Abstract
Copper, as copper sulfate, is increasingly used as an alternative to in-feed antibiotics for growth promotion in weaned piglets. Acquired copper resistance, conferred by a plasmid-borne, transferable copper resistance (tcrB) gene, has been reported in Enterococcus faecium and E. faecalis. A longitudinal field study was undertaken to determine the relationship between copper supplementation and the prevalence of tcrB-positive enterococci in piglets. The study was done with weaned piglets, housed in 10 pens with 6 piglets per pen, fed diets supplemented with a normal (16.5 ppm; control) or an elevated (125 ppm) level of copper. Fecal samples were randomly collected from three piglets per pen on days 0, 14, 28, and 42 and plated on M-Enterococcus agar, and three enterococcal isolates were obtained from each sample. The overall prevalence of tcrB-positive enterococci was 21.1% (38/180) in piglets fed elevated copper and 2.8% (5/180) in the control. Among the 43 tcrB-positive isolates, 35 were E. faecium and 8 were E. faecalis. The mean MICs of copper for tcrB-negative and tcrB-positive enterococci were 6.2 and 22.2 mM, respectively. The restriction digestion of the genomic DNA of E. faecium or E. faecalis with S1 nuclease yielded a band of ∼194-kbp size to which both tcrB and the erm(B) gene probes hybridized. A conjugation assay demonstrated cotransfer of tcrB and erm(B) genes between E. faecium and E. faecalis strains. The higher prevalence of tcrB-positive enterococci in piglets fed elevated copper compared to that in piglets fed normal copper suggests that supplementation of copper in swine diets selected for resistance.
INTRODUCTION
Copper is an essential trace element required for vital biological functions in both prokaryotic and eukaryotic cells (29). The growth response in piglets to copper is independent of—and in addition to—the response to commonly used antibiotics in the feed (16). Copper in excess concentrations is toxic to cells because it induces the production of intracellular reactive oxygen radicals, which inactivate cell components such as nucleic acids, lipids, and proteins (32). Therefore, cells tightly regulate intracellular copper concentrations to avoid toxicity (43). The copper homeostasis mechanism has been well studied in certain Gram-positive bacteria, such as Enterococcus hirae, Lactococcus lactis, and Bacillus subtilis (45). The normal intracellular copper concentration is maintained by the copYZAB operon, where copA and copB encode copper transport ATPases responsible for the influx and efflux of copper, respectively. The copY gene acts as a copper-responsive repressor and copZ encodes a copper chaperone (46).
Some bacteria acquire resistance to copper, which may be either chromosome mediated (17) or plasmid mediated (31). A plasmid-borne gene, designated as transferable copper resistance (tcrB) and homologous to copB of the copYZAB operon, was reported in E. faecium and E. faecalis isolated from piglets, calves, poultry, and humans in Denmark (2). In Denmark, the plasmid also carries the genes erm(B) and vanA, which encode resistance to macrolides and glycopeptides, respectively (2, 20), suggesting a potential link between copper resistance and antibiotic resistance. We have confirmed the occurrence of the tcrB gene in fecal enterococci of piglets fed diets with normal or elevated levels of copper (4). Whether copper supplementation in pigs exerts selection pressure for copper-resistant enterococci, which in turn could lead to coselection for macrolide or glycopeptide resistance, has not been documented. Because of the limited number of isolates tested in our previous study, we were unable to relate copper supplementation to the prevalence of tcrB-positive enterococci. Therefore, the present longitudinal study was undertaken to determine the relationship between supplementation of copper at an elevated concentration (125 ppm) and the prevalence of fecal tcrB-mediated copper-resistant and erm(B)-mediated macrolide-resistant enterococci in piglets. We performed Southern blot hybridization to show that the tcrB gene is carried on a transferable plasmid and provide evidence for the interspecies cotransfer of tcrB and erm(B) genes in enterococci.
(Part of this work was presented at the Second American Society for Microbiology Conference on Antimicrobial Resistance in Zoonotic Bacteria and Foodborne Pathogens, Toronto, Canada, 8 to 11 June 2010, and at the Third American Society for Microbiology Conference on Enterococci, Portland, Oregon, 30 July to 2 August 2010.)
MATERIALS AND METHODS
Animals, experimental design, and sampling.
The use of animals and the experimental procedure followed were approved by the Kansas State University Animal Care and Use Committee. Sixty newly weaned piglets (21 days old) with an average body weight of 7.0 kg (± 1 kg) were randomly allocated to two dietary treatments. The two dietary treatments were as follows: basal diet with 16.5 ppm of copper (control group) or basal diet supplemented with 125 ppm of copper as copper sulfate (copper group). The basal diet consisted of corn, soybean meal, vitamins, amino acids, and trace mineral supplements, and piglets were housed in an environmentally controlled nursery facility. Each treatment group had a total of 30 piglets assigned to 5 pens with 6 piglets per pen. Each pen was provided with a self-feeder containing four holes and a water nipple such that animals had ad libitum access to feed and water. Each pen had a wire-mesh floor that allowed for 0.3 m2 per piglet. Piglets were fed treatment diets for 42 days. Fecal samples were collected randomly from 3 pigs in each pen on days 0, 14, 28, and 42, placed in individual bags, and transported to the laboratory on ice.
Isolation of enterococci.
Unless otherwise mentioned, all culture media used were from Difco (Becton Dickinson, Sparks, MD). Fecal samples were processed by diluting 1 g of feces in 10 ml of phosphate-buffered saline and spread plating 50 μl of the suspension onto M-Enterococcus agar. The plates were incubated for 24 h at 37°C. Five colonies (pinpoint red, pink, or metallic pink) were picked, streaked onto blood agar plates, and incubated overnight at 37°C. All isolates were tested for esculin hydrolysis by inoculating them into 100 μl of Enterococcosel broth in a 96-well microtiter plate (Becton Dickinson, Franklin Lakes, NJ) and incubating at 37°C for 4 h. Three esculin hydrolysis-positive isolates per fecal sample (9 per pen and 45 per treatment at each sampling time) were stored using Protect beads (Cryocare; Key Scientific Products, Stamford, TX) at −80°C until further use.
