Abstract
Salmonella genomic island 1 (SGI1) contains a multidrug resistance region conferring the ampicillin-chloramphenicol-streptomycin-sulfamethoxazole-tetracycline resistance phenotype encoded by blaPSE-1, floR, aadA2, sul1, and tet(G). Its increasing spread via interbacterial transfer and the emergence of new variants are important public health concerns. We investigated the molecular properties of SGI1-carrying Salmonella enterica serovars selected from a European strain collection. A total of 38 strains belonging to S. enterica serovar Agona, S. enterica serovar Albany, S. enterica serovar Derby, S. enterica serovar Kentucky, S. enterica serovar Newport, S. enterica serovar Paratyphi B dT+, and S. enterica serovar Typhimurium, isolated between 2002 and 2006 in eight European countries from humans, animals, and food, were subjected to antimicrobial susceptibility testing, molecular typing methods (XbaI pulsed-field gel electrophoresis [PFGE], plasmid analysis, and multilocus variable-number tandem-repeat analysis [MLVA]), as well as detection of resistance and virulence determinants (PCR/sequencing and DNA microarray analysis). Typing experiments revealed wide heterogeneity inside the strain collection and even within serovars. PFGE analysis distinguished a total of 26 different patterns. In contrast, the characterization of the phenotypic and genotypic antimicrobial resistance revealed serovar-specific features. Apart from the classical SGI1 organization found in 61% of the strains, seven different variants were identified with antimicrobial resistance properties associated with SGI1-A (S. Derby), SGI1-C (S. Derby), SGI1-F (S. Albany), SGI1-L (S. Newport), SGI1-K (S. Kentucky), SGI1-M (S. Typhimurium), and, eventually, a novel variant similar to SGI1-C with additional gentamicin resistance encoded by aadB. Only minor serovar-specific differences among virulence patterns were detected. In conclusion, the SGI1 carriers exhibited pathogenetic backgrounds comparable to the ones published for susceptible isolates. However, because of their multidrug resistance, they may be more relevant in clinical settings.
INTRODUCTION
Salmonella genomic island 1 (SGI1) is a 43-kb mobilizable chromosomal region that is located in Salmonella enterica serovars between the genes thdF and yidY or between thdF and int2 in the retron sequence of S. enterica serovar Typhimurium (7). In 2008, an SGI1 secondary chromosomal attachment site for integration of the island between the sodB and purR genes was described (17). SGI1 contains an antimicrobial resistance gene cluster (7, 16) located on a 13-kb complex class 1 integron designated In104 (31), which confers pentaresistance (Penta-R) to ampicillin (blaPSE-1), chloramphenicol/florfenicol (floR), streptomycin/spectinomycin (aadA2), sulfamethoxazole (sul1), and tetracycline [tet(G)] (8). The genes blaPSE-1 and aadA2 are located on class 1 integrons, named InC and InD, with variable regions of 1.2 kb and 1.0 kb, respectively (3, 24). The five resistance determinants are characteristic of the pandemic S. Typhimurium definite phage type 104 (DT104), which was globally most predominant in causing infections in humans since the 1980s (16, 21, 38). SGI1 was first identified in 2000 in a Canadian multidrug-resistant S. Typhimurium DT104 strain with the Penta-R phenotype (7). Subsequently SGI1 and several variant SGI1 antimicrobial resistance gene clusters were detected worldwide in different Salmonella serovars and in Proteus mirabilis (1, 2, 15, 31, 39, 40).
Within the European Union (EU) Network of Excellence Med-Vet-Net, an investigation on the molecular epidemiology of SGI1 in enteric bacteria other than S. Typhimurium DT104 was conducted (http://www.medvetnet.org). For this study, a collection of 445 multidrug-resistant strains (S. enterica, 277; Escherichia coli, 116; Shigella spp., 43, and Proteus spp., 9), isolated from animals, food, and humans from eight European countries between the years 2002 and 2006, was investigated (3). By conventional gel-based PCR assays, 53% of the Salmonella isolates were SGI1 positive, showing fragments of both typical left and right SGI1 junctions (3, 8). Application of real-time PCR targeting open reading frame (ORF) S004 of SGI1 detected the island in five additional Salmonella strains. These strains lacked both SGI1 junctions (one S. enterica serovar Agona and three S. enterica serovar Derby) or showed the left junction only (one S. Typhimurium) (3).
The spread of multidrug resistance (MDR) among bacterial populations is alarming, and the detection of SGI1 is regarded as important for public health (3). Therefore, the characterization of SGI1-carrying bacteria as presented in this study is a useful tool to support efforts in the health care sector. Taking this into account, we selected from the collection of Amar et al. (3) all strains that acquired SGI1 and belonged to serovars other than S. Typhimurium or showed phage types unrelated to DT104. In order to gain more knowledge on their phenotypic and molecular backgrounds, all isolates were analyzed for their antimicrobial resistance phenotypes and the underlying molecular mechanisms, screened for selected virulence genes, and subjected to molecular typing. In this way, a well-characterized collection of strains was set up to be used as controls for several molecular-epidemiological approaches to antimicrobial resistance and virulence.
MATERIALS AND METHODS
Participating institutes.
Institutes from eight European countries provided SGI1-positive Salmonella enterica subsp. enterica isolates for this study: Agence Nationale de Sécurité Sanitaire de L'alimentation de L'environment et du Travail (ANSES), Maisons-Alfort, France; Central Veterinary Institute (CVI), Lelystad, The Netherlands; Federal Institute for Risk Assessment (BfR), Berlin, Germany; Istituto Superiore di Sanità (ISS), Rome, Italy; Statens Serum Institute (SSI), Copenhagen, Denmark; Panstwowy Zaklad Higieny (PZH), Warsaw, Poland; Veterinary Laboratories Agency (VLA), Weybridge, United Kingdom; and Veterinary Medical Research Institute (VMRI), Budapest, Hungary.
Bacterial isolates.
