Abstract
In the dairy cattle industry, Holstein and Jersey are the breeds most commonly used for production. They differ in performance by various traits, such as body size, milk production, and milk composition. With increased concerns about the impact of agriculture on climate change, potential differences in other traits, such as methane emission, also need to be characterized further. Since methane is produced in the rumen by methanogenic archaea, we investigated whether the population structure of methanogen communities would differ between Holsteins and Jerseys. Breed-specific rumen methanogen 16S rRNA gene clone libraries were constructed from pooled PCR products obtained from lactating Holstein and Jersey cows, generating 180 and 185 clones, respectively. The combined 365 sequences were assigned to 55 species-level operational taxonomic units (OTUs). Twenty OTUs, representing 85% of the combined library sequences, were common to both breeds, while 23 OTUs (36 sequences) were found only in the Holstein library and 12 OTUs (18 sequences) were found only in the Jersey library, highlighting increased diversity in the Holstein library. Other differences included the observation that sequences with species-like sequence identity to Methanobrevibacter millerae were represented more highly in the Jersey breed, while Methanosphaera-related sequences and novel uncultured methanogen clones were more frequent in the Holstein library. In contrast, OTU sequences with species-level sequence identity to Methanobrevibacter ruminantium were represented similarly in both libraries. Since the sampled animals were from a single herd consisting of two breeds which were fed the same diet and maintained under the same environmental conditions, the differences we observed may be due to differences in host breed genetics.
INTRODUCTION
Because of their specific production attributes, the Holstein and Jersey breeds account for the vast majority of animals used in the dairy cattle industry within the United States and worldwide. For instance, a Holstein cow, on average, is larger and produces larger amounts of milk, while a Jersey cow has better fertility, is more efficient at maintaining its body condition during lactation, and produces milk with a higher content of solids (13, 17–19, 26). The relative importance given to certain traits is greatly influenced by circumstances that change priorities. For instance, while Holsteins have been the most popular breed by far because of their higher levels of milk production, there is increased interest in Jersey and Holstein-Jersey crosses as a strategy to improve the decreased fertility associated with the current Holstein genetic stocks.
Methane emission is emerging as an important efficiency trait due to its negative impact on animal production and its contribution to climate change (6). Methane is produced in the rumen by methanogenic archaea, which use the hydrogen and carbon dioxide produced by enteric fermentation as substrates (9). Once released into the environment, mostly through eructation (16), methane acts as a potent greenhouse gas, with a much greater effect on climate change than that of carbon dioxide. Since the continuous growth of the human population is expected to result in an increase in the number of domesticated ruminants, reducing methane emissions by livestock has become a priority and an integral part of climate control policies (25).
A number of reports have compared methane emissions between the Holstein and Jersey breeds, with conflicting results on whether differences exist or not (12, 14, 15). A major difficulty is the high variability and low repeatability of direct methane measurements. Since methane is produced specifically by methanogens, an alternative approach is to identify and compare the population structures of rumen methanogen communities between dairy cattle breeds by using culture-independent detection methods. A great variety of methanogens populate the rumen, as estimated by 16S rRNA gene public data sets, and it is predicted that more remain to be identified (10). Each individual report on rumen methanogens in dairy cattle so far has focused on a specific breed (7, 8, 23, 27). Since methanogen populations vary in response to diet and other environmental conditions (6), it is difficult to reach valid conclusions on differences due to breed when relying on comparisons between independent studies.
In this context, we present results of a comparative analysis of rumen methanogen population structures between lactating animals from a single herd consisting of Holstein and Jersey cows which were fed the same diet and maintained under the same environmental conditions. We investigated potential differences between breeds by sequencing methanogen 16S rRNA genes from breed-specific clone libraries to assess their molecular diversity. The specific objectives of our study were (i) to identify methanogens that reside in the rumens of Jersey and Holstein cattle and to determine their phylogeny, (ii) to investigate whether methanogen population structures vary significantly between breeds, and (iii) to compare our findings with previously published reports.
MATERIALS AND METHODS
Animal sampling.