PCR for detection of the tcrB gene.
The tcrB gene was detected according to the procedure described by Hasman et al. (20). Each isolate from a single Protect bead was streaked onto a blood agar plate and incubated overnight at 37°C. Bacterial DNA was extracted by suspending a single colony in nuclease-free water with Chelex 100 Resin (Bio-Rad Laboratories, Hercules, CA) and boiling it for 10 min (4). A tcrB-positive E. faecium strain (7430275-4 or 7430272-6; kindly provided by Henrik Hasman, National Food Institute, Technical University of Denmark) served as the positive control.
Speciation of tcrB-positive enterococci.
Species identification of the enterococcal isolates that were tcrB positive and an equal number of tcrB-negative isolates, randomly chosen from the control and treatment groups, was performed by a multiplex PCR that identifies E. faecium, E. faecalis, E. gallinarum, and E. casseliflavus (24). Additionally, superoxide dismutase (sodA) gene sequence analysis (42) was used for species confirmation. The DNA template was prepared as mentioned before. The ATCC (Manassas, VA) strains of E. faecium (ATCC 19434), E. faecalis (ATCC 19433), E. gallinarum (ATCC 49579), and E. casseliflavus (ATCC 25788) served as positive controls. Master mixes, primers, and running conditions for the multiplex PCR and sodA gene PCR were as described by Jackson et al. (24) and Poyart et al. (42), respectively. The sodA gene products were purified by the Wizard SV gel and PCR cleanup system (Promega, Madison, WI). The purified PCR products were sequenced at Genomics Core, Institute for Integrative Genome Biology, University of California at Riverside. The sequences were analyzed by a BLAST search in the NCBI GenBank database.
Detection of erm(B) and vanA genes.
The primers and PCR conditions for detection of erm(B) and vanA genes were as per the work of Jacob et al. (26) and Kariyama et al. (27), respectively. Enterococcus faecalis MMH 594 (provided by Lynn Hancock, Division of Biology, Kansas State University) and E. faecium (ATCC 51559) served as positive controls for the erm(B) and vanA genes, respectively.
Copper susceptibility determinations.
Copper susceptibility determination for tcrB-positive and an equal number of tcrB-negative enterococcal isolates was performed by the agar dilution method (19, 20). The two strains from Denmark (7430162-6 and 7430275-4) were included as positive controls. Mueller-Hinton (MH) agar plates prepared with concentrations of copper, added as copper sulfate (Fischer Scientific, Fair Lawn, NJ), at 0, 2, 4, 8, 12, 16, 20, 24, 28, 32, 36, or 40 mM and with pH adjusted to 7.0 were used. The plates were spot inoculated with 20 μl of bacterial culture that was adjusted to McFarland turbidity standard no. 0.5 (Remel, Lenexa, KS) and incubated at 37°C for 48 h to determine growth or lack of growth. The susceptibility determinations were repeated on another day with different inoculum preparations.
Antibiotic susceptibility determinations.
The microbroth dilution method was used to determine the MICs for erythromycin and vancomycin (Sigma-Aldrich, St. Louis, MO) per CLSI guidelines (11). Antibiotic stock solutions, each containing a final concentration of 1,000 μg/ml based on potency, were prepared in sterile distilled water. The bacterial cultures were prepared by inoculating a single colony in 10 ml MH broth, incubating for 6 h, and then diluting 100-fold with sterile MH broth. Antibiotics were tested at concentrations of 100, 50, 25, 12.5, 6.25, 3.125, 1.56, 0.78, 0.39, 0.195, and 0.098 μg/ml. The antimicrobial susceptibility test was performed in 96-well microtiter plates (Becton Dickinson), and inoculated plates were incubated at 37°C for 24 h, and results were recorded as growth or no growth. The MIC determinations were repeated on another day with different inoculum preparations.
Pulsed-field gel electrophoresis.
The pulsed-field gel electrophoresis (PFGE) analysis of tcrB-positive isolates was performed as per the work of Murray et al. (36) with minor modifications. A strain of Salmonella enterica serotype Braenderup H9812 was used as the standard (B. Murray, University of Texas Medical School, personal communication). A single colony of the isolate was inoculated into 5 ml of brain heart infusion (BHI) broth and incubated overnight at 37°C. One milliliter of overnight culture was transferred into a 1.5-ml Eppendorf tube, and cells were pelleted by centrifugation at 9,300 × g for 1 min. The supernatant was discarded and the pellet was suspended in 200 μl of 0.85% NaCl. The plugs were prepared by mixing 200 μl of the cell suspension with 200 μl of 1.6% SeaKem gold agarose. The plugs were lysed by transferring them into a 10-ml lysis solution (6 mM Tris-HCl, pH 7.4, 100 mM EDTA, 1 M NaCl, 0.5% sodium lauroyl sarcosine, 0.5% Brij, 0.2% deoxycholate, lysozyme [500 μg/ml] and RNaseA [20 μg/ml]) for 4 h at 37°C with gentle shaking. The plugs were then transferred to ESP buffer (10 mM Tris-HCl, pH 7.4, 1 mM EDTA, 1% SDS, and 50 μg/ml proteinase K) and incubated overnight at 50°C with gentle shaking. The plugs were washed three times at room temperature for 10 min each with TE dilute (10 mM Tris-HCl, pH-7.4, 0.1 mM EDTA) and stored at 4°C until used.
