Abstract
Phytosanitary regulations and the provision of plant health certificates still rely mainly on long and laborious culture-based methods of diagnosis, which are frequently inconclusive. DNA-based methods of detection can circumvent many of the limitations of currently used screening methods, allowing a fast and accurate monitoring of samples. The genus Xanthomonas includes 13 phytopathogenic quarantine organisms for which improved methods of diagnosis are needed. In this work, we propose 21 new Xanthomonas-specific molecular markers, within loci coding for Xanthomonas-specific protein domains, useful for DNA-based methods of identification of xanthomonads. The specificity of these markers was assessed by a dot blot hybridization array using 23 non-Xanthomonas species, mostly soil dwelling and/or phytopathogens for the same host plants. In addition, the validation of these markers on 15 Xanthomonas spp. suggested species-specific hybridization patterns, which allowed discrimination among the different Xanthomonas species. Having in mind that DNA-based methods of diagnosis are particularly hampered for unsequenced species, namely, Xanthomonas fragariae, Xanthomonas axonopodis pv. phaseoli, and Xanthomonas fuscans subsp. fuscans, for which comparative genomics tools to search for DNA signatures are not yet applicable, emphasis was given to the selection of informative markers able to identify X. fragariae, X. axonopodis pv. phaseoli, and X. fuscans subsp. fuscans strains. In order to avoid inconsistencies due to operator-dependent interpretation of dot blot data, an image-processing algorithm was developed to analyze automatically the dot blot patterns. Ultimately, the proposed markers and the dot blot platform, coupled with automatic data analyses, have the potential to foster a thorough monitoring of phytopathogenic xanthomonads.
INTRODUCTION
Xanthomonas is a genus of Gammaproteobacteria that includes numerous phytopathogenic species, each characterized by a narrow host range. However, as a whole, the genus members are able to infect a broad range of plants, distributed over 124 monocotyledonous and 268 dicotyledonous plant species (15). The nomenclature of this complex genus is still under debate, and the taxonomic rank of many previously described pathovars has been revised (28, 41, 48). At the moment, the European and Mediterranean Plant Protection Organization (EPPO) recommends that 13 members of the genus Xanthomonas be considered quarantine pests. Therefore, reliable, fast, and technically and commercially accessible screening methods of detection and identification are needed to allow the survey of a large number of samples. This would ensure the phytosanitary certification of plants, prevent the spread of contaminated plant material, and facilitate the implementation of timely phytosanitation and quarantine measures (4).
The current certified methods of bacterial detection rely mainly on culture-based approaches and plant bioassays (35). While these methods allow for a presumptive identification, they lack resolution of detection to the species or pathovar level, are often exceedingly time-consuming and costly for routine usage in quarantine procedures, or require specific biocontainment facilities, such as greenhouses or growth chambers (17). To circumvent these limitations, molecularly based detection methodologies have been proposed as more accurate and efficient alternatives. Particularly, DNA-based detection methods, some of which have already been validated in ring tests, have had their potential acknowledged for application in routine surveys (18, 25, 43).
The selection of DNA signatures, i.e., taxon-specific markers with discriminatory resolution for the target organism(s), and the optimization of a sensitive and suitable detection technique (PCR or hybridization based) are key premises for the development of a specific and reliable DNA-based method of bacterial detection and identification. For identification of xanthomonads, the selection of DNA signatures has been made mostly within specific regions of functional genes (7, 11, 14, 27). However, apart from the low number of markers provided by these approaches, these loci are frequently characterized by a low infrageneric resolution. Furthermore, the identification of other genes coding for specific functional traits is dependent on a comprehensive knowledge of bacterial metabolism, including the bacterium-specific infection mechanisms, such as virulence factors. This extensive knowledge, required to search for new markers, is poor or missing for most phytopathogenic bacteria. Other approaches for selection of DNA signatures have been described for Xanthomonas species, based on random specific regions discovered through repetitive sequence-based PCR (rep-PCR) (23), randomly amplified polymorphic DNA (RAPD)-PCR (12, 21), or subtractive hybridization (13, 34). Even though such approaches potentially allow the design of primers and probes for poorly characterized organisms, they require an extensive and laborious specificity validation (16, 44). Furthermore, the number of specific markers obtained with such approaches is low, and their genomic stability or intraspecific variability is generally not known (20).
Presently, the more than 1,200 fully sequenced bacterial genomes and the overall genomic information available in public databases allow access to a large spectrum of bacterial taxa and genomic information facilitating the selection of DNA signatures to any sequenced target bacteria, using the increasingly resourceful bioinformatics applications (2). However, these workflows are dependent on comparative genomics and thus are mainly restricted to fully sequenced organisms, which undermines their utility concerning unsequenced bacterial species. Therefore, new strategies are required to select markers for unsequenced phytopathogenic species. To date, most of the EPPO-recommended quarantine Xanthomonas species do not have a fully sequenced representative, among which are Xanthomonas fragariae, Xanthomonas axonopodis pv. phaseoli, and Xanthomonas fuscans subsp. fuscans, phytopathogens responsible for considerable losses in the agricultural production of strawberry and bean plants. X. fuscans subsp. fuscans strains are responsible for disease symptoms on bean plants identical to the common bacterial blight caused by X. axonopodis pv. phaseoli (24) and, until recently, were considered to be a variety of X. axonopodis pv. phaseoli (X. axonopodis pv. phaseoli variant fuscans). Although both the EPPO and the International Seed Testing Association (ISTA) still do not take into account this updated nomenclature (10, 35), the work of Schaad et al. (33) and subsequent research (1, 28) have helped to establish the taxonomic distinctiveness of X. fuscans subsp. fuscans.
The use of disease-free propagating material is considered the best control method for these phytopathogens, as chemical treatment of infected plants and use of resistant cultivars are considered secondary disease management procedures (35), which emphasizes the importance of developing rapid and effective detection methods. For X. fragariae, a few loci were identified as suitable for the design of species-specific primers: three RAPD-specific regions (29) and within the hrp (31) and gyrB (45) genes. Further research has mainly been focused in technological improvements of PCR-based detection methods, using the mentioned DNA regions (36, 39, 40, 49). In the case of X. axonopodis pv. phaseoli and X. fuscans subsp. fuscans, DNA-based detection methods are limited to conventional PCR-based methodologies (5, 38), with the classical methods remaining the standard detection procedures.
In this work, a comprehensive screening of Xanthomonas-specific molecular markers was validated by PCR and dot blot hybridization, in order to select and validate markers for the unsequenced xanthomonads X. fragariae, X. axonopodis pv. phaseoli, and X. fuscans subsp. fuscans. In addition, a dot blot platform coupled with automatic image analysis software was optimized to allow the fast detection of these bacteria over a large number of isolates.
MATERIALS AND METHODS
Selection of Xanthomonas-specific protein domains.
