Abstract
The stringent response (SR) enables bacteria to adapt to nutrient limitation through production of the nucleotides guanosine tetraphosphate and guanosine pentaphosphate, collectively known as (p)ppGpp. Two enzymes are responsible for the intracellular pools of (p)ppGpp: RelA acts as a synthetase, while SpoT can function as either a synthetase or a hydrolase. We investigated how the SR affects the ability of the biological control agent Pseudomonas sp. strain DF41 to inhibit the fungal pathogen Sclerotinia sclerotiorum (Lib.) de Bary. Strain DF41 relA and relA spoT mutants were generated and found to exhibit increased antifungal activity. Strain DF41 produces a lipopeptide (LP) molecule that is essential for Sclerotinia biocontrol. LP production and protease activity were both elevated in the relA and relA spoT mutants. Addition of relA but not spoT in trans restored the mutant phenotype to that of the parent. Next, we investigated whether an association exists between the SR and known regulators of biocontrol, including the Gac system and RpoS. A gacS mutant of strain DF41 produced less (p)ppGpp and exhibited a 1.7-fold decrease in relA expression compared to the wild type, suggesting that relA forms part of the Gac regulon. We discovered that rpoS transcription was reduced significantly in the SR mutants. Furthermore, rpoS provided in trans restored protease activity to wild-type levels but did not attenuate antifungal activity. Finally, relA expression was decreased in the mutants, indicating that the SR is required for maximum expression of relA.
INTRODUCTION
A number of bacteria are able to antagonize the effects of fungal pathogens through a process known as biological control. Pseudomonas sp. strain DF41 is one such bacterium that has demonstrated excellent antifungal activity against Sclerotinia sclerotiorum (Lib.) de Bary in both greenhouse and field assays (2, 32). Strain DF41 produces several secondary metabolites that are believed to contribute to biocontrol, including hydrogen cyanide, protease, and a novel lipopeptide (LP) molecule (1, 2). LP production has been shown to be essential for strain DF41 biocontrol, as an LP-deficient mutant, DF41-1278, demonstrated greatly reduced S. sclerotiorum inhibition (1, 2). LP synthesis is somewhat unique in that it does not involve ribosome-generated proteins; instead, these compounds are synthesized on large, multimodular enzymes termed nonribosomal peptide synthetases (NRPS) (24).
Several regulators have been found to govern secondary metabolite production in biocontrol strains of Pseudomonas. For example, the Gac two-component signal transduction system, comprised of the sensor kinase GacS and the cognate response regulator GacA, is essential for biocontrol. A mutation in either gacS or gacA results in a loss of biocontrol activity in several pseudomonads, including strain DF41 (2, 13). The alternative stationary-phase sigma factor, RpoS, has also been implicated in secondary metabolite production; however, regulation by RpoS appears to differ among species of pseudomonads. For example, an rpoS mutant of Pseudomonas chlororaphis PCL1391 exhibited decreased production of phenazine-1-carboxamide compared to the wild type (10), whereas in Pseudomonas fluorescens Pf-5, an rpoS mutation resulted in enhanced pyoluteorin and 2,4-diacetylphloroglucinol expression (31). Several other global and pathway-specific regulators have been identified that influence the biocontrol properties of pseudomonads (12).
In addition to these regulatory elements, environmental conditions can have a significant impact on bacterial metabolism. When bacteria such as strain DF41 colonize the plant environment, they experience dramatic fluctuations in many environmental conditions, including nutrient availability. One means by which bacteria are able to survive starvation is through induction of a global stress mechanism known as the stringent response (SR). During the SR, cells accumulate the nucleotides guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp), collectively referred to as (p)ppGpp. In gamma- and betaproteobacteria, (p)ppGpp accumulation is controlled by two enzymes, RelA and SpoT (see reference 27 and references therein). RelA is a synthetase that generates (p)ppGpp when available amino acids are in limiting amounts. SpoT is a bifunctional enzyme that can act as either a hydrolase or a synthetase depending on the conditions present (27). (p)ppGpp exerts its influence on cell physiology by binding RNA polymerase (RNAP) near the catalytic site. This leads to increased transcription of certain genes, for example, those involved in amino acid biosynthesis, and decreased transcription of others, for instance, tRNA and rRNA genes (27). Accordingly, the SR enables bacteria to alter their gene expression to favor activities that promote survival under nutrient-limiting conditions.
Although the SR has been shown to affect antibiotic production in Streptomyces species (4, 11, 15, 18, 19), there is a paucity of knowledge regarding how this global stress response influences biocontrol traits in pseudomonads. The purpose of this study was to investigate the impact of the SR on strain DF41 antifungal compound production. In addition, we examined whether there was a link between the SR and other known regulators of biocontrol, including the GacS/GacA two-component regulatory system and RpoS.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were cultured on Lennox Luria-Bertani (LB) agar (Difco Laboratories, Detroit, MI). Pseudomonas sp. DF41 and its derivatives were routinely cultured at 28°C on King's B agar plates supplemented with 2% glycerol (21). (p)ppGpp analysis was performed using cells grown in morpholinepropanesulfonic acid (MOPS; Sigma-Aldrich Canada, Oakville, Ontario, Canada) medium, as described by Cashel (3), supplemented with 1% Casamino Acids (Difco) and 400 μg/ml of dl-serine hydroxamate (Sigma). For LP analysis, cells were grown in M9 minimal medium (Difco) supplemented with 1% Casamino Acids, 1 mM MgSO4, and 0.2% glycerol. For β-galactosidase assays, strains were grown in M9 medium supplemented with 1 mM MgSO4 and 0.2% glucose. S. sclerotiorum was maintained on potato dextrose agar (PDA; Difco). As required, media were supplemented with the following antibiotics from Research Products International Corp. (Mt. Prospect, IL): gentamicin (Gm; 20 μg/ml), tetracycline (Tc; 15 μg/ml), piperacillin (Pip; 100 μg/ml), and rifampin (Rif; 100 μg/ml) for strain DF41 and ampicillin (Amp; 100 μg/ml), Gm (15 μg/ml), and Tc (15 μg/ml) for E. coli.