Restriction digestion of the plugs was performed by placing a small slice of agarose plug in a 1.5-ml microcentrifuge tube with 2 μl of SmaI in 10 μl of 10× buffer; the volume was increased to 100 μl with double distilled water and the mixture was held for 4 h at 25°C. The digested plugs were transferred onto a gel (1% SeaKem Gold Agarose with 0.5× Tris-borate-EDTA [TBE]) and the gel was placed in an electrical field device, CHEF-DR II (Bio-Rad, Richmond, CA), at 200 V. Pulse times were as follows: for block 1, an initial time of 3.5 s and a final time of 25 s for 12 h; and for block 2, an initial time of 1 s and a final time of 5 s for 8 h. Then the gel was stained with 0.0001% (or 1 μg/ml) ethidium bromide for 30 min followed by destaining in distilled water 3 times for 20 min each. Gel images were captured using a Gel Doc 2000 system (Bio-Rad), and band patterns were analyzed and compared by using BioNumerics version 3.0 (Applied Maths, Austin, TX). For clustering with a position tolerance setting of 1.5% for optimization and a position tolerance of 1.5% for band comparison, we used the band-based Dice similarity coefficient and the unweighted-pair group mathematical average algorithm method (unweighted-pair group method using average linkages [UPGMA]). Isolates were grouped based on identical banding patterns (100% Dice similarity).
Southern blot hybridization of tcrB and erm(B) genes.
The Southern blot hybridization was performed on two tcrB-positive E. faecium isolates, one tcrB-positive E. faecalis isolate, and an equal number of tcrB-negative isolates by using digoxigenin (DIG) High Prime labeling and detection starter kit II (Roche Diagnostics, Indianapolis, IN). The procedure was performed with slight modifications to the instructions provided in the commercial kit. The genomic DNA was used to prepare plugs as in the PFGE protocol, which was followed by restriction digestion with S1 nuclease enzyme (New England BioLabs Inc., Beverly, MA). The restriction digestions were performed using 10 U of enzyme in 25 μl of buffer (44). The digested DNA samples were subjected to electrophoresis using the CHEF-DRII apparatus with Midrange II PFG molecular marker (New England BioLabs, Inc.). The staining and destaining procedures were carried out as in the PFGE protocol. After PFGE, each band was considered a unit length of linear plasmid. The gel was then subjected to depurination (0.25 N HCl), denaturation (1.5 M NaCl, 0.5 N NaOH), and neutralization (Tris base, NaCl, pH 7.4) buffers. The gel containing linearized and separated plasmid DNA was transferred onto a positively charged nylon membrane (Genescreen Plus, Boston, MA) and left overnight for blotting at room temperature by capillary transfer with 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) transfer buffer (pH 7.0). After blotting, DNA was immobilized by UV cross-linking for 2 min (UV Cross-linker, FB-UVXL-1000). The tcrB and erm(B) probes were prepared by random primer labeling using DIG High Prime. The immobilized DNA on the membrane was subjected to prehybridization using DIG Easy Hyb solution followed by hybridization with DIG-labeled DNA probe. The hybridization was carried out overnight at 37°C with gentle agitation. After hybridization, the membrane was developed using CL-XPosure X-ray film (Thermo Scientific, Rockford, IL).
Interspecies transferability of tcrB gene.
A conjugation assay was performed using the filter mating procedure (47). The tcrB-positive donor strains (5 E. faecium and 5 E. faecalis strains) were resistant to tetracycline [MIC > 100 μg/ml; tet(M) positive] and susceptible to spectinomycin [MIC=12.5 μg/ml]. E. faecium strain TX 5034 (provided by Barbara Murray, University of Texas Medical School), resistant to spectinomycin (MIC > 100 μg/ml) and tet(M) negative and susceptible to tetracycline (MIC=0.78 μg/ml), and E. faecalis strain OG1SSp (provided by Ludek Zurek, Kansas State University), resistant to spectinomycin (MIC > 100 μg/ml) and tet(M) negative and sensitive to tetracycline (MIC=0.39 μg/ml), were used as recipient strains. The donors and recipients were grown on BHI agar plates containing tetracycline (40 μg/ml) and spectinomycin (500 μg/ml), respectively. The resultant transconjugants were selected on BHI agar plates containing both tetracycline and spectinomycin. The transconjugants were tested for tcrB and erm(B) genes by PCR and their susceptibilities to copper were determined as described above. The transfer frequency for each strain was calculated as the CFU of transconjugants per recipient CFU.
Statistical analysis.
The tcrB gene prevalence in treatment groups was analyzed using a generalized mixed model (PROC GLIMMIX, SAS, Version 9.2). The statistical model included the fixed effect of dietary treatment (control or elevated copper) with pen as a random effect. The analysis was performed to determine the effects both of sampling days and copper supplementation. Copper and vancomycin MIC values were analyzed for evidence of normal distribution (PROC UNIVARIATE) within and between tcrB-positive and -negative isolates. The MIC values were transformed based on rank (PROC RANK), and analysis of variance (PROC GLIMMIX) was performed on the ranked values since the MIC values were not normally distributed (P < 0.05).
RESULTS
Prevalence of the tcrB, erm(B), and vanA genes.
A total of 360 enterococcal isolates consisting of 45 isolates (3 isolates per fecal sample, 3 piglets sampled per pen, and 5 pens per treatment) per treatment group (control or copper group) and sampling day (days 0, 14, 28, and 42) were obtained. All 360 enterococcal isolates were screened for the tcrB gene and 43 (11.9%) isolates were positive (amplicon size of 663 bp). Out of 43 tcrB-positive isolates, 5 (5/180; 2.8%) were from piglets fed the basal diet with a normal amount of copper (16.5 ppm) and 38 isolates (38/180; 21.1%) were from piglets fed the basal diet supplemented with an elevated amount of copper (Table 1). The overall prevalence of tcrB-positive enterococcal isolates was higher (P < 0.05) in the copper-supplemented group than in the control group. The prevalence of tcrB was affected by sampling day (P < 0.05) and also exhibited a significant treatment and sampling time interaction (P < 0.05). The 43 tcrB-positive and a subset of tcrB-negative isolates (n=44) randomly chosen from the control group were used for the species identification. Based on multiplex PCR and sodA gene sequence analyses, 35 tcrB-positive isolates were E. faecium and 8 isolates were E. faecalis. Among 44 tcrB-negative isolates, 25 isolates were E. faecium and 19 isolates were E. faecalis. All tcrB-positive (n=43) and tcrB-negative (n=44) isolates were positive for the erm(B) gene and negative for the vanA gene.
