Abstract
Nonulosonic acids (NulOs) encompass a large group of structurally diverse nine-carbon backbone α-keto sugars widely distributed among the three domains of life. Mammals express a specialized version of NulOs called sialic acids, which are displayed in prominent terminal positions of cell surface and secreted glycoconjugates. Within bacteria, the ability to synthesize NulOs has been demonstrated in a number of human pathogens and is phylogenetically widespread. Here we examine the distribution, diversity, evolution, and function of NulO biosynthesis pathways in members of the family Vibrionaceae. Among 27 species of Vibrionaceae examined at the genomic level, 12 species contained nab gene clusters. We document examples of duplication, divergence, horizontal transfer, and recombination of nab gene clusters in different Vibrionaceae lineages. Biochemical analyses, including mass spectrometry, confirmed that many species do, in fact, produce di-N-acetylated NulOs. A library of clinical and environmental isolates of Vibrio vulnificus served as a model for further investigation of nab allele genotypes and levels of NulO expression. The data show that lineage I isolates produce about 20-fold higher levels of NulOs than lineage II isolates. Moreover, nab gene alleles found in a subset of V. vulnificus clinical isolates express 40-fold higher levels of NulOs than nab alleles associated with environmental isolates. Taken together, the data implicate the family Vibrionaceae as a “hot spot” of NulO evolution and suggest that these molecules may have diverse roles in environmental persistence and/or animal virulence.
INTRODUCTION
Nonulosonic acids (NulOs) are a family of negatively charged nine-carbon backbone α-keto sugars that include the neuraminic (also known as sialic), legionaminic, and pseudaminic acids (2, 35). The sialic acids are the best-understood NulOs and are found in prominent outermost positions on the surfaces of all vertebrate cells (55). In mammals, the most common NulO is sialic acid, a molecule found at particularly high levels at mucosal surfaces of mammals. Pseudaminic and legionaminic acids are not expressed in animals. In their various locations on microbial surfaces, different NulO structures have been implicated in a variety of host-microbe interactions. NulOs of the sialic, legionaminic, and pseudaminic acid types are involved in bacterial behaviors like biofilm formation, autoagglutination, and motility (1, 15, 18, 45, 52), as well as direct protein-carbohydrate interactions between hosts and pathogens (8, 9, 26, 30, 52). In particular, sialic acid-containing bacterial glycans participate in strategies of immune suppression and subversion, likely contributing to clinical conditions ranging from urogenital, airway, and ear infections to systemic bacteremia, meningitis, and the induction of autoimmunity (1, 24, 32, 56, 57, 63, 64).
Sialic acids were once thought to be unique to the deuterostome lineage of “higher” animals and absent from most protostomes, fungi, plants, and protists (61). In fact, the biosynthetic pathways for sialic acids appear to be quite ancient, likely predating the divergence of the three domains of life (35). Recent studies show that NulO biosynthetic (nab) gene clusters are found in a surprising variety of bacterial strains and species, including a large number of gammaproteobacteria, including the family Vibrionaceae (35). Different proteobacteria have been shown to express NulOs as modifications of polymerized cell surface molecules such as capsular polysaccharides (16, 34), lipopolysaccharides (LPSs) (37, 48, 51), and flagella (40–43, 50). Among the members of the family Vibrionaceae, strains of Vibrio parahaemolyticus, V. vulnificus, Aliivibrio salmonicida, and Photobacterium profundum have also been shown to express NulOs (7, 13, 35, 58). However, little is known about the larger distribution patterns, the natural history of NulOs in Vibrionaceae, or the biology of these molecules in aquatic or host-associated niche environments.
As a group, the members of the family Vibrionaceae engage in a full spectrum of lifestyles, from free-living states to colonization or infection of both aquatic and terrestrial hosts (3, 17). V. vulnificus is an excellent example of the range of environmental and host niches that can be occupied by different members of the same Vibrio species. V. vulnificus is an obligate halophile found in estuarine and marine coastal environments worldwide (28, 29, 46, 53). It is found in association with zooplankton, crabs, and various filter feeders, such as oysters and mussels (12, 21, 22, 44). V. vulnificus is also a highly invasive pathogen of both fish and humans, and in humans, infection is characterized by primary septicemia and wound infections with mortality rates of greater than 50% among susceptible individuals (27, 38, 47). Multilocus sequence typing analysis of six housekeeping genes and genotyping data have previously divided V. vulnificus isolates into at least three distinct clusters or lineages; lineage I is composed exclusively of biotype 1 isolates recovered mainly from clinical sources, lineage II contains biotype 1 and all biotype 2 isolates recovered from environmental sources, including diseased fish, and the third lineage is composed of biotype 3 (4–6, 11, 59, 60). Biotype 3 isolates, which are recovered from one geographic region and associated with one fish species, were shown to be genetically identical and distinct from lineage I and II isolates (4, 5).
We hypothesize that the family Vibrionaceae may be a particularly active lineage of NulO evolution. Here we combine genomic and biochemical approaches to more systematically investigate and document the distribution, phylogeny, and functional activity of homologous NulO biosynthetic (NAB) pathways in members of the family Vibrionaceae.
(A portion of this work was presented by S.A. at the 2009 INTEL International Science and Engineering Fair.)
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Tables 1 and 2. Unless otherwise noted, all strains were grown aerobically (250 rpm) at 37°C in Luria-Bertani broth (Fisher Scientific, Fair Lawn, NJ) with a final NaCl concentration of 2% (Fisher Scientific). The 67 V. vulnificus isolates examined in this study were temporally (1980 to 2005) and geographically (Asia, Europe, and North America) widespread, encompassing all 3 biotype groups as previously reported (11). Of the 67 isolates examined, 27 were recovered from clinical sources (wound infections and blood) and 40 were from environmental sources (clams, oysters, mussels, fish, seawater, and sea sediment). All V. vulnificus strains were grown in LB supplemented with 2% NaCl and stored at −80°C in LB broth with 20% (vol/vol) glycerol.
Table 1.
Species and strains examined in this study
| Species | Isolate name |
|---|---|
| Listonella anguillarum | B559 |
| Listonella anguillarum | B561 |
| Listonella anguillarum | B563 |
| Vibrio angustum | B70 |
| Vibrio fischeri | ATCC 33983 |
| Vibrio fischeri | 394 |
| Vibrio fischeri | 63 |
| Vibrio fischeri | ES114 |
| Vibrio harveyi | ATCC 33843 |
| Vibrio harveyi | L222 |
| Vibrio logei | B583 |
| Vibrio ordalii | B572 |
| Vibrio orientalis | B717 |
| Vibrio pelagius | B98 |
| Vibrio splendidus | B12 |
| Vibrio parahaemolyticus | 428/01 |
| Vibrio parahaemolyticus | 30824 |
| Vibrio parahaemolyticus | 9808/1 |
| Vibrio parahaemolyticus | UCMA178 |
| Vibrio parahaemolyticus | 090-96 |
| Vibrio parahaemolyticus | 906-97 |
| Vibrio parahaemolyticus | 190-2004 |
| Vibrio parahaemolyticus | 155-05 |
| Vibrio parahaemolyticus | 357-99 |
| Vibrio parahaemolyticus | 518-01 |
| Vibrio parahaemolyticus | VP81 |
| Vibrio parahaemolyticus | Vphy191 |
| Vibrio parahaemolyticus | V0441 |
| Vibrio parahaemolyticus | V0586 |
Table 2.
