Abstract
To understand the mechanisms by which CDKs regulate cell cycle progression, it is necessary to identify and characterize the physiological substrates of these kinases. We have developed a screening method to identify novel CDK substrates. One of the cDNAs identified in the screen is identical to the recently isolated NPAT gene. Here we show that NPAT associates with cyclin E–CDK2 in vivo and can be phosphorylated by this CDK. The protein level of NPAT peaks at the G1/S boundary. Overexpression of NPAT accelerates S-phase entry, and this effect is enhanced by coexpression of cyclin E–CDK2. These results suggest that NPAT is a substrate of cyclin E–CDK2 and plays a role in S-phase entry.
Keywords: Cyclin, CDK, substrates, NPAT, S-phase entry
Progression through the eukaryotic cell cycle requires the activation of cyclin-dependent kinases (CDKs). The activities of these kinases are tightly regulated by both positive and negative factors in normal cells, and perturbation of CDK activities can result in tumorigenesis (Hunter and Pines 1994; Sherr 1996). Although the regulation of CDK activation has been studied intensively in recent years, the mechanisms by which these CDKs regulate cell cycle progression are not fully understood partly because few substrates of these kinases have been identified.
Previous studies have shown that some protein kinases, including CDKs, form stable complexes with their respective substrates in vivo (Kato et al. 1993; Dynlacht et al. 1994, 1997; Krek et al. 1994; Karin and Hunter 1995; Zhu et al. 1995; Adams et al. 1996; Kallunki et al. 1996). This physical interaction has been suggested as a mechanism by which the specificity and efficiency of the kinase reactions are achieved. Thus, a possible approach for identifying substrates of a protein kinase is to search for proteins that interact physically with the kinase. We have developed a method to screen expression libraries for potential CDK substrates and identified a novel substrate of cyclin E–CDK2, a kinase complex that regulates the G1/S-phase transition and is required for the initiation of DNA replication in higher eukaryotes (Fang and Newport 1991; Ohtsubo and Roberts 1993; Tsai et al. 1993; Resnitzky et al. 1994).
Results and Discussion
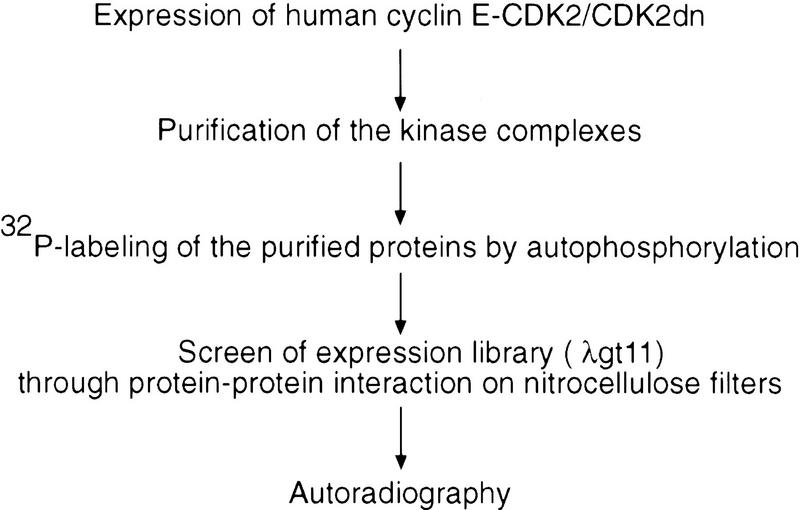
To identify cDNAs that encode new cyclin E–CDK2-interacting proteins and thus potential substrates, we screened a phage expression library by a binding assay (Fig. 1). Unlike previously reported yeast two-hybrid screens in which only CDK2 was used as the probe for interacting proteins (Hannon et al. 1993; Harper et al. 1993), we used a preparation of purified cyclin E–CDK2 complex as the probe for two reasons: (1) Active cyclin E–CDK2 complex was used, so the protein complexes could easily be labeled with [γ-32P]ATP by the active cyclin E–CDK2; and (2) more importantly, it has been shown that cyclins are involved in determining the substrate specificity of CDKs (Peeper et al. 1993; Dynlacht et al. 1994, 1997; Krek et al. 1994; Zhu etal. 1995). Thus, the presence of cyclin E may be required in a screen to identify important substrates of cyclin E–CDK2 complex. The kinase-inactive CDK2 (CDK2dn) was also used in the screen to maximize the binding to the substrates in case the kinase activity of the wild-type CDK2 weakened its binding to the substrates. From 1.2 × 106 clones screened, we isolated 34 positive clones. Of these, 27 encode proteins that have been demonstrated previously to be associated with cyclin E–CDK2 in vivo (8 clones for p27, 1 clone for p107, 18 clones for p130) (Lees et al. 1992; Hannon et al. 1993; Li et al. 1993; Polyak et al. 1994; Toyoshma and Hunter 1994). Isolation of p27, p107, and p130 from the screen demonstrates that the method can identify authentic cyclin E–CDK2-associated proteins. The other six cDNAs (one cDNA was isolated twice) encode fragments of proteins that have not been shown previously to interact with cyclin E–CDK2 in vivo. The deduced peptide sequence from one clone contained residues similar to the RRLFG motif shown previously to be involved in the interaction with cyclin A/E–CDK2 complexes (Zhu et al. 1995; Adams et al. 1996; Russo et al. 1996). Recombinant protein made from this cDNA clone bound cyclin E–CDK2, but not cyclin D1/CDK6, and could be phosphorylated by cyclin E–CDK2, but not cyclin D1/CDK4, in vitro (data not shown). Therefore, this clone was chosen for further studies. While our work was in progress, the gene was also isolated independently, under the names NPAT, E14, and CAN3, through positional cloning by virtue of the fact that the gene is located next to ATM on chromosome 11q22–q23 and may share a bidirectional promoter with ATM (Byrd et al. 1996; Imai et al. 1996; Chen et al. 1997).