Table 1.
Prevalence of tcrB-positive fecal enterococci in piglets fed diets supplemented with or without copper
| Treatment (copper level in the feed) | No. of isolates positive for tcrB gene/total isolates tested |
||||
|---|---|---|---|---|---|
| Sampling day |
Total (%) | ||||
| 0 | 14 | 28 | 42 | ||
| Control (16 ppm) | 0/45 | 3/45 | 1/45 | 1/45 | 5/180 (2.8) |
| Copper (125 ppm) | 0/45 | 19/45 | 8/45 | 11/45 | 38/180 (21.1) |
MICs of copper, erythromycin, and vancomycin.
All tcrB-positive isolates (35 E. faecium and 8 E. faecalis) grew on MH agar containing copper at 16 or 20 mM. However, the tcrB-negative isolates (25 E. faecium and 19 E. faecalis isolates) grew only at a 2 to 8 mM copper concentration. The mean MIC values of copper for tcrB-negative and tcrB-positive isolates were 6.2 and 22.2 mM, respectively, and the difference was significant at P < 0.001. The two reference strains obtained from Denmark (7430162-6 and 7430275-4) had MICs of 23 and 24 mM, respectively.
All tcrB-positive and tcrB-negative isolates were resistant to erythromycin (MIC > 100 μg/ml) and susceptible to vancomycin. The MIC values for vancomycin were higher (P < 0.001) for the tcrB-positive isolates (0.42 μg/ml) than for the tcrB-negative isolates (0.22 μg/ml); however, none of the isolates can be considered “resistant” using the breakpoint criterion (11).
PFGE typing.
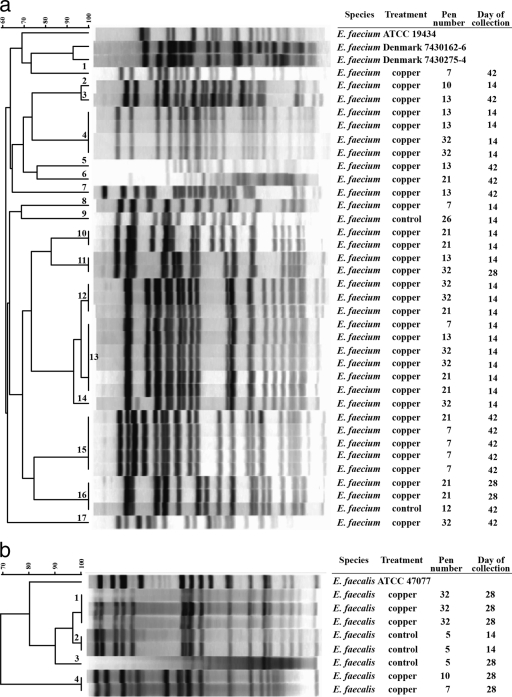
The clonal relationship among the 43 tcrB-positive isolates was determined (Fig. 1) by PFGE typing. Based on the species-specific distribution of PFGE patterns within and between the copper group and the control group, a total of 17 PFGE types were obtained (<95% Dice similarity) among the 35 tcrB-positive E. faecium isolates while the 8 tcrB-positive E. faecalis isolates showed 4 PFGE types. Among the five tcrB-positive isolates from the control group, three E. faecalis isolates belonged to two different PFGE types and the two E. faecium isolates were of different PFGE types. Among the 38 tcrB-positive isolates obtained from the copper group, 33 E. faecium isolates belonged to 15 different PFGE types and five E. faecalis isolates shared two PFGE patterns. Six E. faecium isolates (type 13) with identical banding patterns (100% Dice similarity) were obtained from four pens in the copper group on sampling day 14. Similarly, four E. faecium isolates (type 4) with identical banding patterns (100% Dice similarity) were obtained from two different pens in the copper group on sampling day 14. On day 28, three E. faecalis isolates obtained from the same pen had identical banding patterns. Among the two tcrB-positive E. faecium isolates in the control group, one isolate had a PFGE pattern identical to that of two tcrB-positive isolates (from the same pen) in the copper group. All tcrB-positive isolates obtained on three different sampling days (14, 28, and 42) belonged to different PFGE types except for two E. faecium isolates obtained from the same pen on day 28 matched with another isolate (from a different pen) obtained on day 42. Few of the tcrB-positive isolates obtained within each pen over three different sampling days (14, 28, and 42) belonged to different PFGE types. Among the isolates obtained from day 14, one PFGE type was distributed among three different pens, which was matched with other two different PFGE types shared between pens. On day 28, we observed only one PFGE that was common between two pens. In one pen (pen 32) that had 12 tcrB-positive isolates obtained on three different sampling days, there were six different PFGE types. In pens 7 and 13, which had seven tcrB-positive isolates each, there were five and six different PFGE types, respectively. Pen 21 with nine tcrB-positive isolates had five different PFGE types. Pen 10 with two tcrB-positive isolates had two PFGE types. In one pen of the copper group that had five tcrB-positive E. faecium isolates on sampling day 42, four had identical PFGE banding patterns.
Fig. 1.
(a) Pulsed-field gel electrophoresis banding patterns of SmaI-digested genomic DNA of tcrB-positive Enterococcus faecium isolates from piglets fed diets supplemented with or without copper. (b) Pulsed-field gel electrophoresis banding patterns of SmaI-digested genomic DNA of tcrB-positive Enterococcus faecalis isolates from piglets fed diets supplemented with or without copper.
Southern hybridization.