V. vulnificus strains used in this study and their nab alleles
| Strain name | nab1 allele | nab2 allele | Place of isolation | Source | Yr of isolation |
|---|---|---|---|---|---|
| YJ002 | YJ016-like | YJ016-like | Taiwan | Clinical | 1993 |
| JJ068 | Taiwan | Clinical | 1993 | ||
| JJ072 | Taiwan | Clinical | 1993 | ||
| JJ067 | YJ016-like | Taiwan | Clinical | 1993 | |
| YJ016 | YJ016-like | YJ016-like | Taiwan | Clinical | 1993 |
| L-180 | Japan | Clinical | 1980 | ||
| N-87 | CMCP6-like | CMCP6-like | Japan | Clinical | 1987 |
| KH-03 | YJ016-like | YJ016-like | Japan | Clinical | 2003 |
| SPRC 10143 | YJ016-like | YJ016-like | NKa | Clinical | NK |
| CMCP6 | CMCP6-like | CMCP6-like | Korea | Clinical | NK |
| CDC 9038-96 | CMCP6-like | CMCP6-like | Texas | Clinical | 1996 |
| CDC 9062-96 | CMCP6-like | YJ016-like | Louisiana | Clinical | 1996 |
| CDC 9005-97 | YJ016-like | Louisiana | Clinical | 1997 | |
| CDC 9030-95 | YJ016-like | YJ016-like | Florida | Clinical | 1995 |
| 313-98 | Israel | Clinical | 1998 | ||
| 11028 | Israel | Clinical | NK | ||
| CIP 81.90 | YJ016-like | YJ016-like | France | Clinical | 1980 |
| LSU 1866 | YJ016-like | NK | Clinical | NK | |
| M06 | CMCP6-like | CMCP6-like | United States | Clinical | NK |
| M06-24/O | CMCP6-like | CMCP6-like | California | Clinical | NK |
| 6353/O | YJ016-like | Maryland | Clinical | NK | |
| 85A667/O | YJ016-like | California | Clinical | NK | |
| K2637 | CMCP6-like | Louisiana | Clinical | 2005 | |
| K2667 | CMCP6-like | CMCP6-like | Louisiana | Clinical | 2005 |
| K2719 | YJ016-like | YJ016-like | Louisiana | Clinical | 2005 |
| C-7184 | United States | Clinical | NK | ||
| IFVv8 | Florida | Clinical | NK | ||
| CG27 | CMCP6-like | CMCP6-like | Taiwan | Oyster | 1993 |
| 98-640 DP-B9 | YJ016-like | YJ016-like | Louisiana | Oyster | 1998 |
| 99-609 DP-A4 | YJ016-like | YJ016-like | Oregon | Oyster | 1999 |
| 99-779 DP-D2 | YJ016-like | Louisiana | Oyster | 1999 | |
| 99-509 DP-A6 | YJ016-like | YJ016-like | Texas | Oyster | 1999 |
| 99-540 DP-B6 | YJ016-like | YJ016-like | Texas | Oyster | 1998 |
| 300-1C1 | YJ016-like | YJ016-like | NK | Oyster | NK |
| JY1701 | YJ016-like | Louisiana | Oyster | NK | |
| Env1 | YJ016-like | YJ016-like | Louisiana | Oyster | NK |
| SS108A-3A | YJ016-like | YJ016-like | NK | Oyster | NK |
| b60 | CMCP6-like | CMCP6-like | India | Oyster | NK |
| b122 | CMCP6-like | CMCP6-like | India | Oyster | NK |
| IFVv10 | France | Mussel | NK | ||
| IFVv11 | France | Mussel | NK | ||
| 72M4 | CMCP6-like | CMCP6-like | India | Clam | NK |
| 79M4 | YJ016-like | YJ016-like | India | Clam | NK |
| 76M3 | YJ016-like | YJ016-like | India | Fish | NK |
| 80M4 | CMCP6-like | YJ016-like | India | Fish | NK |
| G-83 | YJ016-like | YJ016-like | Korea | Fish | NK |
| NCIMB 2136 | YJ016-like | YJ016-like | Japan | Eel | NK |
| 90-2-11 | YJ016-like | YJ016-like | Norway | Eel | NK |
| ATCC 33149 | YJ016-like | YJ016-like | Japan | Eel | NK |
| NCIMB 2137 | YJ016-like | YJ016-like | Japan | Eel | NK |
| E86 | YJ016-like | YJ016-like | Spain | Eel | 1990 |
| M79 | CMCP6-like | CMCP6-like | Spain | Eel | NK |
| IFVv18 | France | Seawater | NK | ||
| MLT362 | CMCP6-like | CMCP6-like | Hawaii | Seawater | 1991 |
| MLT364 | CMCP6-like | CMCP6-like | Hawaii | Seawater | 1991 |
| MLT406 | YJ016-like | Florida | Seawater | 1991 | |
| SPRC 10215 | NK | Seawater | NK | ||
| CG62 | YJ016-like | Taiwan | Seawater | 1993 | |
| CG63 | YJ016-like | YJ016-like | Taiwan | Seawater | 1993 |
| CG123 | YJ016-like | YJ016-like | Taiwan | Seawater | 1993 |
| 96-9-114s | Denmark | Sediment | NK | ||
| L-49 | Japan | Environmental | 1988 | ||
| JY1305 | YJ016-like | YJ016-like | Louisiana | Environmental | NK |
| UNCC 1015 | CMCP6-like | CMCP6-like | North Carolina | Environmental | NK |
| MLT 365 | CMCP6-like | CMCP6-like | Florida | Environmental | NK |
| UNCC 913 | YJ016-like | YJ016-like | North Carolina | Environmental | NK |
| 345/O | CMCP6-like | CMCP6-like | Louisiana | Environmental | NK |
NK, not known.
Molecular analysis.
Chromosomal DNA was extracted from each of the V. vulnificus isolates using the Genome DNA isolation kit from Bio 101 as previously described (11) (MP Biomedicals, Solon, OH). The PCR primers used to amplify the nab1 and nab2 genes from V. vulnificus YJ016 and CMCP6 were designed based on unique sequences of each strain and purchased from Integrated DNA Technologies (Coralville, IA) (Table 3). PCRs were performed in a 25-μl reaction mixture using the following program: an initial denaturation step of 94°C for 1 min, followed by 30 cycles of denaturation at 94°C for 30 s, 30 s of primer annealing at 57 to 61°C, and 60 s of primer extension at 72°C. PCR products were visualized on agarose gels.
Table 3.
Primers used for PCR assays in this study
| Oligonucleotide name | Sequence (5′-3′) | Product size (bp) | Tm (°C) |
|---|---|---|---|
| VV0312F | CGA CGA AGC ACT GGC GTT TAA A | ||
| VV0312R | GCT CGA GCA TCT CCC AAT ACT | 986 | 61 |
| VV0316F | GGC CAC CCC TTC AAT TGA G | ||
| VV0316R | GTC GCA TAC ACA ACC GTG G | 435 | 60 |
| VV10808F | TAT TCG TTT AGC CAA ACA GTT GA | ||
| VV10808R | CCA CTT CAT CCC AAC GCG TT | 902 | 57 |
| VV10803F | TTA TCG GCG ACA AGG TGA | ||
| VV10803R | ATC CAT TAC ATA GGC AAA TAT G | 346 | 60 |
Bioinformatic and phylogenetic analysis of nab genes.