In a previous work, the Health Protection Agency (HPA), Laboratory of Enteric Pathogens, United Kingdom, analyzed between May 2006 and March 2007 a European selection of 445 bacterial isolates for the presence of SGI1. In this study, a total of 152 Salmonella isolates tested positive for SGI1 (3). Of these, 38 strains were sent to the BfR, Berlin, Germany, for further molecular analyses. This collection of SGI1-positive isolates included (i) all isolates belonging to serovars other than S. Typhimurium, i.e., S. Agona (1 isolate), S. enterica serovar Kentucky (1), S. enterica serovar Paratyphi B dT+ (S. enterica serovar Java) (1), S. enterica serovar Albany (3), S. enterica serovar Newport (4), and S. Derby (5); (ii) S. Typhimurium non-DT104 (18); and, for comparison, (iii) S. Typhimurium DT104 (5). The strains were isolated between 2002 and 2006 from humans (19 isolates), animals (10), and food (9) in the eight European countries (Table 1). We included in this series all isolates (15) that showed atypical SGI1 properties (lack of SGI1 markers) in the collection described by Amar et al. (3). These 15 isolates were designated “unconventional” in the present study (Table 2).
Table 1.
Molecular typing of Salmonella isolates used in the study
| Countrya | NRL-Salm no. (isolation yr) | Serotype | Origin | Phage type | Resistance phenotypeb | PFGE profile | Plasmid size(s) (kb) | MLVA type (STTR 9-5-6-10-3) |
|---|---|---|---|---|---|---|---|---|
| NL | 08-02864 (04) | S. Agona | Human | NDh | [Penta]c | XAg1 | ≤6 | ND |
| D | 08-00310 (02) | S. Albany | Fungi | ND | AMP/AMC-CHL/FLO-STR(i)d-SUL-TET-TMP/SXT-NAL | XAl1 | ≤6 | ND |
| F | 08-00265 (06) | S. Albany | Food | ND | AMP/AMC-CHL/FLO-STR(i)d-SUL-TET-TMP/SXT-NAL | XAl2 | ≤6 | ND |
| F | 08-00309 (06) | S. Albany | Food | ND | AMP/AMC-CHL/FLO-STR(i)d-SUL-TET-TMP/SXT-NAL | XAl2 | ≤6 | ND |
| NL | 08-00263 (04) | S. Derby | Human | ND | [Penta]-TMP/SXT-NAL | XD1 | 110 | ND |
| NL | 08-00311 (04) | S. Derby | Human | ND | [Penta]-TMP/SXT | XD2 | None | ND |
| NL | 08-02866 (05) | S. Derby | Human | ND | STR/SPE-SUL-TET | XD3 | 90, 30 | ND |
| NL | 08-02867 (05) | S. Derby | Human | ND | STR/SPE-SUL-TET | XD3a | 15, ≤6 | ND |
| NL | 08-02868 (05) | S. Derby | Human | ND | STR/SPE-SUL-TET | XD3a | 30, 8 | ND |
| UK | 08-00267 (04) | S. Paratyphi B dT+ | Cattle | ND | [Penta] | XJ1 | 110 | ND |
| PL | 08-00266 (03) | S. Kentucky | Human | ND | AMP/AMC-STR/SPE-SUL-TET-GEN-NAL | XK1 | None | ND |
| DK | 08-00268 (05) | S. Newport | Human | ND | [Penta]-GEN-KAN/NEO-TMP/SXT-NAL | XN1 | None | ND |
| DK | 08-00269 (05) | S. Newport | Human | ND | [Penta]-GEN-KAN/NEO-TMP/SXT-NAL | XN2 | None | ND |
| DK | 08-00313 (05) | S. Newport | Human | ND | [Penta]-GEN-TMP/SXT-NAL | XN3 | ≤6 | ND |
| DK | 08-00314 (05) | S. Newport | Human | ND | [Penta]-GEN-KAN/NEO-TMP/SXT-NAL | XN3 | None | ND |
| D | 08-00325 (04) | S. Typhimurium | Sausage | DT193 | [Penta] | XT1 | 90, 10, 7, ≤6 | 3-14-15-21-311 |
| D | 08-00326 (05) | S. Typhimurium | Swine | RDNCe | [Penta]-TMP/SXT | XT2a | 90, 100, 9, 7, ≤6 | 3-14-15-13-311 |
| D | 08-00272 (05) | S. Typhimurium | Swine | 206 | [Penta]-GEN-TMP/SXT | XT2 | 90, 100, 150 | 3-14-13-14-311 |
| D | 08-02885 (05) | S. Typhimurium | Rabbit | DT104L | GEN-STR/SPE-SUL | XT3 | 90, ≤6 | 3-13-8-24-311 |
| D | 08-02886 (05) | S. Typhimurium | Rodent | DT120 | AMP/AMC-CHL/FLO-SUL-TET-GEN-TMP/SXT | XT3 | 90, 55, ≤6, ≤6 | 3-13-8-24-311 |
| NL | 08-00264 (05) | S. Typhimurium | Chicken | DT104L | [Penta]-NAL | XT3 | 90, 7, ≤ 6 | 3-12-10-24-311 |
| NL | 08-02865 (05) | S. Typhimurium | Human | DT104B low | [Penta]-GEN | XT3 | 90, 20, ≤6 | 3-14-13-18-311 |
| NL | 08-00315 (05) | S. Typhimurium | Rapeseed | DT104L | [Penta] | XT7 | 90, 180 | 3-16-17-22-311 |
| NL | 08-00312 (05) | S. Typhimurium | Human | DT104L | [Penta] | XT8 | 90, ≤6 | 3-11-18-17-311 |
| NL | 08-00316 (04) | S. Typhimurium | Human | NTf | [Penta]-NAL(i) | XT3 | 90, ≤6, ≤6, ≤6 | 3-17-6-23-311 |
| NL | 08-00270 (04) | S. Typhimurium | Pig | U309 | [Penta]-TMP/SXT | XT4 | 90, 50, ≤6 | 3-15-12-15-311 |
| NL | 08-00317 (05) | S. Typhimurium | Pig | U309 | [Penta] | XT9 | 130, 90, 9, 7, ≤6 | 3-10-14-26-311 |
| NL | 08-00318 (05) | S. Typhimurium | Human | 206 | [Penta]-GEN | XT2 | 90, 150 | 3-14-14-13-311 |
| NL | 08-00319 (05) | S. Typhimurium | Pig | RDNC | [Penta] | XT2a | 90, 9, 7, ≤6 | 3-14-16-23-311 |
| I | 08-00322 (04) | S. Typhimurium | Human | NT | [Penta]-TMP/SXT | XT3 | 90, 25, ≤6, ≤6 | 3-10-17-14-311 |
| I | 08-00271 (04) | S. Typhimurium | Human | U309 | [Penta]-KAN/NEO-NAL | XT3a | 90 | 3-14-16-21-311 |
| DK | 08-00323 (05) | S. Typhimurium | Human | NT | [Penta]-GEN | XT5 | 95, 90, ≤6 | 3-15-14-16-311 |
| DK | 08-00324 (05) | S. Typhimurium | Human | DT92 | [Penta]-GEN | XT6 | 170, 90 | 3-14-15-14-311 |
| UK | 08-00320 (04) | S. Typhimurium | Sheep | DT193 | [Penta] | XT10 | 90 | 3-14-12-24-311 |
| UK | 08-00321 (05) | S. Typhimurium | Cattle | DT151 | [Penta] | XT2a | 90, ≤6 | 3-14-16-13-311 |
| H | 08-00327 (06) | S. Typhimurium | Goose | RDNC | [Penta] | XT11 | 90, 45, 30, 10, ≤6 | 3-11-5-NAg-311 |
| H | 08-00328 (06) | S. Typhimurium | Goose | RDNC | [Penta] | XT11 | 90, 30, ≤6 | 3-11-5-NA-311 |
| H | 08-00329 (06) | S. Typhimurium | Pork sausage | RDNC | [Penta]-GEN-TMP/SXT | XT2b | 90, 100, 150 | 3-14-14-14-311 |
Country abbreviations: D, Germany; DK, Denmark; F, France; H, Hungary; I, Italy; NL, Netherlands; PL, Poland; UK, United Kingdom.