Identification of Xanthomonas-specific protein domains was carried out using the “Compare Genomes” feature of the Pfam online database (release 22.0) (9), as previously described (42). Six fully sequenced Xanthomonas strains and 16 soil-dwelling or phytopathogenic strains were compared: Xanthomonas axonopodis pv. citri strain 306, Xanthomonas campestris pv. campestris strain 8004, X. campestris pv. campestris strain ATCC 33913, X. campestris pv. vesicatoria strain 85-10, Xanthomonas oryzae pv. oryzae KACC 10331, Xanthomonas oryzae pv. oryzae MAFF 311018, Agrobacterium tumefaciens Cereon, Aster yellows witches' broom phytoplasma, Chromobacterium violaceum, Frankia sp., Nocardia farcinica, Pseudomonas fluorescens Pf-5, Pseudomonas fluorescens PfO-1, Pseudomonas putida, Pseudomonas syringae pv. phaseolicola, Pseudomonas syringae pv. syringae, Pseudomonas syringae pv. tomato, Ralstonia solanacearum, Rhizobium etli, Rhizobium loti, Rhizobium meliloti, and Xylella fastidiosa 9a5c. The Pfam analysis allowed filtering out the protein domains that were not exclusive to Xanthomonas, and the nucleotide sequences coding for the remaining proteins' domains (a total of 48), i.e., xanthomonad specific, were retrieved. These sequences were checked for specificity using the BLAST (blastn) utility (3), and 21 were further chosen for experimental validation (Table 1).
Table 1.
Xanthomonas-specific protein domains selected for molecular marker designa
| Protein domain | Locus | No. of domains (no. of proteins) inb: |
Corresponding marker | |||||
|---|---|---|---|---|---|---|---|---|
| X. axonopodis pv. citri strain 306 |
X. campestris pv. campestris |
X. campestris pv. vesicatoria strain 85-10 |
X. oryzae pv. oryzae |
|||||
| Strain 8004 | ATCC 33913 | KACC 10331 | MAFF 311018 | |||||
| NAGLU | XAC0709 | 1 (1) | 0 | 0 | 0 | 1 (1) | 2 (2) | XA1 |
| Peptidase_M35 | XAC2763 | 1 (1) | 0 | 0 | 1 (1) | 0 | 0 | XA2 |
| DUF239 | XAC3314 | 1 (1) | 0 | 0 | 1 (1) | 0 | 0 | XA3 |
| TFR_dimer | XAC3611 | 1 (1) | 0 | 0 | 0 | 0 | 0 | XA4 |
| Sigma70_ECF | XAC4128 | 1 (1) | 0 | 0 | 1 (1) | 0 | 0 | XA5 |
| DUF938 | XC_0087 | 1 (1) | 1 (1) | 1 (1) | 1 (1) | 1 (1) | 0 | XC1 |
| DUF1105 | XC_0608 | 1 (1) | 1 (1) | 1 (1) | 1 (1) | 0 | 0 | XC2 |
| Glyco_hydro_12 | XC_0783 | 1 (1) | 1 (1) | 1 (1) | 1 (1) | 1 (1) | 1 (1) | XC3 |
| DUF819 | XC_1392 | 1 (1) | 1 (1) | 1 (1) | 1 (1) | 0 | 0 | XC4 |
| CelD_N | XC_1727 | 1 (1) | 1 (1) | 1 (1) | 1 (1) | 0 | 0 | XC5 |
| Avidin | XC_2088 | 0 | 1 (1) | 0 | 0 | 0 | 1 (1) | XC6 |
| DUF1130 | XC_2584 | 1 (1) | 1 (1) | 1 (1) | 1 (1) | 0 | 0 | XC7 |
| 3-HAO | XC_2679 | 1 (1) | 1 (1) | 1 (1) | 1 (1) | 1 (1) | 1 (1) | XC8 |
| PLA1 | XC_2818 | 1 (1) | 1 (1) | 1 (1) | 1 (1) | 1 (1) | 1 (1) | XC9 |
| Peptidase_M2 | XC_3130 | 1 (1) | 1 (1) | 1 (1) | 1 (1) | 1 (1) | 1 (1) | XC10 |
| Glyco_hydro_39 | XC_4065 | 1 (1) | 1 (1) | 1 (1) | 0 | 0 | 0 | XC11 |
| Glyco_hydro_67C | XC_4193 | 1 (1) | 1 (1) | 1 (1) | 1 (1) | 1 (1) | 1 (1) | XC12 |
| LEA_4 | XOO_0116 | 0 | 0 | 0 | 0 | 1 (1) | 1 (1) | XO1 |
| NTase_sub_bind | XOO_3261 | 0 | 0 | 0 | 0 | 1 (1) | 1 (1) | XO2 |
| CBM_6 | XOO_3566 | 1 (1) | 0 | 0 | 1 (1) | 1 (1) | 1 (1) | XO3 |
| BsuBI_PstI_RE | XOO_3728 | 0 | 0 | 0 | 0 | 0 | 1 (1) | XO4 |
The corresponding gene identifier (locus) and distribution of each domain, across the six analyzed Xanthomonas proteomes, are shown.
The numbers shown represent the number of domains, with the number of proteins in which the domain is present shown in parentheses.
Bacterial strains, culture conditions, and DNA extraction.
The bacterial strains used in this study are listed in Table S1 in the supplemental material. Xanthomonas spp. and Stenotrophomonas maltophilia were cultured in YGC medium (glucose, 10 g liter−1; yeast extract, 5 g liter−1; CaCO3, 30 g liter−1; agar, 15 g liter−1) at 28°C, except for X. fragariae, which was cultured in YPGA medium (yeast extract, 5 g liter−1; Bacto peptone, 5 g liter−1; glucose, 10 g liter−1; agar, 15 g liter−1) at 20°C. Non-Xanthomonas strains were cultured in nutrient agar (beef extract, 1 g liter−1; yeast extract, 2 g liter−1; peptone, 5 g liter−1; NaCl, 5 g liter−1; KH2PO4, 0.45 g liter−1; Na2HPO4 ·12H2O, 2.39 g liter−1; agar, 15 g liter−1), except for Xylella fastidiosa, which was cultured in BCYE medium (46). DNA was extracted from pure bacterial cultures using the EZNA bacterial DNA purification kit (Omega Bio-Tek, Norcross, GA), following the manufacturer's instructions, and quantified by NanoDrop (Thermo Scientific, Wilmington, DE). Escherichia coli was cultured on Luria-Bertani medium at 37°C. Standard E. coli manipulation and in vitro DNA manipulations were carried out as described by Sambrook and Russell (32).
Primers and PCR validation.
Primer pairs (see Table S2 in the supplemental material) were designed for each of the 21 selected loci, using the Vector NTI software (Invitrogen, Carlsbad, CA). In order to allow PCR assays to be performed using identical reaction conditions, all primer pairs were chosen in order to have a predicted amplicon size of 150 to 350 bp and a calculated optimal annealing temperature of around 60°C. Primer pairs were designed having as the template the sequence of X. axonopodis pv. citri strain 306 for primers XA1F/R to XA5F/R; Xanthomonas campestris pv. campestris strain 8004 for primers XC1F/R to XC12F/R, and Xanthomonas oryzae pv. oryzae MAFF 311018 for primers XO1F/R to XO4F/R (see Table S2 in the supplemental material). Moreover, based on a BLAST analysis of all predicted amplicons, primers were designed to anneal to the sites of each locus that showed higher specificity toward Xanthomonas. Most of the predicted amplicons exhibited specificity to Xanthomonas, as shown by the high E values obtained with non-Xanthomonas strains. Markers XC1, XC2, XC4, XC8, XC9, and XC10 revealed similarity with the member of the Xanthomonadaceae S. maltophilia, and the lowest E value was obtained for marker XC9 (E-value, 2e−51). Concerning marker XC12, the best BLAST hit for non-Xanthomonas was with the Brassicaceae pathogen Hyaloperonospora parasitica (E value, 8e−71) (see Table S2 in the supplemental material).