We performed BLAST searches (blastp) against the sequenced genome database using as seeds the sequences of proteins encoded by nab1, nab2, nab3, and nab4 from YJ016 and CMCP6, respectively. Phylogenetic analysis was performed on Nab1 and Nab2 from 21 strains encompassing 12 species. The nab1 gene alleles from 2 of the 3 nab clusters in A. salmonicida are pseudogenes and were not included in the analysis of Nab1 proteins. Complete protein sequences were aligned using ClustalW 2.0 or MUSCLE V3.8, and the alignment was manually checked for invariant and conserved regions using GENEDOC (version 2.7; National Resource for Biomedical Supercomputing (Pittsburgh, PA) (14, 33). We used neighbor joining (NJ) as implemented in MEGA4 (19, 20, 54). The bootstrap values for NJ trees were obtained after 1,000 generations, and MEGA4 tree viewer was used to visualize the trees and calculate confidence values (25, 54). 16S rRNA sequences were also analyzed as described above for overlay of NAB pathway presence or absence.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequence accession numbers used to construct Vibrio phylogeny were as follows: Vibrio cholerae CECT 514, X76337; Vibrio furnissii ATCC 35016, X76336; Vibrio fluvialis NCTC 11327, X76335; Vibrio brasiliensis LMG 20546, AJ316172; Vibrio mimicus ATCC 33653T, X74713; Vibrio metschnikovii CIP69.14T, X74711; V. vulnificus ATCC 27562, X76333; Vibrio alginolyticus ATCC 17749, X56576; Vibrio splendidus LMG 4042, AJ515230; Vibrio parahaemolyticus ATCC 17802, AF388386; Vibrio harveyi NCIMB1280T, AY750575; Vibrio tubiashii ATCC 19109, X74725; Vibrio orientalis ATCC 33934T, X74719; Vibrio sinaloensis CAIM 797, DQ451211; Vibrio rotiferatus LMG 21460, AJ316187; Vibrio navarrensis, X74715; Vibrio sp. AND4, AF025960; Vibrio sp. EX25, CP001805; Vibrio coralliilyticus LMG 20984, AJ440005; Vibrio shiloni AK1, AF007115.1; Listonella anguillarum, AM235737; Photobacterium damselae ATCC 33539, AB032015; Photobacterium angustum ATCC 25915, D25307; Photobacterium profundum DSJ4, D21226; Grimontia hollisae LMG 17719, AJ514909; Aliivibrio fischeri ATCC 774T, X74702; Aliivibrio salmonicida NCMB 2262, X70643; Vibrio ordalii ATCC 33509T, X74718; Shewanella benthica ATCC 43992, X82131. PCR primers for V. vulnificus YJ016 and CMCP6 nab1 and nab2 were designed using accession numbers VV0316 (NP_933109.1), VV0312 (NP_933105.1), VV10803 (NP_759780.1), and VV10808 (NP_759785.1) respectively. The accession numbers for the other Vibrio nab genes used, including those used for BLAST analysis and tree construction, are as follows. The nab1 accession numbers for V. vulnificus (2 strains), Vibrio sp. EX25, V. parahaemolyticus, V. salmonicida, V. fischeri, Photobacterium profundum (2 strains), V. harveyi, V. splendidus, V. coralliilyticus, V. shilonii, V. mimicus, and Grimontia hollisae are NP_759780.1, NP_933109.1, ZP_04922879.1, NP_796582.1, YP_002264336.1, YP_203530.1, YP_002154944.1, ZP_01218688.1, YP_130889.1, ZP_00989910.1, YP_001443906.1, ZP_01867489.1, ZP_06040339.1, ZP_06054134.1, and ZP_06053975.1. The nab2 accession numbers for V. vulnificus (2 strains), Vibrio sp. EX25, V. parahaemolyticus, V. salmonicida, V. fischeri, P. profundum (2 strains), V. harveyi, V. splendidus, V. coralliilyticus, V. shilonii, V. mimicus, and G. hollisae are NP_759785.1, NP_933105.1, ZP_04922882.1, NP_796579.1, YP_002261714.1, YP_002261791.1, YP_002264335.1, YP_203526.1, YP_002154946.1, ZP_01218684.1, YP_130887.1, YP_001443899.1, ZP_00989905.1, ZP_05888524.1, ZP_01867488.1, ZP_06040343.1, ZP_06054132.1, and ZP_06053977.1.
Release of NulOs by mild acid hydrolysis.
NulO sugars were released from extensively washed culture pellets by mild acid hydrolysis using 2 N acetic acid for 3 h at 80°C as previously described (36). Supernatants were filtered over Centricon centrifugal filtration cassettes with a molecular weight cutoff of 10,000 (Millipore, Kankakee, IL). The low-molecular-weight fraction was then lyophilized and stored at −20°C for use in thiobarbituric acid (TBA), 1,2-diamino-4,5-methylene dioxybenzene (DMB) high-performance liquid chromatography (HPLC), or mass spectrometric analyses as described below.
TBA assays.
The TBA assay is relatively simple, fast, and inexpensive and has been used extensively for studies of NulOs such as N-acetylneuraminic acid. Potential NulOs released from vibrios (see above) were treated for 30 min at 37°C with 0.1 N (final concentration) sodium hydroxide, followed by neutralization as previously described (23). The Vibrio strains used in these assays are listed in Table 1. Measurement of TBA-reactive species (TBARs) in this material was performed as previously described (62). Group B and A streptococci were used as positive and negative controls, respectively; the former display high levels of surface sialic acids, while the latter do not. A standard curve for TBARs was generated in each experiment using N-acetylneuraminic acid (Sigma). Results were normalized to the total protein contents of bacterial culture pellet lysates that were set aside prior to NulO hydrolysis. Protein content was measured using the bicinchoninic acid assay (Pierce).
DMB derivatization and HPLC.
NulOs released by mild acid hydrolysis were derivatized with DMB. Reaction mixtures consisted of 7 mM DMB, 18 mM sodium hydrosulfite, 1.4 M acetic acid, and 0.7 M 2-mercaptoethanol, and reactions were carried out for 2 h at 50°C in the dark. DMB-NulO derivatives were resolved by HPLC using a reverse-phase C18 column (Varian) eluted isocratically at a rate of 0.9 ml/min over 50 min using 85% Milli-Q ultrapure water, 7% methanol, and 8% acetonitrile as previously described (35, 36). Detection of fluorescently labeled NulO sugars was achieved at excitation and emission wavelengths of 373 nm and 448 nm, respectively. 3-Deoxy-d-manno-octulosonic acid (Kdo) is an eight-carbon backbone α-keto acid that forms part of the conserved core portion of LPS in virtually all Gram-negative bacteria. Kdo was released and derivatized under the same conditions as the related NulO α-keto acids and served as an internal control in these assays to express relative levels of NulOs. The Mann-Whitney nonparametric test was used for statistical evaluation of NulO/Kdo ratio differences between strains. All of the chemicals used in DMB reactions were from Sigma, St. Louis, MO.