All procedures were approved under The University of Vermont's Institutional Animal Care and Use Committee (IACUC) protocol 07-021 and Institutional Biosafety Committee (IBC) protocol 10-029. From a lactating herd consisting of 17 Holstein and 17 Jersey cows maintained at the Miller Research Center (The University of Vermont [UVM], Burlington, VT), 9 Holsteins and 10 Jerseys were randomly selected for sampling. Approximately 20 ml of rumen digesta was obtained through stomach tubing under the supervision of a licensed veterinarian. Rumen samples were frozen at −20°C until DNA extraction.
Animals were individually offered 44.1 kg of the same ration at 90 to 95% voluntary intake. Feed dry matter content was 35% corn silage, 33% second-cut haylage, 72% hay, 13.2% canola meal, and 19.8% soybean meal.
Microbial DNA isolation and clone library construction and sequencing.
Microbial DNAs from individual rumen samples were isolated as described by Yu and Morrison (34). Methanogen 16S rRNA genomic sequences were amplified from each purified rumen microbial DNA sample by PCRs using the methanogen-specific primers Met86F and Met1340R (28). PCRs were performed with Taq polymerase (Invitrogen) on a C1000 thermal cycler (Bio-Rad) under the following conditions: a hot start (4 min, 95°C) followed by 35 cycles of denaturation (30 s, 95°C), annealing (30 s, 58°C), and extension (2 min, 72°C), with a final extension period of 10 min at 72°C. Amplicons produced from animals of the same breed were pooled and cloned into the pCR2.1-TOPO vector (Invitrogen) to generate Holstein- and Jersey-specific methanogen 16S rRNA gene clone libraries. Recombinant plasmids from white colonies were screened by colony PCR with the M13 forward and M13 reverse primers to confirm the correctly sized inserts for sequencing. PCR products from positive bacterial clones were used directly as templates for Sanger DNA sequencing using the new sequencing primers Met643F (5′-GGA CCA CCW RTG GCG AAG GC-3′) and Met834R (5′-CTT GCG RCC GTA CTT CCC AGG-3′) to generate overlapping sequences. Nucleotide sequencing was performed by the DNA Analysis Core Facility at UVM.
Computational analysis of nucleotide sequences.
ChromasPro (version 1.5; Technelysium Pty. Ltd.) was used to proofread methanogen 16S rRNA gene sequences from positive clones and assemble them into contigs of 1,255 to 1,265 bp. Library clones were designated UVM-H or UVM-J to indicate the breed, followed by the clone number. All nonchimeric sequences from both libraries were pooled in silico and then assigned to operational taxonomic units (OTUs) by the open-source program MOTHUR (22), based on a 98% sequence identity cutoff (33). For OTU assignment, MOTHUR used distance data generated from the combined clone libraries by the Kimura two-parameter model (11) in PHYLIP (version 3.69) (5). OTU coverage (C) was calculated using the equation C = [1 − (n/N)] × 100%, where n is the number of OTUs represented by a single sequence and N is the total number of sequences obtained in a library. Shannon and Libshuff diversity indices were generated using MOTHUR (22). Characterization of representative OTU sequences was performed using the BLAST search engine against the NCBI nucleotide sequence database (1). For phylogenetic reconstruction, we combined one representative sequence from each OTU with 48 methanogen 16S rRNA gene sequences representing major archaeal phylogenetic groups. PHYLIP (version 3.69) (5) was used to construct a neighbor-joining tree (21), which was bootstrap resampled 1,000 times.
Nucleotide sequence accession numbers.
The sequences from this study have been deposited in the GenBank database under accession numbers JF682889 to JF683253 (see Tables S1 and S2 in the supplemental material).
RESULTS
Combined analysis of methanogens in lactating dairy cattle.
A total of 365 nonchimeric sequences were obtained, with 180 and 185 clones isolated from the Holstein and Jersey clone libraries, respectively. Sequences from both libraries were pooled in silico for analysis and assigned to 55 OTUs based on a 98% sequence identity criterion (Table 1). This species-level cutoff was established previously from the sequence identity that exists between 16S rRNA genes from validly characterized Methanobrevibacter species (33).
Table 1.