Table 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Genotype, phenotype, or sequence (5′–3′)a | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa strain | ||
| PAO1 | Wild type | 17 |
| Pseudomonas sp. strains | ||
| DF41 | Rifr; wild type (canola root tip isolate) | 32 |
| DF41 relA | DF41 with a Gmr cassette inserted into the relA gene | This study |
| DF41 relA spoT | DF41 relA mutant with a Tetr cassette inserted into the spoT gene | This study |
| DF41-469 | Rifr; gacS::Tn5-1063 genomic insertion | 2 |
| DF41-1278 | Rifr; lp::Tn5-1063 genomic insertion | 2 |
| 1278 relA | DF41-1278 with Gmr cassette inserted into the relA gene | This study |
| 1278 relA spoT | 1278 relA mutant with Tetr cassette inserted into the spoT gene | This study |
| E. coli strain | ||
| DH5α | supE44 ΔlacU169 φ80dlacZΔM15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Gibco |
| Plasmids | ||
| pLP170 | Promoterless lacZ transcriptional fusion | 26 |
| pRPOS-lacZ | rpoS promoter in pLP170 | This study |
| pSW205 | Promoterless lacZ translational fusion | 9 |
| pSWrelA | relA promoter and first 21 codons of the open reading frame cloned into pSW205 | This study |
| pCR2.1 | TA cloning vector Ampr | Invitrogen |
| pCRrelA-41 | relA in pCR2.1 | This study |
| pCRspoT | spoT in pCR2.1 | This study |
| pEX18Ap | Suicide plasmid Ampr | 16 |
| pEXrelA | relA in pEX18Ap | This study |
| pEXrelA-800 | pEX18Ap with an 800-bp deletion in relA | This study |
| pEXrelA-Gent | pEXrelA-800 Gmr | This study |
| pEXspoT | spoT in pEX18Ap | This study |
| pEXspoT-Tet | pEXspoT with Tetr cassette inserted into spoT | This study |
| pUCP22 | Broad-host-range vector Ampr Gmr | 36 |
| pUCPrelA | relA in pUCP22 | This study |
| pUCPspoT | spoT in pUCP22 | This study |
| pUCP22-rpoS | rpoS in pUCP22 | 26 |
| pUCP23-gacS | gacS in pUCP23 | 26 |
| pME-rpoS | rpoS in pME6010 | This study |
| pME6010 | pVS1-p15A shuttle cloning vector; Tetr | 14 |
| pME3219 | pME6010 containing an hcnA-lacZ translational fusion | 22 |
| pRK600 | Contains tra genes for mobilization; Chlr | 8 |
| pUCGm | Source of Gmr cassette | 33 |
| pFTC1 | Source of Tetr cassette | 5 |
| Primers | ||
| relA-FOR | ACCGTGGTAAAGGGTAGGCAAG | This study |
| relA-REV | GGGAAATCCCCCTGCTCTATG | This study |
| relAtransl-FRW | GGAATTCCCGCTTTTTTCAAGCCGAT | This study |
| relAtransl-REV | GAGGATCTCGGCGATCTCCA | This study |
| spoT-FOR | GCGTCACCGTTGAAGACTG | This study |
| spoT-REV | TTACTCGAGGACGACGATGG | This study |
| rpoSF | TACGTCAGTGCTTACGGCCA | This study |
| rsmZR | TATGACCCGCCCACATTTTT | This study |
| P170fecorpoS | TGTGAATTCGGGAGGGACA | This study |
| P170rxbarpoS | AGTCTAGAATCACCACTTCCCATTGCTT | This study |
Amp, ampicillin; Chl, chloramphenicol; Gm, gentamicin; Rif, rifampin; Tet, tetracycline.
Nucleic acid manipulation.
Standard techniques were used for purification, cloning, and other DNA manipulations (30). PCR was performed under standard conditions suggested by Invitrogen Life Technologies data sheets supplied with the Taq polymerase.
Sequence analysis.
PCR products were sequenced at the Centre for Applied Genomics at The Hospital for Sick Children (Toronto, Ontario, Canada), and the sequences were analyzed with the BLASTN and BLASTX databases.
Generating relA and relA spoT mutants of strains DF41 and DF41-1278.
All primers used for the construction of mutant strains are listed in Table 1. The DF41 relA mutant strain was generated as follows. The relA gene of strain DF41 was amplified using primers relA-FOR and relA-REV. The resulting 2.4-kb fragment was then cloned into pCR2.1-TOPO (Invitrogen) to generate pCRrelA-41. To liberate the insert, pCRrelA-41 was digested with BamHI and EcoRI. The 2.4-kb fragment was subcloned into the same sites of the suicide vector pEX18Ap (16). Next, an 865-bp SalI fragment containing the Gmr cassette from pUCGm (33) was cloned into the SalI site of pEXrelA to create pEXrelA-Gent. Allelic exchange through triparental mating between the donor [E. coli DH5α(pEXrelA-Gent)], helper [E. coli DH5α(pRK600)], and recipient (either DF41 or DF41-1278) strains was used to replace the wild-type relA gene with the mutated copy of the gene. Transconjugants were screened on LB agar supplemented with 100 μg/ml Rif and 20 μg/ml Gm. Bacteria that had undergone a double-crossover event were selected on LB agar containing 20% sucrose and 20 μg/ml Gm. Verification of the mutation was achieved by PCR using the same primers. For creation of the DF41 relA spoT mutant, a 2.7-kb fragment containing the spoT gene was PCR amplified using primers spoT-FOR and spoT-REV. The resulting PCR product was subsequently cloned into pCR2.1-TOPO to generate pCRspoT. The spoT gene was then subcloned into pEX18Ap via SacI and XbaI sites. The resulting plasmid, pEXspoT, was digested with SmaI and ligated with a 2.0-kb SmaI fragment containing the Tcr marker from pFTC1 (5) to generate pEXspoT-Tet. Triparental mating was performed using E. coli DH5α(pEXspoT-Tet), E. coli DH5α(pRK600), and the DF41 relA or DF41-1278 relA strain. Transconjugants were screened on LB agar supplemented with 15 μg/ml Tc and 100 μg/ml Rif. Sucrose plates containing Tc were used to identify bacteria that had undergone a double-crossover event. To confirm the insertion of the Tc marker into the spoT gene, Southern blot analysis was performed.
Construction of plasmids.