For abbreviations, see Materials and Methods. Resistance phenotypes were assessed by the broth microdilution method, as described by Schroeter et al. (37), by following the guidelines of the CLSI (11). The breakpoints used for AMP, AMC, XNL, CHL, CIP, GEN, KAN, SUL, and TET were the CLSI breakpoints (12). For CIP, the MIC breakpoints were as follows: resistant, ≥4 μg/ml; intermediate, 2 μg/ml; and susceptible, ≤1 μg/ml. The breakpoints used for COL (resistant, ≥16 μg/ml; susceptible, ≤8 μg/ml), FLO (resistant, ≥32 μg/ml; susceptible, ≤8 μg/ml), NAL (resistant, ≥32 μg/ml; susceptible, ≤16 μg/ml), SPE (resistant, ≥128 μg/ml; susceptible, ≤64 μg/ml), and TMP (resistant, ≥16 μg/ml; susceptible, ≤8 μg/ml) were the DANMAP breakpoints (19). Finally, the breakpoints used for NEO (resistant, ≥16 μg/ml; susceptible, ≤4 μg/ml), STR (resistant, ≥32 μg/ml; susceptible, ≤8 μg/ml), and SXT (resistant, ≥4 and ≥76 μg/ml respectively; susceptible, ≤2 and ≤38 μg/ml, respectively) were taken from Schroeter et al. (37).
[Penta], SGI-1 characteristic pentaresistance phenotype [AMP/AMC-CHL/FLO-STR/SPE-SUL-TET].
Intermediate (i) resistance phenotype to STR (MIC, 16 μg/ml) (37).
RDNC, reacts with phages but does not conform to definite or provisional types.
NT, nontypeable.
NA, locus not present.
ND, not done.
Table 2.
Detection of SGI1 and virulence and resistance determinants
| Countrya | NRL-Salm no. (isolation yr) | Serotype | Origin | SGI1b | Resistance phenotypec | Resistance genotype | Class 1 integron | Virulence genotype |
|---|---|---|---|---|---|---|---|---|
| NL | 08-02864 (04) | S. Agona (U)m | Human | S004 | [Penta]d | [Penta-R]e | [I]f | [V-core]g-avrA |
| D | 08-00310 (02) | S. Albany (U) | Fungi | LJ/RJ | AMP/AMC-CHL/FLO-STR(i)h-SUL-TET-TMP/SXT-NAL | blaPSE-1-floR-sul1-tet(G)-dfrA1-gyrAAsp87→Asn87 | 1,200 bp/blaPSE-1 + 1,250 bp/dfrA1-orf | [V-core]-avrA |
| F | 08-00265 (06) | S. Albany (U) | Food | LJ/RJ | AMP/AMC-CHL/FLO-STR(i)-SUL-TET-TMP/SXT-NAL | blaPSE-1-floR-sul1-tet(G)-dfrA1-gyrAAsp87→Asn87 | 1,200 bp/blaPSE-1 + 1,250 bp/dfrA1-orf | [V-core]-avrA |
| F | 08-00309 (06) | S. Albany (U) | Food | LJ/RJ | AMP/AMC-CHL/FLO-STR(i)-SUL-TET-TMP/SXT-NAL | blaPSE-1-floR-sul1-tet(G)-dfrA1-gyrAAsp87→Asn87 | 1,200 bp/blaPSE-1 + 1,250 bp/dfrA1-orf | [V-core]-avrA |
| NL | 08-00263 (04) | S. Derby | Human | LJ/RJ | [Penta] -TMP/SXT-NAL | [Penta-R]-dfrA10-gyrASer83→Ala83 | [I] | [V-core]-avrA |
| NL | 08-00311 (04) | S. Derby | Human | LJ/RJ | [Penta] -TMP/SXT | [Penta-R]-dfrA10-gyrASer83→Ala83 | [I] | [V-core]-avrA |
| NL | 08-02866 (05) | S. Derby (U) | Human | S004 | STR/SPE-SUL-TET | aadA2-sul1-tet(A) | 1,000 bp/aadA2 | [V-core]-avrA-sopE1 |
| NL | 08-02867 (05) | S. Derby (U) | Human | S004 | STR/SPE-SUL-TET | aadA2-sul1-tet(A) | 1,000 bp/aadA2 | [V-core]-avrA-sopE1 |
| NL | 08-02868 (05) | S. Derby (U) | Human | S004 | STR/SPE-SUL-TET | aadA2-sul1-tet(A) | 1,000 bp/aadA2 | [V-core]-avrA-sopE1 |
| UK | 08-00267 (04) | S. Paratyphi B dT+ | Cattle | LJ/RJ | [Penta] | [Penta-R]-sul2 | [I] | [V-core]-sodC1 |
| PL | 08-00266 (03) | S. Kentucky (U) | Human | LJ/RJ | AMP/AMC-STR/SPE-SUL-TET-GEN-NAL | blaTEM-1-strA/aadA7-sul1-tet(A)-aac(3)-Ie-gyrASer83→Phe83 | 1,600 bp/aac(3)-Ie-aadA7 | [V-core]-avrA |
| DK | 08-00268 (05) | S. Newport (U) | Human | LJ/RJ | [Penta] -GEN-KAN/NEO-TMP/SXT-NAL | blaPSE-1-floR-strA/aadA7-sul1-tet(A)-tet(G)-aac(3)-Ie-aphA1-dfrA15-gyrAAsp87→Gly87 | 750 bp/dfrA15 + 1,200 bp/blaPSE-1 + 1,600 bp/aac(3)-Ie-aadA7 | [V-core]-avrA-sodC1-sopE1 |
| DK | 08-00269 (05) | S. Newport (U) | Human | LJ/RJ | [Penta] -GEN-KAN/NEO-TMP/SXT-NAL | blaPSE-1-floR-strA/aadA7-sul1-tet(A)-tet(G)-aac(3)-Ie-aphA1-dfrA15-gyrAAsp87→Gly87 | 750 bp/dfrA15 + 1,200 bp/blaPSE-1 + 1,600 bp/aac(3)-Ie-aadA7 | [V-core]-avrA-sodC1-sopE1 |