Comparison of OTUs between Holstein and Jersey dairy cows
| OTU | No. of sequences |
Nearest valid taxon | % Sequence identity | |
|---|---|---|---|---|
| Holstein | Jersey | |||
| 1 | 48 | 52 | Methanobrevibacter ruminantium | 98.9 |
| 2 | 28 | 46 | Methanobrevibacter millerae | 98.5 |
| 3 | 19 | 1 | Methanobrevibacter ruminantium | 97.4 |
| 4 | 7 | 10 | Methanobrevibacter millerae | 97.5 |
| 5 | 6 | 0 | Methanobrevibacter ruminantium | 97.1 |
| 6 | 2 | 1 | Methanobrevibacter millerae | 95.4 |
| 7 | 4 | 1 | Methanobrevibacter olleyae | 95.7 |
| 8 | 4 | 0 | Methanobrevibacter ruminantium | 97.5 |
| 9 | 9 | 10 | Methanobrevibacter olleyae | 96.3 |
| 10 | 2 | 18 | Methanobrevibacter millerae | 98.7 |
| 11 | 1 | 1 | Methanobrevibacter ruminantium | 97.4 |
| 12 | 2 | 3 | Methanosphaera stadtmanae | 97.4 |
| 13 | 1 | 0 | Aciduliprofundum boonei | 80.0 |
| 14 | 3 | 1 | Methanobrevibacter smithii | 96.4 |
| 15 | 1 | 1 | Methanobrevibacter olleyae | 96.3 |
| 16 | 1 | 0 | Methanobrevibacter ruminantium | 97.0 |
| 17 | 1 | 0 | Methanobrevibacter smithii | 96.1 |
| 18 | 2 | 0 | Methanobrevibacter ruminantium | 95.1 |
| 19 | 4 | 1 | Thermoplasma acidophilum | 80.1 |
| 20 | 4 | 1 | Methanobrevibacter gottschalkii | 97.3 |
| 21 | 1 | 1 | Methanobrevibacter gottschalkii | 96.2 |
| 22 | 2 | 0 | Methanobrevibacter ruminantium | 97.3 |
| 23 | 1 | 2 | Methanobrevibacter millerae | 97.5 |
| 24 | 1 | 0 | Methanobrevibacter ruminantium | 94.0 |
| 25 | 1 | 0 | Methanobrevibacter ruminantium | 96.0 |
| 26 | 2 | 0 | Methanobrevibacter millerae | 97.5 |
| 27 | 1 | 0 | Methanobrevibacter thaurei | 96.9 |
| 28 | 1 | 0 | Methanobrevibacter millerae | 97.2 |
| 29 | 3 | 5 | Methanobrevibacter gottschalkii | 97.1 |
| 30 | 2 | 0 | Methanobrevibacter millerae | 96.5 |
| 31 | 1 | 0 | Methanobrevibacter millerae | 95.4 |
| 32 | 1 | 0 | Methanobrevibacter ruminantium | 96.6 |
| 33 | 2 | 0 | Methanobrevibacter ruminantium | 93.2 |
| 34 | 2 | 9 | Methanobrevibacter arboriphilus | 95.1 |
| 35 | 1 | 0 | Methanobrevibacter millerae | 96.4 |
| 36 | 1 | 0 | Methanobrevibacter ruminantium | 94.8 |
| 37 | 1 | 0 | Methanobrevibacter ruminantium | 98.6 |
| 38 | 1 | 0 | Methanobrevibacter millerae | 96.3 |
| 39 | 1 | 0 | Methanobrevibacter millerae | 92.6 |
| 40 | 1 | 2 | Methanobrevibacter smithii | 98.1 |
| 41 | 2 | 1 | Methanobrevibacter olleyae | 96.5 |
| 42 | 1 | 0 | Methanobrevibacter smithii | 96.3 |
| 43 | 1 | 0 | Methanobrevibacter gottschalkii | 96.6 |
| 44 | 0 | 2 | Methanobrevibacter wolinii | 95.5 |
| 45 | 0 | 2 | Methanobrevibacter millerae | 96.3 |
| 46 | 0 | 1 | Methanobrevibacter olleyae | 95.8 |
| 47 | 0 | 1 | Methanobrevibacter millerae | 96.7 |
| 48 | 0 | 1 | Methanobrevibacter ruminantium | 94.0 |
| 49 | 0 | 2 | Methanobrevibacter gottschalkii | 97.1 |
| 50 | 0 | 1 | Methanobrevibacter ruminantium | 96.6 |
| 51 | 0 | 1 | Methanobrevibacter olleyae | 97.0 |
| 52 | 0 | 1 | Methanobrevibacter millerae | 96.3 |
| 53 | 0 | 3 | Methanobrevibacter millerae | 96.7 |
| 54 | 0 | 2 | Methanobrevibacter millerae | 97.7 |
| 55 | 0 | 1 | Methanobrevibacter smithii | 95.8 |
| Total | 180 | 185 | ||
We found that only 5 of 55 OTUs displayed species-like identity to validly defined methanogen species. OTUs 1, 34, and 37, representing 28.2% (101 of 365 sequences) of the sequences in our study, showed ≥98% sequence identity to Methanobrevibacter ruminantium. A similar number of sequences (25.8%) showed species-like sequence identity to Methanobrevibacter millerae and were assigned to OTUs 2 and 10. In contrast, only 0.8% (3 of 365 sequences) of sequences, which were assigned to OTU 40, showed ≥98% sequence identity to Methanobrevibacter smithii.