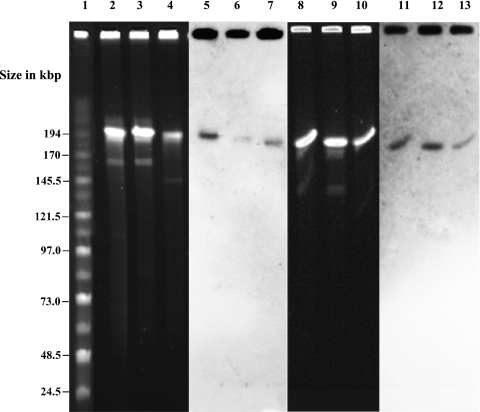
The restriction digestion of the genomic DNA of E. faecium or E. faecalis with S1 nuclease yielded a distinct band of ∼194-kbp size (Fig. 2). The restriction digestion of tcrB-negative E. faecium or E. faecalis with S1 nuclease did not yield 194-kbp bands but showed bands that ranged from 150 to 170 kbp in size (data not shown). In all three tcrB-positive E. faecium and E. faecalis isolates tested, both tcrB and the erm(B) gene probes hybridized with the ∼194-kbp band (Fig. 2). In case of tcrB-negative E. faecium or E. faecalis strains, as expected, no hybridization was observed with tcrB gene probe; however, the erm(B) gene probe hybridized with 150- to 170-kbp bands (data not shown).
Fig. 2.
Hybridization of tcrB and erm(B) probes to S1 nuclease-digested genomic DNA of three enterococcal strains. Lanes: 1, midrange PFG marker II; 2 and 3, genomic DNA of E. faecium isolates digested with S1 nuclease enzyme; 4, genomic DNA of E. faecalis isolate digested with S1 nuclease enzyme; 5, 6, and 7, Southern blot hybridization with tcrB probe; 8 and 9, genomic DNA of E. faecium isolates digested with S1 nuclease enzyme; 10, genomic DNA of E. faecalis isolate digested with S1 nuclease enzyme; 11, 12, and 13, Southern blot hybridization with erm(B) probe.
Interspecies transferability of tcrB gene.
Five each of tcrB-positive E. faecium and E. faecalis isolates were used to demonstrate the interspecies transferability of tcrB gene by conjugation. The 10 resultant transconjugants were positive for tcrB, erm(B), and tet(M) genes. As expected, the transconjugants grew on MH agar containing high copper concentrations (16 or 20 mM). The mean copper MIC of the 10 transconjugants was 18.4 mM. The mean transfer frequencies for tcrB-positive E. faecium and E. faecalis isolates were 9.3 × 10−6 and 8.2 × 10−6, respectively (Table 2).
Table 2.
Transfer frequency of tcrB gene in Enterococcus faecium and Enterococcus faecalis
| Donor strain | Recipient strain | Transfer frequency (no. of transconjugants per recipient) |
|---|---|---|
| E. faecium KSU 143 | E. faecalis OG1SSp | 1.18 × 10−5 |
| E. faecium KSU 150 | E. faecalis OG1SSp | 7.88 × 10−6 |
| E. faecium KSU 153 | E. faecalis OG1SSp | 6.97 × 10−6 |
| E. faecium KSU 160 | E. faecalis OG1SSp | 9.48 × 10−6 |
| E. faecium KSU 167 | E. faecalis OG1SSp | 1.02 × 10−5 |
| E. faecalis KSU 101 | E. faecium TX5034 | 1.05 × 10−5 |
| E. faecalis KSU 102 | E. faecium TX5034 | 8.15 × 10−6 |
| E. faecalis KSU 201 | E. faecium TX5034 | 7.43 × 10−6 |
| E. faecalis KSU 234 | E. faecium TX5034 | 7.67 × 10−6 |
| E. faecalis KSU 269 | E. faecium TX5034 | 6.96 × 10−6 |
DISCUSSION
In the present study, we examined the effect of feeding an elevated concentration of copper on the prevalence of tcrB-mediated, copper-resistant enterococcal isolates in the feces of piglets. Only a small proportion of fecal enterococcal isolates (11.9%; 43/360) obtained from piglets fed diets supplemented with normal or elevated levels of copper harbored the tcrB gene. This suggests that acquired resistance to copper may not be widespread in enterococci. In contrast to our study, Hasman and Aarestrup (19) have reported a prevalence of 76% (45/59) of copper-resistant enterococcal isolates from pigs obtained at the time of slaughter in Denmark. The higher prevalence of tcrB gene in the Danish piglets may be related to higher levels (175 to 250 ppm) or longer durations (birth to slaughter) of copper supplementation (21). In our study, the tcrB gene was detected in only two species, E. faecium and E. faecalis, and the majority of the resistant isolates (35/43; 81%) were E. faecium. Our results are in agreement with Danish studies, where the prevalence of tcrB gene was highest among E. faecium swine isolates; however, the tcrB was also detected in E. gallinarum, E. casseliflavus, and E. mundtii (19, 21). Similar results have been reported from a recent Australian study, in which the highest prevalence of tcrB was observed among E. faecium and E. faecalis isolates; however, tcrB was also detected in E. gallinarum, E. casseliflavus, E. durans, and E. mundtii (33). Even among the randomly chosen tcrB-negative isolates (44 isolates) that were subjected to species identification, we did not detect species other than E. faecium and E. faecalis. Studies conducted in swine in the United States have generally reported E. cecorum, E. durans, E. faecalis, E. faecium, E. hirae, and E. malodoratus as the dominant species (14, 48). Differences in species distribution may be attributable to factors such as age, diet, use of in-feed antibiotics, isolation methods employed, and geographical location (24). Because samples in our study were collected only during the nursery stage, the predominant species may be representative of the population at that stage of production (24).
In piglets, copper is added to diets at concentrations above those physiologically required by the animal because of its growth-promoting effects (38). The exact mechanism of action of copper as a growth promoter has not been elucidated, but suggested mechanisms attributed to the antimicrobial effects of copper include altered gut microbial populations, increased availability of nutrients and energy because of the reduced microbial activity in the gut, and possibly the suppression of enteric bacterial pathogens (18, 23). In European countries, copper is supplemented in swine diets at elevated levels, often as a replacement for in-feed antibiotics, which have been banned for use as growth promotants since the mid-1990s (3, 21).