| DK | 08-00313 (05) | S. Newport (U) | Human | LJ/RJ | [Penta] -GEN-TMP/SXT-NAL | blaPSE-1-floR-strA/aadA7-sul1-tet(G)-aac(3)-Ie-dfrA15-gyrASer83→Phe83 | 750 bp/dfrA15 + 1,200 bp/blaPSE-1 + 1,600 bp/aac(3)-Ie-aadA7 | [V-core]-avrA-sodC1-sopE1 |
| DK | 08-00314 (05) | S. Newport (U) | Human | LJ/RJ | [Penta] -GEN-KAN/NEO-TMP/SXT-NAL | blaPSE-1-floR-strA/aadA7-sul1-tet(A)-tet(G)-aac(3)-Ie-aphA1-dfrA15-gyrAAsp87→Gly87 | 750 bp/dfrA15 + 1,200 bp/blaPSE-1 + 1,600 bp/aac(3)-Ie-aadA7 | [V-core]-avrA-sodC1-sopE1 |
| D | 08-00325 (04) | S. Typhimurium | Sausage | LJ/RJ + R | [Penta] | [Penta-R]-catA1 | [I] | [V-core]-avrA-spvC-sodC1 |
| D | 08-00326 (05) | S. Typhimurium | Swine | LJ/RJ + R | [Penta] -TMP/SXT | [Penta-R]-catA1-sul3-(unknown)i | [I] | [V-core]-avrA-spvC-sodC1 |
| D | 08-00272 (05) | S. Typhimurium | Swine | LJ/RJ + R | [Penta] -GEN-TMP/SXT | [Penta-R]-blaTEM-1-like-catA1-strA/aadA1-sul2-aacC4-dfrA1 | [I] + 1,600 bp/dfrA1-aadA1 | [V-core]-avrA-spvC-sodC1 |
| D | 08-02885 (05) | S. Typhimurium (U) | Rabbit | LJ/RJ + R | GEN-STR/SPE-SUL | aadB-strA/aadA2-sul1 | 800 bp/aadB + integron containing aadA2j | [V-core]-avrA-spvC-sodC1 |
| D | 08-02886 (05) | S. Typhimurium (U) | Rodent | LJ/RJ + R | AMP/AMC-CHL/FLO-SUL-TET-GEN-TMP/SXT | blaPSE-1-floR-sul1-tet(G)-aadB-(unknown) | 800 bp/aadB + 1,200 bp/blaPSE-1 | [V-core]-avrA-spvC-sodC1 |
| NL | 08-00264 (05) | S. Typhimurium | Chicken | LJ/RJ + R | [Penta] -NAL | [Penta-R]-gyrAAsp87→Tyr87 | [I] | [V-core]-avrA-spvC-sodC1 |
| NL | 08-02865 (05) | S. Typhimurium (U) | Human | LJ | [Penta] -GEN | [Penta-R]-(unknown) | [I] | [V-core]-avrA-spvC-sodC1 |
| NL | 08-00315 (05) | S. Typhimurium | Rapeseed | LJ/RJ + R | [Penta] | [Penta-R]-blaTEM-1-like | [I] | [V-core]-avrA-spvC-sodC1 |
| NL | 08-00312 (05) | S. Typhimurium | Human | LJ/RJ + R | [Penta] | [Penta-R] | [I] | [V-core]-avrA-spvC-sodC1 |
| NL | 08-00316 (04) | S. Typhimurium | Human | LJ/RJ + R | [Penta] -(CIP-NAL)k | [Penta-R]-catA1-qnrB19 | [I] | [V-core]-avrA-spvC-gipA-sodC1 |
| NL | 08-00270 (04) | S. Typhimurium | Pig | LJ/RJ + R | [Penta] -TMP/SXT | [Penta-R]-blaTEM-1-like-catA1-sul2-tet(A)-dfrA14 | [I] | [V-core]-avrA-spvC-sodC1 |
| NL | 08-00317 (05) | S. Typhimurium | Pig | LJ/RJ + R | [Penta] | [Penta-R]-blaTEM-1-like-catA1-tet(A) | [I] | [V-core]-avrA-spvC-sodC1 |
| NL | 08-00318 (05) | S. Typhimurium | Human | LJ/RJ + R | [Penta] -GEN | [Penta-R]-catA1-strA-tet(B)-aacC4 | [I] | [V-core]-avrA-spvC-sodC1 |
| NL | 08-00319 (05) | S. Typhimurium | Pig | LJ/RJ + R | [Penta] | [Penta-R]-catA1 | [I] | [V-core]-avrA-spvC-sodC1 |
| I | 08-00322 (04) | S. Typhimurium | Human | LJ/RJ + R | [Penta] -TMP/SXT | [Penta-R]-catA1-sul2-dfrA14 | [I] | [V-core]-avrA-spvC-sodC1 |
| I | 08-00271 (04) | S. Typhimurium | Human | LJ/RJ + R | [Penta] -KAN/NEO-NAL | [Penta-R]-catA1-strA-sul2-tet(B)-aphA1-gyrAAsp87→Asn87 | [I] | [V-core]-avrA-spvC-sodC1 |
| DK | 08-00323 (05) | S. Typhimurium | Human | LJ/RJ + R | [Penta] -GEN | [Penta-R]-catA1-strA-aacC4 | [I] | [V-core]-avrA-spvC-sodC1 |
| DK | 08-00324 (05) | S. Typhimurium | Human | LJ/RJ + R | [Penta] -GEN | [Penta-R]-catA1-strA-aacC4 | [I] | [V-core]-avrA-spvC-sodC1 |
| UK | 08-00320 (04) | S. Typhimurium | Sheep | LJ/RJ + R | [Penta] | [Penta-R]-catA1 | [I] | [V-core]-avrA-spvC-sodC1 |
| UK | 08-00321 (05) | S. Typhimurium | Cattle | LJ/RJ + R | [Penta] | [Penta-R]-catA1 | [I] | [V-core]-avrA-spvC-sodC1 |
| H | 08-00327 (06) | S. Typhimurium | Goose | LJ/RJ + R | [Penta] | [Penta-R]-blaTEM-1-like | [I] | [V-core]-avrA-sodC1 |
| H | 08-00328 (06) | S. Typhimurium | Goose | LJ/RJ + R | [Penta] | [Penta-R] | [I] | [V-core]-avrA-sodC1 |
| H | 08-00329 (06) | S. Typhimurium | Pork sausage | LJ/RJ + R | [Penta] -GEN-TMP/SXT | [Penta-R]-cmlA1-strA-sul3-aacC4-dfrA12 | [I] + integron containing dfrA12-aadA2-cmlA-aadA1l | [V-core]-avrA-spvC-sodC1 |
For country abbreviations, see note a to Table 1.