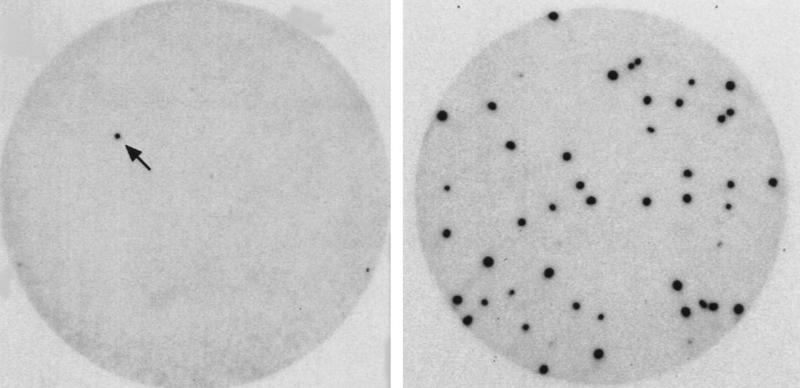
Figure 1.
Binding screen for cyclin E–CDK2-interacting proteins. (Top) Outline of the binding screen (for details, see Materials and Methods). (Bottom) Autoradiographs showing a positive clone identified in primary (left) and tertiary (right) screens, respectively. In the primary screen, the λgt11 phage library was plated at a density of 1.8 × 104 PFU/15-cm plate. In the tertiary screen, 50–100 PFU was plated on a 10-cm plate. The arrow indicates the positive clone in the primary screen.
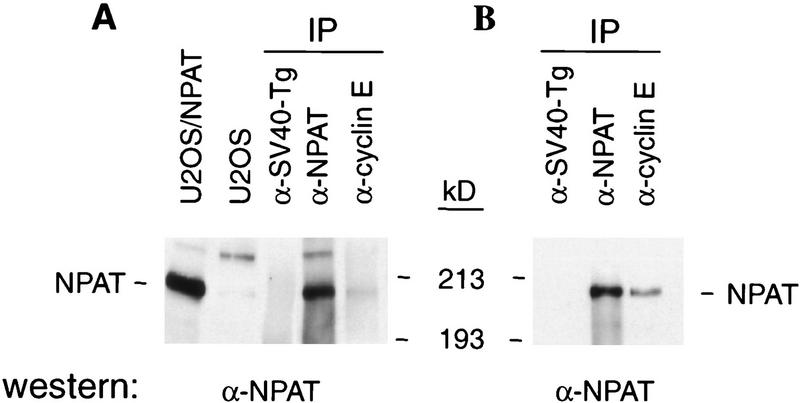
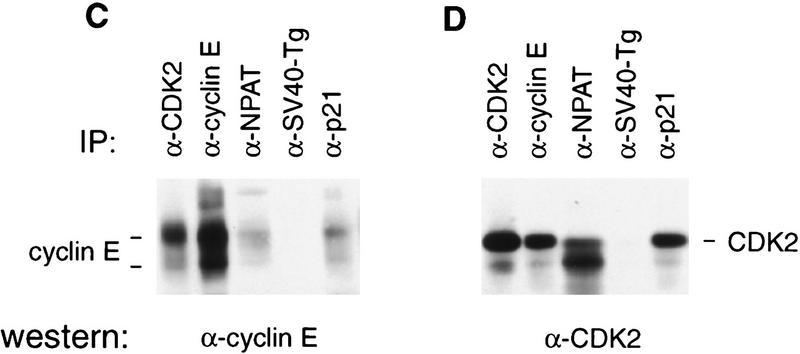
The NPAT (for nuclear protein mapped to the AT locus) protein consists of 1427 amino acids (Imai et al. 1996) and has an apparent molecular mass of 210 kD when analyzed by SDS-PAGE (Fig. 2A; data not shown). To determine whether NPAT is associated with cyclin E–CDK2 in vivo, cell lysates were subjected to immunoprecipitation and Western blotting analysis using specific anti-cyclin E, anti-CDK2, and anti-NPAT antibodies. Although the proteins immunoprecipitated by a control antibody included neither cyclin E–CDK2 nor NPAT (Fig. 2), the protein complex immunoprecipitated by anti-cyclin E monoclonal antibody from human osteosarcoma U2OS cell lysates contained NPAT protein (Fig. 2A). Consistently, the amount of NPAT precipitated by anti-cyclin E antibody was increased in cells transfected with NPAT, cyclin E, and CDK2 expression plasmids (Fig 2B). In reciprocal experiments, analysis of the endogenous protein complex immunoprecipitated by anti-NPAT monoclonal antibody demonstrated the presence of cyclin E (Fig. 2C) and CDK2 (Fig. 2D) in the complex. The amount of cyclin E–CDK2 immunoprecipitated by the anti-NPAT antibody is comparable to that precipitated by an antibody against p21 (Fig. 2C,D), a known cyclin E–CDK2 interacting protein (Harper et al. 1993). These results confirmed physical association of NPAT with cyclin E–CDK2 in vivo. The identity of the 230-kD protein that is recognized by anti-NPAT antibody (Fig. 2A) is not clear at present, but it could be the product of a differentially spliced NPAT mRNA or the product of a related gene. Consistent with this notion, two mRNA species that hybridize to the NPAT sequence have been detected in all human adult tissues examined and in several human cell lines (Byrd et al. 1996; Imai et al. 1996; Chen et al. 1997; data not shown). The intensity of the band that migrates faster than the CDK2 band (Fig. 2D) varied greatly from experiment to experiment. The identity of the band is not clear and is currently being investigated.
Figure 2.
In vivo association of NPAT with cyclin E–CDK2. (A) Lysates (1 mg of total protein) from U2OS cells were immunoprecipitated with an anti-NPAT monoclonal antibody (DH3), anti-cyclin E antibody (HE67) or a control monoclonal antibody (anti-SV40-Tg antibody, PAB419), respectively. The immunoprecipitates were analyzed by Western blotting with a mouse anti-NPAT polyclonal antibody. The first lane was loaded with total protein (25 μg) from U2OS cells transfected with pCMV–NPAT, and the second lane was loaded with 50 μg of total protein from U2OS cells. (B) Same as described in A except that the lysates (0.25 mg of total protein) were prepared from U2OS cells transfected with NPAT (10 μg), cyclin E (2.5 μg), and CDK2 (2.5 μg) expression plasmids. (C, D) Lysates (1 mg of total protein) from U2OS cells were immunoprecipitated with indicated antibodies. The immunoprecipitates were analyzed by Western blotting with anti-cyclin E monoclonal antibody (HE12), (C) or anti-CDK2 antibody (D).