Electrospray ionization mass spectrometry.
DMB-derivatized extracts or individually isolated HPLC peaks were analyzed at the University of California San Diego Glycotechnology Core resource using a ThermoFinnigan LCQ ion trap mass spectrometer with tandem HPLC.
RESULTS
Frequency of nab gene clusters in Vibrionaceae.
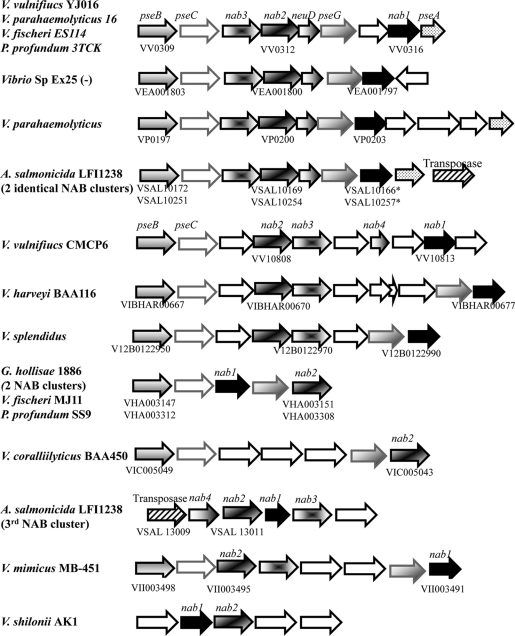
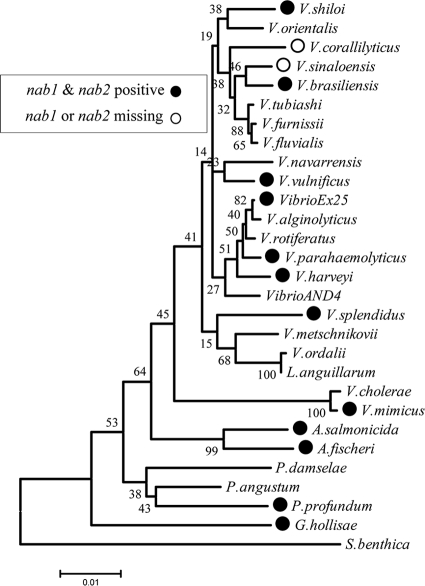
As of September 2010, there were 24 species and 66 strains of the family Vibrionaceae in the genome database (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). This includes 27 strains of V. cholerae, four strains of the closely related species V. mimicus, seven V. parahaemolyticus genome sequences, and 21 additional species with either two genomes or a single genome represented. NAB pathways are encoded by nab gene clusters, which share a core portion composed of nab1, a homolog of CMP-N-acetylneuraminic acid synthetase, nab2, a homolog of N-acetylneuraminic acid synthase, and nab3, a homolog of UDP-N-acetylglucosamine 2 epimerase. Many of these sequenced Vibrio species contained a putative NAB pathway. BLAST analysis of nab genes identified 12 species and a total of 21 strains containing nab gene clusters, i.e., V. vulnificus (2 strains), V. parahaemolyticus (6 strains), V. mimicus (1 strain), V. harveyi (2 strains), V. shilonii (1 strain), V. splendidus (1 strain), V. coralliilyticus (1 strain), V. fischeri (2 strains), V. salmonicida (1 strain), Vibrio sp. EX25 (1 strain), Photobacterium profundum (2 strains), and Grimontia hollisae (1 strain) (Fig. 1). However, about 50% (13/27) of the fully sequenced Vibrionaceae species isolates did not contain homologous pathways for NulO biosynthesis (Fig. 2).
Fig. 1.
Genetic structure of the nab gene cluster among sequenced Vibrionaceae species. Shown is a schematic representation of the gene arrangement of NAB clusters among sequenced isolates of members of the family Vibrionaceae. The annotated homologs of the ORFs are nab1 (CMP-NeuAc synthase homolog), nab2 (N-acetylneuraminic acid synthase homolog), nab3 (UDP-N-acetylglucosamine 2 epimerase homolog), neuD (acetyltransferase homolog), pseA (flagellin modification protein homolog), pseB (polysaccharide biosynthesis homolog), pseC (DegT aminotransferase homolog), and pseG (nucleotidyl/sugar P transferase homolog). Strains with identical gene orders are shown only once with isolate names listed on the left. Accession numbers for nab genes are shown below schematic ORFs.
Fig. 2.
Distribution of nab genes among sequenced members of the family Vibrionaceae. A phylogenetic tree of completely sequenced members of the family Vibrionaceae based on 16S rRNA sequences was constructed. Evolutionary history was inferred by the NJ method (49). The bootstrap consensus tree inferred from 500 replicates is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method and are expressed as the number of base substitutions per site. All positions containing gaps and missing data were eliminated from the data set (complete deletion option). There were a total of 1,191 positions in the final data set. Phylogenetic analyses were conducted in MEGA4 (54). Shewanella benthica 16S rRNA served as the outgroup. Strains that contain nab genes are indicated by circles on the right. Accession numbers for 16S sequences are provided in Materials and Methods.
Evolution of NulO biosynthesis in Vibrionaceae.
Analysis of the distribution and arrangement of nab gene clusters among Vibrionaceae lineages reveals a complex evolutionary history. Within many lineages, there is a conspicuous presence or absence of homologous NAB pathways, with some notable exceptions. For example, among the seven V. parahaemolyticus strains examined, only AN-5034 lacked the nab cluster and the nab genes from 5/6 other strains shared 100% amino acid sequence identity. V. parahaemolyticus strain 16 shared only 90% amino acid identity with the nab genes from the other strains. Among the 27 V. cholerae genomes, none contained nab genes. Yet, within sequenced isolates of V. mimicus, a species closely related to V. cholerae, 1 of the 4 sequenced strains (MB451) contained a nab region with a divergent gene order compared to that of related species (Fig. 1).
For other vibrios, phylogenetic analysis of Nab amino acid sequences revealed that members of the same species sometimes contain NAB pathways that are highly divergent from each other. One of the clearest examples of phylogenetic divergence involved the two sequenced strains of V. vulnificus (YJ016 and CMCP6), both clinical biotype 1 strains from Asia (10). The nab gene cluster within V. vulnificus YJ016 encompasses open reading frames (ORFs) VV0311 to VV0316 on chromosome 1. In strain CMCP6, the NAB cluster encompasses ORFs VV10803 to VV10808 and is also found on chromosome 1 (Fig. 1). Both Nab1 and Nab2 from CMCP6 and YJ106 are found in two divergent lineages (a and b in Fig. 3 A and B). Nab2 from YJ016 is closely related to Nab2 from V. fischeri ES114 and P. profundum 3TCK, whereas Nab2 from CMCP6 is closely related to Nab1 from V. splendidus and V. harveyi (a and b in Fig. 3B).
Fig. 3.