Three different annealing temperatures were tested (57, 59, and 61°C) in order to optimize the PCR conditions for each primer pair. The PCR mastermix contained 1× reaction buffer IV (ABgene, Epsom, United Kingdom), 0.2 mM each deoxynucleoside triphosphate (dNTP; Fermentas, Ontario, Canada), 1.5 mM MgCl2, 0.2 μM each primer, and 1 U of Simple Red DNA polymerase (ABgene). Twenty-five nanograms of genomic DNA was used as the template, and the PCR conditions were as follows: an initial denaturation step of 5 min at 95°C, followed by 35 cycles of 30 s at 95°C, 30 s at 57, 59, or 61°C, and 30 s at 72°C, with a final extension step of 10 min at 72°C. Amplicons were extracted and purified from agarose gels stained with ethidium bromide (Bio-Rad, Hercules, CA), using the GFX PCR and gel band purification kit (GE Healthcare, Buckinghamshire, United Kingdom). Purified amplicons were cloned in pGEM-T Easy vector (Promega, Madison, WI), according to the manufacturer's instructions, and their identity was confirmed by sequencing (STAB Genomica, Portugal).
Dot blot hybridization assays.
For dot blot assays, 100 ng of heat-denatured DNA from each bacteria was spotted into a nylon membrane using a Bio-Dot apparatus (Bio-Rad). DNA probes were obtained from purified PCR amplicons labeled with digoxigenin (DIG), using the DIG-High Prime labeling kit (Roche, Basel, Switzerland) and following the manufacturer's instructions. Hybridization was carried out overnight at 68°C, with a final probe concentration of 100 ng ml−1. Washing and detection steps were carried out according to the DIG system recommendations (Roche). DIG-labeled nucleic acids were detected by chemiluminescence using X-ray films (GE Healthcare) or a Molecular Imager ChemiDoc system (Bio-Rad).
Dot blot analysis using an image-processing algorithm.
In order to ensure an unbiased and automated analysis of the dot blot assays, an algorithm was developed to process the images obtained (22). Briefly, this MATLAB-based algorithm, available upon request, allows the automated rotation of the obtained dot blot images and adjustment of all dots to a user-defined grid. The software then calculates the probability of each dot being a positive (ON) result, using as references the positive and negative controls present in each membrane (6a). To achieve a proper quantification of signal intensities and ON probability, the exposure time of the Chemidoc system was adjusted so that all dots were below pixel saturation. Each probability value was calculated based on the analysis of four independent dots. By doing so, the variation of dot intensities due to different membrane positioning and/or to different hybridization assays was taken into account.
Nucleotide sequence accession number.
DNA sequences have been deposited in the NCBI database under accession no. HQ315628 to HQ315642.
RESULTS
Selection of Xanthomonas-specific protein domains.
The “Compare Genomes” feature of Pfam was used to directly compare all the protein domains from the deduced proteomes of the selected microorganisms. On average, 1,700 different protein domains are present in each Xanthomonas proteome, with around 1,500 domains being shared by the six fully sequenced strains considered in this study. All of the unspecific domains, present in at least one of the 16 nonxanthomonads were filtered out, leaving 48 protein domains exclusive to at least one of the six Xanthomonas species used for this in silico screening. After the Pfam comparison, the nucleotide coding region for each specific protein domain region was retrieved, and a BLAST analysis was carried out, aiming to widen the specificity assessment to the full universe of genomic sequences deposited in the NCBI database. Twenty-one out of the 48 loci, corresponding to the xanthomonad-specific protein domains, showed low similarity toward any other genus and were selected for PCR and hybridization validation. The occurrence of the selected loci was not uniform among Xanthomonas strains, ranging from domains present in only one strain (TFR_dimer and Bsu_PstI_RE) to domains present in all strains (Glyco_hydro_12, 3-HAO, PLA1, Peptidase_M2, and Glyco_hydro_67C) (Table 1).
PCR validation of markers with the nonsequenced X. fragariae and X. axonopodis pv. phaseoli.
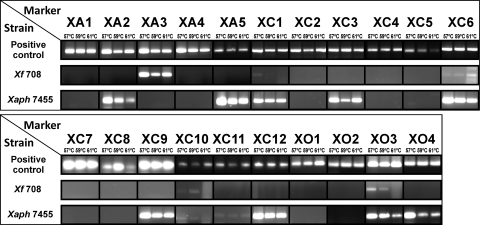
PCRs were initially performed with DNA extracted from Xanthomonas strains corresponding to the three template genome sequences used for primer design. As expected, positive amplification was obtained for markers XA1 to XA5 with DNA from X. axonopodis pv. citri LMG 9322, markers XC1 to XC12 with X. campestris pv. campestris LMG 568, and markers XO1 to XO4 with Xanthomonas oryzae pv. oryzae LMG 5047 (Fig. 1). Moreover, amplification was obtained for the three annealing temperatures assayed, and the amplicons' identity was confirmed by sequencing.
Fig. 1.
PCR analysis results using the selected primer pairs. The template DNA used as positive control was Xanthomonas axonopodis pv. citri LMG 9322 for markers XA1 to XA5, Xanthomonas campestris pv. campestris LMG 568 for markers XC1 to XC12, and Xanthomonas oryzae pv. oryzae LMG 5047 for markers XO1 to XO4. Xf 708, X. fragariae LMG 708; Xaph 7455, X. axonopodis pv. phaseoli LMG 7455.
All 21 primer pairs were then evaluated with DNA from the pathovar reference strain of X. axonopodis pv. phaseoli (LMG 7455) and type strain of X. fragariae (LMG 708), using the same PCR conditions. The results showed that for X. fragariae LMG 708, positive amplification was only consistently achieved with markers XA3, XC6, XC10, and XO3, along with a faint amplification with marker XC1. However, for X. axonopodis pv. phaseoli LMG 7455, a larger number of markers (XA2, XA5, XC1, XC3, XC6, XC9, XC10, XC11, XC12, XO3, and XO4) provided consistent positive amplification under the tested PCR conditions (Fig. 1). The PCR amplicons were sequenced, and high similarity was verified with the template Xanthomonas strains used for primer design (query coverage higher than 98% and E value lower than 1e−60), therefore demonstrating the presence of the selected regions in X. fragariae and X. axonopodis pv. phaseoli.
Dot blot specificity analysis.
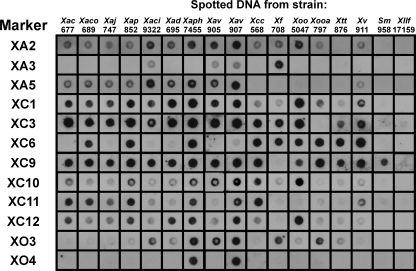
In order to access the specificity of the selected markers, 13 quarantine Xanthomonas strains and 23 non-Xanthomonas strains were analyzed by dot blot hybridization. The PCR products, obtained using template DNA from X. fragariae and X. axonopodis pv. phaseoli, were labeled with digoxigenin and used as X. fragariae- and X. axonopodis pv. phaseoli-specific probes. The tested probes were XA3, XC6, XC10, and XO3 obtained from X. fragariae LMG 708 and XA2, XA5, XC1, XC3, XC9, XC11, XC12, and XO4 obtained from X. axonopodis pv. phaseoli LMG 7455.
Concerning the two target bacteria, seven probes provided positive hybridization signals with X. fragariae, while XA3 was the only marker negative for X. axonopodis pv. phaseoli (Fig. 2). Probes XA2 and XC3 provided positive hybridization signals with X. fragariae LMG 708, although no PCR amplification was observed.
Fig. 2.