All primers are listed in Table 1. To generate the relA overexpression plasmid pUCPrelA, primers relAtranslFRW and relA-REV were used to amplify a 2.9-kb fragment. The PCR product was cloned into pCR2.1-TOPO to yield pCRrelA-41. pCRrelA-41 was then digested with EcoRI, and the 2.8-kb fragment containing the relA gene was cloned into pUCP22, placing the gene under the control of the plasmid-borne lac promoter. The spoT overexpression vector pUCPspoT was created by first amplifying the spoT gene from strain DF41, using primers spoT-FOR and spoT-REV. The 2.8-kb fragment containing the gene was cloned into pCR2.1-TOPO to generate pCRspoT, which was subsequently digested with XbaI. The linearized plasmid was treated with Klenow DNA polymerase (Invitrogen) and digested again with BamHI. The 2.8-kb insert was cloned into the BamHI and SmaI sites of pUCP22 such that the transcription of spoT was dependent on the lac promoter. To create an rpoS-lacZ transcriptional fusion, the rpoS gene was amplified from DF41 genomic DNA by using primers rpoSF and rsmZR. The resulting 1.7-kb fragment was cloned into the pCR2.1-TOPO vector, resulting in pCR2.1-rpoS. Using pCR2.1-rpoS as the template, a 1.1-kb fragment was amplified using primers P170fecorpoS and P170rxbarpoS, which contain EcoRI and XbaI sites, respectively. The PCR product was digested with EcoRI and XbaI and cloned into the same sites of pLP170 (28), generating pRPOS-lacZ. For the relA-lacZ translational fusion pSWrelA, a 644-bp fragment containing 72 nucleotides upstream of the ATG translational start site was PCR amplified from strain DF41 genomic DNA, using primers relAtranslFRW and relAtranslREV. The product was digested with EcoRI and SmaI and cloned into the same sites of pSW205 (9). To create pME-rpoS, containing rpoS constitutively expressed from the kanamycin promoter, a 1.3-kb KpnI-HindIII fragment was isolated from pUCP22-rpoS and ligated into the same sites of pME6010 (14).
(p)ppGpp and relA expression analysis.
Determination of (p)ppGpp levels was performed as described by Cashel (3), with the following modifications. Cells were grown overnight in MOPS minimal medium (3) at 28°C, followed by 1/100 dilution of the culture in MOPS phosphate-free minimal medium containing 1 mg/ml of Casamino Acids and 100 μCi/ml of 32P (Perkin Elmer, Waltham, MA). Three 200-μl aliquots of culture were added to wells of a polystyrene microtiter plate (Costar; Corning Incorporated, Corning, NY) and grown at 28°C for an additional 8 h. dl-Serine hydroxamate was added to each well at a concentration of 400 μg/ml and allowed to incubate for 2 h. To ensure that an equivalent number of cells was extracted for each strain, PA23 and its derivatives were grown on a parallel plate as described above, but excluding the 32P. All of the cultures showed equivalent turbidity levels (optical densities at 600 nm [OD600s]). Thus, the three radiolabeled 200-μl aliquots were pooled for each strain, and nucleotides were extracted with an equal volume of cold 13 M formic acid. A 20-μl aliquot of the nucleotide sample was separated on polyethyleneimine-cellulose chromatography sheets (Sigma), using 1.5 M KH2PO4 as the solvent, which was allowed to ascend the plate for 2.5 h. The spots were visualized by autoradiography. For the (p)ppGpp analysis, Pseudomonas aeruginosa strain PAO1 was included as a positive control, and the experiments were repeated five times. Expression of an relA-lacZ translational fusion was monitored in the DF41, DF41 relA, and DF41 relA spoT strains and in the gacS mutant strain DF41-469. Strains harboring pSWrelA were grown for 4, 8, 16, and 24 h and then assayed for β-galactosidase activity (25).
Antifungal assays.
To assess the ability of strain DF41 and its derivatives to inhibit fungal growth in vitro, radial diffusion assays were performed according to the method of Poritsanos et al. (26). Five replicates for each strain were analyzed, and the experiments were repeated three times.
HPLC analysis.
LP was extracted from cultures and analyzed by high-performance liquid chromatography (HPLC) as described by Berry et al. (2), with the following modifications. To determine the efficiency of the extraction, surfactin (Sigma) was used as an internal control. Cell-free supernatants were spiked with a 500-μl aliquot of surfactin (1-mg/ml stock concentration), which was extracted with the LP. The chromatograms obtained for each strain were normalized using the surfactin peak height. We have previously shown that the peak at 28 min corresponds to the DF41 LP (2); consequently, the amount of LP present in the extracts was determined by measuring the height of the 28-min peak.
Bioluminescence.
Five-milliliter cultures were grown in M9 minimal medium plus 1 mM MgSO4 plus 0.2% glucose and assayed by the OD600 to determine the number of cells present. The cultures were centrifuged and resuspended in an equal volume of 1× phosphate-buffered saline (PBS). A 1-ml aliquot of the cell suspension was mixed with 1 ml of 2 × 523 medium supplemented with 10 mg/liter of sodium citrate (20). A 2.5-μl aliquot of a 10% n-decanal solution was added to each tube, which was mixed for 15 s with a vortex mixer and measured for bioluminescence 90 s later by use of a BG-P luminometer (GEM Biomedical Inc., Hamden, CT). Bioluminescence was expressed in relative light units (RLU) by dividing the total bioluminescence signal by the OD600.
HCN analysis.
Qualitative determination of hydrogen cyanide production was performed using Cyantesmo paper (Macherey-Nagel GmbH & Co., Duren, Germany). Plasmid pME3219, harboring an hcnA-lacZ translational fusion (22), was mobilized into the DF41, DF41 relA, and DF41 relA spoT strains. Strains were grown for 24 h and then assayed for β-galactosidase activity. Samples were analyzed in triplicate, and experiments were repeated three times.
Protease production.
Cultures were grown in M9 minimal medium supplemented with 1 mM MgSO4, 0.2% glucose, and 1.5% skim milk (Difco) for 5 days at 28°C to induce protease production. A 200-μl aliquot of cell-free supernatant was analyzed for the activity of this enzyme in a 0.65% solution of casein according to the method of Cupp-Enyard (7). Tyrosine, which is released upon the hydrolysis of casein by the protease enzyme, is able to react with the Folin-Ciocalteu reagent (Sigma) to produce a blue chromophore (7). This chromophore is measured spectrophotometrically at a wavelength of 660 nm. To determine the amount of tyrosine liberated, a standard curve was generated using pure tyrosine at the following concentrations: 0.055, 0.111, 0.221, 0.442, and 0.553 μM. Each strain was assayed in triplicate, and experiments were performed three times.
rpoS expression in DF41, DF41 relA, and DF41 relA spoT strains.