Forty-one of 55 OTUs (41.1% of sequences) showed genus-level (95 to 97.9%) sequence identity to validly defined methanogen species. To gain further insight, we conducted BLAST searches to determine if similar environmental clones were in the GenBank database. We found, for instance, that UVM-H004, representing OTU 3, was identical to environmental clones 1, 4, and 5 identified in anaerobic fungal cultures of rumen origin. Other findings of interest included UVM-H014 (OTU 7), which was almost identical (1,246 of 1,248 bp) to environmental clone SRmetF2 from Svalbard reindeer (24), and UVM-H024 (OTU 10), which was very similar (1,257 of 1,260 bp) to clone MLR 10 from cattle in Malaysia.
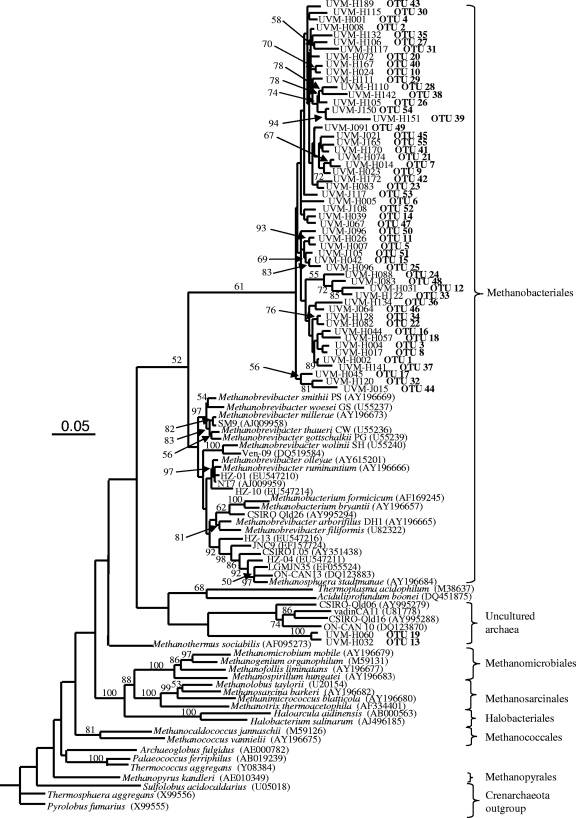
While 47 of 55 OTUs, representing 95.3% (348 of 365 clones) of the combined clones, displayed genus-level sequence identity (≥95%) to several Methanobrevibacter species, the remaining 8 OTUs were found to be distantly (<94.9%) related to Methanobrevibacter. Four OTUs (24, 36, 39, and 48), representing 1.1% of combined clones (4 of 365 sequences), were likely from methanogens representing uncharacterized genera within the Methanobacteriales, since their sequence identity to validly characterized Methanobrevibacter species was between 92.6% and 94.8%. The novelty of OTU 39 can be emphasized further by the observation that its closest environmental sample clone (MLR 10, found in cattle from Malaysia) was only 93.4% identical in sequence. OTUs 12 and 33 (7 sequences) showed genus-level sequence identity to Methanosphaera stadtmanae, at 96.9% and 95%, respectively. Finally, 6 sequences, grouped into OTUs 13 and 19, were found to be phylogenetically very distant from methanogens of the Methanobacteriales order (Fig. 1). They are part of a group of archaea that have consistently been identified in microbial communities of environmental samples, but they have yet to be validly characterized.