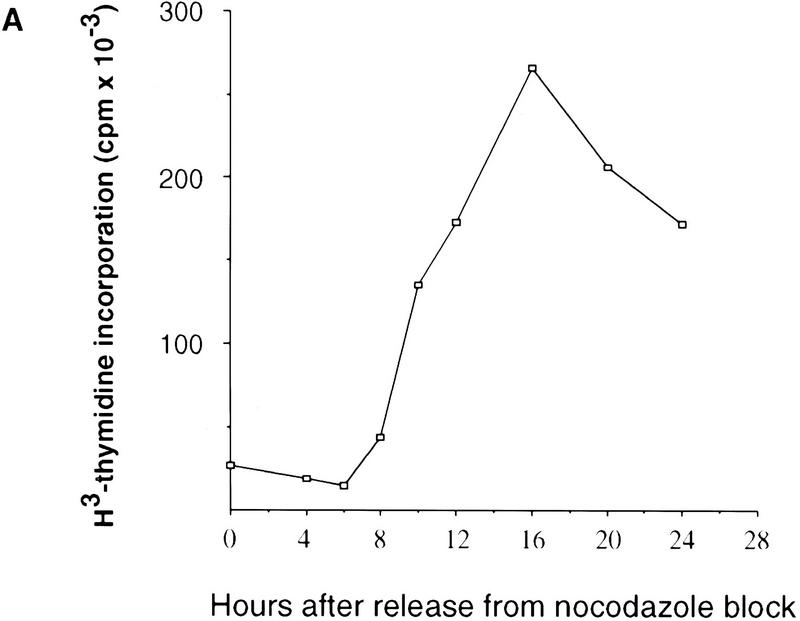
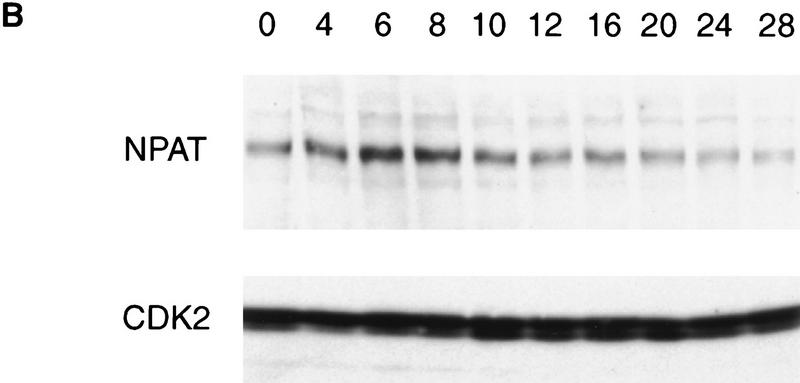
To examine the protein level of NPAT throughout the cell cycle, U2OS cells were synchronized in M phase by nocodazole treatment. The amount of NPAT in the cells at different times after release from the nocodazole block was analyzed by anti-NPAT antibody. Although NPAT appears to be expressed in all phases of the cell cycle, the protein level of NPAT peaks at the G1/S boundary and decreases when cells enter S phase (Fig. 3), suggesting that NPAT may play a role at the G1/S boundary. Consistently, NPAT protein is diminished in serum-starved or contact-inhibited normal human fibroblatst WI38 cells (data not shown).
Figure 3.
The level of NPAT protein peaks at the G1/S boundary. (A) DNA synthesis in U2OS cells after removal of nocodazole was monitored by [3H]thymidine incorporation. (B) Lysates (50 μg of total protein) prepared from cells at the indicated times (hr) after release from nocodazole block were analyzed by Western blotting with a rabbit anti-NPAT polyclonal antibody (top). As a control, the lysates were also subjected to Western blotting analysis with anti-CDK2 antibody (bottom).
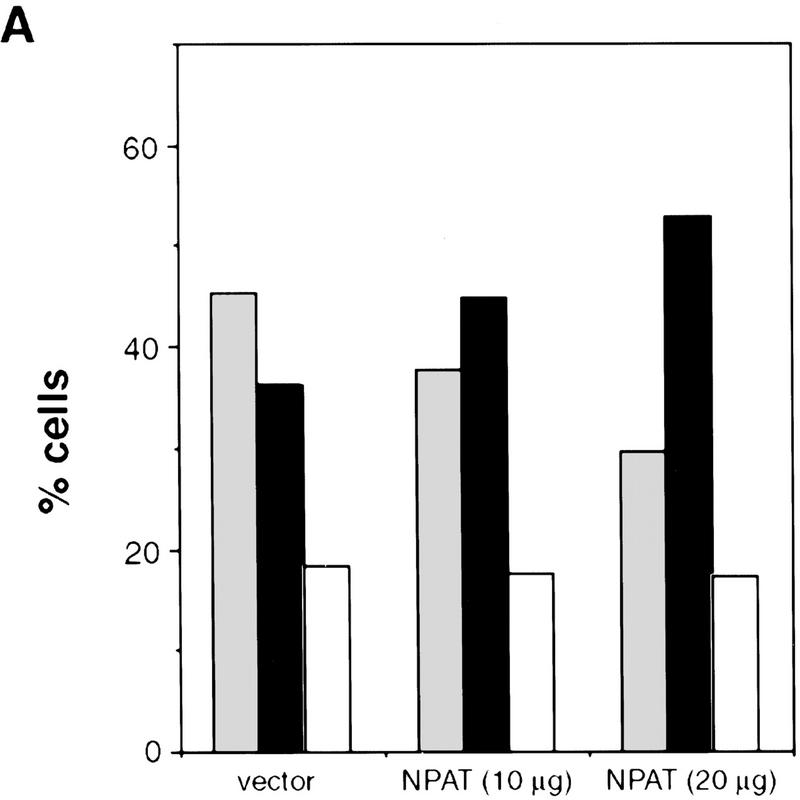
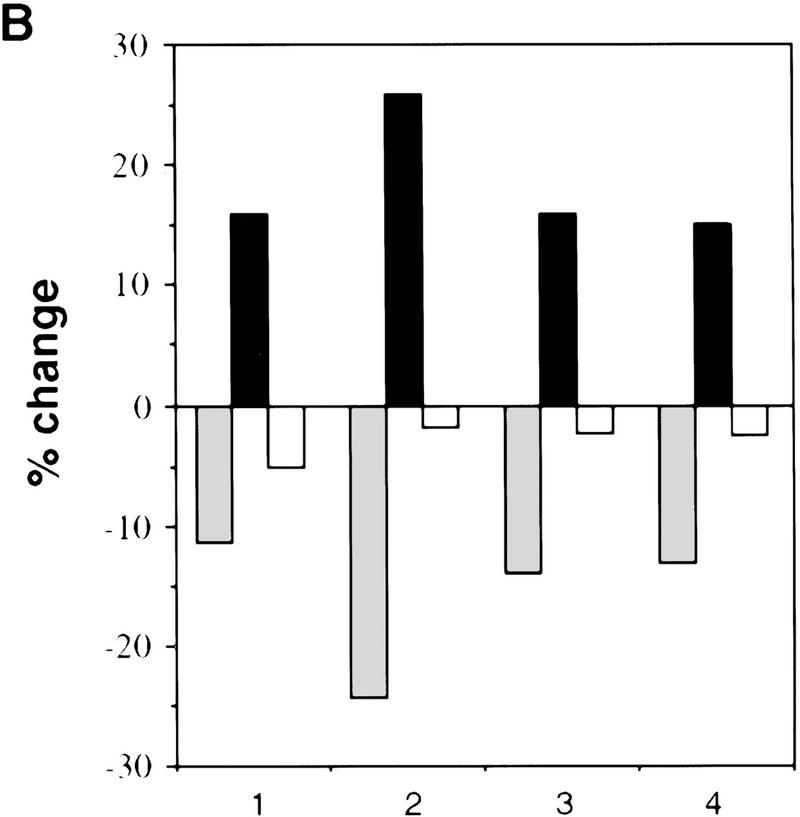
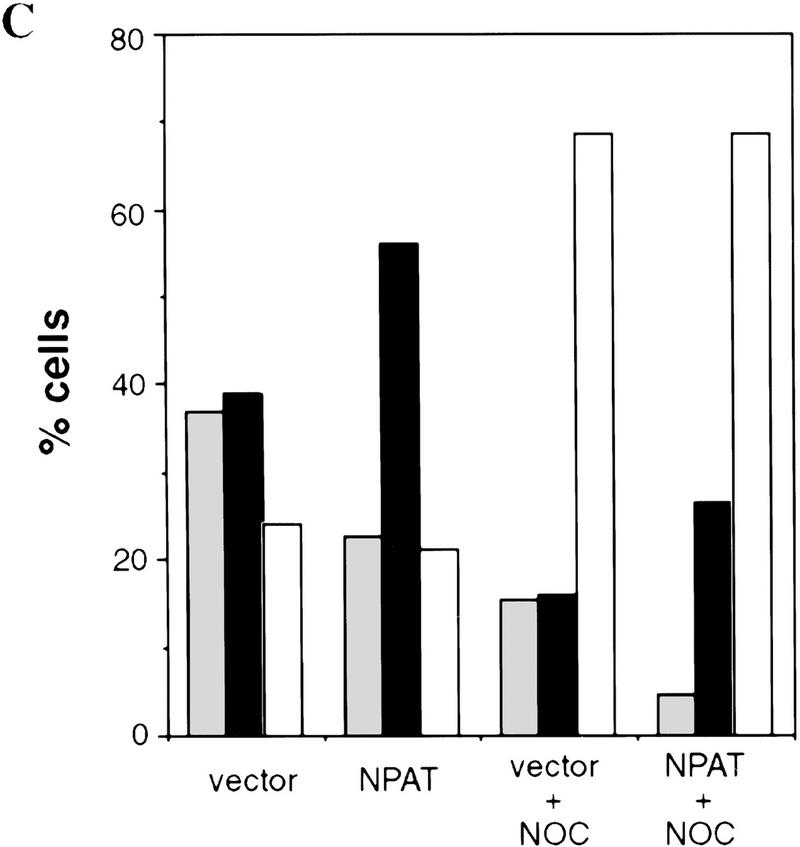
To gain further insight into the function of NPAT, we determined the effect of NPAT overexpression on cell cycle distribution. When transfected into U2OS cells, NPAT caused a large increase in the fraction of cells in S-phase and a concomitant decrease in the G1 population. (Fig. 4A,B). To rule out the possibility that the increase of S-phase population was due to an S phase block caused by overexpression of NPAT, we used nocodazole to block cell cycle at M phase and compared cell cycle distribution of NPAT-transfected cells with that of vector-transfected cells. As shown in Figure 4C, NPAT-transfected cells, like vector-transfected cells, proceeded to the M phase, demonstrating that the increase