Dot blot analysis of digoxigenin-labeled probes using DNA from a collection of phytopathogenic Xanthomonas strains and the closely related Xanthomonadaceae Stenotrophomonas maltophilia LMG 958 and Xylella fastidiosa LMG 17159. Strain abbreviations are defined in Table 2. Markers XA3, XC6, XC10, and XO3 were obtained with DNA template from X. fragariae LMG 708, and markers XA2, XA5, XC1, XC3, XC9, XC11, XC12, and XO4 were obtained with DNA template from Xanthomonas axonopodis pv. phaseoli LMG 7455.
When assessing 13 quarantine Xanthomonas strains, the hybridization results vary from markers present in all tested strains (XC1 and XC10) to markers present only in four strains (XO4). When the full array of 12 probes was considered, the strain-specific hybridization profile allowed us to distinguish the different Xanthomonas strains, with the exception of Xanthomonas arboricola pv. corylina LMG 689, X. arboricola pv. pruni LMG 852, Xanthomonas axonopodis pv. dieffenbachiae LMG 695, and Xanthomonas vesicatoria LMG 911, which shared the same hybridization pattern (Fig. 2).
To dismiss unspecific hybridization to other bacteria and further confirm the unequivocal specificity of the selected probes toward Xanthomonas, 23 nonxanthomonads, comprising other phytopathogens or bacteria with matching hosts or habitats, were assayed by dot blotting (data not shown). Results confirmed the probes' specificity for Xanthomonas, with exception of probe XC9, for which hybridization signals were obtained with the closely related member of the Xanthomonadaceae Stenotrophomonas maltophilia LMG 958, while for Xylella fastidiosa LMG 17159, another phytopathogenic member of the Xanthomonadaceae, no hybridization was observed (Fig. 2).
Identification of X. fragariae, X. axonopodis pv. phaseoli, and X. fuscans subsp. fuscans strains by dot blot hybridization with automatic analysis.
The exploratory dot blot used for marker validation enabled the choice of a combination of three markers (XA3, XA5, and XO4) that provided unique hybridization patterns for X. fragariae and X. axonopodis pv. phaseoli in comparison with the other 13 Xanthomonas species tested. Marker XC1, a broad-spectrum marker present in all tested Xanthomonas species, was also included in this validation as a horizontal marker for xanthomonads.
To avoid operator-dependent analyses of the dot blot results, a key limitation to implement this hybridization technique in routine phytodiagnostics assays, an image analysis algorithm was developed. Essentially, this algorithm allows the determination of the variation in dot intensities among experimental replicates and, therefore, allows us to evaluate the reliability of the hybridization patterns. Furthermore, as the algorithm outputs a probability value, with each dot being a positive hybridization signal based on the measured intensity of the pixels, it was possible to calculate the variation among the signals obtained for each strain and marker tested (6a, 22).
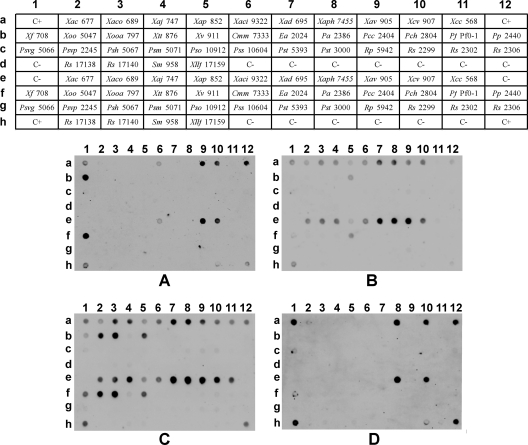
Dot blots using the four markers (XA3, XA5, XC1, and XO4) mentioned above as probes against template DNA of all the bacterial species used in this study confirmed the previous qualitative validation (Fig. 2), strengthening the consistency of the obtained patterns (Fig. 3). Indeed, the computed probability values obtained with the automatic analyses of the dot blots show a high consistency (Table 2). Furthermore, all of the non-Xanthomonas strains presented low probability values, including the Xanthomonadaceae S. maltophilia and Xylella fastidiosa, further emphasizing the specificity of these four markers and the software's reliability to quantify the signals. The results obtained for marker XC1 showed a negligible probability for Xanthomonas translucens pv. translucens LMG 876, which displayed a positive hybridization signal in the previous dot blot analysis (Fig. 2). This result is likely due to the low signal intensity obtained for this strain when chemiluminescence was acquired by a ChemiDoc system below the saturation point of all pixels.
Fig. 3.
Validation of dot blot hybridization patterns. Strain abbreviations are defined in Table 2. (A) Probe XA3; (B) probe XA5; (C) probe XC1; and (D) probe XO4. Tris-EDTA (TE) buffer was used for the negative controls (C−). A mixture of 6 ng of each purified PCR amplicon, corresponding to each of the digoxigenin-labeled probes, was used for the positive controls (C+). Probability values are detailed in Table 2.
Table 2.
Outputted probability values concerning the dot blot validation assays
| Strain no. | Strain name | Species abbreviation in Fig. 1–4 | Calculated ON probabilitya |
|||
|---|---|---|---|---|---|---|
| XA3 | XA5 | XC1 | XO4 | |||
| 1 | Xanthomonas arboricola pv. celebensis LMG 677 | Xac | 0.04 ± 0.05 | 0.86± 0.22 | 0.81± 0.22 | 0.1 ± 0.13 |
| 2 | Xanthomonas arboricola pv. corylina LMG 689 | Xaco | 0.86± 0.12 | 1± 0 | 1± 0.01 | 0 ± 0 |