Fig. 1.
Neighbor-joining distance matrix tree for archaea in the rumens of lactating Holstein and Jersey cows derived from 16S rRNA gene evolutionary distances produced by the Kimura two-parameter correction model (11). Bootstrap supports are indicated as a percentage at the base of each bifurcation. Bootstrap values of <50% are not shown. Evolutionary distances are represented by the horizontal components separating the species in the figure. The scale bar corresponds to 5 changes per 100 positions.
Comparative analysis between Holstein and Jersey libraries.
A comparative analysis first revealed that the Holstein and Jersey clone libraries had 20 OTUs in common, representing 80.0% (144 of 180 sequences) and 90.3% (167 of 185 sequences) of their respective sequences. OTUs 1 and 2 were the most highly represented, comprising 42.2% (76 of 180 sequences) of sequences from the Holstein library and 53.0% (98 of 185 sequences) of those from the Jersey library. BLAST searches revealed that methanogen sequences assigned to OTUs 1 and 2 were almost identical to rumen methanogen sequences from other environmental samples. For instance, sequences from OTU 1 were almost identical to those of environmental clones AK-87 and CSIRO3.04 (1,259 of 1,260 bp), identified in the rumens of sheep and cattle from Australia (20, 32), while OTU 2 was very similar (1,259 of 1,262 bp) to environmental clone M21, isolated from the rumens of sheep sampled in China.
While both methanogen 16S rRNA gene clone libraries shared common features, we also observed a difference in their sequence diversity. Holstein sequences were assigned to 43 OTUs, in contrast to 32 OTUs for Jersey sequences, and exhibited significantly more methanogen diversity (P = 0.05) according to the Shannon diversity index values for these clone libraries (2.81 ± 0.20 and 2.29 ± 0.19, respectively). Another major difference was that sequences with species-like sequence identity to Methanobrevibacter millerae were approximately twice as abundant in the Jersey library as in the Holstein library. In contrast, there was equal representation in both libraries of sequences with species-like sequence identity to Methanobrevibacter ruminantium. Other noticeable differences between the two libraries were the higher representation in the Holstein library of clones with 90 to 97.9% sequence identity to Methanobrevibacter species and of clones from the novel uncultured group of archaea. Libshuff analysis also indicated that differences in the community structure between the two libraries were significant (P < 0.0147). Clone library OTU coverage rates were estimated at 88.3% and 90.8% for Holsteins and Jerseys, respectively, indicating that the libraries were very well sampled for the diversity that they contained.
DISCUSSION
Methane is a by-product of plant biomass digestion by herbivores, and its release comes at an energy cost from the host and contributes to climate change. To investigate potential differences in methane production between the Holstein and Jersey dairy cattle breeds, we compared the structures of rumen methanogen populations of animals from a single herd consisting of Holstein and Jersey cows, which have been kept together for more than 40 years. Previous studies have reported that differences in methanogen population structure likely exist between dairy cattle, but they were limited in scope by sampling animals from only a single breed (7, 8, 23, 27). To the best of our knowledge, there has been only one paper reporting methanogens from Jersey cattle (23). Unfortunately, the data are limited, as only 18 clones from a single Jersey cow were described.
Our study is the first to compare lactating Holstein and Jersey breeds under the same diet and environmental conditions, allowing confirmation of suggested differences between breeds. We found very significant differences in methanogen population structure between them, both in diversity at the OTU level and in sequence representation.