Expression of the rpoS gene was monitored using pRPOS-lacZ. Cultures of the DF41, DF41 relA, and DF41 relA spoT strains harboring pRPOS-lacZ were grown for 4, 8, 16, 24, and 48 h and then measured for β-galactosidase activity.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences of the DF41 rpoS, relA, and spoT genes are EU595545.1, HQ615419, and HQ615420, respectively.
RESULTS
Generation of DF41 relA and relA spoT mutant strains and (p)ppGpp analysis.
To determine the impact of the SR on the biocontrol activity of strain DF41, relA and relA spoT mutants were created. Primers designed from the relA and spoT sequences of Pseudomonas aeruginosa PAO1 were used to amplify these genes from strain DF41 genomic DNA. Sequence analysis revealed the DF41 alleles to be 99% identical to the relA and spoT genes of P. aeruginosa PAO1 (GenBank accession no. AE004091.2), 85% identical to those of P. fluorescens Pf0-1 (GenBank accession no. CP000094.2), and 84% identical to those of P. fluorescens Pf-5 (GenBank accession no. CP000076.1). DF41 relA and relA spoT mutants were generated through allelic exchange. Double-crossover mutations were confirmed by PCR and Southern blot analysis (data not shown). We were unable to isolate an spoT single null mutant, indicating that the absence of SpoT in an relA+ background is likely lethal, similar to what has been reported for other bacteria (27, 37).
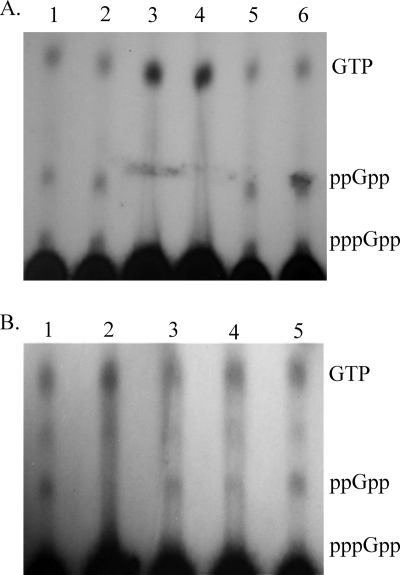
Next, we analyzed (p)ppGpp levels in the aforementioned strains. In the presence of dl-serine hydroxamate, which induces the SR, no (p)ppGpp was detected in the relA and relA spoT mutants, unlike the case in strain DF41, which produced both nucleotides (Fig. 1 A). The presence of pUCPrelA, a vector harboring the strain DF41 relA gene, restored (p)ppGpp production in both mutants (Fig. 1A, lanes 5 and 6). The presence of spoT on plasmid pUCPspoT, however, did not alter (p)ppGpp production in the mutants (data not shown).
Fig. 1.
(p)ppGpp analysis of Pseudomonas sp. strain DF41 and its SR derivatives after serine hydroxamate induction. Cells were labeled with 32P, and nucleotides were extracted and separated by thin-layer chromatography. (A) Lane 1, Pseudomonas aeruginosa strain PAO1; lane 2, Pseudomonas sp. DF41; lane 3, DF41 relA mutant; lane 4, DF41 relA spoT mutant; lane 5, DF41 relA(pUCPrelA) mutant; lane 6, DF41 relA spoT(pUCPrelA) mutant. (B) Lane 1, DF41; lane 2, DF41-469 (gacS mutant); lane 3, DF41-469(pUCP23-gacS); lane 4, DF41-469(pUCPrelA); lane 5, PAO1.
SR affects strain DF41 antifungal activity.
When we assessed the ability of the mutants to inhibit S. sclerotiorum in vitro, we observed 1.5-fold and 1.8-fold increases in antifungal activity for the DF41 relA and DF41 relA spoT strains, respectively, compared to the parent (Table 2). The presence of pUCPrelA was able to restore the antifungal activities of both mutants to that of the wild type (Table 2). Production of many secondary metabolites begins at the transition between the logarithmic and stationary phases, also known as the idiophase. As such, it is important that there were no differences in growth rate between the wild type, the SR mutants, and the complemented strains (see Fig. 4A).
Table 2.
Antifungal activity of Pseudomonas sp. strain DF41 and its derivatives after 5 days of growth
| Strain | Zone of fungal growth inhibition (mm)a |
|---|---|
| DF41(pUCP22) | 4.58 (1.0) |
| DF41 relA(pUCP22) | 7.25 (0.5)b |
| DF41 relA spoT(pUCP22) | 8.30 (1.5)c |
| DF41 relA(pUCPrelA) | 4.75 (1.2)d |
| DF41 relA spoT(pUCPrelA) | 5.50 (1.2)d |
Mean (standard deviation) obtained for five replicates.
Significantly different from the wild type (P < 0.005).
Significantly different from the wild type (P < 0.05).
Not significantly different from the wild type.
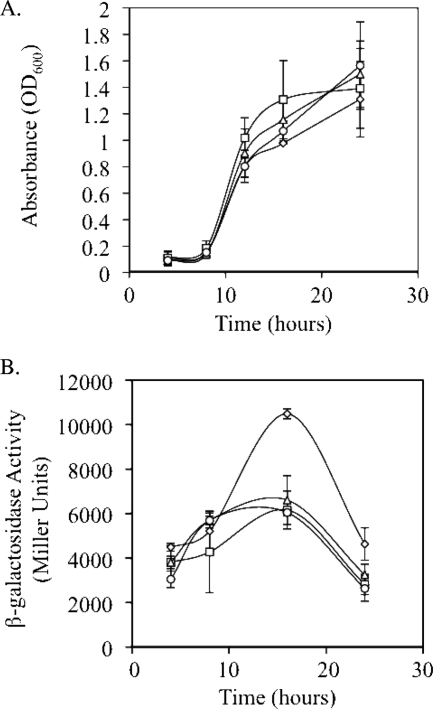
Fig. 4.