| 3 | Xanthomonas arboricola pv. juglandis LMG 747 | Xaj | 0.01 ± 0.01 | 0.99± 0.02 | 0.97± 0.04 | 0.02 ± 0.03 |
| 4 | Xanthomonas arboricola pv. pruni LMG 852 | Xap | 0.01 ± 0.02 | 1± 0 | 1± 0 | 0 ± 0 |
| 5 | Xanthomonas axonopodis pv. citri LMG 9322 | Xaci | 0.01 ± 0.01 | 0.97± 0.06 | 0.92± 0.14 | 0.01 ± 0.01 |
| 6 | Xanthomonas axonopodis pv. diffenbachiae LMG 695 | Xad | 0.02 ± 0.03 | 0.88± 0.13 | 0.87± 0.13 | 0 ± 0 |
| 7 | Xanthomonas axonopodis pv. phaseoli LMG 7455 | Xaph | 0 ± 0 | 1± 0 | 1± 0 | 1± 0 |
| 24 | Xanthomonas axonopodis pv. vesicatoria LMG 905 | Xav | 1± 0 | 1± 0 | 1± 0 | 0 ± 0.01 |
| 25 | Xanthomonas campestris pv. vesicatoria LMG 907 | Xcv | 1± 0.01 | 1± 0 | 1± 0 | 0.99± 0.02 |
| 26 | Xanthomonas campestris pv. campestris LMG 568 | Xcc | 0.03 ± 0.03 | 0.04 ± 0.06 | 0.84± 0.23 | 0.01 ± 0.01 |
| 28 | Xanthomonas fragariae 708 | Xf | 1± 0 | 0.4 ± 0.37 | 0.8± 0.31 | 0.27 ± 0.34 |
| 54 | Xanthomonas oryzae pv. oryzae LMG 5047 | Xoo | 0.02 ± 0.01 | 0.08 ± 0.08 | 1± 0 | 0.05 ± 0.06 |
| 55 | Xanthomonas oryzae pv. oryzicola LMG 797 | Xooa | 0.01 ± 0.02 | 0.06 ± 0.07 | 1± 0 | 0.01 ± 0.01 |
| 56 | Xanthomonas translucens pv. translucens LMG 876 | Xtt | 0 ± 0.01 | 0.13 ± 0.17 | 0.14 ± 0.13 | 0 ± 0 |
| 57 | Xanthomonas vesicatoria LMG 911 | Xv | 0 ± 0.01 | 1± 0 | 0.98± 0.04 | 0 ± 0 |
| 58 | Clavibacter michiganensis subsp. michiganensis LMG 7333 | Cmm | 0 ± 0 | 0.02 ± 0.04 | 0.01 ± 0.01 | 0 ± 0 |
| 59 | Erwinia amylovora LMG 2024 | Ea | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.03 ± 0.02 | 0 ± 0 |
| 60 | Pectobacterium atrosepticum LMG 2386 | Pa | 0 ± 0 | 0 ± 0 | 0.01 ± 0.03 | 0 ± 0 |
| 61 | Pectobacterium carotovorum subsp. carotovorum LMG 2404 | Pcc | 0.01 ± 0.01 | 0.01 ± 0.02 | 0.07 ± 0.05 | 0 ± 0 |
| 62 | Pectobacterium chrysanthemi LMG 2804 | Pch | 0.02 ± 0.02 | 0.01 ± 0.01 | 0.06 ± 0.08 | 0.01 ± 0.01 |
| 63 | Pseudomonas fluorescens Pf0-1 | Pf | 0.03 ± 0.02 | 0.01 ± 0.02 | 0.02 ± 0.02 | 0 ± 0.01 |
| 64 | Pseudomonas putida KT 2440 | Pp | 0.07 ± 0.1 | 0 ± 0 | 0.02 ± 0.02 | 0.01 ± 0.03 |
| 65 | Pseudomonas savastanoi pv. glycinea LMG 5066 | Psvg | 0.01 ± 0.02 | 0.03 ± 0.05 | 0.23 ± 0.35 | 0.1 ± 0.19 |
| 66 | Pseudomonas savastanoi pv. phaseolicola LMG 2245 | Psvp | 0 ± 0.01 | 0.01 ± 0.02 | 0.07 ± 0.09 | 0 ± 0 |
| 67 | Pseudomonas syringae pv. helianthi LMG 5067 | Psh | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0 ± 0 |
| 68 | Pseudomonas syringae pv. maculicola LMG 5071 | Psm | 0 ± 0 | 0.03 ± 0.03 | 0.01 ± 0.01 | 0 ± 0 |
| 69 | Pseudomonas syringae pv. oryzae LMG 10912 | Pso | 0 ± 0 | 0.02 ± 0.04 | 0.05 ± 0.05 | 0 ± 0 |
| 70 | Pseudomonas syringae pv. syringae DSM 10604 | Pss | 0 ± 0 | 0.01 ± 0.02 | 0.01 ± 0.01 | 0 ± 0 |
| 71 | Pseudomonas syringae pv. tabaci LMG 5393 | Pstb | 0.04 ± 0.05 | 0.18 ± 0.2 | 0.09 ± 0.07 | 0.14 ± 0.14 |
| 72 | Pseudomonas syringae pv. tomato DC 3000 | Pst | 0.01 ± 0.01 | 0.02 ± 0.04 | 0 ± 0 | 0.03 ± 0.05 |
| 73 | Ralstonia picketii LMG 5942 | Rp | 0.04 ± 0.06 | 0.32 ± 0.1 | 0.13 ± 0.08 | 0 ± 0.01 |
| 74 | Ralstonia solanacearum LMG 2299 | Rs | 0.06 ± 0.1 | 0.06 ± 0.05 | 0.07 ± 0.09 | 0 ± 0 |
| 75 | Ralstonia solanacearum LMG 2302 | Rs | 0.05 ± 0.08 | 0.02 ± 0.04 | 0.01 ± 0.03 | 0 ± 0.01 |
| 76 | Ralstonia solanacearum LMG 2306 | Rs | 0.1 ± 0.15 | 0.01 ± 0.02 | 0.01 ± 0.01 | 0.04 ± 0.05 |
| 77 | Ralstonia solanacearum LMG 17138 | Rs | 0 ± 0.01 | 0.04 ± 0.08 | 0.01 ± 0.03 | 0 ± 0 |
| 78 | Ralstonia solanacearum LMG 17140 | Rs | 0.01 ± 0.02 | 0.01 ± 0.02 | 0.06 ± 0.05 | 0 ± 0 |
| 79 | Stenotrophomonas maltophilia LMG 958 | Sm | 0.01 ± 0.02 | 0.04 ± 0.06 | 0.06 ± 0.06 | 0 ± 0.01 |
| 80 | Xylella fastidiosa LMG 17159 | Xllf | 0 ± 0.01 | 0.02 ± 0.04 | 0.04 ± 0.03 | 0.03 ± 0.05 |
| 7–19b | Xanthomonas axonopodis pv. phaseoli | Xaph | (0.000-0.094) | (0.873-1.000) | (0.855-0.990) | (0.831-1.000) |
| 20 | Xanthomonas fuscans subsp. fuscans CPBF 507 | Xff | 0.01 ± 0.01 | 1± 0 | 0.95± 0.06 | 0 ± 0.01 |
| 21 | Xanthomonas fuscans subsp. fuscans CPBF 508 | Xff | 0 ± 0 | 1± 0.01 | 0.98± 0.04 | 0 ± 0 |
| 22 | Xanthomonas fuscans subsp. fuscans CPBF 509 | Xff | 0.01 ± 0.01 | 0.96± 0.07 | 0.96± 0.05 | 0 ± 0 |
| 23 | Xanthomonas fuscans subsp. fuscans CPBF 795 | Xff | 0 ± 0 | 0.99± 0.02 | 1± 0 | 0 ± 0 |
| 27–53b | X. fragariae | Xf | (1.000-1.000) | (0.026-0.088) | (0.450-0.630) | (0.006-0.018) |
The values shown represent the average probability ± standard deviation. The values considered as positive signals are highlighted in bold. The 99% confidence intervals for the mean probability values obtained with the Xanthomonas axonopolis pv. phaseoli (strains 7 to 19) and X. fragariae (strains 27 to 53) collections are displayed in parentheses and calculated according to the equation x̄ + z̄[1 − (α/2)](s/), where x= is the average probability value, z is the standard score, s is the standard deviation, and n is the number of replicates.
Strain-specific probability values are detailed in Table S3 in the supplemental material.