in S-phase population was not the result of an S-phase block. The effect of NPAT overexpression on cell cycle distribution is reminiscent of that resulting from overexpression of cyclin E or cyclin D1 in mammalian cells (Ohtsubo and Roberts 1993; Resnitzky et al. 1994). To determine whether overexpression of NPAT, like overexpression of cyclin E or cyclin D1, promotes entry into S phase, we examined the changes in the distribution of cells in the cell cycle at different times after transfection with NPAT or control vector (Table 1). Nocodazole was added at 22 hr after transfection to arrest cells at M phase so that the relative rates at which transfected cells passing through G1 phase into S phase could be measured and compared. At early times (e.g., 22 hr) after transfection, distribution of cells in the cell cycle was virtually identical for both NPAT- and vector-transfected cells. At later times (25, 28, 31, 34 hr), however, the percentage of cells in S phase is much higher in NPAT-transfected cells than in control vector-transfected cells. This increase in S-phase population was accompanied by a decrease in the G1 population of NPAT-transfected cells, whereas there was no significant change in G2/M population. These results show that overexpression of NPAT accelerates S-phase entry in U2OS cells. Taken together, these observations (Figs. 3 and 4; Table 1) suggest that NPAT may function in some step that is rate-limiting in the G1/S-phase transition. Although our results suggest that overexpression of NPAT results in shortening of G1 phase, it is not clear, though possible, whether S phase is lengthened to compensate for the shortened G1 when the cells are pushed into S phase. We note that the effect of overexpression of NPAT on cell cycle distribution varied considerably among different cell lines (data not shown), suggesting that in addition to the expression of NPAT, other unidentified events are also involved in the accelerated S-phase entry in these cells.
Figure 4.