Ability of GacS and the stringent response to regulate relA translation. Growth curves for each strain are depicted in panel A, while translational analysis of relA is shown in panel B. Strains used were as follows: Pseudomonas sp. strain DF41 (diamonds), DF41 relA mutant (squares), DF41 relA spoT mutant (triangles), and DF41-469 (circles). Strains in panel B harbor the relA translation fusion pSWrelA. Strains were grown in M9 minimal medium supplemented with 1 mM MgSO4 and 0.2% glucose. Note that relA expression was markedly reduced in both the SR mutants and the gacS mutant.
Secondary metabolite production by strain DF41 and its SR mutant derivatives.
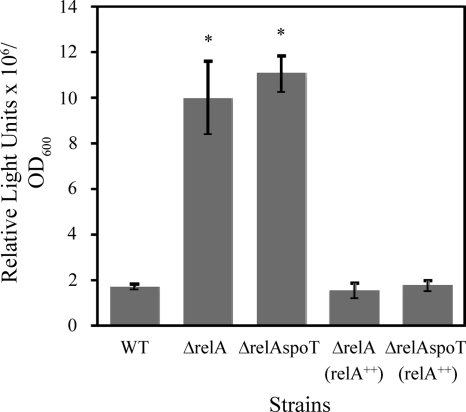
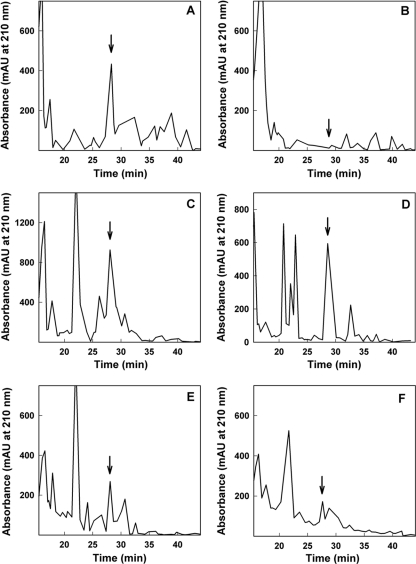
Because LP production is essential for strain DF41 biocontrol (1, 2), we investigated whether the ppGpp-deficient strains exhibited elevated LP expression. A derivative of strain DF41, termed DF41-1278, is LP deficient due to a Tn5-1063 (luxAB) insertion in the NRPS locus responsible for LP synthesis (1, 2). Consequently, it is possible to monitor transcription of the NRPS genes in strain DF41-1278 by using a bioluminescence assay (1). Two SR mutants of strain DF41-1278 were created, designated the 1278 relA and 1278 relA spoT mutants. After 12 h of growth, NRPS transcription in the 1278 relA and 1278 relA spoT strains was elevated 5.6-fold and 6.1-fold, respectively, over that of the wild type (Fig. 2). Addition of relA in trans restored transcription to levels close to wild-type levels (Fig. 2). By 24 h, all strains exhibited equal levels of transcription (data not shown). Next, we determined the level of LP present in DF41, DF41 relA, and DF41 relA spoT culture supernatants through HPLC analysis. A single peak eluting at 28 min (Fig. 3) was observed for all strains except for strain DF41-1278 (Fig. 3B). We demonstrated previously that this 28-min peak corresponds to the DF41 LP molecule (2). For the SR mutants, the 28-min peak was 1.5- to 2.0-fold larger (Fig. 3C and D) than that for the wild type (Fig. 3A). When relA was expressed in trans, LP production by the SR mutants was markedly reduced (Fig. 3E and F).
Fig. 2.
Transcription of NRPS genes in Pseudomonas sp. strain DF41-1278 and in the 1278 relA, 1278 relA spoT, and relA-complemented mutants. Column 1, DF41-1278(pUCP22); column 2, 1278 relA(pUCP22) strain; column 3, 1278 relA spoT(pUCP22) strain; column 4, 1278 relA(pUCPrelA) strain; and column 5, 1278 relA spoT(pUCPrelA) strain. Bioluminescence was monitored after 12 h of growth in M9 minimal medium supplemented with 1 mM MgSO4 and 0.2% glucose. For strains that differ significantly from the wild type, columns are labeled with asterisks (*, P < 0.005).
Fig. 3.
HPLC analysis of LP produced by Pseudomonas sp. strain DF41, the stringent response mutants, and strains harboring pUCPrelA. LP was extracted from cell-free supernatants of 4-day-old cultures and separated by HPLC. Chromatograms depict the following strains: DF41(pUCP22) (A), DF41-1278(pUCP22) (B), DF41 relA(pUCP22) (C), DF41 relA spoT(pUCP22) (D), DF41 relA(pUCPrelA) (E), and DF41 relA spoT(pUCPrelA) (F). Peaks containing the strain DF41 LP molecule are indicated with arrows.
Besides LP molecules, strain DF41 liberates the volatile antibiotic HCN (1, 2). Using Cyantesmo paper, we determined that the wild type and the SR mutants all produced HCN (data not shown). Furthermore, no differences in hcnA-lacZ expression were observed between the DF41 (888 ± 166 Miller units), DF41 relA (1,163 ± 124 Miller units), and DF41 relA spoT (1,152 ± 113 Miller units) strains.
Quantitative analysis of protease production revealed that the DF41 relA and DF41 relA spoT mutants produced 2-fold more protease than the wild type (Table 3). Addition of pUCPrelA restored the protease activity of the SR mutants to wild-type levels (Table 3).
Table 3.
Protease activity of Pseudomonas sp. strain DF41 and derivatives harboring the relA overexpression plasmid pUCPrelA and the rpoS overexpression plasmid pUCP22-rpoS
| Strain | Protease activity (U of enzyme/ml)a |
|---|---|
| DF41(pUCP22) | 0.752 (0.04) |
| DF41 relA(pUCP22) | 1.480 (0.01)b |
| DF41 relA spoT(pUCP22) | 1.487 (0.02)b |
| DF41 relA(pUCPrelA) | 0.882 (0.10)c |
| DF41 relA spoT(pUCPrelA) | 0.914 (0.20)c |
| DF41 relA(pUCP22-rpoS) | 0.677 (0.10)c |
| DF41 relA spoT(pUCP22-rpoS) | 0.735 (0.02)c |
Mean (standard deviation) for five replicates.