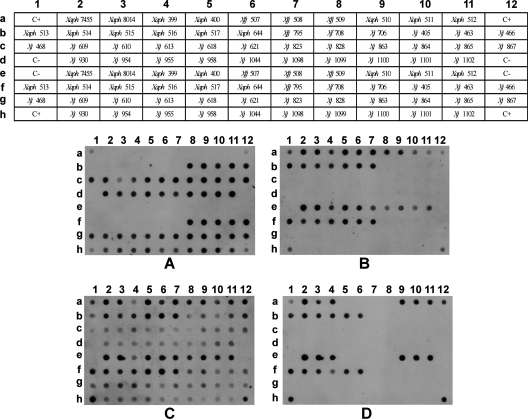
These markers were further validated on 27 X. fragariae strains, 13 X. axonopodis pv. phaseoli strains, and 4 X. fuscans subsp. fuscans strains (Fig. 4) to determine if the hybridization patterns were consistent between different strains of the target Xanthomonas species. All tested X. fragariae strains displayed maximum probability for marker XA3 (1 ± 0) and low probabilities for markers XA5 (≤0.18) and XO4 (≤0.04), while X. axonopodis pv. phaseoli strains had low probability for XA3 (≤0.21) and high probability for XA5 (≥0.75) (see Table S3 in the supplemental material). Furthermore, the standard deviation in the probability values was very low for all the strains tested using these probes. When analyzing the results for marker XO4, positive hybridization results were obtained for all of the tested X. axonopodis pv. phaseoli strains (≥0.99), with the exception of one strain—X. axonopodis pv. phaseoli CPBF 400 (see Table S3 in the supplemental material). Concerning the four X. fuscans subsp. fuscans strains, these presented low probability for XA3 (≤0.01) and high probability for XA5 (≥0.96), similarly to X. axonopodis pv. phaseoli strains. However, and unlike X. axonopodis pv. phaseoli strains, all X. fuscans subsp. fuscans strains presented very low values for XO4 (0.00). The genus-specific marker XC1 presented high probability values for all X. axonopodis pv. phaseoli strains (≥0.87, with the exception of X. axonopodis pv. phaseoli strain CPBF 399, which presents a probability of 0.41). Similarly, all tested X. fuscans subsp. fuscans strains present high probability values for this marker (≥0.95). The probability values obtained with marker XC1 for X. fragariae strains were unexpectedly highly variable (≥0.30 and ≤ 0.81), with the standard deviations of the results from different experiments being much higher than those obtained for any other probe (see Table S3 in the supplemental material). Although XC1 was considered a broad-spectrum marker, the fact that it was obtained from amplification of X. axonopodis pv. phaseoli LMG 7455, coupled with the use of high-stringency conditions of hybridization, might explain the lower consistency obtained for the different X. fragariae strains for this marker. Nevertheless, these values are undoubtedly higher than the values obtained for those considered negative signals (see Table S3 in the supplemental material).
Fig. 4.
Dot blot validation results with a collection of Xanthomonas fragariae, Xanthomonas axonopodis pv. phaseoli, and Xanthomonas fuscans subsp. fuscans strains. Strain abbreviations are defined in Table 2. (A) Probe XA3; (B) probe XA5; (C) probe XC1; and (D) probe XO4. TE buffer was used for the negative controls (C−). A mixture of 6 ng of each purified PCR amplicon, corresponding to each of the digoxigenin-labeled probes, was used for the positive controls (C+). Probability values are detailed in Table S3 in the supplemental material.
DISCUSSION
According to the recommended detection standards from EPPO and ISTA, the current methods for the detection of X. fragariae and X. axonopodis pv. phaseoli are primarily based on serological techniques using polyclonal antibodies and on culture on selective medium, while the DNA-based methods are still largely underrepresented (10, 25). The frequent cross-reactions of the X. fragariae polyclonal antibodies, as highlighted by an assessment study of diagnostics methods for quarantine organisms carried out by several laboratories across Europe, known as DIAGPRO (25), and the need to develop culture-independent diagnostic standards to hasten the detection of these phytopathogens, particularly of the fastidious organism X. fragariae, underlined the importance of specific and reliable DNA-based methods of detection able to provide fast confirmatory diagnostics for X. fragariae and X. axonopodis pv. phaseoli. In this work, we propose several novel detection markers for xanthomonads in general, and for X. fragariae, X. axonopodis pv. phaseoli, and X. fuscans subsp. fuscans in particular, in order to increase diagnostic reliability and contribute to development of single-step DNA-based and culture-independent confirmatory identification of these phytopathogens.
At the moment, the limited number of specific primers described for X. fragariae (29, 31, 45), for X. axonopodis pv. phaseoli by Audy et al. (5) and other works that followed based on the same set of primers (19, 26, 30, 37), and for X. fuscans subsp. fuscans (38) have been hampering the implementation of DNA-based detection protocols as trustworthy alternatives to isolation in pure culture. Furthermore, the unavailability of complete genome sequences for most quarantine phytopatogenic Xanthomonas strains does not allow the direct selection of target-specific DNA signatures using comparative genomics bioinformatics tools. Therefore, to identify novel DNA-specific markers able to detect X. fragariae, X. axonopodis pv. phaseoli, and X. fuscans subsp. fuscans isolates, which are unsequenced bacteria, an indirect in silico-based approach previously described by us was used (42). Essentially, we hypothesized that some of the Pfam protein domains, present exclusively in the sequenced Xanthomonas strains, would also be present in the unsequenced target bacteria X. fragariae, X. axonopodis pv. phaseoli, and X. fuscans subsp. fuscans. Furthermore, having in mind that this marker selection methodology takes into account only putative functional regions, we increased the likelihood of these protein domain-encoding loci to be conserved across different species of the same genus.
From the comparison of the proteomes of Xanthomonas strains with several nontarget bacteria that share hosts or habitats, 48 protein domains were filtered as specific for the genus. The follow-up BLAST analysis of the selected protein domains and their respective encoding regions (i.e., putative markers) enabled confirmation of the specificity of 21 protein domains, emphasizing the importance of including a BLAST analysis as a fine-tuning tool in any marker selection workflow (2). The distribution of the 21 selected protein domains among the sequenced Xanthomonas strains was not uniform, ranging from domains present in only one strain to domains common to all proteomes (Table 1). These data suggested the existence of a specific pattern of markers, determined by the presence or absence of each marker, for the different Xanthomonas species.
Using X. fragariae, X. axonopodis pv. phaseoli, and X. fuscans subsp. fuscans as the target bacteria, we analyzed the potential of the selected markers for detection of nonsequenced Xanthomonas. A preliminary PCR assay showed that eight markers were amplified with X. axonopodis pv. phaseoli LMG 7455, and one marker was amplified with X. fragariae LMG 708, while three markers were consistently amplified for both strains (Fig. 1). These 12 markers were used as probes in dot blot assays. Each probe provided positive hybridization with several Xanthomonas strains, and species-specific hybridization patterns were obtained, with the exception of four Xanthomonas strains that shared the same hybridization pattern (Fig. 2). Interestingly, although markers XA2 and XC3 gave positive hybridization for X. fragariae, the negative amplification obtained in the preliminary PCR assays for these markers (Fig. 1), suggests sequence mismatches at the primers' annealing sites, preventing amplification. These PCR false-negative results, which are predominantly frequent if the sequence differences are located in the 3′-primer region (47), are a favorable argument for the implementation of hybridization detection methods over PCR-based methods.
The specificity of the probes toward Xanthomonas was further strengthened by the fact that no hybridization signal was obtained with the 21 non-Xanthomonadacea tested. The results obtained in the above-mentioned validation assays allowed identification of a combination of three probes (XA3, XA5, and XO4) able to distinguish specifically X. fragariae, X. axonopodis pv. phaseoli, and X. fuscans subsp. fuscans from the other tested Xanthomonas species. In fact, while probe XA3 was shown to be specific for X. fragariae, probe XA5 hybridized to all X. axonopodis pv. phaseoli and X. fuscans subsp. fuscans strains tested, but not to X. fragariae. To distinguish X. axonopodis pv. phaseoli from X. fuscans subsp. fuscans, a xanthomonad species symptomatically indistinguishable from X. axonopodis pv. phaseoli in infected plants and up to recently considered a subspecies of X. axonopodis pv. phaseoli (8, 33), probe XO4 hybridized to X. axonopodis pv. phaseoli, with exception of X. axonopodis pv. phaseoli strain CPBF400, but not to the X. fuscans subsp. fuscans strains used in this study, allowing a presumptive discrimination of these two species. Probe XC1, chosen as a xanthomonad-specific marker, hybridized to all X. fragariae, X. axonopodis pv. phaseoli, and X. fuscans subsp. fuscans strains tested and not to the closely related Xanthomonadaceae S. maltophilia and Xylella fastidiosa, confirming its usefulness as a genus-positive control.
