–572. doi: 10.1093/genetics/140.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S, Levine M. Transcriptional repression in development. Curr Opin Cell Biol. 1996a;8:358–364. doi: 10.1016/s0955-0674(96)80010-x. [DOI] [PubMed] [Google Scholar]
- ————— Short-range transcriptional repressors mediate both quenching and direct repression within complex loci in Drosophila. Genes & Dev. 1996b;10:700–710. doi: 10.1101/gad.10.6.700. [DOI] [PubMed] [Google Scholar]
- Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes & Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- Hansen SK, Takada S, Jacobson RH, Lis JT, Tjian R. Transcription properties of a cell type-specific TATA-binding protein, TRF. Cell. 1997;91:71–83. doi: 10.1016/s0092-8674(01)80010-6. [DOI] [PubMed] [Google Scholar]
- Ip YT, Park RE, Kosman D, Bier E, Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes & Dev. 1992;6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- Jiang J, Kosman D, Ip YT, Levine M. The dorsal morphogen gradient regulates the mesoderm determinant twist in early Drosophila embryos. Genes & Dev. 1991;5:1881–1891. doi: 10.1101/gad.5.10.1881. [DOI] [PubMed] [Google Scholar]
- Kaufman PD, Rio DC. Drosophila P-element transposase is a transcriptional repressor in vitro. Proc Natl Acad Sci. 1991;88:2613–2617. doi: 10.1073/pnas.88.7.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann J, Verrijzer CP, Shao J, Smale ST. CIF, an essential cofactor for TFIID-dependent initiator function. Genes & Dev. 1996;10:873–886. doi: 10.1101/gad.10.7.873. [DOI] [PubMed] [Google Scholar]
- Laughon A, Scott MP. Sequence of a Drosophila segmentation gene: Protein structure homology with DNA-binding proteins. Nature. 1984;310:25–31. doi: 10.1038/310025a0. [DOI] [PubMed] [Google Scholar]
- Lewis EB, Knafels JD, Mathog DR, Celniker SE. Sequence analysis of the cis-regulatory regions of the bithorax complex of Drosophila. Proc Natl Acad Sci. 1995;92:8403–8407. doi: 10.1073/pnas.92.18.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Noll M. Compatibility between enhancers and promoters determines the transcriptional specificity of gooseberry and gooseberry neuro in the Drosophila embryo. EMBO J. 1994;13:400–406. doi: 10.1002/j.1460-2075.1994.tb06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald PM, Ingham P, Struhl G. Isolation, structure, and expression of even-skipped: A second pair-rule gene of Drosophila containing a homeo box. Cell. 1986;47:721–734. doi: 10.1016/0092-8674(86)90515-5. [DOI] [PubMed] [Google Scholar]
- Manley JL, Um M, Li C, Ashali H. Mechanisms of transcriptional activation and repression can both involve TFIID. Philos Trans R Soc Lond B Biol Sci. 1996;351:517–526. doi: 10.1098/rstb.1996.0050. [DOI] [PubMed] [Google Scholar]
- Martin CH, Mayeda CA, Davis CA, Ericsson CL, Knafels JD, Mathog DR, Celniker SE, Lewis EB, Palazzolo MJ. Complete sequence of the bithorax complex of Drosophila. Proc Natl Acad Sci. 1995;92:8398–8402. doi: 10.1073/pnas.92.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merli C, Bergstrom DE, Cygan JA, Blackman RK. Promoter specificity mediates the independent regulation of neighboring genes. Genes & Dev. 1996;10:1260–1270. doi: 10.1101/gad.10.10.1260. [DOI] [PubMed] [Google Scholar]
- Mihaly J, Hogga J, Gausz I, Gyurkovics H, Karch F. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development. 1997;124:1809–1820. doi: 10.1242/dev.124.9.1809. [DOI] [PubMed] [Google Scholar]
- Oelgeschlager T, Chiang CM, Roeder RG. Topology and reorganization of a human TFIID-promoter complex. Nature. 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- Pick L, Schier A, Affolter M, Schmidt-Glenewinkel T, Gehring WJ. Analysis of the ftz upstream element: Germ layer-specific enhancers are independently autoregulated. Genes & Dev. 1990;7:1224–1239. doi: 10.1101/gad.4.7.1224. [DOI] [PubMed] [Google Scholar]
- Pirrotta V, Steller H, Bozzetti MP. Multiple upstream regulatory elements control the expression of the Drosophila white gene. EMBO J. 1985;4:3501–3508. doi: 10.1002/j.1460-2075.1985.tb04109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Rivera-Pomar R, Jackle H. From gradients to stripes in Drosophila embryogenesis: Filling in the gaps. Trends Genet. 1996;12:478–483. doi: 10.1016/0168-9525(96)10044-5. [DOI] [PubMed] [Google Scholar]
- Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- Rusch J, Levine M. Threshold responses to the dorsal regulatory gradient and the subdivision of primary tissue territories in the Drosophila embryo. Curr Opin Genet Dev. 1996;6:416–423. doi: 10.1016/s0959-437x(96)80062-1. [DOI] [PubMed] [Google Scholar]
- Rushlow CA, Levine M. Combinatorial expression of a ftz-zen fusion promoter suggests the occurrence of cis interactions between genes of the ANT-C. EMBO J. 1988;7:3479–3485. doi: 10.1002/j.1460-2075.1988.tb03223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer F, Wassarman DA, Rubin GM, Tjian R. TAF(II)s mediate activation of transcription in the Drosophila embryo. Cell. 1996;87:1271–1284. doi: 10.1016/s0092-8674(00)81822-x. [DOI] [PubMed] [Google Scholar]
- Schier AF, Gehring WJ. Direct homeodomain-DNA interaction in the autoregulation of the fushi tarazu gene. Nature. 1992;356:804–807. doi: 10.1038/356804a0. [DOI] [PubMed] [Google Scholar]
- Scott KS, Geyer PK. Effects of the su(Hw) insulator protein on the expression of the divergently transcribed Drosophila yolk protein genes. EMBO J. 1995;14:6258–6267. doi: 10.1002/j.1460-2075.1995.tb00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Brose M, Treisman J, Desplan C. Synergy between the hunchback and bicoid morphogens is required for anterior patterning in Drosophila. Cell. 1994;78:855–865. doi: 10.1016/s0092-8674(94)90622-x. [DOI] [PubMed] [Google Scholar]
- Smale ST. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stargell LA, Struhl K. Mechanisms of transcriptional activation in vivo: Two steps forward. Trends Genet. 1996;12:311–315. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Verrijzer CP, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–432. [PubMed] [Google Scholar]
- Zenzie-Gregory B, Khachi A, Garraway IP, Smale ST. Mechanism of initiator-mediated transcription: Evidence for a functional interaction between the TATA-binding protein and DNA in the absence of a specific recognition sequence. Mol Cell Biol. 1993;13:3841–3849. doi: 10.1128/mcb.13.7.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Barolo S, Szymanski P, Levine M. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes & Dev. 1996;10:3195–3201. doi: 10.1101/gad.10.24.3195. [DOI] [PubMed] [Google Scholar]