Correlation of phylogenetic branching patterns and analysis of Nab1 and Nab2 among Vibrionaceae species. Evolutionary relationships of Nab1 (A) and Nab2 (B) amino acid sequences from 12 species of the family Vibrionaceae was inferred by the NJ method (49). The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches. The phylogenetic trees are drawn to scale, with branch lengths in the same units as the evolutionary distances used to infer each phylogenetic tree. Evolutionary distances were computed using the Poisson correction method and are expressed as the number of amino acid substitutions per site. All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons (pairwise deletion option). There were a total of 432 positions in the final data set. Phylogenetic analyses were conducted in MEGA4 (54). The lowercase letters a and b refer to similar phylogenetic clades and gene cluster arrangements shown in green and yellow, respectively. Asterisks highlight Vibrio sp. EX25, which exhibits evidence of recombination between the a and b gene cluster alleles. See Fig. 1 for a complete description of gene identifiers and accession numbers.
Representative members of other species (A. salmonicida and G. hollisae) contain multiple paralogous nab gene clusters within the same genome. In A. salmonicida, ORFs VSAL_I0164 to VSAL_I0172 are duplicated at positions VSAL_I0250 to VSAL_I0259, but on the negative strand, and the nab1 gene is a pseudogene. A third copy of the nab cluster is found in this species at ORFs VSAL_I3009 to VSAL_I3013, but the region shows low sequence similarity to the other two clusters and has a unique gene arrangement (Fig. 1). Flanking all three nab regions in A. salmonicida is a transposase. In the G. hollisae genome, two copies of the nab region are present but the regions show less than 65% amino acid identity, ORFs VHA_003148 to VHA_003151 and VHA_003308 to VHA_003011; however, the gene order is identical. Taken together, these observations suggest that nab gene clusters in Vibrionaceae are prone to duplication, divergence, and horizontal transfer.
Gene cluster organization matches phylogenetic signatures of Nab1 and Nab2 in Vibrionaceae.
Initial observations of the nab gene clusters in sequenced strains of Vibrionaceae and other gammaproteobacteria suggested that there are at least three divergent nab gene alleles found within different gene cluster arrangements (Fig. 1). To examine this more carefully, selected nab gene clusters were aligned (Fig. 3C) and compared to the phylogenetic branching patterns of the Nab1 (Fig. 3A) and Nab2 (Fig. 3B) amino acid sequences. There was a strong correlation between the nab gene cluster arrangement and the phylogenetic lineages of the Nab1 and Nab2 amino acid sequences (Fig. 3, lineages a and b). These data also revealed examples of apparent recombination between the different nab alleles. For example, while most of the members of the family Vibrionaceae contained nab1 and nab2 alleles that both cluster in phylogenetic clade a or b, the Vibrio sp. EX25 strain contained nab1 and nab2 alleles that clustered in different phylogenetic clades (b and a, respectively) (Fig. 3).
Biochemical analyses of Vibrionaceae isolates confirm that many species express NulOs.
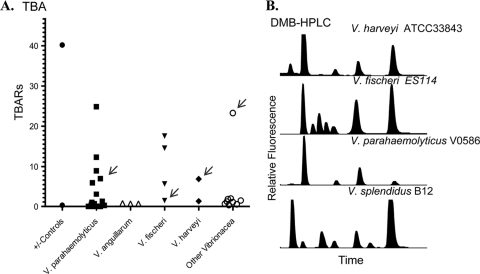
To determine whether NAB pathways are functional in Vibrionaceae, analytical approaches were applied to investigate the potential production of NulOs in different lineages. Initially, Vibrionaceae isolates (28 strains representing 14 species) were subjected to the classic TBA assay, a method originally used to evaluate the distribution and levels of sialic acids expressed by various animal lineages (61). Indeed, many of the vibrios expressed TBARs when grown under standard culture conditions (Fig. 4A). However, as suggested by genomic observations of nab gene clusters in sequenced vibrios, many of the Vibrionaceae isolates did not show TBARs at levels above those observed in negative controls. The method of TBA reactivity has some limitations, including the potential for interfering molecular species that may confound interpretation of the results (62). To further characterize potential NulO expression, selected isolates were analyzed using a method of derivatization that relies specifically on the α-keto acid shared by all types of NulOs. Briefly, mild acid hydrolysates of selected Vibrio isolates were subjected to derivatization (fluorescence tagging) of NulOs with DMB. Molecular species of DMB-NulOs were then resolved by HPLC. Chromatograms revealed multiple peaks of DMB-derivatized molecules in several Vibrio isolates (Fig. 4B).
Fig. 4.
Analysis of putative NulOs in Vibrionaceae. (A) A TBA assay of Vibrio strains listed in Table 1 was carried out as described in Materials and Methods. Results were normalized to total protein content and expressed as TBARs. Controls gave the expected results and included Streptococcus agalactiae (group B), which is known to express sialic acids, and Streptococcus pyogenes (group A), which does not express sialic acids (+ and −, respectively). (B) Putative NulOs released from selected strains (indicated by arrows in panel A) were fluorescently derivatized with DMB as described in Materials and Methods. DMB-derivatized α-keto acids were then resolved by HPLC with reverse-phase separation.
Further verification that HPLC peaks of DMB-derivatized molecules correspond to masses characteristic of NulOs was achieved by mass spectrometry (Fig. 5). For example, experiments with V. fischeri ES114 showed several retention times of DMB-derivatized molecules that correspond to masses of di-N-acetylated NulOs (m/z 450 to 451) (), while V. parahaemolyticus V0586 and V. vulnificus N-87 produced a single dominant NulO species of this mass (note that 473 and 433 correspond to the sodium adduct and dehydrated forms of the same molecule). Due to the multiple epimers and modifications of NulOs that can occur naturally (31), identical masses found within many of the HPLC peaks could not be assigned unambiguously to specific chemical structures. However, it is clear from these studies that a variety of di-N-acetylated NulOs are produced by multiple strains of V. parahaemolyticus, V. fischeri, and V. vulnificus. Taken together, these experiments conclusively show that many members of the family Vibrionaceae contain functional NAB pathways that are capable of producing a variety of NulOs.
Fig. 5.
Mass spectrometry reveals masses of DMB-derivatized di-N-acetylated NulOs in Vibrionaceae. V. fischeri (ES114*) (A), V. parahaemolyticus (518-01 and V0586*) (B), and V. vulnificus (CDC903896, N-87*, MLT362, MO6-24/O, and SS108A3A) (C) were subjected to electrospray ionization mass spectrometry performed in tandem with HPLC separation of DMB-NulOs. Asterisks indicate data representative of the strains tested. The total ion currents (TIC) of selected m/z 450 to 451 during HPLC elution are shown on the left, while mass spectra are shown on the right along with the retention times (RT).
Distribution of nab alleles among diverse V. vulnificus isolates.
To more clearly define potential associations between the different nab alleles, functional output of NulOs, and specific bacterial phylogenetic lineages, a more directed approach within a single species was utilized. V. vulnificus was chosen for these studies due to the phylogenetically divergent nab gene clusters found in the two available reference genome strains (YJ016 and CMCP6) (Fig. 3 and 6 ). Allele typing of nab1 and nab2 was performed on a collection of 67 V. vulnificus strains whose phylogenetic relationships are known, using a set of primer pairs for nab1 and nab2 alleles designed using genomic sequences from reference strains YJ016 and CMCP6 (alleles here referred to as YJ-like and CM-like [Table 2]). Four PCR assays were performed (Table 3) using genomic DNA isolated from the 67 V. vulnificus strains (Table 2), of which 40 were from environmental sources and 27 were from clinical sources. We then mapped the presence of YJ-like or CM-like nab1 and/or nab2 alleles onto the V. vulnificus phylogenetic tree (11) (Fig. 6).
