Abstract
We measured expression and used biochemical characterization of multiple carbohydrate esterases by the xylanolytic rumen bacterium Prevotella ruminicola 23 grown on an ester-enriched substrate to gain insight into the carbohydrate esterase activities of this hemicellulolytic rumen bacterium. The P. ruminicola 23 genome contains 16 genes predicted to encode carbohydrate esterase activity, and based on microarray data, four of these were upregulated >2-fold at the transcriptional level during growth on an ester-enriched oligosaccharide (XOSFA,Ac) from corn relative to a nonesterified fraction of corn oligosaccharides (AXOS). Four of the 16 esterases (Xyn10D-Fae1A, Axe1-6A, AxeA1, and Axe7A), including the two most highly induced esterases (Xyn10D-Fae1A and Axe1-6A), were heterologously expressed in Escherichia coli, purified, and biochemically characterized. All four enzymes showed the highest activity at physiologically relevant pH (6 to 7) and temperature (30 to 40°C) ranges. The P. ruminicola 23 Xyn10D-Fae1A (a carbohydrate esterase [CE] family 1 enzyme) released ferulic acid from methylferulate, wheat bran, corn fiber, and XOSFA,Ac, a corn fiber-derived substrate enriched in O-acetyl and ferulic acid esters, but exhibited negligible activity on sugar acetates. As expected, the P. ruminicola Axe1-6A enzyme, which was predicted to possess two distinct esterase family domains (CE1 and CE6), released ferulic acid from the same substrates as Xyn10D-Fae1 and was also able to cleave O-acetyl ester bonds from various acetylated oligosaccharides (AcXOS). The P. ruminicola 23 AxeA1, which is not assigned to a CE family, and Axe7A (CE7) were found to be acetyl esterases that had activity toward a broad range of mostly nonpolymeric acetylated substrates along with AcXOS. All enzymes were inhibited by the proximal location of other side groups like 4-O-methylglucuronic acid, ferulic acid, or acetyl groups. The unique diversity of carbohydrate esterases in P. ruminicola 23 likely gives it the ability to hydrolyze substituents on the xylan backbone and enhances its capacity to efficiently degrade hemicellulose.
INTRODUCTION
Hemicelluloses comprise a heterogeneous, highly branched mixture of complex polysaccharides. In plant cell walls, these polysaccharides form intimate associations with cellulose, pectin, and lignin and markedly reduce the degradability of plant fiber (20). In a microbial ecosystem such as the rumen, fiber-degrading microorganisms meet this challenge by operating as a consortium. Several hemicellulolytic rumen species, especially the ones belonging to Prevotella and Butyrivibrio, have been shown to synergistically enhance the rate and extent of plant cell wall hydrolysis during co- or sequential culturing with cellulolytic bacteria (12, 18, 31, 32).
The hemicelluloses present in the cell walls of grasses, cereals, and hardwoods largely comprise xylan, a polymer consisting of linear chains of d-xylopyranosyl residues linked through β-(1→4) glycosidic linkages. For grasses and cereals, the xylan backbone is heavily substituted at the O-2 and/or O-3 position(s) with α-l-arabinofuranosyl and to a lesser extent with (4-O-methyl)-α-d-glucuronic acids and O-acetyl esters (6, 9, 42, 46). Ferulic acid esters, linked to the O-5 of the arabinose moieties, may link with other esters, forming various types of di-ester bridges, resulting in the inter- and possibly even intralinking of polymers within plant cell walls (24). The xylan backbone from hardwoods is mainly decorated with O-acetyl esters and 4-O-methyl-α-d-glucuronic acids (17, 39, 44).
Hemicellulose degradation is largely affected by the presence of ferulic acid and O-acetyl esters. In nature, these esters can be hydrolyzed by microbial carbohydrate esterases. Acetyl xylan esterases catalyze the removal of acetyl ester groups from O-2 or O-3 positions of d-xylopyranosyl residues (5), while ferulic acid esterases release ferulic acid from the O-2 or O-5 positions of α-l-arabinofuranosyl side chains (36, 37, 41). Consequently, acetyl and feruloyl esterases play important roles in hemicellulose degradation and, therefore, contribute to the complete hydrolysis of plant polysaccharides.
Various ruminal bacteria, including Prevotella ruminicola 23, have been shown to have esterase activities (21). However, knowledge regarding the diversity of esterases present in the genome, including the regulation, biochemical characteristics, and mechanistic action of these enzymes, is limited. Recently, the complete genome sequence of the ruminal bacterium P. ruminicola 23 was made available (38), therefore providing an opportunity to evaluate the repertoire of enzymes enabling this organism to function as a highly efficient hemicellulose-degrading bacterium.
In order to provide insight into the diversity and complexity of the multiple carbohydrate esterases present in P. ruminicola 23 cells, the two most upregulated esterases were biochemically characterized and their activities were compared with those of two other esterases that were constitutively expressed during growth on either an ester-enriched corn oligosaccharide, nonesterified corn oligosaccharide, or simple sugar substrates.
MATERIALS AND METHODS
Preparation of water-soluble acetylated and feruloylated XOSFA,Ac.
Corn fibers were kindly provided by a corn processing plant. These fibers were milled (<1 mm) by using a Retsch ZM200 mill (Retsch Gmbh, Haan, Germany), and a suspension of milled fibers (231 g dry weight in total) in water (1,869 g) plus sulfuric acid (4.6 g) was heated at 140°C for 30 min, followed by cooling within 5 min to room temperature.
A mixture of hemicellulases with an endoxylanase (Humicola insolens GH10 [49]), β-xylosidase (Trichoderma reesei GH3 [49]), and arabinofuranosidases (Meripilus giganteus GH51 and H. insolens GH43 [49]) (Novozymes, Bagsvaerd, Denmark) and the commercially available amyloglucosidase Spirizyme (Novozymes, Bagsvaerd, Denmark) were added to the suspension in order to degrade the most easily accessible starch and hemicellulose components (enzyme or protein [dry matter] in suspension was 0.025 and 0.05% [by weight] for each of the hemicellulases and amyloglucosidase, respectively). Two yeast strains (Nedalco, Bergen op Zoom, Netherlands) were used to consume the resulting arabinose, xylose, and glucose monosaccharides. The hydrolysate was then filtered, and the filtrate (2 liters in total), which contained mainly xylo-oligosaccharides, was applied in portions of 30 ml to Extract Clean C18 columns (Grace, Deerfield, IL). The bound fraction, named XOSFA,Ac, was eluted with methanol in one fraction. Pure water was added to keep the oligomers in solution. After methanol evaporation, the XOSFA,Ac was lyophilized, yielding 19 g of dry material.
The nonbound C18 fraction, named AXOS, was collected as well (composition based on dry matter: carbohydrates, 27% [arabinose, 8%; xylose, 10%; galactose, 4%; glucose, 3%; mannose plus rhamnose plus glucuronic acid, 2%]; protein, 11%; acetic acid and ferulic acid esters, 0%). AXOS components were separated further on an AKTA explorer instrument (GE Healthcare, Piscataway, NJ) equipped with a BioGel P2-column (900 by 26 mm; 200/400 mesh; Bio-Rad Laboratories, Hercules, CA). The column was thermostated at 60°C, and elution was performed with pure water (2 ml/min). One of the fractions (FA-20) contained only two oligomers: a xylobiose substituted with a single glucuronic acid group and a single acetyl group (X2GlcAAc) and a xylobiose substituted with a single 4-O-methylglucuronic acid group and a single acetyl group (X2GlcAmeAc). This fraction, FA-20, was used as one of the substrates in the esterase enzymatic assays.
Bacterial strain and growth conditions.
P. ruminicola 23 (ATCC 19189) was cultured anaerobically at 39°C in basal medium, consisting of 1.3% (NH4)2SO4, 0.1% hemin, 0.09% each of KH2PO4, NaCl, and methionine, 0.002% each of CaCl2 and MgCl2, 0.001% FeSO4, and 0.0001% CoCl and the volatile fatty acid (VFA) and vitamin solutions described by Pittman and Bryant (34). The medium was supplemented with either 30 mM xylose or glucose or equivalent percentages (wt/vol) of XOSFA,Ac and AXOS. Each culture was grown to midexponential phase (optical density at 600 nm [OD600], ∼0.5) and then harvested by centrifugation.
RNA extraction and cDNA generation.