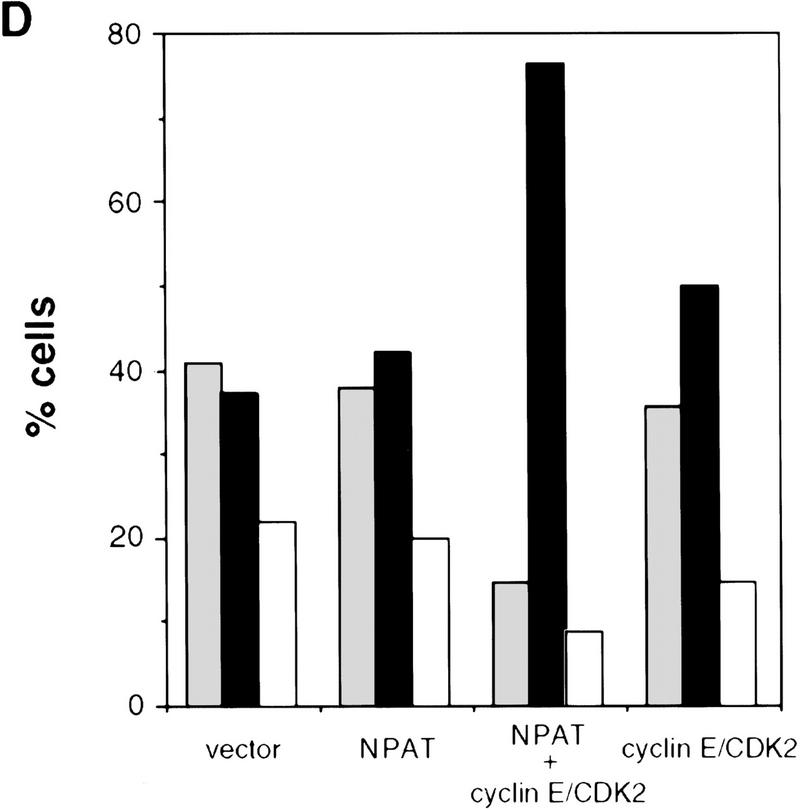
Effects of overexpression of NPAT on U2OS cell cycle distribution. (A) cells were transfected with control pCMV vector alone or with indicated amounts of pCMV–NPAT, and were analyzed by FACS. The results represent the means from triplicate samples. (B) Cells were transfected with 20 μg of vector or 20 μg of pCMV–NPAT and analyzed as in A. The absolute percentage differences between NPAT-transfected cells and vector-transfected cells (% NPAT-transfected cells-% vector-transfected cells) were calculated. The mean results from duplicate samples in four independent experiments (1–4) are depicted. (C) Cells were transfected with 20 μg of vector or pCMV–NPAT in the presence (vector + NOC; NPAT + NOC) or absence of nocodazole and were analyzed as in A. Shown are the mean results from duplicate samples; similar results have also been obtained in four other independent experiments. (D) Cells were transfected with the indicated expression plasmids (10 μg of NPAT; 2.5 μg of cyclin E; and 2.5 μg of CDK2) and analyzed as in A. The mean results from duplicate samples are presented. (░⃞) G1 phase; (▪) S phase; (□) G2/M phase.
Table 1.
Overexpression of NPAT in U2OS cells accelerates S-phase entry
|
|
22
hr
|
25 hr
|
28 hr
|
31 hr
|
34 hr
|
36 hr
|
46
hr
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| vector
|
NPAT
|
vector
|
NPAT
|
vector
|
NPAT
|
vector
|
NPAT
|
vector
|
NPAT
|
vector
|
NPAT
|
vector
|
NPAT
|
|
| G1 (%) | 62.9 | 60.8 | 57.4 | 48.3 | 43.6 | 31.5 | 35.7 | 17.5 | 29.6 | 11.6 | 34.8 | 12.6 | 27.7 | 9.9 |
| S (%) | 31.0 | 31.1 | 32.8 | 41.9 | 44.2 | 56.8 | 47.0 | 64.9 | 35.4 | 54.9 | 30.2 | 44.3 | 16.7 | 23.5 |
| G2M (%) | 6.1 | 8.1 | 9.8 | 9.8 | 12.2 | 11.7 | 17.3 | 17.6 | 35.0 | 33.5 | 35.0 | 43.1 | 55.6 | 66.6 |
U2OS cells were transfected with pCMV-neo (vector) or pCMV-NPAT. Nocodazole (75 ng/ml) was added to the culture medium at 22 hr post-transfection. Cells were harvested at the indicated times post-transfection, and the distribution of transfected cells in the cell cycle was analyzed by FACS. The data representing the mean results from two independent experiments deviated < 3% from each other.
Because NPAT and cyclin E–CDK2 associate in vivo and expression of each accelerates S-phase entry, we examined the effect of coexpression of NPAT and cyclin E–CDK2 on the cell cycle distribution. Coexpression of NPAT and cyclin E–CDK2 together has a cooperative effect on the increase in S-phase population (Fig. 4D), whereas coexpression of NPAT with cyclin A–CDK2 or with cyclin D1–CDK4 does not have this effect (data not shown), indicating that NPAT and cyclin E–CDK2 are not only associated physically but related functionally as well.