Although the dot blot validation of these probes confirmed the consistency and specificity of the obtained hybridization profiles toward numerous strains of X. fragariae, X. axonopodis pv. phaseoli, and X. fuscans subsp. fuscans, the ambiguity inherent in operator-dependent analysis of dot blots' hybridization data is still a major weakness to the implementation of macroarrays for microbial detection assays and likely a reason why PCR-based protocols are generally favored. In order to overcome this limitation, we developed and optimized an innovative automated image analysis algorithm to ensure the numerical analysis of dot blot data (6a, 22). By converting each hybridization signal into probability values, the software enables comparisons of data from different independent experiments, which allows us to validate the data statistically.
Overall, this work proposes 21 novel markers useful for the identification of Xanthomonas, particularly for those species in which the number of markers for DNA-based methods of detection is limited. The proposed detection dot blots might complement the established PCR methods that do not possess the throughput of dot blotting, by narrowing down the samples for confirmatory PCR-based detection. It is further shown that dot blots coupled with automatic data analysis are convenient platforms for fast and easy screening of dozens of isolates simultaneously, contrary to microarrays that only allow the assay of a single isolate at a time and are economically unsustainable for routine phytosanitary analysis (6, 18). Most importantly, while the complex microarray data sets require extended expertise to interpret the results, the image-processing software developed here allows a reliable and user-friendly analysis of dot blot hybridization data, which ultimately might facilitate the use of macroarrays by plant diagnostic laboratories.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the Fundação para a Ciência e Tecnologia—FCT (POCTI/AGG/39216/2001). Pedro Albuquerque and Cristina Caridade were supported by FCT fellowships SFRH/BD/37249/2007 and SFRH/BD/32150/2006, respectively. Marta V. Mendes was supported by the “Ciência 2007” FCT program, sponsored by the POPH(QREN) program subsidized by the European Social Fund and by the MCTES.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 24 June 2011.
REFERENCES
- 1. Alavi S. M., et al. 2008. Assessment of the genetic diversity of Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans as a basis to identify putative pathogenicity genes and a type III secretion system of the SPI-1 family by multiple suppression subtractive hybridizations. Appl. Environ. Microbiol. 74:3295–3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Albuquerque P., Mendes M. V., Santos C. L., Moradas-Ferreira P., Tavares F. 2009. DNA signature-based approaches for bacterial detection and identification. Sci. Total Environ. 407:3641–3651 [DOI] [PubMed] [Google Scholar]
- 3. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 4. Alvarez A. M. 2004. Integrated approaches for detection of plant pathogenic bacteria and diagnosis of bacterial diseases. Annu. Rev. Phytopathol. 42:339–366 [DOI] [PubMed] [Google Scholar]
- 5. Audy P., Laroche A., Saindon G., Huang H. C., Gilbertson R. L. 1994. Detection of the bean common blight bacteria, Xanthomonas campestris pv. phaseoli and X. c. phaseoli var fuscans, using the polymerase chain-reaction. Phytopathology 84:1185–1192 [Google Scholar]
- 6. Call D. R. 2005. Challenges and opportunities for pathogen detection using DNA microarrays. Crit. Rev. Microbiol. 31:91–99 [DOI] [PubMed] [Google Scholar]
- 6a. Caridade C., et al. 2010. Automatic analysis of macroarray images, p. 6122–6125 In Proceedings of the 32nd Annual International Conference of the IEEE EMBS-EMBC [DOI] [PubMed] [Google Scholar]
- 7. Cubero J., Graham J. H. 2002. Genetic relationship among worldwide strains of Xanthomonas causing canker in citrus species and design of new primers for their identification by PCR. Appl. Environ. Microbiol. 68:1257–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Darrasse A., Bureau C., Samson R., Morris C., Jacques M.-A. 2007. Contamination of bean seeds by Xanthomonas axonopodis pv. phaseoli associated with low bacterial densities in the phyllosphere under field and greenhouse conditions. Eur. J. Plant Pathol. 119:203–215 [Google Scholar]
- 9. Finn R. D., et al. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247–D251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. International Seed Testing Association, 2007. International rules for seed testing: annex to chapter 7. International Seed Testing Association. https://www.seedtest.org/upload/cms/user/7-021.pdf
- 11. Kang M. J., et al. 2008. Specific detection of Xanthomonas oryzae pv. oryzicola in infected rice plant by use of PCR assay targeting a membrane fusion protein gene. J. Microbiol. Biotechnol. 18:1492–1495 [PubMed] [Google Scholar]
- 12. Khoodoo M. H. R., Sahin F., Jaufeerally-Fakim Y. 2005. Sensitive detection of Xanthomonas axonopodis pv. dieffenbachiae on Anthurium andreanum by immunocapture-PCR (IC-PCR) using primers designed from sequence characterized amplified regions (SCAR) of the blight pathogen. Eur. J. Plant Pathol. 112:379–390 [Google Scholar]
- 13. Kuflu K. M., Cuppels D. A. 1997. Development of a diagnostic DNA probe for xanthomonads causing bacterial spot of peppers and tomatoes. Appl. Environ. Microbiol. 63:4462–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Leite R. P., Minsavage G. V., Bonas U., Stall R. E. 1994. Detection and identification of phytopathogenic Xanthomonas strains by amplifcation of DNA sequences related to the hrp genes of Xanthomonas campestris pv. vesicatoria. Appl. Environ. Microbiol. 60:1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leyns F., Decleene M., Swings J. G., Deley J. 1984. The host range of the genus Xanthomonas. Bot. Rev. 50:308–356 [Google Scholar]
- 16. Lievens B., Thomma B. 2005. Recent developments in pathogen detection arrays: implications for fungal plant pathogens and use in practice. Phytopathology 95:1374–1380 [DOI] [PubMed] [Google Scholar]
- 17. Lopez M. M., et al. 2003. Innovative tools for detection of plant pathogenic viruses and bacteria. Int. Microbiol. 6:233–243 [DOI] [PubMed] [Google Scholar]
- 18. Lopez M. M., et al. 2009. Are molecular tools solving the challenges posed by detection of plant pathogenic bacteria and viruses? Curr. Issues Mol. Biol. 11:13–45 [PubMed] [Google Scholar]
- 19. Lopez R., Asensio C., Gilbertson R. L. 2006. Phenotypic and genetic diversity in strains of common blight bacteria (Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscans) in a secondary center of diversity of the common bean host suggests multiple introduction events. Phytopathology 96:1204–1213 [DOI] [PubMed] [Google Scholar]
- 20. Louws F. J., Rademaker J. L. W., de Bruijn F. J. 1999. The three Ds of PCR-based genomic analysis of phytobacteria: diversity, detection, and disease diagnosis. Annu. Rev. Phytopathol. 37:81–125 [DOI] [PubMed] [Google Scholar]