The higher prevalence of tcrB in enterococci from piglets fed an elevated level of copper compared to the control group (21.1 versus 2.8%) suggests the exertion of selection pressure for copper resistance. The presence of tcrB-positive isolates in the control group fed normal level of copper in the diet suggests the occurrence of naturally resistant isolates not favored by a high level of copper in the diet. Only one isolate (among five) of E. faecium in the control group was clonally identical to an isolate from the copper group, suggesting that the occurrence was not because of the transmission of tcrB-positive strains from the copper-treated group. We obtained three enterococcal isolates per sample (nine per pen), which allowed us to screen 45 isolates per treatment group and sampling day. Although the presence of tcrB-positive isolates was detected only after the initiation of copper supplementation, we did not find a linear increase in the prevalence with continued supplementation of an elevated level of copper. In fact, the highest prevalence was on day 14 of copper supplementation. The PFGE analysis of tcrB-positive isolates revealed that there was no dominant or persistent PFGE type in the copper-supplemented group. Almost all isolates obtained on different sampling days (14 days apart) belonged to different PFGE types. Based on an in vivo pig challenge study, Hasman et al. (21) reported that feeding elevated level of copper resulted in the selection of tcrB-mediated copper-resistant E. faecium. The in vivo challenge study was done with piglets fed low-copper (6 ppm) or high-copper diets (175 ppm), which were orally inoculated with tcrB-positive (copper-resistant) and tcrB-negative (copper-sensitive) E. faecium strains. The fecal prevalence of tcrB-positive E. faecium was significantly higher in the copper-supplemented group (175 ppm copper) than in the control (6 ppm copper).
Hasman and Aarestrup (19) have reported on the isolation of a large plasmid (∼175 kb) from a copper-resistant E. faecium isolate from a pig in Denmark. The plasmid was transferable and carried genes for copper, macrolide, and glycopeptide resistance. Because the isolation of a large plasmid with a low copy number from a Gram-positive bacterium is difficult, we used S1 nuclease to facilitate the detection of the large plasmid DNA (8). The S1 nuclease attacks only a single-stranded DNA (50, 41), leading to the conversion of supercoiled plasmid DNA into a full-length linear molecule (8). The S1 nuclease digestion of genomic DNA of three randomly chosen tcrB-positive strains of E. faecium or E. faecalis yielded bands of approximately 194-kb size to which both tcrB and erm(B) gene probes hybridized, suggesting their colocation on the same plasmid. The tcrB-negative strains of E. faecium or E. faecalis that we tested did not carry the 194-kbp plasmid but had plasmids that ranged from 150 to 170 kbp in size, which carried the erm(B) gene. The presence of the erm(B) gene on plasmids of different sizes have been reported (22, 25). In enterococci of human origin, vanA-type glycopeptide resistance and other antibiotic resistance determinants have been shown to be located on a plasmid of about 100 kb in size (49). The large plasmids of enterococci may play a major role in the horizontal transfer of resistance determinants in enterococci (30).
An interesting aspect of copper resistance in enterococci of piglets is the possible linkage to macrolide and glycopeptide resistance because of colocalization of tcrB and erm(B) genes on the same plasmid (19). Hasman et al. (21) have shown that elevated levels of copper fed to piglets coselected for erythromycin and vancomycin resistance in enterococcal isolates obtained in Denmark and Spain. More often, the antibiotic and heavy metal resistance genes are located on the same mobile genetic elements, such as plasmids, transposons, and integrons (5, 39). Therefore, there is a possibility that the natural selection pressure imposed by heavy metals in the feed may indirectly coselect for antibiotic resistance. We were unable to assess the coselection because in our study, both tcrB-positive and tcrB-negative isolates were phenotypically resistant to erythromycin (MIC > 100 μg/ml) and contained the erm(B) gene. The erm(B) gene is the most common determinant associated with enterococcal resistance to macrolides (25) and tylosin, an erythromycin derivative, is used in the feed of piglets in the United States to treat enteric infections and also for growth promotion (25). Not surprisingly, our tcrB-positive isolates did not carry the vanA gene and were susceptible to vancomycin (MIC=0.42 μg/ml). However, the vancomycin MIC values were higher for tcrB-positive isolates than for tcrB-negative isolates. The reason for the difference is not known: possibly a nonspecific shared resistance mechanism(s) imparted by the tcrB gene is likely responsible for the difference in susceptibility to vancomycin (7). However, the biological implication of the difference in susceptibility to vancomycin is not known. The prevalence of vanA in enterococcal isolates of piglets and chickens in Europe was likely because of the use of avoparcin, a glycopeptide, for growth promotion (6). The use of avoparcin was banned in Denmark in 1995 and in the European Union in 1997 (1, 3). The absence of glycopeptide resistance in swine enterococcal isolates in our study is likely because avoparcin or other related glycopeptide derivatives have never been used in the swine industry in the United States (12, 34). However, isolation of vancomycin-resistant E. faecium (VRE) containing vanA has recently been reported from swine in Michigan (15). The source of VRE acquisition was not determined in the study, but there was no association between the occurrence of VRE and antibiotic administration in the pigs.