Harvested cells were ground under liquid N2 using an oven-sterilized pestle and mortar. The cell homogenate was incubated in 1 ml of Trizol (Invitrogen, Carlsbad, CA) for 5 min at room temperature and then transferred to a sterile Eppendorf tube. The homogenate was mixed with 200 μl chloroform, and the mixture was centrifuged at 12,000 × g for 15 min at 4°C. The upper aqueous layer, containing total RNA, was collected, and RNA was precipitated using 0.5 ml isopropanol. The total RNA was subsequently pelleted, washed in 70% ethanol, and resuspended in sterile diethyl pyrocarbonate (DEPC)-treated double-distilled water (ddH2O). The resuspended RNA was then further purified using an RNeasy kit (Qiagen, Hilden, Germany) per the manufacturer's instructions. Purified RNAs were analyzed for quality using a Bioanalyzer RNA 6000 nano assay. All samples used gave an RNA index number (RIN) of 9 or greater. Total RNA was transcribed to cDNA, labeled, and purified using the Fairplay III system (Stratagene, La Jolla, CA) and Cy3 and Cy5 dyes (GE Healthcare, Piscataway, NJ), per the manufacturers' instructions.
Genome analysis and sequence availability.
The P. ruminicola 23 genome (accession number CP002006), along with the protein sequences of the three putative carbohydrate esterases designated Axe1-6A (YP_003575954), AxeA1 (YP_003575473), and Axe7A (YP_003575925) and that of the biochemically characterized xyn10D-fae1A (YP_003575973), are available in the NCBI GenBank database. Amino acid sequences were analyzed and classified into gene families using BlastP (1) and Interpro (23). The cellular localization of proteins was predicted with GnegPloc (10, 45) and by the presence of transmembrane (30) or signal peptide (2) hidden Markov models (HMMs). HMMs were predicted using HMMer (15).
Microarray analysis.
Transcriptional profiles were obtained using custom-designed 8- by 15-K microarrays supplied by Agilent (Santa Clara, CA). Each array possessed five replicate probes for each of the 2,875 open reading frames (ORFs) and 12 structural RNAs identified in the P. ruminicola 23 genome. Microarray slides were scanned using a GenePix Professional 4200 scanner and GenePix Pro 6.0 software (Molecular Devices, Sunnyvale, CA). Each condition (growth on glucose, xylose, XOSFA,Ac, or AXOS) was examined in triplicate, including a single dye swap. The resulting data were normalized and then analyzed in Bioconductor (19, 48) using a single-channel analysis. Genes with a transcriptional difference of 2-fold or greater and a false discovery rate (FDR) value of <0.05 were considered to be statistically significant. Since our interest was in carbohydrate esterase diversity, to validate the microarray analysis the transcriptional responses of 12 of the esterase genes were further examined by reverse transcription-quantitative PCR (RT qPCR). All RT qPCR assays were performed relative to three constitutively expressed genes, encoding the translation initiation factor IF-2 (infB), the beta subunit of the ATP-synthase F1 (atpD), and the beta subunit of the RNA polymerase (rpoB). Unlabeled cDNAs, prepared as described above from cells grown on either XOSFA,Ac or glucose, were amplified using Power SYBR green PCR master mix (Applied Biosystems, Carlsbad, CA) per the manufacturer's instructions. The 20-μl reactions were carried out in 384-well plates using an ABI 7900HT fast real-time PCR system (Applied Biosystems). Reaction mixtures were heated to 95°C for 10 min and then underwent 40 cycles of 95°C for 15 s followed by 62°C for 1 min. Reactions were performed in triplicate, and those with an amplification efficiency of ≥1.98 were analyzed using SDS software version 2.2.1 (Applied Biosystems). All primers used were designed using VectorNTI (Invitrogen, Carlsbad, CA) and are listed in Table 1.
Table 1.
Oligonucleotide primers used in this study
| Targeta | Primerb | Sequencec |
|---|---|---|
| RT qPCR | ||
| infB | infBf | 5′-TCAGAGCTGGCCACCATGA-3′ |
| infBr | 5′-CAGCATCCAGACGCTGGTT-3′ | |
| atpD | atpDf | 5′-TGGGTATCTATCCCGCTGTTG-3′ |
| atpDr | 5′-TTGACACGCTGGGCACAAT-3′ | |
| rpoB | rpoBf | 5′-GAAGACCTTGCTGAGTGGACTGA-3′ |
| rpoBr | 5′-TAGCAGGCTGGTCGAAACG-3′ | |
| PRU_0012 | 12f | 5′-TACTGCTATTACTCGCCATCAT-3′ |
| 12r | 5′-ATCCATAAATGCACGGCA-3′ | |
| PRU_1101 | 1101f | 5′-ACTGATTTTTGCACTGGTGATT-3′ |
| 1101r | 5′-AGTTTGCCCGATTGTTTAT-3′ | |
| PRU_1396 | 1396f | 5′-ACTAATTGTAGTGCTGGTGAGT-3′ |
| 1396r | 5′-TGGCAAAACGTGTAACGTA-3′ | |
| PRU_1726 | 1726f | 5′-GACTGGATGTGACTGCCCAT-3′ |
| 1726r | 5′-GGCAGCAAACGCTTGGTA-3′ | |
| PRU_2033 | 2033f | 5′-TGGTTCGCGCTCAGATTT-3′ |
| 2033r | 5′-CCGTTAGCATCCTTCTTCAT-3′ | |
| PRU_2212 | 2212f | 5′-CTGATGCTGGCTATGACCAT-3′ |
| 2212r | 5′-AACAATGGCTCTGCCTGA-3′ | |
| PRU_2584 | 2584f | 5′-AAGCCGAACTTTAGGTAAGA-3′ |
| 2584r | 5′-TTCTGTCTGCAACGGTTT-3′ | |
| PRU_2630 | 2630f | 5′-TCCTATTATCAGCAGCATTGCT-3′ |
| 2630r | 5′-CGGGCAGATAAACAGTTAGTT-3′ | |
| PRU_2678 | 2678f | 5′-TGTTTAACTTTGCACCCAAA-3′ |
| 2678r | 5′-CCACAAATAATCAGCACGATA-3′ | |
| PRU_2707 | 2707f | 5′-CTACCGAACAGGTTGGCAA-3′ |
| 2707r | 5′-ATCAGGAGGAAGCGTGGA-3′ | |
| PRU_2728 | 2728f | 5′-ACGTCCTGTAGATTATGCAGCA-3′ |
| 2728r | 5′-GCAGGATCAGAGATGTTGAA-3′ | |
| Heterologous | ||
| PRU_2212 | 2212_HEf | 5′-catatgCAGACCGCAAAGAAGTTTACGTTGAACCTGTC-3′ |
| 2212_HEr | 5′-ctcgagTTATTTGATGGCAGCCGCAATGATCTCGGCC-3′ | |
| PRU_2678 | 2678_HEf | 5′-catatgGAGAATTATCCCTATCGTGCTGATTATTTG-3′ |
| 2678_HEr | 5′-ctcgagTTATTTTACTTGTTTTTTGAGCCATAGC-3′ | |
| PRU_2707 | 2707_HEf | 5′-gacgacgacaagatgGCCCAGAAACGTAAGGC-3′ |
| 2707_HEr | 5′-gaggagaagcccggTTATTTTTTGAACAGATGTGG-3′ | |
| PRU_2728 (14) | 2728_HEf | 5′-catatgAAGAAACTATTAGTAGCGTTATCG-3′ |
| 2728_HEr | 5′-ctcgagTTACTTAAAGAGACTCTGAGCCATCTTTTC-3′ |
Primers were designed and used for the purposes of either RT qPCR or heterologous expression and characterization.
f, forward; r, reverse.
Primers were synthesized by Integrated DNA Technologies (Coralville, IA), with restriction enzyme sites indicated by lowercase type.
Cloning, expression, and purification of Xyn10D-Fae1A, Axe1-6A, AxeA1, and Axe7A.