We observed that clones with species-like identity to Methanobrevibacter ruminantium and Methanobrevibacter millerae were present at very similar frequencies in lactating Jersey cows. In contrast, methanogen 16S rRNA gene clones with 95 to 97.9% identity to Methanobrevibacter species were found to be the most abundant (47.8%) in Holsteins, followed by clones with species-like identity to Methanobrevibacter ruminantium. A high representation of methanogens related to Methanobrevibacter ruminantium was also observed in lactating Holstein cattle from Alberta, Canada (27), in corn-fed cattle from Ontario, Canada (30), in beef cattle fed a low-energy diet (36), in the hoatzin (33), and in yak (2). In contrast, different methanogens were found to be dominant in other studies. For instance, 16S rRNA gene clones showing species-like identity to Methanobrevibacter gottschalkii were dominant in sheep from Venezuela (31) and in wallabies in November (4), while the majority of clones in the Murrah buffalo (India) were from the genus Methanomicrobium (3). Moreover, Zhou et al. (35) found a higher prevalence of Methanosphaera stadtmanae and Methanobrevibacter sp. AbM4 in beef cattle from Alberta, Canada. Other studies reported that clones belonging to the uncultured archaeal group were dominant in sheep from Queensland, Australia (29), in wallabies in May (4), in reindeer (24), and in potato-fed cattle from Prince Edward Island, Canada (30).
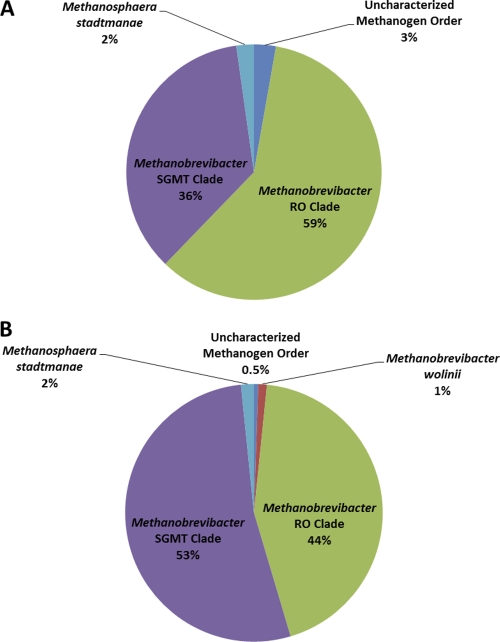
Like the case in this study, environmental clones that show at least genus-level sequence identity (≥95%) to Methanobrevibacter species are often the most abundant in gastrointestinal samples from herbivores (6). We recently proposed elsewhere to divide Methanobrevibacter-related environmental clones into two categories based on their phylogenetic distribution and representation. One phylogenetic grouping consists of sequences related to Methanobrevibacter smithii, Methanobrevibacter gottschalkii, Methanobrevibacter millerae, or Methanobrevibacter thaurei, which we refer to as the smithii-gottschalkii-millerae-thaurei (SGMT) group. The other phylogenetic grouping consists of Methanobrevibacter ruminantium and Methanobrevibacter olleyae-like sequences, which we refer to as the ruminantium-olleyae (RO) group. When unassigned Methanobrevibacter sequences were divided between the SGMT and RO groups, we found that the Holstein library had a higher representation of unassigned RO sequences than the Jersey library but that both libraries had similar numbers of unassigned SGMT sequences. This distribution pattern is opposite to the distribution of sequences with species-like identity to Methanobrevibacter ruminantium or Methanobrevibacter millerae in either library. When all sequences with at least genus-level sequence identity were included and divided between the SGMT and RO groups, each breed exhibited a distinct population structure (Fig. 2). Since SGMT and RO sequences tend to show opposite distribution patterns, each group perhaps represents methanogens that thrive under conditions that are not optimal for members of the opposite group. While this model still remains to be validated, it may potentially provide a means of estimating the methane synthesis potential of a methanogen community.
Fig. 2.
Pie chart representation of methanogen 16S rRNA gene clone distributions in lactating Holstein (A) and Jersey (B) cows. Methanobrevibacter sequences that phylogenetically group within the major clade consisting of Methanobrevibacter smithii, Methanobrevibacter gottschalkii, Methanobrevibacter millerae, and Methanobrevibacter thaurei sequences are represented in the smithii-gottschalkii-millerae-thaurei clade (SGMT clade). Similarly, the ruminantium-olleyae or RO clade consists of sequences that phylogenetically group within the major clade consisting of Methanobrevibacter ruminantium and Methanobrevibacter olleyae sequences.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to John Barlow and the Cooperative for Real Education in Agricultural Management (CREAM) students for access to the lactating cows.