Markers for the presence of SGI1: S004, SGI1 ORF outside the MDR region; LJ, left junction of SGI1; RJ, right junction of SGI1; R, retron phage element.
For abbreviations, see Materials and Methods.
See note c to Table 1.
[Penta-R], SGI1 characteristic pentaresistance genotype [blaPSE-1-floR-aadA2-sul1-tet(G)].
[I], SGI1 characteristic class 1 integron [1,000 bp/aadA2 + 1,200 bp/blaPSE-1].
[V-core], virulence genes present throughout all 38 isolates: ssaQ, mgtC, spi4_D, sopB, and bcfC.
Intermediate (i) resistance phenotype to STR (MIC, 16 μg/ml) (37).
Unknown, none of the genes tested were present.
Defective integron, undetectable by 5′CS/3′CS primers. PCR products were observed using the following primer sets: 5′CS/aadA2-B, int1-R/aadA2-R, and int1-R/qacΔE-R. No PCR product was obtained using aadA2-F/sul1-B and aadA2-F/3′CS primers (see Table SA in the supplemental material).
Decreased susceptibility phenotype to CIP (MIC, 0.5 μg/ml) and NAL (MIC, 8 μg/ml) (12).
Defective integron, undetectable by 5′CS/3′CS primers, as described by Antunes et al. (5).
(U), unconventional isolate.
Phenotypic analysis.
Serotyping was done following the White-Kauffmann-Le Minor scheme (22). Phage typing of S. Typhimurium isolates was performed according to the method of Anderson et al. (4). All isolates were tested by broth microdilution, according to the guidelines of the CLSI (11), for their susceptibility to 17 antimicrobial agents: ampicillin (AMP), amoxicillin-clavulanic acid (AMC), ceftiofur (XNL), chloramphenicol (CHL), ciprofloxacin (CIP), colistin (COL), florfenicol (FLO), gentamicin (GEN), kanamycin (KAN), nalidixic acid (NAL), neomycin (NEO), spectinomycin (SPE), streptomycin (STR), sulfamethoxazole (SUL), tetracycline (TET), trimethoprim (TMP), and sulfamethoxazole/trimethoprim (SXT), using custom-defined microtiter plates (Trek Diagnostic Systems, United Kingdom). The antimicrobial concentrations tested were described by Schroeter et al. (37). The breakpoints used are shown in Table 1.
Molecular typing.
Plasmid profiling was performed by alkaline denaturation according to the method of Kado and Liu (28). Vertical electrophoresis was carried out on 0.8% agarose (standard low melting point; Bio-Rad Laboratories, Munich, Germany) gels for 3 to 4 h at 100 V. The E. coli reference plasmids R27 (169 kb), R1 (94 kb), RP4 (55 kb), and ColE1 (6 kb) were used as standards for the determination of plasmid sizes. Genomic DNA from the SGI1 isolates was subjected to macrorestriction analysis with the XbaI endonuclease (Roche Diagnostics, Mannheim, Germany). The generated fragments were separated by pulsed-field gel electrophoresis (PFGE) using the CHEF-DRIII SYS220/240 system (Bio-Rad Laboratories, Munich, Germany). Agarose plug preparation and PFGE running conditions followed the PulseNet standardized protocol (http://www.pulsenet-europe.org). The resulting profiles were analyzed by recording the presence or absence of fragments larger than 33 kb. Profiles with differences in two or more bands were designated using numbers (e.g., X1, X2, etc.). Similar patterns in which only one band, or rather, the intensity of a band, was different were designated using lowercase letters (e.g., X3 and X3a). Multilocus variable-number tandem-repeat analysis (MLVA) of S. Typhimurium isolates was performed as described by Lindstedt et al. (32) using an ABI 310 DNA Sequencer (Applied Biosystems, Darmstadt, Germany). MLVA nomenclature according to Larsson et al. (29) was used.
Detection of resistance and virulence determinants.
In a first approach, detection of resistance and virulence genes in all isolates was undertaken by PCR amplification (primers and conditions are given in Table SA in the supplemental material). The isolates were analyzed for the presence of resistance genes related to their resistance phenotypes. Overall, genes encoding resistance to ampicillin (blaPSE1, blaTEM1-like, and blaOXA1-like), chloramphenicol (catA1, cmlA, and floR), gentamicin [aacC2, aacC4, aac(3)-Ie (aacC5), aadB, and armA], kanamycin [aphA1, aphA2, and aac(6)-1b], fluoroquinolones (qnrA, qnrB, and qnrS), streptomycin (aadA1-like, aadA2, aadA7, and strA-strB), sulfamethoxazole (sul1, sul2, and sul3), tetracycline [tet(A), tet(B), and tet(G)], and trimethoprim (dfrA1-like, dfrA5-14, dfrA7-17, dfrA10, dfrA12, and dfrA17) were screened as previously described (23, 35). The presence of class 1 and 2 integrons was investigated by using the 5′-CS/3′-CS and Hep74/Hep51 primers to amplify their variable regions (30, 41). Gene cassettes inserted therein were identified by DNA sequencing of the amplicons performed by Agowa GmbH (Berlin, Germany). The sequences obtained were compared using the BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
For virulotyping analysis, isolates were screened for the presence of 10 virulence genes (avrA, bcfC, gipA, mgtC, ssaQ, sopB, sopC1, sopE1, spvC, and spi4_D). PCR amplifications were performed as described by Huehn et al. (27).