The genes encoding the esterases, designated the Axe1-6A (PRU_2707), AxeA1 (PRU_2212), and Axe7A (PRU_2678) genes, were amplified from P. ruminicola 23 genomic DNA by PCR using primers with 5′ modifications to contain NdeI and XhoI restriction sites in the forward and reverse primers, respectively (primers are listed in Table 1). The amplified genes were cloned into the TA cloning vector pGEM-T (Promega, Madison, WI), transformed into Escherichia coli JM109 competent cells (Stratagene, La Jolla, CA), and sequenced to confirm the integrity of the coding sequence. The correct inserts were subcloned into a modified pET28a vector (Novagen, San Diego, CA). The modification was a replacement of the kanamycin resistance gene present in the plasmid with the ampicillin resistance gene. The sequencing of each coding sequence was carried out by the W. M. Keck Center for Comparative and Functional Genomics, University of Illinois. The cloning of the gene for xyn10D-fae1A into the modified pET28a vector was described in our earlier publication (14). Each resulting pET28a-gene construct was introduced into E. coli BL21-CodonPlus (DE3) RIL competent cells (Strategene, La Jolla, CA), and protein was expressed as described by Dodd et al. (14). The cells were pelleted, resuspended in 25 ml of lysis buffer (50 mM Tris-HCl, 300 mM NaCl [pH 7.5]), and then ruptured by using a French pressure cell (American Instrument Company Inc., Silver Spring, MD). The ruptured cell suspension was centrifuged (30,000 × g, 20 min, 4°C), and the supernatant was subjected to purification by immobilized-metal affinity chromatography (IMAC) (Talon polyhistidine tag purification resin; Clontech, Mountain View, CA) as described by the manufacturer. Aliquots of the collected fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Purification of Xyn10D-Fae1A followed the procedure described by Dodd et al. (14). For the proteins Axe1-6A, AxeA1, and Axe7A, IMAC fractions were pooled and subjected to size exclusion chromatography with an AKTA purifier system equipped with a Superdex 200 10/300 GL column and UV detector (GE Healthcare, Piscataway, NJ). Protein solutions (100 μl) with a concentration in the range of 3 to 10 mg/ml were subjected to the Superdex 200 column. The buffer used was 50 mM citrate (pH 6.0), and the flow rate was 0.8 ml/min. Fractions (100 μl) were collected and analyzed by SDS-PAGE, and highly purified fractions were assayed for esterase activity using β-d-glucose-pentaacetate as a substrate as described below. Fractions active in esterase activity were pooled, and enzyme concentrations were determined with the Pierce bicinchoninic acid (BCA) microplate procedure (Thermo Scientific, Rockford, IL).
Protein content and sugar composition analysis of XOSFA,Ac.
The nitrogen content of XOSFA,Ac was quantified using a Dumas-based FlashEA1112 nitrogen analyzer (Thermo Scientific, Rockford, IL) with methionine as a calibration standard (47). The protein content (percentage [wt/wt]) was then calculated by multiplying the nitrogen content (percentage [wt/wt]) by a factor of 6.25. Sugar composition was determined using methanolysis as previously described (13).
Ester content.
The amount of alkali-labile acetic acid was determined using a high-pressure liquid chromatography (HPLC) (Thermo Separation Products) instrument equipped with an Aminex HPX column (54). Ferulic and coumaric acids were released under N2 in alkali methanol (2% [vol/vol] methanol in 0.5 M KOH) in the absence of UV light for 16 h at room temperature using p-hydroxybenzoic acid as an internal standard for quantification purposes. Subsequently, the ferulic and coumaric acids were extracted with ethyl acetate, redissolved in methanol, and analyzed. The analysis was performed on an Acella ultraperformance liquid chromatography (UPLC) system (Thermo Scientific, Rockford, IL) coupled to a photodiode array (PDA) detector and an LTQ XL mass detector equipped with an electrospray ionization matrix (ESI) source (Thermo Scientific, Rockford, IL). Separation was performed on a Hypersyl GOLD column (1.9 by 150 mm; 1.9 μm; Thermo Scientific, Rockford, IL) at a flow rate of 0.4 ml per minute at 30°C. The mobile phase was composed of H2O plus 1% (vol/vol) acetonitrile plus 0.2% (vol/vol) acetic acid (phase A) and acetonitrile plus 0.2% (vol/vol) acetic acid (phase B). The elution profile was as follows: the first 5 min isocratic on 0% phase B; 5 to 23 min, phase B (linearly 0% to 50%); 23 to 24 min, phase B (linearly 50% to 100%); followed by cleaning and reconditioning of the column. Spectral data were collected from 200 to 600 nm, and quantification was performed at 320 nm. Mass spectrometry (MS) data were collected in the negative mode with an ion spray voltage of 3.5 kV, a capillary voltage of −20 V, and a capillary temperature of 350°C. Full MS scans were made within the range m/z 150 to 1,500, and MS2 data of the most intense ions were obtained.
MALDI-TOF MS.
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) was performed using an Ultraflex instrument (Bruker Daltonics, Bremen, Germany) equipped with a nitrogen laser of 337 nm. The sample preparation and choice of matrix were described previously (26). Oligosaccharides were detected using the reflector mode. Proteins were detected in the linear mode.
Capillary electrophoresis (CE) of 9-aminopyrene-1,4,6-trisulfonate (APTS)-derivatized oligosaccharides was performed as described previously (27). Both CE coupled to a laser-induced fluorescence (LIF) detector and CE coupled to an electrospray ion-trap mass spectrometry (ESI-MSn) detector were used for the identification and characterization of the oligosaccharides (28).
Enzyme assays.
Feruloyl esterase activity was assayed by evaluating the release of ferulic acid from methylferulate (Sigma-Aldrich, St. Louis, MO), wheat bran (25), and corn fiber isolated in the current study. Ferulic acid released was analyzed as described above under “Ester content.” Various acetylated substrates were used to evaluate the acetyl esterase activity. The 4-O-methylumbelliferylacetate, xylose-tetraacetate, β-d-glucose-pentaacetate, cephalosporin C zinc salt, and N-acetyl-neuraminic acid type IV-S were obtained from Sigma-Aldrich (St. Louis, MO). The N-acetyl-d-glucosamine was purchased from Merck (Whitehouse Station, NJ), and the xanthan-acetylated CX911 was obtained from Degussa (Baupte, France). Other acetylated substrates, i.e., sugar beet pectin (33), arabinofuranosidase B-treated apple MHR-B (43), acetylated neutral xylo-oligosaccharides (DP 2-20) (Euc NI A) (26), acetylated GlcAme-xylo-oligosaccharides (DP 4-12) (Euc AII plus AIII) (26), and acetylated xylan (29), were prepared as described in earlier reports. The XOSFA,Ac and FA-20 (X2GlcAAc plus X2GlcAmeAc) were prepared as described above.
Substrate and enzyme concentrations used were as shown (see Table 4), except those used in the hydrolysis of AXOS, where 4 g/liter substrate was assayed with 0.4 μM enzyme. Acetic acid released was assayed with the K-ACETAF kit from Megazyme (Bray, Ireland).
Table 4.
Activity of P. ruminicola 23 esterases tested on various acetylated substrates
| Substrate | Reaction conditions |
Activity (mmol HAc/μmol protein) at 20 min; 2 h; 20 ha |
||||
|---|---|---|---|---|---|---|
| Substrate (g/liter) | Protein concn (μM), AxeA1; Axe7A; Axe1-6A; Xyl10D-Fae1A | AxeA1 | Axe7A | Axe1-6A | Xyn10D-Fae1A | |
| Glucose-pentaacetate | 0.47 (1.2 mM) | 0.05; 0.05; 0.08; 0.25 | 27; 63; ND | 4; 12; ND | 9; 19; ND | <0.5b |
| Cephalosporin C | 0.68 (1.7 mM) | 0.05; 0.05 | 11; 13; ND | 7; 8; ND | ND | ND |
| N-Acetyl-d-glucosamine | 0.64 (2.9 mM) | 0.33; 0.33 | <0.5b | <0.5b | ND | ND |
| N-Acetyl-neuraminic acid (type IV-S) | 0.60 (1.9 mM) | 0.33; 0.33 | <0.5b | <0.5b | ND | ND |
| Acetylated xanthan | 2.0 | 0.04; 0.37 | ND; 1; 1 | <0.5b | ND | ND |
| Sugar beet pectin (acetylated homogalacturonan) | 4.0 | 0.04; 0.37 | <0.5b | <0.5b | ND | ND |
| Apple MHR (branched pectin) | 4.0 | 0.04; 0.37 | ND; 5; 7 | <0.5b | ND | ND |
| Acetylated xylo-oligosaccharides (DP 2-20; wood) | 4.0 | 0.04; 0.37 | ND; 79; 116 | ND; <0.5; 4 | ND | ND |
| Acetylated xylan (polymer) | 4.0 | 0.04; 0.37 | ND; 26; 29 | <0.5b | ND | ND |
| Acetylated GlcAme-xylo-oligosaccharides (DP 4-12) | 4.0 | 0.04; 0.37 | ND; 125; 125 | ND; <0.5; 3 | ND | ND |
| X2GlcAAc + X2GlcAmeAc (FA-20) | 1.0 | 0.04; 0.37 | ND; 8; 11 | <0.5b | ND | ND |
ND, not determined.
Amount of acetic acid released below the level of detection (0.5 μM) in 20 h.
Temperature and pH optima.
The pH and temperature ranges for AxeA1, Axe7A, and Axe1-6A were determined by analyzing the amount of acetic acid released from 1.2 mM glucose-pentaacetate in 30 min using 0.05 μM the appropriate enzyme. Buffers used for pHs in the range 3.0 to 6.5 were 50 mM citrate plus 150 mM NaCl, and those used in the range pH 7.0 to 8.0 were 50 mM HEPES plus 150 mM NaCl.
Microarray data accession number.
Microarray results have been deposited in the Gene Expression Omnibus (GEO accession number GSE19088) in accordance with Minimum Information about a Microarray Experiment (MIAME) standards.