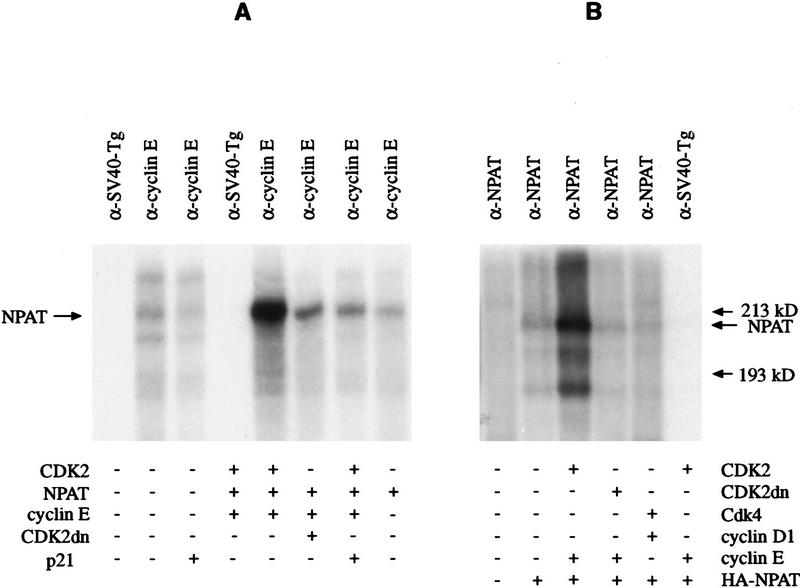
The observation that NPAT interacts physically and functionally with cyclin E–CDK2 in vivo and that recombinant NPAT can be phosphorylated by purified cyclin E–CDK2 in vitro (data not shown) prompted us to examine whether NPAT complexed with cyclin E–CDK2 in cells can be phosphorylated by this kinase in vitro. Cell lysates were immunoprecipitated with anti-cyclin E antibody, and the immunocomplexes were incubated with [γ-32P]ATP. As shown in Figure 5A, both endogenous and transfected NPAT in anti-cyclin E immunoprecipitations were phosphorylated. The fact that phosphorylation of NPAT is inhibited by purified p21 protein and that the phosphorylation of transfected NPAT is diminished when a kinase-inactive CDK2 is cotransfected (Fig. 5A) indicates that phosphorylation of NPAT in the anti-cyclin E immunoprecipitations depends on active cyclin E–CDK2 in the complex. To determine whether NPAT can be phosphorylated by cyclin E–CDK2 in vivo, we examined phosphorylation of NPAT in cells transfected with NPAT alone or together with cyclin E–CDK2 expression constructs. NPAT was phosphorylated at basal level in U2OS cells when transfected alone (Fig. 5B). When coexpressed with active cyclin E–CDK2, phosphorylation of NPAT was greatly increased. In contrast, coexpression of a kinase-inactive CDK2 with cyclin E did not increase the phosphorylation of NPAT. Increased phosphorylation of NPAT by cyclin E–CDK2 is not merely a consequence of accelerated S-phase entry, as overexpression of cyclin D1/CDK4, which also promotes S-phase entry (data not shown), did not result in increased phosphorylation of NPAT (Fig. 5B). These results suggest that NPAT is a substrate of cyclin E–CDK2 in human cells and that the cooperative effect of coexpression of NPAT and cyclin E–CDK2 on the cell cycle distribution (Fig. 4D) may result from the phosphorylation of NPAT by the cyclin E–CDK2 complex. Because cotransfection of NPAT with cyclin A/CDK2 did not show the cooperative effect on the cell cycle progression and the recombinant GST fusion protein encoded by the original isolated NPAT cDNA did not bind cyclin A/CDK2 in vitro (data not shown), we did not investigate further whether NPAT is associated with cyclin A/CDK2 in vivo or whether it is also a substrate of this kinase complex.
Figure 5.
Phosphorylation of NPAT by cyclin E–CDK2. (A) Lysates from U2OS cells (1 mg of total protein) or from U2OS cells (0.25 mg of total protein) transfected with plasmids expressing indicated proteins were precipitated with anti-cyclin E (HE 67) or a control antibody (pAb 419) as indicated. The immunoprecipitates were incubated with [γ-32p]ATP in the absence (−) or presence (+) of purified CDK inhibitor p21 protein (20 ng) as indicated. The phosphorylation of NPAT was determined by reimmunoprecipitation of the phosphorylated protein with anti-NPAT antibody (DH3). The amounts of expression plasmids used in the transfection experiments were 15 μg of NPAT, 4 μg of cyclin E, and 6 μg of CDK2 and CDK2dn, respectively. (B) U2OS cells were transfected with indicated expression plasmids, and the cellular phosphoproteins were labeled with [32P]orthophosphate. Lysates were immunoprecipitated with anti-NPAT antibody (DH3) or a control antibody as indicated. The immunoprecipitates were denatured and reimmunoprecipitated with anti-HA antibody (12CA5). Fifteen micrograms of HA-tagged NPAT, 4 μg of cyclin E, 6 μg of CDK2/CDK2dn, 4 μg of cyclin D1, and 6 μg of CDK4 expression plasmids were used in the transfection experiments.
Identification of cyclin E–CDK2 substrates is crucial for elucidating the mechanism by which this kinase regulates the G1/S-phase transition and initiation of DNA replication. Several cell cycle-related proteins have been proposed as substrates of cyclin E–CDK2 (Hinds et al. 1992; Li et al. 1993; Zhu et al. 1995; Herrera et al. 1996; Sheaff et al. 1997). Using the method we developed, we have identified NPAT as a novel substrate of cyclin E–CDK2. Here we show that NPAT appears to play a role at the G1/S boundary where cyclin E–CDK2 executes its function. Unlike the other potential substrate described, overexpression of NPAT promotes S-phase entry and this effect is enhanced by the expression of cyclin E–CDK2. Therefore, it appears likely that NPAT is one of the critical targets of cyclin E–CDK2 in promoting S-phase entry. This notion is consistent with the observation that inactivation of the mouse NPAT/NPAT gene by proviral insertion resulted in embryonic arrest at the eight-cell stage (Fruscio et al. 1997).
This work is the first characterization of the function of NPAT biochemically. Additional studies, employing different experimental approaches, are needed to elucidate the mechanism by which overexpression of NPAT promotes S-phase entry. One of the interesting questions that needs to be addressed is whether the function of NPAT protein is required for the G1/S phase transition. These important issues are currently under investigation. The method described in this report should also be applicable to the identification of substrates for other protein kinases. Thus, it provides a new approach for the study of not only cyclin-dependent kinases but also other protein kinases.
Materials and methods
Preparation of 32P-labeled human cyclin E–CDK2/cyclin E–CDK2dn
Expression and purification of human cyclin E–CDK2 and cyclin E–CDK2dn from recombinant baculoviruses in insect cells were carried out as described previously (Dynlacht et al. 1994). To label the protein complexes, purified cyclin E–CDK2 and cyclin E–CDK2dn were mixed and incubated in the presence of [γ-32P]ATP at 30°C for 30–60 min. The unincorporated free [γ-32P]ATP was removed from the labeled proteins using a Centricon 10 (Amicon). The specificities of labeled protein complexes were 5 × 106–1 × 107 cpm/μg of protein.