In a second approach, nine isolates (4 S. Typhimurium, 1 S. Newport, 1 S. Paratyphi B dT+, 1 S. Albany, 1 S. Kentucky, and 1 S. Derby), representing different serovars, phage types, countries, and/or origins, were selected for an extensive analysis of the presence of virulence determinants using DNA microarrays (see Fig. 2). Probes, microarray production, and performance of whole-genome DNA labeling, hybridization, analysis, and validation were as previously described (26). Virulence determinants for each strain analyzed (101 genes) were categorized according to their locations on the Salmonella genome: SPIs, prophages, plasmids, islets, and fimbrial clusters. Signals that were assigned as “uncertain” (cutoff between 0.25 and 0.4) by microarray analysis were verified by PCR as described previously (26), and an individual decision was made for the presence or absence of each target.
Fig. 2.
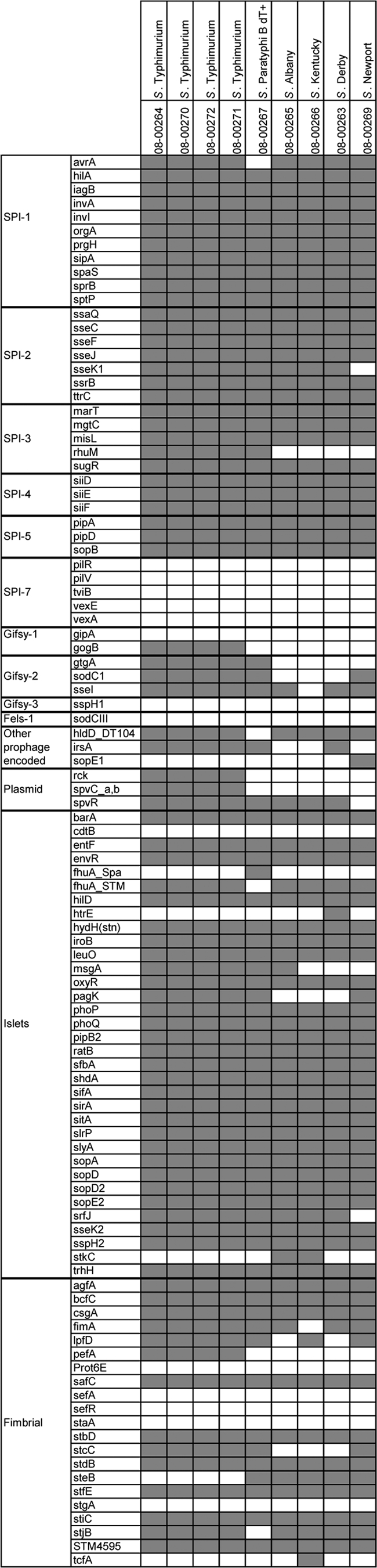
Virulence determinant microarray data for 9 SGI1-positive Salmonella strains. The analyzed genes are grouped according to their genomic locations (SPI-1 to SPI-7; prophages Gifsy-1, Gifsy-2, Gifsy-3, and Fels-1; plasmids; and islets) or function (fimbrial).
Additional analysis of unconventional isolates lacking a classical chromosomal attachment site.
Mapping of the regions flanking the primary chromosomal integration site of SGI1 was performed for a selected group of five isolates already described as unconventional by Amar et al. (3): S. Agona 08-02864 (MDR region and S004 present; other markers outside the MDR region and left and right junctions absent); S. Derby 08-02866, 08-02867, and 08-02868 (only S004 present); and S. Typhimurium 08-02865 (right junction absent). For this, PCR amplifications were carried out targeting genes framing the SGI1 region: upstream (dnaN, dnaA, rpmH, rnpA, yidC, and thdF) and downstream (int2, urt, rt, yidY, yidZ, yieE, yieF, yieG, and yieH). The primers for the genes cited above were designed from the sequence of S. Typhimurium LT2 (accession number AE008879) (see Table SA in the supplemental material).
To detect a secondary integration site of SGI1, previously reported in the intergenic region between the chromosomal genes sodB and purR, primers sodB-F and purR-B were used (17). Only in case of a nonintegration of SGI1 was an amplicon size of 508 bp expected.
Data analysis.
Results were analyzed with BioNumerics software (version 5.1; Applied Maths, Sint-Martens-Latem, Belgium).
RESULTS
Molecular typing.
In 33 of the 38 isolates (87%), one to five plasmids were detected, ranging from approximately ≤6 to 180 kb. All S. Typhimurium isolates (n = 23) carried a serovar-specific virulence plasmid of 94 kb (33, 36). No plasmids were identified in five isolates belonging to the serovars S. Newport (3 isolates), S. Kentucky (1), and S. Derby (1).
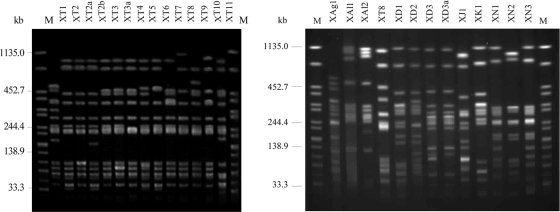
PFGE analysis using XbaI generated 26 patterns. The 23 S. Typhimurium isolates showed 14 different patterns, the 5 S. Derby isolates 4 patterns, the 4 S. Newport isolates 3 patterns, and the 3 S. Albany isolates 2 different patterns (Fig. 1).
Fig. 1.