RESULTS
Characteristics of XOSFA,Ac.
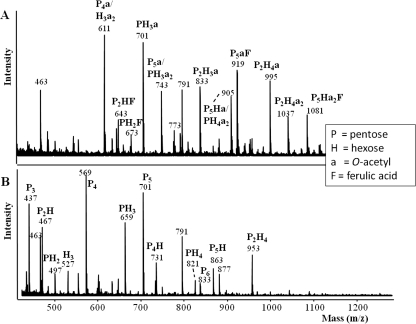
The main compositional characteristics of fraction XOSFA,Ac are described in Table 2. The fraction XOSFA,Ac was found to be soluble and also to contain large amounts of esterified O-acetyl and ferulic acid groups. The mass spectrum of fraction XOSFA,Ac (Fig. 1 A) revealed a range of oligosaccharide structures that either contain a ferulic acid, an O-acetyl group, or both. The addition of NaOH caused a shift in the mass spectra (Fig. 1B). Mass decreases of 42, 176, or 218 Da were observed for oligosaccharides, consistent with the expected mass decreases resulting from the loss of an O-acetyl or ferulic acid moiety or both, respectively, confirming the presence of alkali-labile ester groups. The mass spectra also revealed the oligosaccharides present to be mostly pentose-based oligomers (Fig. 1), and this was confirmed by the sugar composition (Table 2), which suggested that the main pentose present in XOSFA,Ac was xylose. Based on 100 xylosyl residues (molar ratio), the relative amount of side chains was calculated to be 32 arabinosyl, 10 galactosyl, 6 glucuronic acid, 24 (bound) ferulic acid, 5 (bound) coumaric acid, and 50 (bound) O-acetyl residues. Based on 100 arabinosyl residues (molar ratio), the relative amounts of ferulic acid and coumaric acid were calculated to be 75 and 17, respectively. Collectively, these results show that the corn fraction used as a substrate was composed of an arabinoxylo-oligosaccharide rich in O-acetyl and ferulic acid ester linkages.
Table 2.
Compositional data of the corn fiber fraction XOSFA,Ac (based on dry matter)
| Component | Content (% [wt/wt] XOSFA,Ac) |
|---|---|
| Proteina | 13 |
| Carbohydrates (total) | 45 |
| Monomeric sugars | <0.5 |
| Oligosaccharides | 44 |
| Arabinose | 8 |
| Xylose | 25 |
| Galactose | 3 |
| Glucose | 6 |
| Mannose | 0.3 |
| Rhamnose | 0 |
| Glucuronic acid | 2 |
| Ferulic acid | 17 (9, 8)b |
| Coumaric acid | 2.5 (1, 1.5)b |
| Acetic acid | 4 (0, 4)b |
%N × 6.25.
Parenthetical values: (free acid, acid bound in oligosaccharides).
Fig. 1.
MALDI-TOF mass spectra of fraction XOSFA,Ac (A) and fraction XOSFA,Ac (B) after removal of the esterified ferulic acid and O-acetyl groups with NaOH. (Fraction XOSFA,Ac was dissolved in 50 mM NaOH at 4°C overnight to remove the ester groups.) Masses represent sodium-adducted oligosaccharides.
Growth characteristics on corn fiber fractions.
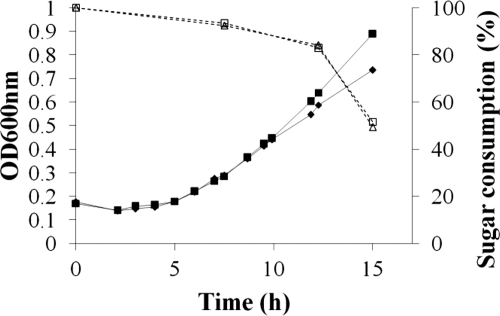
The growth dynamics of P. ruminicola 23 cells on XOSFA,Ac were determined. The growth rate of P. ruminicola was 0.12 h−1, and the bacterium attained a maximum cell density (OD600) for growth of 0.997 on this substrate (Fig. 2). Measures of sugar consumption across the growth curve showed that as much as 50% of the available sugars were consumed upon entering stationary phase (Fig. 2).
Fig. 2.
Growth curves of Prevotella ruminicola 23 grown on a minimal medium with addition of XOSFA,Ac (in duplicates). The data marked by the solid lines represent the OD600, while the dotted lines represent the sugar consumption related to the amount of total sugar present at the start of growth. Sugar consumption was analyzed based on the orcinol-sulfuric acid reaction (7) with an autoanalyzer (Skalar Analytical BV, Breda, Netherlands).
Bioinformatic analysis of the esterases of P. ruminicola 23.
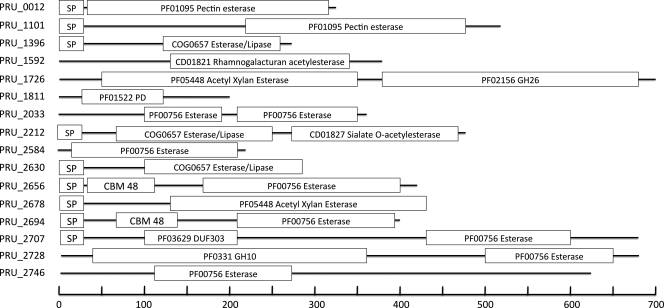
Using BlastP alignments and hidden Markov model searches, a total of 16 putative carbohydrate esterases (Table 3; Fig. 3) were identified. Eight of these carbohydrate esterases (PRU_1592, PRU_1811, PRU_2656, PRU_2678, PRU_2694, PRU_2707, PRU_2728, and PRU_2746) were clustered with one or more glycoside hydrolase-encoding genes. Additionally, PRU_1726 was found proximal to a predicted GPS family sugar transporter, and PRU_1396 was contiguous with a group 2 glycosyl transferase. Three of the putative esterases (PRU_2678, PRU_2707, and PRU_2728), in particular, were contiguous with a large number of genes likely to be involved in polysaccharide hydrolysis. For example, PRU_2678 was surrounded by six putative glycoside hydrolase (2 endo-1,5-α-l-arabinosidases, a β-1,4-xylosidase, an α-amylase, an endo-1,4-β-galactanase, and a GH 43 family protein)-encoding genes, as well as two glycosyl transferases (GTs) (GT families 1 and 2).
Table 3.
Predicted location and expression of the carbohydrate esterases of P. ruminicola 23a
| Locus tag | Product | CE familyb | Cellular locationc | RT qPCRd | Microarraye (Glu; Xyl; AXOS) | REf |
|---|---|---|---|---|---|---|
| PRU_0012 | Pectin esterase, PecE1 | 8 | Extracellular | 0.93 | 0.61; 0.83; 0.79 | 8.7; 0.07 |
| PRU_1101 | Pectin esterase, PecE2 | 8 | Extracellular | 2.18 | 1.37; 1.14; 1.35 | 6.1; 0.12 |
| PRU_1396 | Pectin acetyl esterase, PacE1 | Extracellular | 0.94 | 0.26; 0.37; 0.96 | 1.4; 0.01 | |
| PRU_1592 | Conserved hypothetical protein | 12 | Cytoplasm | ND | 2.25; 1.77; 0.72 | ND |
| PRU_1726 | Carbohydrate esterase | 7 | Periplasm | 11.5 | 2.36; 0.6; 1.26 | 2.7; 0.27 |
| PRU_1811 | Polysaccharide deacetylase | 4 | Cytoplasm | ND | 0.61; 0.86; 0.87 | ND |
| PRU_2033 | Carbohydrate esterase | Cytoplasm | 7.19 | 6.16; 3.60; 3.14 | 3.8; 0.23 | |
| PRU_2212 | Carbohydrate acetyl esterase, AxeA1 | Extracellular | 1.15 | 0.66; 0.58; 1.40 | 13.2; 0.13 | |
| PRU_2584 | Carbohydrate esterase | Cytoplasm | 3.17 | 1.02; 1.26; 1.61 | 0.9; 0.02 | |
| PRU_2630 | Pectin acetyl esterase, PacE2 | Periplasm | 1.86 | 1.58; 1.65; 1.25 | 5.4; 0.09 | |
| PRU_2656 | Isoamylase/esterase | 1 | Periplasm | ND | 1.68; 1.11; 0.90 | ND |
| PRU_2678 | Carbohydrate esterase, Axe7A | 7 | Periplasm | 1.71 | 1.81; 1.26; 1.05 | 1.1; 0.02 |
| PRU_2694 | Isoamylase/esterase | 1 | Extracellular | ND | 1.62; 1.26; 0.85 | ND |
| PRU_2707 | Acetyl-xylan esterase, Axe1-6A | 1/6 | Extracellular | 48.77 | 41.72; 36.01; 4.67 | 1.0; 0.38 |
| PRU_2728 | Xylanase/ferulic acid esterase, Xyn10D-Fae1A | 1 | Periplasm | 119.54 | 41.73; 38.10; 4.35 | 1.0; 1.00 |
| PRU_2746 | Carbohydrate esterase | 1 | Cytoplasm | 20.36 | 24.97; 18.00; 2.45 | 0.9; 0.15 |
Biochemically characterized proteins are distinguished in boldface type. ND, not determined.