We thank the College of Agriculture and Life Sciences (CALS) for a Distinguished Undergraduate Research (DUR) award granted to Erin King. This study was supported in part by a University of Vermont Undergraduate Research Endeavors Competitive Award (URECA!) provided to Erin King.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 24 June 2011.
REFERENCES
- 1. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. An D., Dong X., Dong Z. 2005. Prokaryote diversity in the rumen of yak (Bos grunniens) and Jinnan cattle (Bos taurus) estimated by 16S rDNA homology analyses. Anaerobe 11:207–215 [DOI] [PubMed] [Google Scholar]
- 3. Chaudhary P. P., Sirohi S. K. 2009. Dominance of Methanomicrobium phylotype in methanogen population present in Murrah buffaloes (Bubalus bubalis). Lett. Appl. Microbiol. 49:274–277 [DOI] [PubMed] [Google Scholar]
- 4. Evans P. N., et al. 2009. Community composition and density of methanogens in the foregut of the Tammar wallaby (Macropus eugenii). Appl. Environ. Microbiol. 75:2598–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Felsenstein J.2006. PHYLIP (Phylogeny Inference Package) documentation files, version 3.66. Department of Genome Sciences, University of Washington, Seattle, WA [Google Scholar]
- 6. Hook S. E., Wright A.-D. G., McBride B. W. 2010. Methanogens: methane producers of the rumen and mitigation strategies. Archaea 2010:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hook S. E., Northwood K. S., Wright A.-D. G., McBride B. W. 2009. Long-term monensin supplementation does not significantly affect the quantity or diversity of methanogens in the rumen of the lactating dairy cow. Appl. Environ. Microbiol. 75:374–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jeyanathan J., Kirs M., Ronimus R. S., Hoskin S. O., Janssen P. H. 2011. Methanogen community structure in the rumens of farmed sheep, cattle and red deer fed different diets. FEMS Microbiol. Ecol. doi:10.1111/j.1574-6941.2011.01056.x [DOI] [PubMed] [Google Scholar]
- 9. Johnson K. A., Johnson D. E. 1995. Methane emissions from cattle. J. Anim. Sci. 73:2483–2492 [DOI] [PubMed] [Google Scholar]
- 10. Kim M., Morrison M., Yu Z. 2011. Status of the phylogenetic diversity census of ruminal microbiomes. FEMS Microbiol. Ecol. 76:49–63 [DOI] [PubMed] [Google Scholar]
- 11. Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111–120 [DOI] [PubMed] [Google Scholar]
- 12. Lassen J., Lovendahl P., Madsen J. 2010. Experiences with large scale individual measurements of methane emission in dairy cattle using a Fourier transform infrared (FTIR) measuring unit, paper 247. In CD communication proceedings. Gesellschaft für Tierzuchtwissenschaften, Giessen, Germany [Google Scholar]
- 13. Mackle T. R., Parr C. R., Stakelum G. K., Bryant A. M., Macmillan K. L. 1996. Feed conversion efficiency, daily pasture intake, and milk production of primiparous Friesian and Jersey cows calved at two different liveweights. N. Z. J. Agric. Res. 39:357–370 [Google Scholar]
- 14. Münger A., Kreuzer M. 2006. Methane emission as determined in contrasting dairy cattle breeds over the reproduction cycle. Int. Cong. Ser. 1293:119–122 [Google Scholar]
- 15. Münger A., Kreuzer M. 2007. Absence of persistent methane emission differences in three breeds of dairy cows. Aust. J. Exp. Agric. 48:77–82 [Google Scholar]
- 16. Murray R. M., Byrant A. M., Leng R. A. 1976. Rates of production of methane in the rumen and large intestine in sheep. Br. J. Nutr. 36:1–14 [DOI] [PubMed] [Google Scholar]
- 17. Oldenbroek J. K. 1988. The performance of Jersey cows and cows of larger dairy breeds on two complete diets with different roughage contents. Livestock Prod. Sci. 18:1–17 [Google Scholar]