Binding screen of phage expression library
A λgt11 cDNA library prepared from human T-cell leukemia Molt-4 cells (Clontech) was screened. Phage plating, incubation, and protein transfer were performed essentially as suggested by the manufacturer based on the method described by Young and Davis (1983). After protein transfer, the filters were blocked in 5% milk in PBS with 0.5 mm PMSF, 0.2 mm benzamindine–HCl at 4°C overnight. The blocked filters were incubated with the labeled protein probes at a concentration of 0.12–0.2 μg of protein/ml of binding solution [25 mm Tris-HCl at pH 7.5, 100 mm KCl, 5 mm MgCl2, 1 mm DTT, 0.1% Tween 20, 5% glycerol, 1% milk, 10 mm NaF, 0.1 mm Na3VO4, 0.4 mm AEBSF (Calbiochem), 0.2 mm benzamidine-HCl, 1 μg/ml each of leupeptin and aprotinin] at room temperature for 1–3 hr. Filters were then washed four times for 15 min each in a washing solution (25 mm Tris-HCl at pH 7.5, 100 mm KCl, 5 mm MgCl2, 0.1 mm DTT, 0.1% Tween 20, 5% glycerol, 0.1 mm Na3VO4, 1 mm NaF, 0.2 mm PMSF, 0.2 mm benzamidine-HCl). Washed filters were air-dried and exposed to X-ray films at 80°C for 4–20 hr.
Antibodies, Western blotting, and immunoprecipitation
Anti-CDK2 antibody (M2) was from Santa Cruz. Anti-cyclin E (HE12, HE67) antibodies, anti-p21 antibody (CP68), and anti-HA antibody (12CA5) have been described previously (Zhu et al. 1995). Mouse anti-NPAT polyclonal and monoclonal antibodies were prepared according to Harlow and Lane (1988), using a GST fusion protein containing amino acids 782–1081 of NPAT as the immunogen. Rabbit anti-NPAT polyclonal antibodies were generated against His-tagged protein consisting of amino acids 1082–1427 of NPAT. The monoclonal antibodies used for immunoprecipitation were covalently coupled to protein A or protein G–Sepharose beads as described (Harlow and Lane 1988). Immunoprecipitation and Western blot analysis were performed according to Harlow and Lane (1988), except that the lysis and wash buffer was 50 mm Tris-HCl (pH 8.0), 250 mm NaCl, 5 mm EDTA, 1 mm DTT, 0.2% NP-40, 5% glycerol, 0.4 mm AEBSF, 0.5 mm benzamidine-HCl, 5 μg/ml of leupeptin, 5 μg/ml of aprotinin, 5 μg/ml of pepstatin A, 10 mm NaF, 0.1 mm Na3VO4, and 50 mm β-glycerophosphate.
Expression plasmids
The cyclin D, cyclin E, CDK2, and CDK2dn and the vector (pCMVneo) plasmids have been described previously (Hinds et al. 1992; van den Heuvel and Harlow 1993). The NPAT expression plasmid (pCMV–NPAT) was made by cloning the full-length NPAT cDNA sequence including both 5′ and 3′ noncoding sequence (Imai et al. 1996) into the pCMV–neo vector at the BamHI site. The HA-tagged NPAT was made by PCR. A 10-amino-acid epitope tag (YPYDVPDYAS) was inserted at the amino terminus of the protein. The sequence of the PCR product was verified by DNA sequencing. The DNA sequence encoding HA-tagged NPAT was cloned into the pCMV–neo vector.
Transient transfection and FACS analysis
Transient transfection and FACS analysis were performed essentially as described previously (van den Heuvel and Harlow 1993). Unless otherwise stated, cells were cultured in DMEM supplemented with 10% FBS. Transfections were done on 100-mm dishes using indicated plasmid DNA, together with pCMV–CD20 plasmid for the selection of transfected cells. Cells were harvested at 40–48 hr after transfection or as indicated. In the experiments described in Figure 4C, nocodazole (70 ng/ml) was added to half of the samples at 26 hr after transfection and the cells were harvested for analysis 22 hr later.
Cell cycle synchronization
U2OS cells were incubated with nocodazole (75 ng/ml) for 22 hr in DMEM supplemented with 10% FBS. Then, the cells were washed twice with PBS and incubated in the fresh medium without nocodazole.
[3H]thymidine incorporation
U2OS cells released from nocodazole block were incubated at a concentration of 5 × 105 cells/60-mm dish in DMEM with 10% FBS. At the indicated times 10μCi of [3H]thymidine was added to 3 ml of culture medium. After 1 hr, cells were washed with PBS and lysed with 0.3 n NaOH. DNA was precipitated with TCA (10%), and incorporation of [3H]thymidine into DNA was determined by liquid scintillation counting.
Phosphorylation assay of immunoprecipitates
Following immunoprecipitation with indicated antibodies, the immunoprecipitates were incubated with or without p21 in 50 μl of kinase buffer (50 mm HEPES at pH 7.0, 10 mm MgCl2, 4 mm MnCl2, 1 mm DTT, 0.1 mm BSA, 2 μCi of [γ-32P]ATP) at 30°C for 30 min. The reaction was terminated with SDS-PAGE loading buffer. The mixtures were diluted 20-fold with 2% BSA in PBS and reimmunoprecipitated with anti-NPAT monoclonal antibody (DH3). The immunoprecipitates were analyzed by autoradiography following SDS-PAGE. Recombinant human p21 protein was purified as described (Zhu et al. 1995).
In vivo [32P]phosphate labeling
U2OS cells were transfected with indicated plasmids. Thirty-two hours after transfection, the cells were washed twice with phosphate-free DMEM and incubated in phosphate-free DMEM for 1 hr. Then the cells were incubated in phosphate-free DMEM containing 0.5 mCi/ml of [32P]orthophosphate (ICN) for 4 hr. The lysates were immunoprecipitated and analyzed by autoradiography following SDS-PAGE.
Acknowledgments
We are grateful to Drs. Marie Classon, Brian Kennedy, Nick Dyson, and Hubert Chou for critical reading of the manuscript and suggestions. We thank Dr. Jim Koh for technical help, Chidi Ngwu for help with preparation of anti-NPAT monoclonal antibodies, David Dombkowski for assistance with flow cytometry analysis, Dr. Joshua LaBaer for suggestions, and members of Molecular Oncology Laboratory and MGH Cancer Center for stimulating discussions. This study was supported by an American Cancer Society postdoctoral fellowship to J.Z., a Damon Runyon postdoctoral fellowship to B.D., and grants from the National Institutes of Health to E.H.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL zhao@helix.mgh.harvard.edu; FAX (617) 726-7808.