Classified according to carbohydrate esterase (CE) families described in the CAZy database (8) (www.cazy.org).
RT qPCR analysis of the relative expression of each esterase gene during growth on XOSFA,Ac relative to growth on glucose.
Expression of each esterase during growth on XOSFA,Ac relative to that during growth on glucose, xylose, or AXOS.
Expression of each esterase relative to that of xyn10D-fae1A during growth on xylose or XOSFA,Ac.
Fig. 3.
Putative esterase genes from the genome of P. ruminicola 23, illustrating the size and domain structure variability as predicted from their amino acid sequence. HMM identifiers are given, with the exceptions of CBM (carbohydrate-binding domain) and SP (signal peptide).
Nine of the putative esterases were predicted to possess signal peptides (Fig. 3). Using GnegPloc (45), three of the signal peptide-possessing esterases (PRU_2630, PRU_2656, and PRU2678) and two esterases devoid of signal peptides (PRU_1726 and PRU_2728) were predicted to be targeted to the periplasm (Table 3). Six esterases (PRU_0012, PRU_1101, PRU_1396, PRU_2212, PRU_2694, and PRU_2707) were predicted to be secreted out of the cell, and all other esterases were predicted to be located in the cytoplasm (Table 3).
Microarray analysis of P. ruminicola 23 grown on XOSFA,Ac.
The results of the transcriptional analyses are summarized in Table 3. The data from the microarray measurements revealed that 4 of 16 putative esterases in P. ruminicola 23 were significantly upregulated (>2 fold) during growth on XOSFA,Ac, compared to growth on the nonesterified corn arabinoxylo-oligosaccharide fraction (AXOS). The upregulated esterase-encoding genes included xyn10D-fae1A (PRU_2728) and genes predicted to encode an acetyl-xylan esterase, the Axe1-6A gene (PRU_2707), and two esterases (PRU_2033 and PRU_2746), whose specificities were unclear from bioinformatic evidence. Comparing this transcriptional response to that obtained during growth on XOSFA,Ac relative to that on xylose, revealed much of the transcriptional induction observed in the comparison between XOSFA,Ac and AXOS was influenced by the presence of the corn xylo-oligomers rather than the ester groups themselves, which caused a much smaller, albeit significant transcriptional induction. Two additional esterases were upregulated when glucose was used as the comparator to XOSFA,Ac, suggesting that their induction was purely attributable to the presence of xylose, or perhaps a pentameric sugar as opposed to glucose, and their oligomerized derivatives (PRU_1592, PRU_1726). Reverse transcription-quantitative PCR (RT qPCR) largely supported the microarray results, although the data also suggested that the true induction of xyn10D-fae1A exceeded the measurable dynamic range of the microarray analysis. The RT qPCR data also suggested that two esterase-encoding genes, including a pectin esterase, pecE2 (PRU_1101), and PRU_2584, were also induced greater than 2-fold. In both the microarray analysis and RT qPCR analysis, the most significantly upregulated esterase gene was xyn10D-fae1A, which was determined by RT qPCR to be induced 120-fold during growth on XOSFA,Ac relative to that on glucose. All other esterases were constitutively expressed under each of the conditions examined. Despite its constitutive expression, the AxeA1 gene (PRU_2212) was found to be highly expressed, exceeding all others during growth on glucose and having just a 7.5-fold-lower growth rate than xyn10D-fae1A during growth on XOSFA,Ac. This finding suggested, despite its constitutive expression, the AxeA1 gene could make significant contributions to the esterase activities of P. ruminicola 23.
Purification and general characteristics of esterases Xyn10D-Fae1A, Axe1-6A, AxeA1, and Axe7A.
To gain further insights into the diverse esterase activities present in P. ruminicola 23, the two most upregulated esterases (Xyn10D-fae1A and Axe1-6A), the esterase (axeA1) with the highest basal expression on xylose relative to xyn10D-fae1A, and a fourth esterase (axe7A; PRU_2678) that appeared to be neither upregulated nor strongly expressed were selected for biochemical studies. The esterase genes were cloned, expressed heterologously in E. coli, and purified to homogeneity. Protein concentrations of purified Xyn10D-Fae1A, Axe1-6A, AxeA1, and Axe7A were 2.08, 0.24, 1.82 and 0.11 g/liter, respectively. The molecular masses of Xyn10D-Fae1A, Axe1-6A, AxeA1, and Axe7A were determined by MALDI-TOF mass spectrometry to be 83.0, 74.1, 54.5, and 49.2 kDa, respectively, which were in agreement with the predicted masses (84, 74, 53, and 48 kDa, respectively) based on their polypeptide sequences.
Enzymatic activities of Xyn10D-Fae1A, Axe1-6A, AxeA1, and Axe7A.
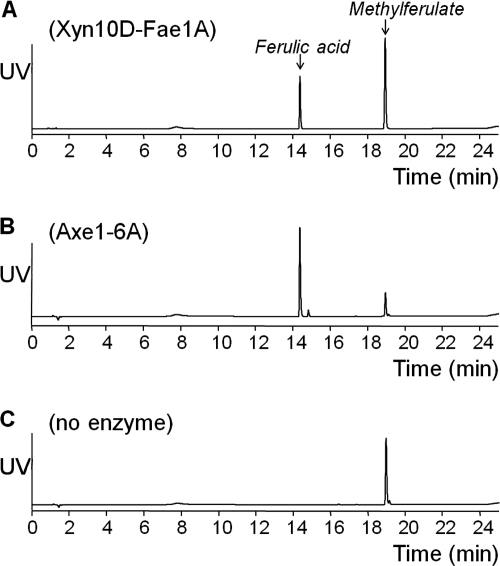
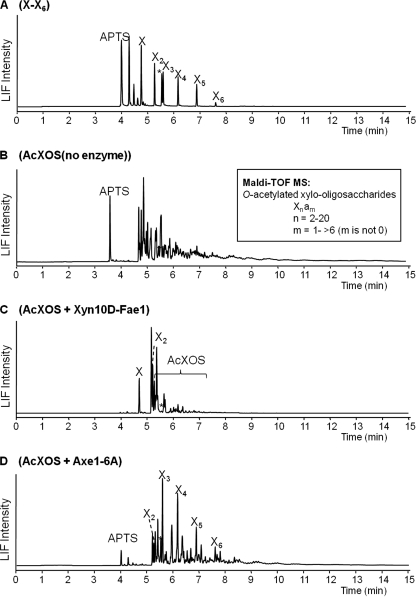
The most highly induced gene, xyn10D-fae1A, was biochemically characterized previously by Dodd et al. (14). These authors demonstrated that Xyn10D-Fae1A possesses both endo-β-1,4-xylanase and ferulic acid esterase activities. In the current study, the esterase activity of Xyn10D-Fae1A was evaluated in more detail. The reported ferulic acid esterase activity was confirmed by the release of ferulic acid from methylferulate (Fig. 4 A), and from the more natural substrates wheat bran and corn fiber. Only mono-ferulic acids were released, as di- and tri-ferulic acids were not identified in the enzyme digests. The enzymatic release of acetic acid from glucose pentaacetate (Table 4), acetylated xylo-oligosaccharides (Fig. 5), and acetylated xylan was negligible. The xylanase activity resulted in a decreased size of acetylated xylo-oligosaccharides (Fig. 5), and acetylated xylan (not shown).
Fig. 4.
UV chromatograms of methylferulate with XynD-Fae1A digest (A), methylferulate with Axe1-6A digest (B), and methylferulate without enzymes (C). Methylferulate in 50 mM citrate buffer, pH 6 (4 mg/ml), was digested overnight at 37°C, with 0.12% (wt/wt) (enzyme protein/substrate) for Axe1-6A or 1.5% (wt/wt) for XynD-Fae1A.
Fig. 5.
LIF electropherograms of a series of APTS-derivatized xylose until release of xylohexaose (A), APTS-derivatized AcXOS (B), APTS-derivatized AcXOS from digestion with Xyn10D-Fae1 until the endpoint (C), and APTS-derivatized AcXOS from digestion with Axe1-6A until the endpoint (D).