Significantly different from the wild type (P < 0.005).
Not significantly different from the wild type.
relA expression and (p)ppGpp production are reduced in a gacS mutant of strain DF41.
To ascertain whether a link exists between the Gac system and the SR, (p)ppGpp production was assessed in a gacS mutant of strain DF41 called DF41-469. As observed in Fig. 1B, (p)ppGpp levels were markedly reduced in the gacS mutant. Production of these nucleotides was increased by the addition of either gacS or relA in trans (Fig. 1B). Next, we generated an relA translational fusion and monitored its activity in strains DF41 and DF41-469 (gacS). Although there were no differences in growth rate between the two strains (Fig. 4 A), a 1.7-fold reduction in relA expression was observed in the gacS mutant compared to the wild type at 16 h (Fig. 4B). Addition of pUCP23-gacS in trans was unable to complement the DF41 relA and DF41 relA spoT mutants for any of the aforementioned phenotypes, including LP production and antifungal and protease activities (data not shown).
The stringent response is required for maximal relA expression.
Expression of an relA translational fusion was monitored in the DF41, DF41 relA, and DF41 relA spoT strains. In strain DF41, relA expression reached its maximum at 16 h, followed by a sharp decline by 24 h (Fig. 4B). In the SR mutants, relA expression peaked at 16 h; however, expression was reduced 1.6-fold in these strains. Thus, it appears that in strain DF41, the SR is required for maximal relA expression.
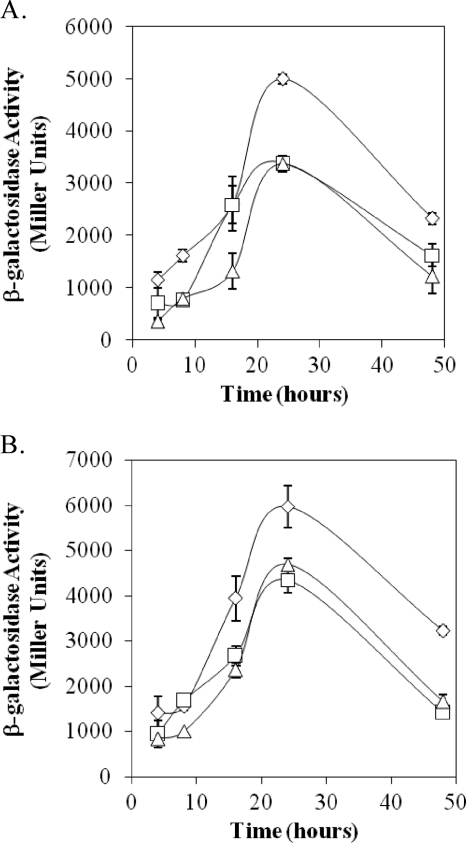
RpoS overexpression complements rpoS transcription and protease activity in SR mutants.
In strain DF41, rpoS was found to be under the control of GacS (2). To determine the impact of the SR on the transcription of this sigma factor, the β-galactosidase activity of an rpoS-lacZ transcriptional fusion was monitored in strain DF41 and its derivatives. At 24 h, rpoS transcription was reduced almost 1.5-fold in the ppGpp-deficient strains compared to the wild type (Fig. 5 A). To determine the effect of constitutively expressed rpoS on transcription of this sigma factor, pME-rpoS was mobilized into the DF41, DF41 relA, and DF41 relA spoT strains, all of which carry the rpoS-lacZ fusion plasmid. As shown in Fig. 5B, the presence of pME-rpoS in the SR mutants elevated rpoS-lacZ expression to a level close to that of DF41 carrying the empty vector (Fig. 5A). Conversely, when pME-rpoS was introduced into the DF41-1278, 1278 relA, and 1278 relA spoT strains, no change in NRPS gene transcription was observed (data not shown). To see if constitutively expressed rpoS would alter phenotypic traits displayed by the SR mutants, the DF41 relA(pME-rpoS) and DF41 relA spoT(pME-rpoS) strains were analyzed for LP production and antifungal and protease activities. No difference in LP production and antifungal activity was observed (data not shown); however, protease activity exhibited by the ppGpp-deficient strains was restored to wild-type levels upon the addition of rpoS in trans (Table 3).
Fig. 5.
Expression of rpoS-lacZ in strain DF41 and its derivatives. Strains used were as follows: Pseudomonas sp. strain DF41 (diamonds), DF41 relA mutant (squares), and DF41 relA spoT mutant (triangles). All strains harbored the rpoS-lacZ reporter plasmid pRPOS-lacZ together with empty vector pME6010 (A) or pME-rpoS (B). The results shown in panels A and B are from the same experiment. Note that the presence of pME-rpoS in the SR mutants (B) restored rpoS activity to a level close to that of DF41(pME6010) (A).
DISCUSSION
The SR is a global regulatory mechanism that enables bacteria to alter a broad range of physiological traits, including growth rate, motility, persistence, biofilm formation, virulence, and secondary metabolite production, in response to nutrient limitation (27). In light of this, we were interested to learn how the SR affects the antifungal activity of DF41. This is particularly important because when DF41 is used as a biocontrol agent against Sclerotinia stem rot of canola, it is applied as a foliar spray (2, 32). The aerial surfaces of the plant represent a harsh environment for bacteria, as they must endure exposure to high and low temperatures, UV radiation, and desiccation (23). Additionally, nutrients are scarce, so bacteria on the plant surface are expected to undergo the SR (23).