- 21. Manulis S., Valinsky L., Lichter A., Gabriel D. W. 1994. Sensitive and specific detection of Xanthomonas campestris pv. pelargonii with DNA primers and probes identified by random amplified polymorphic DNA analysis. Appl. Environ. Microbiol. 60:4094–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marcal A. R. S., Caridade C. M. R., Albuquerque P., Mendes M. V., Tavares F. 2009. Automatic detection of molecular markers in digital images, p. 6710–6713 In Proceedings of the 31st Annual International Conference of the IEEE EMBS-EMBC [DOI] [PubMed] [Google Scholar]
- 23. Moretti C., Amatulli M. T., Buonaurio R. 2009. PCR-based assay for the detection of Xanthomonas euvesicatoria causing pepper and tomato bacterial spot. Lett. Appl. Microbiol. 49:466–471 [DOI] [PubMed] [Google Scholar]
- 24. Mutlu N., et al. 2008. Differential pathogenicity of Xanthomonas campestris pv. phaseoli and X. fuscans subsp fuscans strains on bean genotypes with common blight resistance. Plant Dis. 92:546–554 [DOI] [PubMed] [Google Scholar]
- 25. OEPP/EPPO 2006. Diagnostics: Xanthomonas fragariae. EPPO Bull. 36:135–144 [Google Scholar]
- 26. Osdaghi E., Alizadeh A., Shams-Bakhsh M., Law M. R. 2009. Evaluation of common bean lines for their reaction to the common bacterial blight pathogen. Phytopathol. Mediterr. 48:461–468 [Google Scholar]
- 27. Park D. S., et al. 2009. Sensitive and specific detection of Xanthomonas campestris pv. vesicatoria by PCR using pathovar-specific primers based on rhs family gene sequences. Microbiol. Res. 164:36–42 [DOI] [PubMed] [Google Scholar]
- 28. Parkinson N., et al. 2007. Phylogenetic analysis of Xanthomonas species by comparison of partial gyrase B gene sequences. Int. J. Syst. Evol. Microbiol. 57:2881–2887 [DOI] [PubMed] [Google Scholar]
- 29. Pooler M. R., Ritchie D. F., Hartung J. S. 1996. Genetic relationships among strains of Xanthomonas fragariae based on random amplified polymorphic DNA PCR, repetitive extragenic palindromic PCR, and enterobacterial repetitive intergenic consensus PCR data and generation of multiplexed PCR primers useful for the identification of this phytopathogen. Appl. Environ. Microbiol. 62:3121–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Popovic T., et al. 2010. Detection and identification of Xanthomonas axonopodis pv. phaseoli on bean seed collected in Serbia. Afr. J. Agric. Res. 5:2730–2736 [Google Scholar]
- 31. Roberts P. D., Jones J. B., Chandler C. K., Stall R. E., Berger R. D. 1996. Survival of Xanthomonas fragariae on strawberry in summer nurseries in Florida detected by specific primers and nested PCR. Plant Dis. 80:1283–1288 [Google Scholar]
- 32. Sambrook J., Russell D. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33. Schaad N. W., et al. 2005. Reclassification of Xanthomonas campestris pv. citri (ex Hasse 1915) Dye 1978 forms A, B/C/D, and E as X. smithii subsp citri (ex Hasse) sp nov nom. rev. comb. nov., X. fuscans subsp aurantifolii (ex Gabriel 1989) sp nov nom. rev. comb. nov., and X. alfalfae subsp citrumelo (ex Riker and Jones) Gabriel et al., 1989 sp nov nom. rev. comb. nov.; X. campestris pv. malvacearum (ex Smith 1901) Dye 1978 as X. smithii subsp smithii nov comb. nov nom. nov.; X. campestris pv. alfalfae (ex Riker and Jones, 1935) Dye 1978 as X. alfalfae subsp alfalfae (ex Riker et al.,. 1935 sp nov nom. rev.; and “var. fuscans” of X. campestris pv. phaseoli (ex Smith, 1987) Dye 1978 as X. fuscans subsp fuscans sp nov. Syst. Appl. Microbiol. 28:494–518 [DOI] [PubMed] [Google Scholar]
- 34. Shih H. D., Lin Y. C., Huang H. C., Tzeng K. C., Hsu S. T. 2000. A DNA probe for identification of Xanthomonas campestris pv. campestris, the causal organism of black rot of crucifers in Taiwan. Bot. Bull. Acad. Sin. (Taiwan) 41:113–120 [Google Scholar]
- 35. Smith I. M., McNamara D. G., Scott P. R., Holderness M. (ed.) 1997. Quarantine pests for Europe, 2nd ed. EPPO/CABI International, Wallingford, United Kingdom: http://www.eppo.org/QUARANTINE/listA2.htm [Google Scholar]
- 36. Stoger A., Ruppitsch W. 2004. A rapid and sensitive method for the detection of Xanthomonas fragariae, causal agent of angular leafspot disease in strawberry plants. J. Microbiol. Methods 58:281–284 [DOI] [PubMed] [Google Scholar]
- 37. Tebaldi N. D., et al. 2010. Detection of Xanthomonas axonopodis pv. phaseoli in bean seeds by flow cytometry, immunostaining and direct viable counting. Trop. Plant Pathol. 35:213–222 [Google Scholar]
- 38. Toth I. K., Hyman L. J., Taylor R., Birch P. R. J. 1998. PCR-based detection of Xanthomonas campestris pv. phaseoli var. fuscans in plant material and its differentiation from X. c. pv. phaseoli. J. Appl. Microbiol. 85:327–336 [Google Scholar]
- 39. Turechek W. W., Hartung J. S., McCallister J. 2008. Development and optimization of a real-time detection assay for Xanthomonas fragariae in strawberry crown tissue with receiver operating characteristic curve analysis. Phytopathology 98:359–368 [DOI] [PubMed] [Google Scholar]
- 40. Vandroemme J., Baeyen S., Van Vaerenbergh J., De Vos P., Maes M. 2008. Sensitive real-time PCR detection of Xanthomonas fragariae in strawberry plants. Plant Pathol. 57:438–444 [Google Scholar]
- 41. Vauterin L., Rademaker J., Swings J. 2000. Synopsis on the taxonomy of the genus Xanthomonas. Phytopathology 90:677–682 [DOI] [PubMed] [Google Scholar]
- 42. Vieira J., Mendes M. V., Albuquerque P., Moradas-Ferreira P., Tavares F. 2007. A novel approach for the identification of bacterial taxa-specific molecular markers. Lett. Appl. Microbiol. 44:506–512 [DOI] [PubMed] [Google Scholar]
- 43. Vincelli P., Tisserat N. 2008. Nucleic acid-based pathogen detection in applied plant pathology. Plant Dis. 92:660–669 [DOI] [PubMed] [Google Scholar]
- 44. Ward E., Foster S. J., Fraaije B. A., McCartney H. A. 2004. Plant pathogen diagnostics: immunological and nucleic acid-based approaches. Ann. Appl. Biol. 145:1–16 [Google Scholar]
- 45. Weller S. A., et al. 2007. Detection of Xanthomonas fragariae and presumptive detection of Xanthomonas arboricola pv. fragariae, from strawberry leaves, by real-time PCR. J. Microbiol. Methods 70:379–383 [DOI] [PubMed] [Google Scholar]
- 46. Wells J. M., Raju B. C., Nyland G., Lowe S. K. 1981. Medium for isolation and growth of bacteria associated with plum leaf scald and phony peach diseases. Appl. Environ. Microbiol. 42:357–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wilson I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Young J. M., Park D. C., Shearman H. M., Fargier E. 2008. A multilocus sequence analysis of the genus Xanthomonas. Syst. Appl. Microbiol. 31:366–377 [DOI] [PubMed] [Google Scholar]
- 49. Zimmermann C., Hinrichs-Berger J., Moltmann E., Buchenauer H. 2004. Nested PCR (PCR) for detection of Xanthomonas fragariae in symptomless strawberry plants. J. Plant Dis. Prot. 111:39–51 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.