The esterase Axe1-6A released all of the acetic acid esters present in 1.2 mM glucose-pentaacetate in 2 h (Table 4). Axe1-6A also released part of the acetyl esters from the acetylated xylo-oligosaccharides, resulting in a completely deacetylated series of oligosaccharides, including xylobiose and xylotriose up to xylohexaose (Fig. 5). The protein possesses both acetyl esterase activity and ferulic acid esterase activity, as evidenced by the release of ferulic acid from methyl ferulate (Fig. 4), and from the more natural substrates wheat bran and corn fiber (not shown). The Axe1-6A enzyme is, therefore, a bifunctional esterase.
Analysis of enzymatic activity revealed AxeA1 to be an acetyl esterase with broad substrate specificity, releasing acetic acid from acetylated xylo-oligosaccharides and acetylated xylan as well as xylose-tetra-acetate, 4-O-methylumbelliferyl acetate, glucose-pentaacetate, and cephalosporin C (Table 4). AxeA1 released 100% and 88%, respectively, of the acetic acid esters present in 1.2 mM glucose-pentaacetate and 1.7 mM cephalosporin C in 2 h. AxeA1 appeared to have greater activity on oligosaccharides than on the polymeric substrates, releasing 58% of the acetic acid esters present in 0.4% (wt/vol) acetylated xylo-oligosaccharides in 20 h, compared to just 2% of the acetic acid esters present in 0.4% (wt/vol) acetylated xylan. Based on the relative amount (percentage) of acetic acid released, AxeA1 was also able to release acetic acid from xylo-oligosaccharides with 4-O-methylglucuronic acid side groups proximally located to O-acetyl esters. However, the amount of acetic acid released, in 20 h, from these oligosaccharides was only 15% of the O-acetyl groups present, which suggested that the release of O-acetyl groups by AxeA1 was to some extent hindered by the proximal location of 4-O-methylglucuronic acid side groups.
Axe7A was also able to release acetic acid from xylose-tetraacetate, 4-O-methylumbelliferyl acetate, glucose-pentaacetate, cephalosporin C, and acetylated xylo-oligosaccharides (Table 4), although compared to that of AxeA1, this esterase displayed a reduced enzymatic activity. Axe7A released 19%, 54%, and 2% of the acetic acid present in glucose-pentaacetate (2 h), cephalosporin C (2 h), and acetylated xylo-oligosaccharides (20 h), respectively. Axe7A displayed no detectable activity on polymeric xylan.
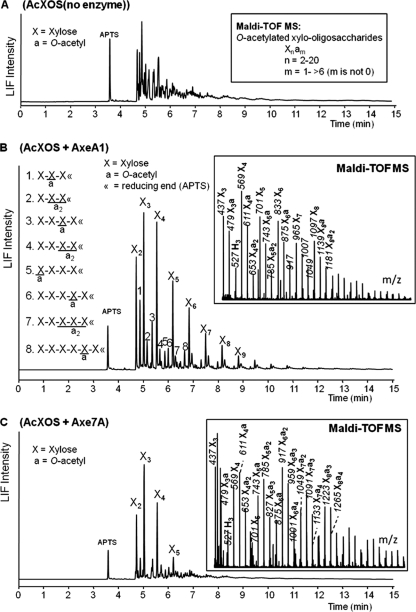
To gain further insight into the mode of action of the acetyl esterase activities of AxeA1 and Axe7A, we collected MALDI-TOF mass spectra during 24 h of incubation on acetylated xylo-oligosaccharides (Fig. 6 A) (26, 28). The results revealed AxeA1 to preferentially utilize xylo-oligosaccharides containing three or more O-acetyl groups over xylo-oligosaccharides containing one or two O-acetyl groups. It was only following the depletion of xylo-oligosaccharides containing three or more O-acetyl groups that AxeA1 began using oligosaccharides with two or fewer O-acetyl groups (see Fig. S1 in the supplemental material). At the conclusion of the incubation, only xylo-oligosaccharides with two O-acetyl esters or fewer remained (Fig. 6B). The residual xylo-oligosaccharides from the 24 h of incubation were labeled with APTS and studied in more detail using capillary electrophoresis coupled to a fluorescence detector and an electrospray ion-trap mass spectrometer (CE-LIF-ESI-MSn). CE-LIF-ESI-MSn results revealed that the remaining esters were located mostly on the second xylosyl residue from the reducing end of the residual xylo-oligosaccharides containing just one O-acetyl group. However, it could not be determined whether these residual O-acetyl groups were located at the O-2 or O-3 position of the xylosyl residue. The position of the O-acetyl groups for xylo-oligosaccharides containing two O-acetyl esters could not be determined, although it was revealed that at least two xylosyl residues from the nonreducing end were not O-acetylated.
Fig. 6.
LIF electropherograms of APTS-derivatized AcXOS (A), APTS-derivatized AcXOS from digestion with AxeA1 until the endpoint (B), and APTS-derivatized AcXOS from digestion with Axe7A until the endpoint (C). Insets show the MALDI-TOF mass data of the corresponding oligosaccharides. Masses represent sodium-adducted oligosaccharides. The suggested structures 1 to 8 (B) were defined by using CE-LIF-ESI-MSn (28).
The corresponding analysis revealed that the acetyl esterase activities of Axe1-6A and Axe7A were limited to acetylated xylo-oligosaccharides smaller than xylo-heptaose (Fig. 5D and 6C); however, the remaining mixture of acetylated oligosaccharides was still too heterogeneous following 24 h of incubation to extract detailed information regarding the deacetylation mechanism of Axe1-6A and Axe7A.
Xyn10D-Fae1 and Axe1-6A were able to release some ferulic acid esters present in XOSFA,Ac. For example, the mass corresponding to an arabino-ferulic acid oligomer disappeared from digests of both enzymes (not shown). AxeA1 and Axe7A were able to release some of the acetic acid esters present in the XOSFA,Ac as revealed by the obtained mass spectra (not shown). In all cases, the masses of the oligosaccharides containing both acetic and ferulic acid esters (in Fig. 1: masses 643, 673, 919, 1,081 Da) remained, suggesting that the proximal location of ferulic acid groups and the O-acetyl group hindered the activities of these two enzymes.
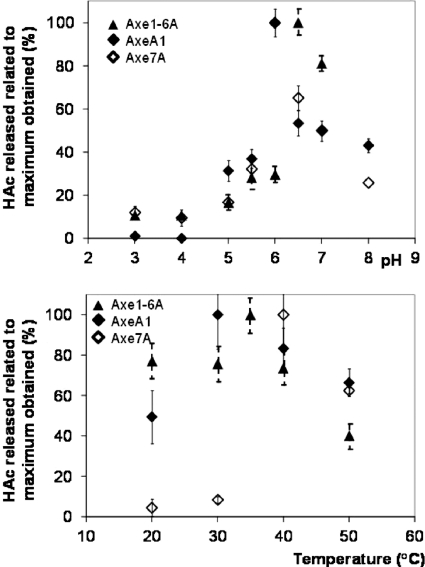
Temperature and pH optima for enzymatic activities of Xyn10D-Fae1A, Axe1-6A, AxeA1, and Axe7A.
The pH and temperature range in which Axe1-6A, AxeA1, and Axe7A were active was determined by the release of acetic acid on glucose-pentaacetate (Fig. 7). Xyn10D-Fae1A had low activity on glucose-pentaacetate. The latter protein was characterized previously (14) and exhibited measurable activity for both the endo-xylanase and the esterase activity at pH 6.0 and 30 to 40°C.
Fig. 7.
Activities of Axe1-6A, AxeA1, and Axe7A on glucose pentaacetate at various pH and temperature values.
Axe1-6A, AxeA1, and Axe7A were active in a pH range 5 to 8. Whereas Axe1-6A displayed maximum activity at pH 6.5, both AxeA1 and Axe7A showed a maximum at pH 6.0. Axe1-6A and AxeA1 were active between 20 and 50°C and still exhibited 40 to 70% of the maximum activity after 20 h of incubation at 50°C. Axe7A was active at temperatures between 30 and 50°C, but at temperatures of 30°C and lower, less than 10% of the maximum activity was left. At 50°C Axe7A retained 60% of its maximum activity. The optimum temperatures for enzymatic activity of Axe1-6A, AxeA1, and Axe7A were 35°C, 30 to 35°C, and 40°C, respectively.
DISCUSSION
The genus Prevotella is one of the more numerically dominant and metabolically versatile bacteria in the rumen (16, 18, 50). Corn, a common ingredient in the diet of U.S. cattle, contains the major hemicellulose xylan and is known to contain a relatively high content of ferulic and coumaric acid esters (22, 40, 41). This observation was further confirmed by our analysis of the corn-derived substrates used in the present study. Microbial capture of nutrients from polysaccharides, including hemicelluloses, followed by their fermentation in the rumen results in the production of short-chain fatty acids that serve as the major energy source for the host ruminant. The enzymatic hydrolysis of ester linkages in hemicelluloses by rumen microbes is, therefore, an important step in the complete degradation and efficient utilization of corn hemicellulose.