References
- Adams PD, Sellers WR, Sharma SK, Wu AD, Nalin CM, Kaelin WG. Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol Cell Biol. 1996;16:6623–6633. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd PJ, Cooper PR, Stankovic T, Kullar HS, Watts GD, Robinson PJ, Taylor MR. A gene transcribed from the bidirectional ATM promoter coding for a serine rich protein: Amino acid sequence, structure and expression studies. Hum Mol Genet. 1996;5:1785–1791. doi: 10.1093/hmg/5.11.1785. [DOI] [PubMed] [Google Scholar]
- Chen X, Yang L, Udar N, Liang T, Uhrhammer N, Xu S, Bay J-O, Wang X, Dandakar S, Chiplunkar S, Klisak I, Telatar M, Yang H, Concannon P, Gatti R. CAND3: A ubiquitously expressed gene immediately adjacent and in opposite transcriptonal orientation to the ATM gene at 11q23.1. Mamm Genome. 1997;8:129–133. [PubMed] [Google Scholar]
- Dynlacht BD, Flores O, Lees JA, Harlow E. Differential regulation of E2F trans-activation by cyclin-cdk2 complexes. Genes & Dev. 1994;8:1772–1786. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- Dynlacht BD, Moberg K, Lees JA, Harlow E, Zhu L. Specific regulation of E2F family members by cyclin-dependent kinase. Mol Cell Biol. 1997;17:3867–3875. doi: 10.1128/mcb.17.7.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Newport JW. Evidence that the G1-S and G2-M transitions are controlled by different cdc2 proteins in higher eukaryotes. Cell. 1991;66:731–742. doi: 10.1016/0092-8674(91)90117-h. [DOI] [PubMed] [Google Scholar]
- Fruscio MD, Weiger H, Vanderhyden BC, Imai T, Shiomi T, Hori T-A, Jaenisch R, Gray DA. Proviral inactivation of the Npat gene of Mpv 20 mice results in early embryonic arrest. Mol Cell Biol. 1997;17:4080–4086. doi: 10.1128/mcb.17.7.4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, Demetrick D, Beach D. Isolation of the Rb-related p130 through its interaction with CDK2 and cyclins. Genes & Dev. 1993;7:2378–2391. doi: 10.1101/gad.7.12a.2378. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Herrera RE, Chen F, Weinberg RA. Increased histone H1 phosphorylation and relaxed chromatin structure in Rb-deficient fibroblasts. Proc Natl Acad Sci. 1996;93:11510–11515. doi: 10.1073/pnas.93.21.11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds PW, Mittnacht S, Dulic V, Arnold A, Reed SI, Weinberg RA. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- Hunter T, Pines J. Cyclins and cancer II: Cyclin D and cdk inhibitors come of age. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- Imai T, Yamauchi M, Seki N, Sugawara T, Saito T, Matsuda Y, Ito H, Nagase T, Nomura N, Hori T. Identification and characterization of a new gene physically linked to the ATM gene. Genome Res. 1996;6:439–447. doi: 10.1101/gr.6.5.439. [DOI] [PubMed] [Google Scholar]
- Kallunki T, Deng T, Hibi M, Karin M. c-Jun can recruit JNK to phosphorylate dimerization pateners via specific docking interactions. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- Karin M, Hunter T. Transcriptional control by protein phosphorylation: Signal transmission from the cell surface to the nucleus. Curr Biol. 1995;5:747–757. doi: 10.1016/s0960-9822(95)00151-5. [DOI] [PubMed] [Google Scholar]
- Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes & Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- Krek W, Ewen M, Shirodkar S, Arany Z, Kaelin WG, Livingston D. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- Lees E, Faha B, Dulic V, Reed SI, Harlow E. Cyclin E/cdk2 and cyclin A/cdk2 kinases associate with p107 and E2F in a temporally distinct manner. Genes & Dev. 1992;6:1874–1885. doi: 10.1101/gad.6.10.1874. [DOI] [PubMed] [Google Scholar]
- Li Y, Graham C, Lacy S, Duncan AM, Whyte P. The adenovirus E1A-associated 130-kD protein is encoded by a member of the retinoblastoma gene family and physically interacts with cyclins A and E. Genes & Dev. 1993;7:2366–2377. doi: 10.1101/gad.7.12a.2366. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M, Roberts JM. Cyclin-dependent regulation of G1 in mammalian fibroblasts. Science. 1993;259:1908–1912. doi: 10.1126/science.8384376. [DOI] [PubMed] [Google Scholar]
- Peeper DS, Parker LL, Ewen ME, Toebes M, Hall FL, Xu M, Zantema A, van der Eb AJ, Piwnica-Worms H. A- and B-type cyclins differentially modulate substrate specificity of cyclin-cdk complexes. EMBO J. 1993;12:1947–1954. doi: 10.1002/j.1460-2075.1993.tb05844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Lee M-H, Erdjument-Bromage H, Koff A, Roberts JM, Tempst P, Massague J. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78:59–66. doi: 10.1016/0092-8674(94)90572-x. [DOI] [PubMed] [Google Scholar]
- Resnitzky D, Gossen M, Bujard H, Reed SI. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol Cell Biol. 1994;14:1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletich NP. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- Sheaff RJ, Groudine M, Gordon M, Roberts JM, Clurman BE. Cyclin E–CDK2 is a regulator of p27kip1. Genes & Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- Tsai L-H, Lees E, Faha B, Harlow E, Riabowol K. The cdk2 kinase is required for the G1-to-S transition in mammalian cells. Oncogene. 1993;8:1593–1602. [PubMed] [Google Scholar]
- van den Heuvel S, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- Young RA, Davis RW. Efficient isolation of genes using antibody probes. Proc Natl Acad Sci. 1983;80:1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Harlow E, Dynlacht BD. p107 uses a p21CIP1-related domain to bind cyclin/cdk2 and regulate interactions with E2F. Genes & Dev. 1995;9:1740–1752. doi: 10.1101/gad.9.14.1740. [DOI] [PubMed] [Google Scholar]