Characterization of DF41 relA and relA spoT SR mutants revealed a lack of (p)ppGpp production (Fig. 1) and enhanced inhibition of S. sclerotiorum (Table 2), indicating that the SR negatively regulates strain DF41 biocontrol. We next addressed whether the SR affects LP expression. HPLC analysis of culture extracts showed that LP levels were elevated between 1.5- and 2.0-fold in the DF41 relA and relA spoT mutants compared to those in strain DF41 (Fig. 3). Moreover, at 12 h, transcription of the NRPS biosynthetic genes was increased over 5 times in the SR mutant background relative to that in the wild type (Fig. 2). These findings suggest that in the absence of (p)ppGpp, the NRPS genes are induced earlier, leading to enhanced LP production. A similar finding was reported for Streptomyces clavuligerus, in which an relA mutant produced elevated levels of the antibiotics clavulanic acid and cephamycin C (11). Moreover, transcription of the clavulanic acid and cephamycin C biosynthetic genes was increased substantially in the relA-deficient strain (11). With respect to SR control over antibiotic production, Streptomyces species are probably the most well-studied group of bacteria (4, 11, 15, 18, 19). Depending on the strain of Streptomyces in question, the antibiotics produced, and the growth media employed, the SR has been found to either positively (4, 15, 18, 19) or negatively (11) regulate antibiotic production. Hence, for Streptomyces spp., the SR appears to have a variable effect on antibiotic production. For biocontrol strains of Pseudomonas, little is known about SR control over secondary metabolite production and fungal antagonism. One exception, however, is Pseudomonas sp. strain MIS38. Therein, a transposon insertion in spoT resulted in elevated (p)ppGpp levels and reduced production of the LP arthrofactin (35), consistent with the inverse relationship between (p)ppGpp levels and LP expression observed for strain DF41.
In pseudomonads, regulation of biocontrol factors is governed by a complex network that functions at both the transcriptional and posttranscriptional levels. At the top of the hierarchy, the GacS/GacA two-component signal transduction system is essential for biological control in Pseudomonas spp. (12, 13). Mutation of either component typically leads to a complete loss of antagonistic activity (13), which was found to be the case for strain DF41 (2). Because the SR inhibits expression of factors that are also under Gac control, we were interested in determining whether a connection exists between the two systems in strain DF41. In this study, we observed a 1.7-fold decrease in relA expression in the gacS-negative background (Fig. 5). (p)ppGpp analysis also revealed a dramatic reduction in the level of this nucleotide in the gacS mutant, which was increased upon the addition of gacS in trans (Fig. 1B). These findings indicate that the expression of relA is influenced by the Gac system, although the molecular mechanism(s) underlying this regulation has yet to be uncovered. In other pseudomonads, the Rsm system forms part of the Gac regulatory cascade (12). After Gac activation, a series of regulatory RNAs are produced that antagonize the effects of RsmA-like proteins, which bind to and block translation of biocontrol mRNAs (12). The only Rsm component that has been identified in strain DF41 thus far is a homolog of the regulatory RNA RsmZ (T. R. de Kievit and C. Berry, unpublished data). The presence of RsmZ suggests that a cognate repressor protein(s) and additional regulatory RNAs likely exist, some or all of which may play a role in relA regulation.
During the SR, RNAP binding to (p)ppGpp and additional effectors, such as DksA, leads to activation and repression of different subsets of genes (27). For genes that are activated, increased expression is believed to occur by both direct and indirect mechanisms (see reference 27 and references therein). The direct effect involves RNAP in complex with (p)ppGpp increasing transcription from a given promoter. Indirect effects arise because (p)ppGpp facilitates RNAP binding to alternative sigma factors, leading to induction of these regulons. In this study, the sigma factor gene rpoS was found to be upregulated during the SR, while the NRPS genes were repressed. To better understand how the SR affects transcription of rpoS and the NRPS genes, we expressed rpoS constitutively from the kanamycin promoter in the SR mutants. The presence of plasmid pME-rpoS in the relA and relA spoT mutants increased rpoS transcription to near wild-type levels (Fig. 5). In strain DF41, rpoS is positively autoregulated, as evidenced by the increased transcription brought about by pME-rpoS (Fig. 5B). It is possible that in the SR mutants, the elevated level of plasmid-encoded RpoS enabled it to bind RNAP in the absence of (p)ppGpp, resulting in increased rpoS transcription. On the other hand, addition of rpoS had no effect on NRPS gene transcription. In this situation, (p)ppGpp may affect transcription from these promoters directly by decreasing NRPS transcription. Alternatively, the SR may exert its effects indirectly, through an as yet unidentified regulator of NRPS gene expression. Clearly, a great deal of work remains in order for us to understand exactly how the SR affects expression of these and other genes in strain DF41. When we examined whether the presence of rpoS in trans altered the SR mutant phenotype, the only trait that was affected was protease activity, which was restored to wild-type levels (Table 3). A protease-encoding gene(s) has not yet been identified in strain DF41; however, the results of the present study suggest that this gene(s) is under RpoS control.
In summary, we demonstrated that the SR negatively affects the antifungal activity of strain DF41 in vitro. Whether SR mutants would demonstrate enhanced biocontrol in planta has not been established. When an organism encounters a stress, several mechanisms may be induced to mount an appropriate response. For instance, bacteria frequently produce long-chain polymers of phosphate residues, termed poly(P)s, when starved for nutrients (29). These molecules are synthesized by polyphosphate kinase, the product of the ppk gene, while their hydrolysis to Pi residues depends upon the ppx-encoded exopolyphosphatase (29). It was reported that in Streptomyces lividans, expression of the antibiotics actinorhodin and undecylprodigiosin was enhanced in the ppk mutant under phosphate-limiting conditions, presumably as a result of reduced endogenous Pi levels (6). Furthermore, Pi-mediated inhibition is believed to occur independently of (p)ppGpp, as relA transcription in the poly(P)-deficient strain was similar to that in the wild type (6). Taken together, these findings suggest that the regulation of antibiotic production is complex and requires multiple pathways to respond appropriately to the prevailing conditions. Synthesis of antifungal factors, including LP and protease, is an energetically costly process. When nutrients are scarce, the SR enables strain DF41 to shift resources from secondary metabolite production to activities required for coping with starvation. Because biocontrol bacteria are often forced to survive under nutrient-depleted conditions, it is essential to understand the impact of the SR on secondary metabolism, as this directly impacts antagonism.
ACKNOWLEDGMENTS
We gratefully acknowledge financial support for this work through grants awarded to T.R.D.K. and W.G.D.F. from the Natural Sciences and Engineering Research Council (NSERC) discovery grant program.
We are indebted to Ivan Oresnik for his assistance with the (p)ppGpp analysis and to Peter Loewen and Tom Ward for help with the HPLC analysis.
Footnotes
Published ahead of print on 24 June 2011.