Previously, it was shown that Xyn10D-Fae1A possessed both endo-xylanase activity on oat spelt xylan and esterase activity on methylferulate (14). In the present study, Xyn10D-Fae1A was also tested on other ester-containing substrates, including wheat bran, corn fiber, and the ester-rich arabinoxylo-oligosaccharide fraction from corn (XOSFA,Ac). For all substrates the release of ferulic acid was observed, although the activity was clearly hindered by the presence of neighboring ester groups such as acetic acid esters. Negligible activity was observed when Xyn10D-Fae1A was incubated with sugar acetates, indicating that the enzyme behaved as a true ferulic acid esterase belonging to the carbohydrate esterase family 1 (8) (http://www.cazy.org).
Axe1-6A was found to release acetic acid from acetylated xylo-oligosaccharides, as well as from glucose-pentaacetate. This esterase was also able to release ferulic acid from methylferulate, wheat bran, corn fiber, and XOSFA,Ac. The protein Axe1-6A was predicted to have two esterase domains corresponding to distinct carbohydrate esterase families, CE1 and CE6 (8). It can be hypothesized that the release of acetic acid is related to the CE6 domain, an esterase family that exclusively contains acetyl xylan esterases (EC 3.1.1.72), while the release of ferulic acid may relate to the CE1 domain. Family 1 carbohydrate esterases include enzymes of numerous specificities, including both acetyl- and feruloyl-linked esterases. Obtaining a clearer understanding of the functions of these two CE domains will be an important and interesting focus of future work.
In addition to Axe1-6A, AxeA1 and Axe7A were also found to possess acetyl esterase activity, making these three esterases the first biochemically characterized acetyl esterases from a Prevotella sp. Axe7A was predicted to belong to CE family 7 (8). The CE-7 family enzymes have previously been suggested to comprise a single class of proteins with multifunctional deacetylase activity against a range of small substrates (11, 53). This is consistent with our findings, with Axe7A demonstrating deacetylase activity toward a variety of substrates, including xylose-tetraacetate, 4-O-methylumbelliferylacetate, glucose-pentaacetate, cephalosporin C, and acetylated xylo-oligosaccharides (DP 2-20), but not toward acetylated xylo-oligosaccharides larger than xylo-heptaose. AxeA1 was active toward a similar range of substrates but, in addition, displayed some activity toward polymeric acetyl-xylan. The high basal expression of AxeA1, relative to that of Xyn10D-Fae1A, suggests that this esterase is critical for P. ruminicola 23, a bacterium with a hemicellulose-degrading lifestyle in the rumen. The preference of both AxeA1 and Axe7A for xylo-oligosaccharides over polymeric xylan illustrates the need for these enzymes to work in synergy with xylan-depolymerizing enzymes, such as xylanases and β-xylosidases. This has previously been reported for other acetyl-xylan esterases, which alone have shown low activities on acetylated xylan (3).
Insight into the mechanistic action of AxeA1 revealed that the enzyme preferentially targeted xylo-oligosaccharides possessing three or more O-acetyl esters, but following their depletion the enzyme was active on the less acetylated portion of the substrate. It also revealed several factors that limited the enzyme's esterase activity. The residual xylo-oligosaccharides following incubation with AxeA1 were enriched for those possessing O-acetyl groups on the second xylosyl residue from the reducing end (X2), suggesting an inability to remove O-acetyl groups at this position. The deacetylase activities of various acetyl-xylan esterases have been described to possess positional specificity (4, 35, 52). This limitation is likely conferred through their binding cleft or catalytic site, and this may be applicable to AxeA1. In addition to xylo-oligosaccharides with an O-acetyl group on X2, a xylo-tetraose having an O-acetyl ester on the nonreducing end xylosyl was observed following incubation with AxeA1. We cannot rule out the possibility that this O-acetyl is present on the O-4 position of the nonreducing end xylosyl residue and not on the O-2 or O-3 position typical of xylan (39, 51). The O-4 position could be the result of internal migration, as has been proposed previously (27). Biely et al. (4) reported that acetyl-xylan esterases from Streptomyces lividans, Trichoderma reesei, and Schizophyllum commune were able to release O-acetyl esters from the O-2 and O-3 positions of methyl 2,3,4-tri-O-acetyl-β-d-xylopyranoside but not from the O-4 position.
The deacetylase activity of Axe1-6A, AxeA1, and Axe7A also appeared to be hindered by the proximal localization of 4-O-methylglucuronic or ferulic acid esters. Likewise, the ferulic acid esterase activity of Axe1-6A and Xyn10D-Fae1A appeared to be hindered by the proximal localization of acetyl esters. As almost all previous investigations into the mechanistic action of other esterases have been performed on substrates possessing only ferulic acid or 4-O-methyl glucuronic or O-acetyl esters and not on substrates with combinations of these esters, it is unclear if such inhibition is a widespread feature of esterase enzymes. However, our results clearly provide significant insights into the biochemical activities of these four esterases from P. ruminicola 23. In addition, this report begins to illuminate the importance of multiple or diverse esterase activities that support the hemicellulose-degrading lifestyle of this important ruminal bacterium.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by National Research Initiative Competitive Grant no. 2008-35206-18784 from the USDA National Institute of Food and Agriculture (to R.I.M., I.K.O.C., and M.M.) and by the Dutch Ministry of Economic Affairs via a grant supporting the short-term research of M.A.K. (EOS-KTO grant).
We thank David van Eylen, Femke van Dongen, and Maaike Appeldoorn for their valuable contributions to this work.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 8 July 2011.
REFERENCES
- 1. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 3. Biely P., MacKenzie C. R., Puls J., Schneider H. 1986. Cooperativity of esterases and xylanases in the enzymatic degradation of acetyl xylan. Nat. Biotechnol. 4:731–733 [Google Scholar]
- 4. Biely P., Cote G. L., Kremnicky L., Weisleder D., Greene R. V. 1996. Substrate specificity of acetylxylan esterase from Schizophyllum commune: mode of action on acetylated carbohydrates. Biochim. Biophys. Acta 1298:209–222 [DOI] [PubMed] [Google Scholar]
- 5. Biely P. 2003. Xylanolytic enzymes, p. 879–916 In Whitaker J. R., Voragen A. G. J., Wong D. W. S. (ed.), Handbook of food enzymology. Marcel Dekker, New York, NY [Google Scholar]
- 6. Brillouet J. M., Joseleau J. P., Utille J. P., Lelievre D. 1982. Isolation, purification and characterization of a complex heteroxylan from industrial wheat bran. J. Agric. Food Chem. 30:488–494 [DOI] [PubMed] [Google Scholar]
- 7. Bruckner J. 1955. Estimation of monosaccharides by the orcinol-sulphuric acid reaction. Biochem. J. 60:200–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cantarel B. L., et al. 2008. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carpita N. C. 1996. Structure and biogenesis of the cell walls of grasses. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47:445–476 [DOI] [PubMed] [Google Scholar]
- 10. Chou K. C., Shen H. B. 2008. Cell-PLoc: a package of web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 3:153–162 [DOI] [PubMed] [Google Scholar]
- 11. Degrassi G., Kojic M., Ljubijankic G., Venturi V. 2000. The acetyl xylan esterase of Bacillus pumilus belongs to a family of esterases with broad substrate specificity. Microbiology 146:1585–1591 [DOI] [PubMed] [Google Scholar]
- 12. Dehority B. A., Scott H. W. 1967. Extent of cellulose and hemicellulose digestion in various forages by pure cultures of rumen bacteria. J. Dairy Sci. 50:1136–1141 [Google Scholar]
- 13. de Ruiter G. A., Schols H. A., Voragen A. G. J., Rombouts F. 1992. Carbohydrate analysis of water-soluble uronic acid-containing polysaccharides with HPAEC using methanolysis combined with TFA hydrolysis is superior to four other methods. Anal. Biochem. 207:176–185 [DOI] [PubMed] [Google Scholar]