Fig. 6.
Distribution of nab1 and nab2 allele types among V. vulnificus. PCR diagnosis of nab alleles was carried out using genomic DNA template from 67 strains of V. vulnificus and primers specific to the nab1 and nab2 alleles from reference strains YJ016 and CMCP6. If only one gene allele was positive, a minus sign was used to indicate a negative PCR result for the other allele. Strains that were negative in all PCR assays are indicated as “untypeable” here (−/−), since biochemical investigations revealed that they do, in fact, express NulOs (see Fig. 7). The phylogenetic tree of V. vulnificus is based on a phylogenetic analysis of six housekeeping genes as previously described (11).
PCR assays were negative for 13 of the 67 isolates examined, including biotype 3 isolates, suggesting that these strains may either lack the genes or contain unique untypeable nab genes (Fig. 6). YJ-like alleles of nab1 and nab2 were identified in 26 strains, while 16 strains contained CM-like alleles of both genes. Among the lineage I isolates, which are mostly from clinical sources, both YJ-like and CM-like alleles of nab1 and nab2 were present. However, among the lineage II isolates, which are predominantly environmental and fish isolates, the YJ-like allele was by far the most common (Fig. 6). Interestingly, two V. vulnificus isolates contained a CM-like nab1 allele with a YJ-like nab2 allele (Fig. 6). Taken together with data presented in Fig. 3, these observations further support the conclusion that members of the family Vibrionaceae sometimes engage in horizontal exchange and recombination of different nab gene cluster alleles.
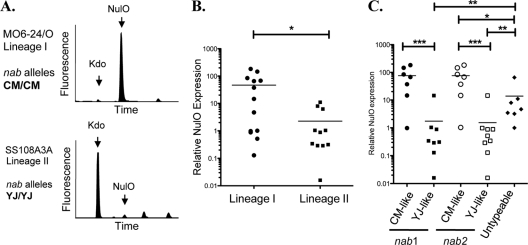
NulOs are expressed at higher levels in lineage I clinical isolates with CMCP6-like alleles.
We hypothesized that allelic differences in nab genes may predict functional differences in the level of NulO production by isolates of V. vulnificus. To test this hypothesis, we subjected isolates of V. vulnificus representing all three lineages and two nab allele types to DMB HPLC analysis. The structurally related 8-carbon α-keto acid known as Kdo served as a convenient internal standard for normalization of NulO expression, since it is present as part of the conserved core structure of LPS in Gram-negative bacteria and also reacts with DMB. Relative NulO production was determined for the different isolates by comparing HPLC peak areas at characteristic retention times of NulO and Kdo in V. vulnificus as described in Materials and Methods (Fig. 7A). Relative NulO levels were compared among lineage I and lineage II isolates (Fig. 7B) and among isolates having YJ-like and CM-like nab1 and nab2 alleles (Fig. 7C). These data show that NulO expression levels are much higher in isolates that contain CM-like nab alleles (Fig. 7C) and that these strains tend to belong to V. vulnificus phylogenetic lineage I (mostly clinical isolates) (Fig. 7B). Alternatively, isolates that contained YJ-like nab alleles expressed much lower levels of NulOs. Nearly all of the lineage II (mostly environmental) isolates contained YJ-like nab alleles and had low levels of NulOs (Fig. 7C). Interestingly, the data also show that most of the isolates that were “untypeable” by these methods did, in fact, express NulOs at levels that were significantly higher than those of strains with YJ-like nab genes but significantly lower than those of strains with CM-like nab genes. Taken together, these data strongly suggest that there are at least 3 alleles of nab genes that correspond to low (YJ-like), intermediate (untypeable), and high (CM-like) levels of NulO expression.
Fig. 7.
CMCP6-like nab alleles are associated with high levels of NulO expression in mostly clinical strains. DMB HPLC analysis of diverse V. vulnificus isolates was carried out, and NulO expression levels were normalized to an internal control monosaccharide (Kdo) that is a part of the conserved core portion of LPS. (A) Raw HPLC data from strains representing the major V. vulnificus lineages (I and II) and common nab allele types (CMCP6 and YJ016). (B) Relative NulO expression levels in lineage I and II strains. (C) V. vulnificus isolates with CM-like alleles of nab genes have significantly higher levels of NulOs than isolates with YJ-like alleles. Most of the “untypeable” isolates do, in fact, express detectable NulOs but at levels that are intermediate compared to those of isolates with YJ-like or CM-like alleles. (B and C) The Mann-Whitney test was used for statistical evaluation (*, P < 0.05; * *, P < 0.01; * * *, P < 0.0001). Note the log scales.
DISCUSSION
These studies illustrate processes of evolution within the family Vibrionaceae by analysis of genetic and phenotypic patterns related to the nine-carbon backbone α-keto acid sugars (NulOs). We show that biosynthetic pathways for NulOs are widespread in members of the family Vibrionaceae but by no means universal (Fig. 1 and 2). The distribution and phylogeny of nab pathways in vibrios suggest that some lineages or strains have lost, while others have apparently duplicated, these gene clusters (Fig. 1 and 2 and Results). Multiple biochemical approaches confirm that NAB pathways are indeed active in a variety of Vibrio isolates (Fig. 4 and 5) and can participate in the biosynthesis of multiple structural variations of NulOs within a given strain (Fig. 5A). Genomic comparisons revealed multiple allele types of NAB pathways which are reflected by similar patterns of gene arrangement and phylogeny (Fig. 3) and correspond to functional differences in NulO expression levels (Fig. 7). In particular, clinical isolates of the CMCP6-like allele expressed, on average, nearly 100-fold higher levels of NulOs than environmental isolates with the YJ016 allele. Moreover, the data show several examples of apparent recombination between nab gene cluster alleles (Fig. 3 and 6). Taken together, these studies indicate a relatively plastic state of NAB pathway evolution in Vibrionaceae. These observations highlight the family Vibrionaceae and the species V. vulnificus as interesting models for investigations of the biological functions of NulOs.
During the completion of this study, the genome sequence of a third V. vulnificus strain, MO6-24/O, a clinical isolate from the United States, became available. We found that strain MO6-24/O contains a NAB cluster identical to CMCP6 and produces high levels of NulOs, confirming our genomic predictions (Fig. 6 and 7). Lastly, our data also strongly suggest the presence of a third NAB cluster type (and possibly more cluster types) that is present among natural V. vulnificus isolates. We identified a total of 13 strains that were untypeable by our PCR method; however, our analysis of NulO levels demonstrates that these strains produce intermediate levels of NulO, indicating the presence of nab genes unrelated to YJ016 or CMCP6. Given the limited number of V. vulnificus isolates that we examined, it is tempting to speculate that there may be even more heterogeneity in nab gene content within this species and the challenge going forward will be to determine the functional role of this diversity in survival and fitness.