XbaI PFGE profiles (X) of representative isolates. To define the profiles, only bands at >33 kb were considered. Similar profiles with only a one-band difference are designated by letters. Profiles with differences in two or more bands are designated by numbers. Lanes M contained XbaI-digested DNA of S. enterica serovar Braenderup H9812, which was used as a size standard. T, S. Typhimurium; Ag, S. Agona; Al, S. Albany; D, S. Derby; J, S. Paratyphi B dT+ (S. Java); K, S. Kentucky; N, S. Newport.
By MLVA analysis, the 23 S. Typhimurium isolates were assigned to 21 MLVA types (Table 1). Two of the types (allele numbers 3-13-8-24-311 and 3-11-5-NA-311) were represented by two strains each, isolated in Germany and Hungary. All isolates showed identical alleles for loci STTR-9 and -3. The highest numbers of alleles were found in loci STTR-6 (12 variants) and STTR-10 (13 variants).
Phenotypic and genotypic characterization of antimicrobial resistance.
Fourteen different resistance patterns, conferring resistance to at least four antimicrobial substances, were identified among the isolates tested (Tables 1 and 2). The SGI1 characteristic Penta-R phenotype was detected in 29 isolates throughout all serotypes. Variations within this phenotype were observed in several isolates (Tables 1 and 2).
The prevalence of resistance genes and their distributions in the different Salmonella serovars are shown in Table 2. Twenty-five isolates were positive for the five characteristic SGI1 resistance genes, blaPSE-1, floR, aadA2, sul1, and tet(G), but again, variations were found (Table 2). The sul1 gene was present in all 38 isolates. The plasmid-mediated-quinolone-resistance (PMQR) gene, qnrB19, was detected on a 2.7-kb ColE-like plasmid in a Dutch S. Typhimurium isolate (CIP MIC = 0.5 μg/ml; NAL MIC = 8 μg/ml) of human origin (25).
All isolates tested in this study harbored class 1 integrons (Table 2), whereas none carried class 2 integrons. Based on the gene cassette content of the variable region and the amplicon size, the nine different types of class 1 integrons shown in Table 3 were observed. The two class 1 integrons typically linked to the presence of SGI1, generating PCR amplicons of 1,200 bp and 1,000 bp and carrying the blaPSE-1 and aadA2 gene cassettes, respectively, were present in 25 (66%) of the isolates. Two S. Typhimurium isolates carried 3′-CS-defective integrons that could not be detected using the 5′-CS/3′-CS primers but were identified by linking the integrase gene intI1 and the different gene cassettes. One isolate carried the aadA2 and blaPSE-1 variable regions characteristic of the SGI1 integron, together with an additional defective integron with the dfrA12-aadA2-cmlA1-aadA1 variable region (5), while the aadB variable region and a defective integron containing aadA2 were present in the other (Tables 2 and 3).
Table 3.
Class 1 integrons detected in SGI1-positive isolates (n = 38)
| Variable-region size(s)a (bp) | Gene cassette(s)a | n | Serovar(s) |
|---|---|---|---|
| 1,000 + 1,200 | aadA2 + blaPSE-1 | 23 | S. Typhimurium (19), S. Derby (2), S. Agona, S. Paratyphi B dT+ |
| 1,000 + 1,200 + 1,600 | aadA2 + blaPSE-1 + dfrA1-aadA1 | 1 | S. Typhimurium |
| 1,000 + 1,200 + defective integronb | aadA2 + blaPSE-1 + dfrA12-aadA2-cmlA-aadA1b | 1 | S. Typhimurium |
| 1,200 + 1,250 | blaPSE-1 + dfrA1-orf | 3 | S. Albany |
| 1,000 | aadA2 | 3 | S. Derby |
| 1,600 | aac(3)-Ie-aadA7 | 1 | S. Kentucky |
| 1,600 + 1,200 + 750 | aac(3)-Ie-aadA7 + blaPSE-1 + dfrA15 | 4 | S. Newport |
| 1,200 + 800 | blaPSE-1 + aadB | 1 | S. Typhimurium |
| 800 + defective integronc | aadB + aadA2c | 1 | S. Typhimurium |
PCR/sequencing was performed using the 5′CS/3′CS primers (30).
Defective integron, undetectable by 5′CS/3′CS primers, as described by Antunes et al. (5).
Defective integron, undetectable by 5′CS/3′CS primers. PCR products were observed using the following primer sets: 5′CS/aadA2-B, int1-R/aadA2-R, and int1-R/qacΔE-R.
Due to the differences in the SGI1 MDR gene content, the S. Albany (three isolates), S. Kentucky (1), S. Newport (4), and S. Typhimurium (2) isolates were considered part of the unconventional isolates (Table 2).
Analyses of SGI1 flanking regions on unconventional isolates lacking a classical chromosomal attachment site.
In the other five unconventional isolates, one S. Agona (08-02864), three S. Derby (08-02866, 08-02867, and 08-02868), and one S. Typhimurium (08-02865), one or both junctions were absent. The flanking regions of these isolates were analyzed by PCR screening. In all five isolates, the normal SGI1 attachment sites thdF and yidY, as well as all analyzed up- and downstream flanking genes (dnaN, dnaA, rpmH, rnpA, yidC, thdF, int2, urt, rt, yidY, yidZ, yieE, yieF, yieG, and yieH), were present.
Integration of SGI1 into a secondary chromosomal attachment site between the genes sodB and purR was not observed, because in all isolates, a PCR product of 518 bp was generated.
Virulotyping.
All isolates were screened by PCR analysis for the presence or absence of 10 selected virulence genes. The ssaQ, mgtC, spi4_D, sopB (carried by Salmonella pathogenicity islands [SPIs]), and bcfC (fimbria-related) genes were present in all the isolates tested. Except for the S. Paratyphi B dT+ strain, all isolates also carried the avrA gene located in SPI1. The spvC gene, carried by the Salmonella virulence plasmid, was present in all S. Typhimurium isolates except two. Likewise, the sodC1 gene, located on a bacteriophage, was found in all isolates belonging to S. Typhimurium, S. Newport, and S. Paratyphi B dT+, whereas for sopE1, only the four S. Newport and three S. Derby isolates tested positive. The gipA gene was detected in only one S. Typhimurium isolate. Altogether, in this strain collection, seven different combinations of virulence genes were detected (Table 2).