Antibiotic-resistant enterococci are opportunistic and nosocomial pathogens in humans (35). Because of the widespread occurrence of resistant enterococci in animals, it has been suggested that these enterococci may serve as a reservoir of potential resistant genes capable of transferring from animal to human bacteria (28). The importance of enterococci is related to their propensity for conjugal transfer of resistance genes, mostly associated with the plasmids or transposons. In our previous study (4), we demonstrated the transferability of the tcrB gene between strains within the same species of enterococci. In the present study, we have demonstrated the interspecies transferability of the tcrB gene between E. faecium and E. faecalis. The efficient transfer of genes by conjugation indicates the presence of conjugative plasmids and or other mobile genetic elements (13, 40). The transconjugants possessing erm(B) and tcrB genes were indicative of their presence on a conjugative plasmid. The resistance determinants tet(M) and erm(B) are frequently carried on conjugative transposons Tn916 and Tn916/Tn1545 (10). These conjugative transposons have a broad bacterial host range and are capable of being transferred horizontally to a variety of Gram-positive and Gram-negative bacteria in the gut bacterial community (9). A critical issue is the coselection of metal and antibiotic resistance on the same genetic determinant, often on a plasmid or a transposon (37). These plasmids or transposons are of concern because they harbor antibiotic resistance genes and have the potential to spread between species (37). The existence of a metal-associated coselection mechanism could potentially be a major issue relative to public health, and further studies are needed to determine the magnitude of such an association. In conclusion, our longitudinal study showed a relationship between elevated levels of copper supplementation and the prevalence of copper-resistant enterococci in piglets. Also, we have confirmed the presence of the tcrB and erm(B) genes on a plasmid, suggesting potential transferability and coselection.
Footnotes
Published ahead of print on 24 June 2011.
Contribution 11-243-J from the Kansas Agricultural Experiment Station.
REFERENCES
- 1. Aarestrup F. M. 1995. Occurrence of glycopeptide resistance among Enterococcus faecium isolated from conventional and ecological poultry farms. Microb. Drug Resist. 1:255–257 [DOI] [PubMed] [Google Scholar]
- 2. Aarestrup F. M., et al. 2002. Antimicrobial resistance among enterococci from pigs in three European countries. Appl. Environ. Microbiol. 68:4127–4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aarestrup F. M., et al. 2001. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 45:2054–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amachawadi R. G., et al. 2010. Occurrence of tcrB, a transferable copper resistance gene, in fecal enterococci of swine. Foodborne Pathog. Dis. 7:1089–1097 [DOI] [PubMed] [Google Scholar]
- 5. Aminov R. I., Mackie R. I. 2007. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 271:147–161 [DOI] [PubMed] [Google Scholar]
- 6. Bager F., Madsen M., Christensen J., Aarestrup F. M. 1997. Avoparcin used as a growth promoter is associated with the occurrence of vancomycin resistant Enterococcus faecium on Danish poultry and pig farms. Prev. Vet. Med. 31:95–112 [DOI] [PubMed] [Google Scholar]
- 7. Baker-Austin C., Wright M. S., Stepanauskas R., McArthur J. V. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol. 14:176–182 [DOI] [PubMed] [Google Scholar]
- 8. Barton B. M., Harding G. P., Zuccarelli A. J. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235–240 [DOI] [PubMed] [Google Scholar]
- 9. Bertram J., Stratz M., Durre P. 1991. Natural transfer of conjugative transposon Tn916 between gram-positive and gram-negative bacteria. J. Bacteriol. 173:443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clewell D. B. 1995. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 3:229–236 [DOI] [PubMed] [Google Scholar]
- 11. Clinical Laboratory Standards Institute (CLSI). 2002. Performance standard for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. M31-A2. Approved Guideline-Second Edition. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 12. Coque T. M., Tomayko J. F., Ricke S. C., Okhyusen P. C., Murray B. E. 1996. Vancomycin-resistant enterococci from nosocomial, community, and animal sources in the United States. Antimicrob. Agents Chemother. 40:2605–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Devirgiliis C., Barile S., Caravelli A., Coppola D., Perozzi G. 2010. Identification of tetracycline and erythromycin resistant gram positive cocci within the fermenting microflora of an Italian dairy food product. J. Appl. Microbiol. 109:313–323 [DOI] [PubMed] [Google Scholar]
- 14. Devriese L. A., Hommez J., Pot B., Haesebrouck F. 1994. Identification and composition of the streptococcal and enterococcal flora of tonsils, intestines and feces of pigs. J. Appl. Bacteriol. 77:31–36 [DOI] [PubMed] [Google Scholar]
- 15. Donabedian S. M., et al. 2010. Characterization of vancomycin-resistant Enterococcus faecium isolated from swine in three Michigan counties. J. Clin. Microbiol. 48:4156–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Edmonds M. S., Izquiredo O. A., Baker D. H. 1985. Feed additive studies with newly weaned pigs: efficacy of supplemental copper, antibiotics, and organic acids. J. Anim. Sci. 60:462–469 [DOI] [PubMed] [Google Scholar]
- 17. Franke S., Grass G., Rensing C., Nies D. H. 2003. Molecular analysis of the copper transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 185:3804–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gould S. M. J., et al. 2009. The antimicrobial properties of copper surfaces against a range of important nosocomial pathogens. Ann. Microbiol. 59:151–156 [Google Scholar]
- 19. Hasman H., Aarestrup F. M. 2002. tcrB, a gene conferring transferable copper resistance in Enterococcus faecium: occurrence, transferability, and linkage to macrolide and glycopeptide resistance. Antimicrob. Agents Chemother. 46:1410–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hasman H., Franke S., Rensing C. 2006. Resistance to metals used in agricultural production, p. 99–114 In Aarestrup F. M. (ed.), Antimicrobial resistance in bacteria of animal origin, ASM Press, Washington, DC [Google Scholar]