NulOs are generally expressed on the surfaces of cells—both in “higher” animals and on bacteria. Vibrionaceae species likely express NulOs as part of one or more surface structures such as capsular polysaccharides, LPSs, or flagella (16, 34, 37, 40–43, 48, 50, 51). For example, strains of Campylobacter jejuni can simultaneously express pseudaminic and legionaminic acids as flagellar modifications and sialic acids as LPS modifications (39, 40). It is possible that different nab alleles in Vibrionaceae encode slightly different NulO structures and/or that NulO sugars may be intended for different surface molecules (i.e., capsule, LPS, flagella). Alternatively, the different NAB alleles in V. vulnificus could be responsible for NulO modifications at different densities on the same molecule. Vibrio NulOs may be involved in motility, biofilm formation, relative phage susceptibility, or other phenotypes relevant to the marine ecosystem niche. In addition, Vibrio NulOs may have functional implications during opportunistic pathogenesis in aquatic or terrestrial animals.
In summary, the further study of NulO biology in Vibrionaceae will require careful analysis of the genetics and biochemistry of cell surface NulO modifications. Taken together with the many known roles of NulOs in host-microbe interactions, these studies provide a basis for further investigations of NulOs in bacterial behaviors and host-pathogen interactions involving the members of the family Vibrionaceae.
Acknowledgements
We thank Sandra Diaz and Sulabha Argade for initial technical support and Doug Bartlett, who kindly provided many of the Vibrio strains (other than V. vulnificus) used in these studies (Table 1). Mass spectrometry was performed at the University of California San Diego Glycotechnology Core.
This research was supported by University of California President's Postdoctoral Fellowship funds to A.L.L. and startup funds to A.L.L. and National Science Foundation CAREER award DEB-0844409 to E.F.B. J.-B.L. is supported by the Chemistry-Biology Interface graduate program at the University of Delaware.
Footnotes
Published ahead of print on 1 July 2011.
REFERENCES
- 1. Anderson G. G., Goller C. C., Justice S., Hultgren S. J., Seed P. C. 2010. Polysaccharide capsule and sialic acid-mediated regulation promote biofilm-like intracellular bacterial communities during cystitis. Infect. Immun. 78:963–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Angata T., Varki A. 2002. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev. 102:439–469 [DOI] [PubMed] [Google Scholar]
- 3. Austin B. 2010. Vibrios as causal agents of zoonoses. Vet. Microbiol. 140:310–317 [DOI] [PubMed] [Google Scholar]
- 4. Bisharat N., Amaro C., Fouz B., Llorens A., Cohen D. I. 2007. Serological and molecular characteristics of Vibrio vulnificus biotype 3: evidence for high clonality. Microbiology 153:847–856 [DOI] [PubMed] [Google Scholar]
- 5. Bisharat N., et al. 2005. Hybrid Vibrio vulnificus. Emerg. Infect. Dis. 11:30–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bisharat N., et al. 2007. The evolution of genetic structure in the marine pathogen, Vibrio vulnificus. Infect. Genet. Evol. 7:685–693 [DOI] [PubMed] [Google Scholar]
- 7. Bøgwald J., Hoffman J. 2006. Structural studies of the O-antigenic oligosaccharide from Vibrio salmonicida strain C2 isolated from Atlantic cod, Gadus morhua L. Carbohydr. Res. 341:1965–1968 [DOI] [PubMed] [Google Scholar]
- 8. Carlin A. F., Lewis A. L., Varki A., Nizet V. 2007. Group B streptococcal capsular sialic acids interact with siglecs (immunoglobulin-like lectins) on human leukocytes. J. Bacteriol. 189:1231–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carlin A. F., et al. 2009. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood 113:3333–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen C. Y., et al. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cohen A. L., Oliver J. D., DePaola A., Feil E. J., Boyd E. F. 2007. Emergence of a virulent clade of Vibrio vulnificus and correlation with the presence of a 33-kilobase genomic island. Appl. Environ. Microbiol. 73:5553–5565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DePaola A., Capers G. M., Alexander D. 1994. Densities of Vibrio vulnificus in the intestines of fish from the U.S. Gulf Coast. Appl. Environ. Microbiol. 60:984–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edebrink P., Jansson P. E., Bøgwald J., Hoffman J. 1996. Structural studies of the Vibrio salmonicida lipopolysaccharide. Carbohydr. Res. 287:225–245 [DOI] [PubMed] [Google Scholar]
- 14. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ewing C. P., Andreishcheva E., Guerry P. 2009. Functional characterization of flagellin glycosylation in Campylobacter jejuni 81-176. J. Bacteriol. 191:7086–7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gil-Serrano A. M., et al. 1999. Structural determination of a 5-acetamido-3,5,7,9-tetradeoxy-7-(3-hydroxybutyramido)-l-glycero-l-manno-nonulosonic acid-containing homopolysaccharide isolated from Sinorhizobium fredii HH103. Biochem. J. 342(Pt. 3): 527–535 [PMC free article] [PubMed] [Google Scholar]
- 17. Grimes D. J., et al. 2009. What genomic sequence information has revealed about Vibrio ecology in the ocean—a review. Microb. Ecol. 58:447–460 [DOI] [PubMed] [Google Scholar]
- 18. Guerry P., et al. 2006. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol. Microbiol. 60:299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 20. Guindon S., Lethiec F., Duroux P., Gascuel O. 2005. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557–W559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heidelberg J. F., Heidelberg K. B., Colwell R. R. 2002. Bacteria of the gamma-subclass Proteobacteria associated with zooplankton in Chesapeake Bay. Appl. Environ. Microbiol. 68:5498–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heidelberg J. F., Heidelberg K. B., Colwell R. R. 2002. Seasonality of Chesapeake Bay bacterioplankton species. Appl. Environ. Microbiol. 68:5488–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Higa H. H., Butor C., Diaz S., Varki A. 1989. O-acetylation and de-O-acetylation of sialic acids. O-acetylation of sialic acids in the rat liver Golgi apparatus involves an acetyl intermediate and essential histidine and lysine residues—a transmembrane reaction? J. Biol. Chem. 264:19427–19434 [PubMed] [Google Scholar]
- 24. Houliston R. S., et al. 2011. The lipooligosaccharide of Campylobacter jejuni: similarity with multiple types of mammalian glycans beyond gangliosides. J. Biol. Chem. 286:12361–12370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huson D. H. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68–73 [DOI] [PubMed] [Google Scholar]
- 26. Jones C., Virji M., Crocker P. R. 2003. Recognition of sialylated meningococcal lipopolysaccharide by siglecs expressed on myeloid cells leads to enhanced bacterial uptake. Mol. Microbiol. 49:1213–1225 [DOI] [PubMed] [Google Scholar]