REFERENCES
- 1. Berry C. L. 2010. An investigation of the mechanisms underlying biocontrol activity of a novel canola-associated bacterial isolate, Pseudomonas species DF41. Ph.D. thesis. University of Manitoba, Winnipeg, Manitoba, Canada [Google Scholar]
- 2. Berry C., Fernando W. G. D., Loewen P. C., de Kievit T. R. 2010. Lipopeptides are essential for Pseudomonas sp. DF41 biocontrol of Sclerotinia sclerotiorum. Biol. Control 55:211–218 [Google Scholar]
- 3. Cashel M. 1994. Detection of ppGpp and pppGpp accumulation patterns in Escherichia coli mutants, p. 341–356 In Adolph K. W. (ed.), Methods in molecular genetics, vol. 3, part A. Academic Press, New York, NY [Google Scholar]
- 4. Chakraburtty R., Bibb M. 1997. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J. Bacteriol. 179:5854–5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choi K. H., et al. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2:443–448 [DOI] [PubMed] [Google Scholar]
- 6. Chouayekh H., Virolle M. J. 2002. The poly-phosphate kinase plays a negative role in the control of antibiotic production in Streptomyces lividans. Mol. Microbiol. 43:919–930 [DOI] [PubMed] [Google Scholar]
- 7. Cupp-Enyard C. 17 September 2008. Sigma's non-specific protease activity assay: casein as a substrate. J. Vis. Exp. doi:10.3791/899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Finan T. M., Kunkel B., Vos G. F. D., Signer E. R. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gambello M. J., Iglewski B. H. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173:3000–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Girard G., Rij E., Lugtenberg J., Bloomberg G. 2006. Regulatory roles of psrA and rpoS in phenazine-1-carboxamide synthesis by Pseudomonas chlororaphis PCL1391. Microbiology 152:43–58 [DOI] [PubMed] [Google Scholar]
- 11. Gomez-Escribano J. P., Martin J. F., Hesketh A., Bibb M. J., Liras P. 2008. Streptomyces clavulingerus relA-null mutants overproduce clavulanic acid and cephamycin C: negative regulation of secondary metabolism by (p)ppGpp. Microbiology 154:744–755 [DOI] [PubMed] [Google Scholar]
- 12. Haas D., Défago G. 2005. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3:307–319 [DOI] [PubMed] [Google Scholar]
- 13. Heeb S., Haas D. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other Gram-negative bacteria. Mol. Plant Microbe Interact. 14:1351–1363 [DOI] [PubMed] [Google Scholar]
- 14. Heeb S., et al. 2000. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram negative, plant-associated bacteria. Mol. Plant Microbe Interact. 13:232–237 [DOI] [PubMed] [Google Scholar]
- 15. Hesketh A., Sun J., Bibb M. 2001. Induction of ppGpp synthesis in Streptomyces coelicolor A3(2) grown under conditions of nutritional sufficiency elicits actII-ORF4 transcription and actinorhodin biosynthesis. Mol. Microbiol. 39:136–144 [DOI] [PubMed] [Google Scholar]
- 16. Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86 [DOI] [PubMed] [Google Scholar]
- 17. Holloway B. W., Krishnapillai V., Morgan A. F. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoyt S., Jones G. H. 1999. relA is required for actinomycin production in Streptomyces antibioticus. J. Bacteriol. 181:3824–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jin W., et al. 2004. Two relA/spoT homologous genes are involved in the morphological and physiological differentiation of Streptomyces clavuligerus. Microbiology 150:1485–1493 [DOI] [PubMed] [Google Scholar]
- 20. Kado C. I., Keskett M. G. 1970. Selective media for isolation of Agrobacterium, Corynebacterium, Erwinia, Pseudomonas, Xanthomonas. Phytopathology 60:969–976 [DOI] [PubMed] [Google Scholar]
- 21. King E. O., Ward M. K., Raney D. E. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301–307 [PubMed] [Google Scholar]
- 22. Laville J., et al. 1998. Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHAO. J. Bacteriol. 180:3187–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lindow S. E., Brandl M. T. 2003. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69:1875–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marahiel M. A., Stachelhaus T., Mootz H. D. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97:2651–2674 [DOI] [PubMed] [Google Scholar]
- 25. Miller J. H. 1972. Experiments in molecular genetics, p. 351–355 Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 26. Poritsanos N., Selin C., Fernando W. G. D., Nakkeeran S., de Kievit T. R. 2006. A GacS deficiency does not affect Pseudomonas chlororaphis PA23 fitness when growing on canola, in aged batch culture or as a biofilm. Can. J. Microbiol. 52:1177–1188 [DOI] [PubMed] [Google Scholar]
- 27. Potrykus K., Cashel M. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62:6235–6251 [DOI] [PubMed] [Google Scholar]
- 28. Preston M., et al. 1997. Contribution of proteases and LasR to the virulence of Pseudomonas aeruginosa during corneal infections. Infect. Immun. 65:3086–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rao N. N., Gómez-Garcia M. R., Kornberg A. 2009. Inorganic polyphosphate: essential for growth and survival. Annu. Rev. Biochem. 78:605–647 [DOI] [PubMed] [Google Scholar]
- 30. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 31. Sarniguet A., Kraus J., Henkels M. D., Muehlchen A. M., Loper J. E. 1995. The sigma factor σs affects antibiotic production and biological control activity of Pseudomonas fluorescens Pf-5. Proc. Natl. Acad. Sci. U. S. A. 92:12255–12259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Savchuk S. C., Fernando W. G. D. 2004. Effect of timing of application and population dynamics on the degree of biological control of Sclerotinia sclerotiorum by bacterial antagonists. FEMS Microbiol. Ecol. 49:379–388 [DOI] [PubMed] [Google Scholar]
- 33. Schweizer H. P. 1993. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques 15:831–833 [PubMed] [Google Scholar]
- 34. Reference deleted.
- 35. Washio K., Lim S. P., Roongsawang N., Morikawa M. 2010. Identification and characterization of the genes responsible for the production of the cyclic lipopeptide arthrofactin by Pseudomonas sp. MIS38. Biosci. Biotechnol. Biochem. 74:992–999 [DOI] [PubMed] [Google Scholar]
- 36. West S. E., Schweizer H. P., Dall C., Sample A. K., Runyen-Janecky L. J. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their application in Pseudomonas aeruginosa. Gene 148:81–86 [DOI] [PubMed] [Google Scholar]
- 37. Xiao H., et al. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980–5990 [PubMed] [Google Scholar]