- 21. Hasman H., et al. 2006. Copper resistance in Enterococcus faecium, mediated by the tcrB gene, is selected by supplementation of pig feed with copper sulfate. Appl. Environ. Microbiol. 72:5784–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haug M. C., Tanner S. A., Lacroix C., Meile L., Stevens M. J. A. 2010. Construction and characterization of Enterococcus faecalis CG110/gfp/pRE25*, a tool for monitoring horizontal gene transfer in complex microbial ecosystems. FEMS Microbiol. Lett. 313:111–119 [DOI] [PubMed] [Google Scholar]
- 23. Højberg O., Canibe N., Poulsen H. D., Hedemann M. S., Jensen B. B. 2005. Influence of dietary zinc oxide and copper sulfate on the gastrointestinal ecosystem in newly weaned piglets. Appl. Environ. Microbiol. 71:2267–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jackson C. R., Fedorka-Cray P. J., Barret J. B. 2004. Use of a genus- and species-specific multiplex PCR for the identification of enterococci. J. Clin. Microbiol. 42:3558–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jackson C. R., Fedorka-Cray P. J., Barret J. B., Ladely S. C. 2004. Effects of tylosin use on erythromycin resistance in enterococci isolated from swine. Appl. Environ. Microbiol. 70:4205–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jacob M. E., et al. 2008. Effects of feeding wet corn distillers grains with solubles with or without monensin and tylosin on the prevalence and antimicrobial susceptibilities of fecal foodborne pathogenic and commensal bacteria in feedlot cattle. J. Anim. Sci. 86:1182–1190 [DOI] [PubMed] [Google Scholar]
- 27. Kariyama R., Mitsuhata R., Chow J. W., Clewell D. B., Kumon H. 2000. Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. J. Clin. Microbiol. 38:3092–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kojima A., et al. 2010. Classification and antimicrobial susceptibilities of Enterococcus species isolated from apparently healthy food-producing animals in Japan. Zoonoses Public Health 57:137–141 [DOI] [PubMed] [Google Scholar]
- 29. Krupanidhi S., Sreekumar A., Sanjeevi C. B. 2008. Copper and biological health. Indian J. Med. Res. 128:448–461 [PubMed] [Google Scholar]
- 30. Leavis H. L., et al. 2007. Insertion sequence driven diversification creates a globally dispersed emerging multiresistant subspecies of Enterococcus faecium. PLoS Pathog. 3:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lim C. K., Cooksey D. A. 1993. Characterization of chromosomal homologs of the plasmid-borne copper resistance operon of Pseudomonas syringae. J. Bacteriol. 175:4492–4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Macomber L., Imlay J. A. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U. S. A. 106:8344–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mazaheri R., Heuzenroeder M. W., Barton M. D. 2010. Antimicrobial and heavy metal resistance in commensal enterococci isolated from pigs. Vet. Microbiol. doi: 10.1016/j.vetmic.2010.09.002 [DOI] [PubMed] [Google Scholar]
- 34. McDonald C. L., Kuehnert M. J., Tenover F. C., Jarvis W. R. 1997. Vancomycin-resistant enterococci outside the health care setting: prevalence, sources, and public health. Emerg. Infect. Dis. 3:311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Murray B. E. 2000. Vancomycin resistant enterococcal infections. N. Engl. J. Med. 342:710–721 [DOI] [PubMed] [Google Scholar]
- 36. Murray B. E., Singh K. V., Heath J. D., Sharma B. R., Weinstock G. M. 1990. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent cutting sites. J. Clin. Microbiol. 28:2059–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nandi S., Maurer J. J., Hofacre C., Summers A. O. 2004. Gram-positive bacteria are a major reservoir of class 1 antibiotic resistance integrons in poultry litter. Proc. Natl. Acad. Sci. U. S. A. 101:7118–7122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. National Research Council. 1998. Nutrient requirements of swine, 10th ed, p. 47–70 National Academy Press, Washington, DC [Google Scholar]
- 39. Nemergut D. R., Martin A. P., Schmidt S. K. 2004. Integron diversity in heavy metal contaminated mine tailings and inferences about integron evolution. Appl. Environ. Microbiol. 70:1160–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pembroke J. T., MacMahon C., McGrath B. 2002. The role of conjugative transposons in the Enterobacteriaceae. Cell. Mol. Life Sci. 59:2055–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pimkin M., et al. 2006. Characterization of a periplasmic S1-like nuclease coded by the Mesorhizobium loti symbiosis island. Biochem. Biophys. Res. Commun. 343:77–84 [DOI] [PubMed] [Google Scholar]
- 42. Poyart C., Quesnes G., Trieu-Cuot P. 2000. Sequencing the gene encoding manganese-dependent superoxide dismutase for rapid species identification of enterococci. J. Clin. Microbiol. 38:415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ridge P. G., Zhang Y., Gladyshev V. N. 2008. Comparative genomic analyses of copper transporters and cuproproteomes reveal evolutionary dynamics of copper utilization and its link to oxygen. PLoS One 3:e1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rosvoll T. C., et al. 2010. PCR-based plasmid typing in Enterococcus faecium strains reveals widely distributed pRE25-, pRUM-, pIP501-, and pHTbeta-related replicons associated with glycopeptides resistance and stabilizing toxin-antitoxin systems. FEMS Immunol. Med. Microbiol. 58:254–268 [DOI] [PubMed] [Google Scholar]
- 45. Solioz M., Abicht H. K., Mermod M. 2010. Response of gram-positive bacteria to copper stress. J. Biol. Inorg. Chem. 15:3–14 [DOI] [PubMed] [Google Scholar]
- 46. Solioz M., Stoyanov J. V. 2003. Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 27:183–195 [DOI] [PubMed] [Google Scholar]
- 47. Tendolkar P. M., Baghdayan A. S., Shankar N. 2006. Putative surface proteins encoded within a novel transferable locus confer a high-biofilm phenotype to Enterococcus faecalis. J. Bacteriol. 188:2063–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thal L. A., et al. 1995. Characterization of antimicrobial resistance in enterococci of animal origin. Antimicrob. Agents Chemother. 40:2112–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Werner G., Klare I., Witte W. 1999. Large conjugative vanA plasmids in vancomycin-resistant Enterococcus faecium. J. Clin. Microbiol. 37:2383–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wiegand R. C., Godson G. N., Radding C. M. 1975. Specificity of S1 nuclease from Asperigillus oryzae. J. Biol. Chem. 250:8848–8855 [PubMed] [Google Scholar]