- 14. Dodd D., et al. 2009. Biochemical analysis of a β-d-xylosidase and a bifunctional xylanase-ferulic acid esterase from a xylanolytic gene cluster in Prevotella ruminicola 23. J. Bacteriol. 191:3328–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eddy S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755–763 [DOI] [PubMed] [Google Scholar]
- 16. Edwards J. E., Travis A. J., Wallace R. J. 2004. 16S rDNA library-based analysis of ruminal bacterial diversity. Antonie Van Leeuwenhoek 86:263–281 [DOI] [PubMed] [Google Scholar]
- 17. Evtuguin D. V., Tomas J. L., Silva A. M. S., Neto C. P. 2003. Characterization of an acetylated heteroxylan from Eucalyptus globulus Labill. Carbohydr. Res. 338:597–604 [DOI] [PubMed] [Google Scholar]
- 18. Fondevila M., Dehority B. A. 1996. Interactions between Fibrobacter succinogenes, Prevotella ruminicola, and Ruminococcus flavefaciens in the digestion of cellulose from forages. J. Anim. Sci. 74:678–684 [DOI] [PubMed] [Google Scholar]
- 19. Gentleman R., Carey V., Huber W., Irazarry R. R., Dudoit S. (ed.). 2005. Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY [Google Scholar]
- 20. Grabber J. H. 2005. How do lignin composition, structure, and cross-linking affect degradability? A review of cell wall model studies. Crop Sci. 45:820–831 [Google Scholar]
- 21. Hespell R. B., O'Bryan-Shah P. J. 1988. Esterase activities in Butyrivibrio fibrisolvens strains. Appl. Environ. Microbiol. 54:1917–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huisman M. M. H., Schols H. A., Voragen A. G. J. 2000. Glucuronoarabinoxylans from maize kernel cell walls are more complex than those from sorghum kernel cell walls. Carbohydr. Polym. 43:269–279 [Google Scholar]
- 23. Hunter S., et al. 2009. InterPro: the integrative protein signature database. Nucleic Acids Res. 37:D224–D228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iiyama K., Lam T., Stone B. A. 1994. Covalent cross-links in the cell wall. Plant Physiol. 104:315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kabel M. A., et al. 2002. Hydrothermally treated xylan rich by-products yield different classes of xylo-oligosaccharides. Carbohydr. Polym. 50:47–56 [Google Scholar]
- 26. Kabel M. A., Schols H. A., Voragen A. G. J. 2002. Complex xylo-oligosaccharides identified from hydrothermally treated Eucalyptus wood and brewery's spent grain. Carbohydr. Polym. 50:191–200 [Google Scholar]
- 27. Kabel M. A., de Waard P., Schols H. A., Voragen A. G. J. 2003. Location of O-acetyl substituents in xylo-oligosaccharides obtained from hydrothermally treated Eucalyptus wood. Carbohydr. Res. 338:69–77 [DOI] [PubMed] [Google Scholar]
- 28. Kabel M. A., et al. 2006. Capillary electrophoresis fingerprinting, quantification and mass-identification of various 9-aminopyrene-1,4,6-trisulfonate-derivatized oligomers derived from plant polysaccharides. J. Chromatogr. A 1137:119–126 [DOI] [PubMed] [Google Scholar]
- 29. Kabel M. A., Van den Borne H., Vincken J.-P., Voragen A. G. J., Schols H. A. 2007. Structural differences of xylans affect their interaction with cellulose. Carbohydr. Polym. 69:91–105 [Google Scholar]
- 30. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 31. Kudo H., Cheng K. J., Costerton J. W. 1987. Interactions between Treponema bryantii and cellulolytic bacteria in the in-vitro degradation of straw cellulose. Can. J. Microbiol. 33:244–248 [DOI] [PubMed] [Google Scholar]
- 32. Miron J. 1991. The hydrolysis of lucerne cell-wall monosaccharide components by monocultures or pair combinations of defined ruminal bacteria. J. Appl. Bacteriol. 70:245–252 [DOI] [PubMed] [Google Scholar]
- 33. Oosterveld A., Beldman G., Searle-van Leeuwen M. J. F., Voragen A. G. J. 2000. Effect of enzymatic deacetylation on gelation of sugar beet pectin in the presence of calcium. Carbohydr. Polym. 43:249–256 [Google Scholar]
- 34. Pittman K. A., Bryant M. P. 1964. Peptides and other nitrogen sources for growth of Bacteroides ruminicola. J. Bacteriol. 88:401–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Poutanen K., Sundberg M., Korte H., Puls J. 1990. Deacetylation of xylans by acetyl esterases of Trichoderma reesei. Appl. Microbiol. Biotechnol. 33:506–510 [Google Scholar]
- 36. Puchart V., et al. 2007. Substrate and positional specificity of feruloyl esterases for monoferuloylated and monoacetylated 4-notrophenyl glycosides. J. Biotechnol. 127:235–243 [DOI] [PubMed] [Google Scholar]
- 37. Puls J., Poutanen K. 1989. Mechanisms of enzymatic hydrolysis of hemicelluloses (xylans) and procedures for determination of the enzyme activities involved, p. 151–165 In Coughlan M. P. (ed.), Enzyme systems for lignocellulose degradation. Elsevier Applied Science, Amsterdam, The Netherlands [Google Scholar]
- 38. Purushe J., et al. 2010. Comparative genome analysis of Prevotella ruminicola and Prevotella bryantii: insights into their environmental niche. Microb. Ecol. 60:721–729 [DOI] [PubMed] [Google Scholar]
- 39. Reicher F., Correa J. B. C. 1984. Location of O-acetyl groups in the acidic D-xylan of Mimosa scabrella (bracatinga). A study of O-acetyl group migration. Carbohydr. Research 135:129–140 [Google Scholar]
- 40. Saulnier L., Thibault J. F. 1999. Ferulic acid and diferulic acid as components of sugar-beet pectins and maize bran heteroxylans. J. Sci. Food Agric. 79:396–402 [Google Scholar]
- 41. Saulnier L., Vigouroux J., Thibault J. F. 1995. Isolation and partial characterization of feruloylated oligosaccharides from maize bran. Carbohydr. Res. 272:241–253 [DOI] [PubMed] [Google Scholar]
- 42. Saulnier L., Sado P.-E., Branlard G., Charmet G., Guillon F. 2007. Wheat arabinoxylans: exploiting variation in amount and composition to develop enhanced varieties. J. Cereal Sci. 46:261–281 [Google Scholar]
- 43. Schols H. A., Posthumus M. A., Voragen A. G. J. 1990. Structural features of hairy regions of pectins isolated from apple juice produced by the liquefaction process. Carbohydr. Res. 206:117–129 [Google Scholar]
- 44. Shatalov A. A., Evtuguin D. V., Neto C. P. 1999. (2-O-α-d-galactopyranosyl-4-O-methyl-α-d-glucurono)-d-xylan from Eucalyptus globulus labill. Carbohydr. Res. 320:93–99 [DOI] [PubMed] [Google Scholar]
- 45. Shen H., Chou K. 2007. Gneg-PLoc: an ensemble classifier for predicting subcellular localization of Gram-negative bacterial proteins. Protein Eng. Des. Sel. 20:39–46 [DOI] [PubMed] [Google Scholar]
- 46. Shibuya N., Iwasaki T. 1985. Structural features of rice bran hemicellulose. Phytochemistry 24:285–289 [Google Scholar]
- 47. Simonne A. H., Simonne E. H., Eitenmiller R. R., Mills H. A., Cresman C. P. 1997. Could the Dumas method replace the Kjeldahl digestion for nitrogen and crude protein determinations in foods? J. Sci. Food Agric. 73:39–45 [Google Scholar]
- 48. Smyth G. K. 2005. Limma: linear models for microarray data, p. 397–420 In Gentleman R., Carey V., Huber W., Irazarry R. R., Dudoit S. (ed.), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY [Google Scholar]
- 49. Sorensen H. R., Pedersen S., Jorgensen C. T., Meyer A. S. 2007. Enzymatic hydrolysis of wheat arabinoxylan by a recombinant “minimal” enzyme cocktail containing beta-xylosidase and novel endo-1,4-beta-xylanase and alpha-(L)-arabinofuranosidase activities. Biotechnol. Prog. 23:100–107 [DOI] [PubMed] [Google Scholar]
- 50. Stevenson D. M., Weimer P. J. 2007. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 75:165–174 [DOI] [PubMed] [Google Scholar]
- 51. Teleman A., Tenkanen M., Jacobs A., Dahlman O. 2002. Characterization of O-acetyl-(4-O-methylglucurono)xylan isolated from birch and beech. Carbohydr. Res. 337:373–377 [DOI] [PubMed] [Google Scholar]
- 52. Tenkanen M., Eyzaguirre J., Isoniemi R., Faulds C. B., Biely P. 2003. Comparison of catalytic properties of acetyl xylan esterases from three carbohydrate esterase families. ACS Symp. Ser. 855:211–229 [Google Scholar]
- 53. Vincent F., et al. 2003. Multifunctional xylooligosaccharide/cephalosporin C deacetylase revealed by the hexameric structure of the Bacillus subtilis enzyme at 1.9 Å resolution. J. Mol. Biol. 330:593–606 [DOI] [PubMed] [Google Scholar]
- 54. Voragen A. G. J., Schols H. A., Pilnik W. 1986. Determination of the degree of methylation and acetylation of pectins by HPLC. Food Hydrocolloids 1:65–70 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.