- 18. Prendiville R., Pierce K. M., Buckley F. 2009. An evaluation of production efficiencies among lactating Holstein-Friesian, Jersey, and Jersey × Holstein-Friesian cows at pasture. J. Dairy Sci. 92:6176–6185 [DOI] [PubMed] [Google Scholar]
- 19. Prendiville R., Pierce K. M., Delaby L., Buckley F. 6 January 2011, posting date Animal performance and production efficiencies of Holstein-Friesian, Jersey and Jersey × Holstein-Friesian cows throughout lactation. Livestock Sci. doi:10.1016/j.livsci.2010.11.023 [Google Scholar]
- 20. Rea S., Bowman J. P., Popovski S., Pimm C., Wright A.-D. G. 2007. Methanobrevibacter millerae sp. nov. and Methanobrevibacter olleyae sp. nov., methanogens from the ovine and bovine rumen that can utilize formate for growth. Int. J. Syst. Evol. Microbiol. 57:450–456 [DOI] [PubMed] [Google Scholar]
- 21. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 22. Schloss P. D., et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Skillman L. C., Evans P. N., Strompl C., Joblin K. N. 2006. 16S rDNA directed PCR primers and detection of methanogens in the bovine rumen. Lett. Appl. Microbiol. 42:222–228 [DOI] [PubMed] [Google Scholar]
- 24. Sundset M. A., et al. 2009. Rumen microbial diversity in Svalbard reindeer, with particular emphasis on methanogenic archaea. FEMS Microbiol. Ecol. 70:553–562 [DOI] [PubMed] [Google Scholar]
- 25. Thorpe A. 2008. Enteric fermentation and ruminant eructation: the role (and control?) of methane in the climate change debate. Clim. Change 93:407–431 [Google Scholar]
- 26. Washburn S. P., White S. L., Green J. T., Jr., Benson G. A. 2002. Reproduction, mastitis, and body condition of seasonally calved Holstein and Jersey cows in confinement or pasture systems. J. Dairy Sci. 85:105–111 [DOI] [PubMed] [Google Scholar]
- 27. Whitford M. F., Teather R. M., Forster R. J. 2001. Phylogenetic analysis of methanogens from the bovine rumen. BMC Microbiol. 1:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wright A.-D. G., Pimm C. 2003. Improved strategy for presumptive identification of methanogens using 16S riboprinting. J. Microbiol. Methods 55:337–349 [DOI] [PubMed] [Google Scholar]
- 29. Wright A.-D. G., Toovey A. F., Pimm C. L. 2006. Molecular identification of methanogenic archaea from sheep in Queensland, Australia reveal more uncultured novel archaea. Anaerobe 12:134–139 [DOI] [PubMed] [Google Scholar]
- 30. Wright A.-D. G., Auckland C. H., Lynn D. H. 2007. Molecular diversity of methanogens in feedlot cattle from Ontario and Prince Edward Island, Canada. Appl. Environ. Microbiol. 73:4206–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wright A.-D. G., Ma X., Obispo N. E. 2008. Methanobrevibacter phylotypes are the dominant methanogens in sheep from Venezuela. Microb. Ecol. 56:390–394 [DOI] [PubMed] [Google Scholar]
- 32. Wright A.-D. G., et al. 2004. Molecular diversity of rumen methanogens from sheep in Western Australia. Appl. Environ. Microbiol. 70:1263–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wright A.-D. G., Northwood K. S., Obispo N. E. 2009. Rumen-like methanogens identified from the crop of the folivorous South American bird, the hoatzin (Opisthocomus hoazin). ISME J. 3:1120–1126 [DOI] [PubMed] [Google Scholar]
- 34. Yu Z., Morrison M. 2004. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36:808–812 [DOI] [PubMed] [Google Scholar]
- 35. Zhou M., Hernandez-Sanabria E., Guan L. L. 2009. Assessment of the microbial ecology of ruminal methanogens in cattle with different feed efficiencies. Appl. Environ. Microbiol. 75:6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou M., Hernandez-Sanabria E., Guan L. L. 2010. Characterization of variation in rumen methanogenic communities under different dietary and host feed efficiency conditions, as determined by PCR-denaturing gradient gel electrophoresis analysis. Appl. Environ. Microbiol. 76:3776–3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.