- 27. Jones M. K., Oliver J. D. 2009. Vibrio vulnificus: disease and pathogenesis. Infect. Immun. 77:1723–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaysner C. A., et al. 1987. Virulent strains of Vibrio vulnificus isolated from estuaries of the United States West Coast. Appl. Environ. Microbiol. 53:1349–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kaysner C. A., Tamplin M. L., Wekell M. M., Stott R. F., Colburn K. G. 1989. Survival of Vibrio vulnificus in shellstock and shucked oysters (Crassostrea gigas and Crassostrea virginica) and effects of isolation medium on recovery. Appl. Environ. Microbiol. 55:3072–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Khatua B., et al. 2010. Sialic acids acquired by Pseudomonas aeruginosa are involved in reduced complement deposition and siglec mediated host-cell recognition. FEBS Lett. 584:555–561 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31. Knirel Y. A., Shashkov A. S., Tsvetkov Y. E., Jansson P. E., Zahringer U. 2003. 5,7-Diamino-3,5,7,9-tetradeoxynon-2-ulosonic acids in bacterial glycopolymers: chemistry and biochemistry. Adv. Carbohydr. Chem. Biochem. 58:371–417 [DOI] [PubMed] [Google Scholar]
- 32. Komagamine T., Yuki N. 2006. Ganglioside mimicry as a cause of Guillain-Barré syndrome. CNS Neurol. Disord. Drug Targets 5:391–400 [DOI] [PubMed] [Google Scholar]
- 33. Larkin M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 34. Le Quéré A. J., et al. 2006. Structural characterization of a K-antigen capsular polysaccharide essential for normal symbiotic infection in Rhizobium sp. NGR234: deletion of the rkpMNO locus prevents synthesis of 5,7-diacetamido-3,5,7,9-tetradeoxy-non-2-ulosonic acid. J. Biol. Chem. 281:28981–28992 [DOI] [PubMed] [Google Scholar]
- 35. Lewis A. L., et al. 2009. Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc. Natl. Acad. Sci. U. S. A. 106:13552–13557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lewis A. L., Nizet V., Varki A. 2004. Discovery and characterization of sialic acid O-acetylation in group B Streptococcus. Proc. Natl. Acad. Sci. U. S. A. 101:11123–11128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li X., et al. 2010. Structural and genetic characterization of the O-antigen of Escherichia coli O161 containing a derivative of a higher acidic diamino sugar, legionaminic acid. Carbohydr. Res. 345:1581–1587 [DOI] [PubMed] [Google Scholar]
- 38. Linkous D. A., Oliver J. D. 1999. Pathogenesis of Vibrio vulnificus. FEMS Microbiol. Lett. 174:207–214 [DOI] [PubMed] [Google Scholar]
- 39. Linton D., et al. 2000. Multiple N-acetyl neuraminic acid synthetase (neuB) genes in Campylobacter jejuni: identification and characterization of the gene involved in sialylation of lipo-oligosaccharide. Mol. Microbiol. 35:1120–1134 [DOI] [PubMed] [Google Scholar]
- 40. Logan S. M., et al. 2009. Identification of novel carbohydrate modifications on Campylobacter jejuni 11168 flagellin using metabolomics-based approaches. FEBS J. 276:1014–1023 [DOI] [PubMed] [Google Scholar]
- 41. Logan S. M., Kelly J. F., Thibault P., Ewing C. P., Guerry P. 2002. Structural heterogeneity of carbohydrate modifications affects serospecificity of Campylobacter flagellins. Mol. Microbiol. 46:587–597 [DOI] [PubMed] [Google Scholar]
- 42. McNally D. J., et al. 2007. Targeted metabolomics analysis of Campylobacter coli VC167 reveals legionaminic acid derivatives as novel flagellar glycans. J. Biol. Chem. 282:14463–14475 [DOI] [PubMed] [Google Scholar]
- 43. McNally D. J., et al. 2006. Functional characterization of the flagellar glycosylation locus in Campylobacter jejuni 81-176 using a focused metabolomics approach. J. Biol. Chem. 281:18489–18498 [DOI] [PubMed] [Google Scholar]
- 44. Motes M. L., et al. 1998. Influence of water temperature and salinity on Vibrio vulnificus in Northern Gulf and Atlantic Coast oysters (Crassostrea virginica). Appl. Environ. Microbiol. 64:1459–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Naito M., et al. 2010. Effects of sequential Campylobacter jejuni 81-176 lipooligosaccharide core truncations on biofilm formation, stress survival, and pathogenesis. J. Bacteriol. 192:2182–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Oliver J. D. 1995. The viable but non-culturable state in the human pathogen Vibrio vulnificus. FEMS Microbiol. Lett. 133:203–208 [DOI] [PubMed] [Google Scholar]
- 47. Oliver J. D. 2005. Wound infections caused by Vibrio vulnificus and other marine bacteria. Epidemiol. Infect. 133:383–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Perepelov A. V., et al. 2010. Structure of the O-antigen and characterization of the O-antigen gene cluster of Escherichia coli O108 containing 5,7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-d-galacto-non-2-ulosonic (8-epilegionaminic) acid. Biochemistry (Mosc.) 75:19–24 [DOI] [PubMed] [Google Scholar]
- 49. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 50. Schirm M., et al. 2003. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 48:1579–1592 [DOI] [PubMed] [Google Scholar]
- 51. Shashkov A. S., et al. 2007. Structure of the O-antigen of Providencia stuartii O20, a new polysaccharide containing 5,7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-d-galacto-non-2-ulosonic acid. Carbohydr. Res. 342:653–658 [DOI] [PubMed] [Google Scholar]
- 52. Swords W. E., et al. 2004. Sialylation of lipooligosaccharides promotes biofilm formation by nontypeable Haemophilus influenzae. Infect. Immun. 72:106–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tamplin M., Rodrick G. E., Blake N. J., Cuba T. 1982. Isolation and characterization of Vibrio vulnificus from two Florida estuaries. Appl. Environ. Microbiol. 44:1466–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 55. Varki A. 2007. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature 446:1023–1029 [DOI] [PubMed] [Google Scholar]
- 56. Vimr E., Lichtensteiger C. 2002. To sialylate, or not to sialylate: that is the question. Trends Microbiol. 10:254–257 [DOI] [PubMed] [Google Scholar]
- 57. Vimr E. R., Kalivoda K. A., Deszo E. L., Steenbergen S. M. 2004. Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 68:132–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vinogradov E., et al. 2009. Structure of the lipopolysaccharide core of Vibrio vulnificus type strain 27562. Carbohydr. Res. 344:484–490 [DOI] [PubMed] [Google Scholar]
- 59. Warner E. B., Oliver J. D. 2008. Multiplex PCR assay for detection and simultaneous differentiation of genotypes of Vibrio vulnificus biotype 1. Foodborne Pathog. Dis. 5:691–693 [DOI] [PubMed] [Google Scholar]
- 60. Warner J. M., Oliver J. D. 1999. Randomly amplified polymorphic DNA analysis of clinical and environmental isolates of Vibrio vulnificus and other vibrio species. Appl. Environ. Microbiol. 65:1141–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Warren L. 1963. The distribution of sialic acids in nature. Comp. Biochem. Physiol. 10:153–171 [DOI] [PubMed] [Google Scholar]
- 62. Warren L. 1959. The thiobarbituric acid assay of sialic acids. J. Biol. Chem. 234:1971–1975 [PubMed] [Google Scholar]
- 63. Wessels M. R., Rubens C. E., Benedi V. J., Kasper D. L. 1989. Definition of a bacterial virulence factor: sialylation of the group B streptococcal capsule. Proc. Natl. Acad. Sci. U. S. A. 86:8983–8987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wu H., Jerse A. E. 2006. Alpha-2,3-sialyltransferase enhances Neisseria gonorrhoeae survival during experimental murine genital tract infection. Infect. Immun. 74:4094–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]