Since major serovar-specific differences were observed, nine representative Salmonella isolates were analyzed by DNA microarray in more detail. All four S. Typhimurium isolates tested had identical virulence gene repertoires (Fig. 2). In contrast to the five isolates of other serovars tested, all S. Typhimurium isolates possessed gogB (encoding a leucine-rich-repeat protein), usually located on Gifsy-1, but lacked gipA (encoding a Peyer's patch-specific virulence factor, GipA). Virulence markers associated with the prophage Gifsy-2 were always present in S. Typhimurium, but no markers of the Gifsy-3 and Fels-1 prophages tested positive. Other prophage genes, like hldD_DT104 (encoding a DT104-specific phage protein) and irsA (encoding a putative transcriptional regulator and internal response element to stress) were present, but sopE1 (encoding a translocated effector protein) was absent. The Salmonella virulence plasmid genes rck, spvC, and spvR were positive, and with the exception of cdtB (encoding a cytolethal distending toxin), fhuA_Spa (encoding an outer membrane protein receptor), htrE (encoding a probable porin/fimbrial assembly protein), and stkC (encoding an outer membrane usher protein), all genes analyzed on virulence islets were present. For the fimbria-coding genes tested, pefA (encoding a virulence plasmid fimbrial protein) was positive and steB (encoding an outer membrane fimbrial usher protein) was negative, in contrast to the isolates of other serovars.
The S. Paratyphi B dT+ isolate was solely negative for avrA (encoding a protein that inhibits activation of NF-κB), hldD_DT104, and the fimbrial stjB (encoding a putative fimbrial usher protein), and it had none of the Gifsy-1 genes tested. For the islet genes, the only difference from S. Typhimurium was fhuA_Spa instead of fhuA_STM.
S. Albany was characterized by a truncated SPI-3 missing rhuM (encoding a putative cytoplasmic protein) and possessed only sseI of Gifsy-2 and hldD_DT104 of the other prophage genes. In contrast to S. Typhimurium and S. Paratyphi B dT+, no pagK (encoding a PhoPQ-activated protein) was detected, but stkC was present. Also, lpfD (encoding a long polar fimbrial operon protein), pefA, and stcC (encoding a putative outer membrane protein) of the fimbrial genes were missing.
For S. Kentucky, five genes were found to be different from those of S. Albany, namely, the islet gene msgA (encoding an ssrB-regulated factor) and fimA (encoding a major type 1 subunit fimbrin) were negative, while lpfD and tcfA (encoding an S. Typhi colonization factor and putative fimbrial protein) were positive. The Gifsy-2 gene sseI was negative only for this isolate.
The S. Derby strain analyzed had the same fimbrial set as S. Albany. In addition, the isolate was positive for irsA and solely for htrE. The genes msgA and pagK were negative.
Also, one S. Newport strain was investigated for its virulence genes. Probes were negative for sseK1 (encoding Salmonella-secreted effector K1) of SPI-2 and rhuM of SPI-3. Of the Gifsy-2 genes, gtgA was missing, but genes of other prophages were positive in the case of hldD_DT104 and, only in this isolate, sopE1. Additionally, it possessed no srfJ gene (encoding a putative virulence factor activated by transcription factor SsrB), which was found only for this isolate.
DISCUSSION
The acquisition of genomic islands has played a central role in bacterial evolution as a mechanism of diversification and adaptation (10). Since the emergence of pandemic multidrug-resistant S. Typhimurium DT104, its antimicrobial resistance-conferring genomic island, SGI1, has increasingly attracted research interest. In particular, the horizontal-transfer potential of SGI1 has been intensively studied (8, 16). In this study, we sought to investigate the molecular properties of SGI1-carrying epidemic Salmonella isolates in a European strain collection of human, animal, and food origin. We specially focused on isolates that belonged to serovars other than S. Typhimurium or to phage types of S. Typhimurium other than DT104. We intended to establish the relationship between virulence, multidrug resistance, and the presence of SGI1.
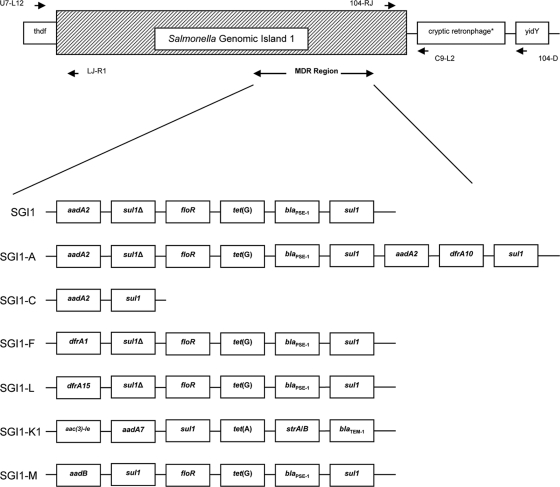
The results of typing showed wide variability within the series (38 isolates from seven serovars). PFGE analysis distinguished within the same serovar several XbaI patterns, with a total of 26 different patterns. MLVA analysis of the S. Typhimurium isolates was also very heterogeneous (21 types). These findings are in line with the supposed epidemiological nonrelatedness of the strains, which were isolated in different countries and from different sources. In contrast, characterization of the phenotypic and genotypic antimicrobial resistance in general displayed certain serovar-specific features. Apart from the resistance genes organized in SGI1, several strains exhibited additional resistance genes with either chromosomal or plasmidic locations. Altogether, we could identify eight possible SGI1 variants in our strain collection, based on the particular resistance genotype and the presence of class 1 integrons. The majority of strains (61%; 23 isolates), belonging to the S. enterica serovars Agona (1 isolate), Paratyphi B dT+ (1), and Typhimurium (21), harbored the classical SGI1 resistance genes (Fig. 3) (3, 34). Isolates with a gene repertoire typical of the variants SGI1-A and SGI1-C (Fig. 3) were found only within S. Derby (one each). The three S. Albany strains harbored genes suggesting the presence of SGI1-F (Fig. 3) (14). SGI1-L was recognized in the four S. Newport isolates and SGI1-K1 in S. Kentucky (Fig. 3), as previously described (13, 18). In the S. Typhimurium isolate 08-02886, the resistance gene cluster found (Fig. 3) suggests the presence of the variant SGI1-M, which was first described for a Dutch S. Typhimurium DT104 horse isolate by Vo et al. (39). To our knowledge, this is the first time that an SGI1-M variant has been detected in a German S. Typhimurium DT120 isolate of rodent origin. The same variant was also detected by Rodríguez et al. (35) in German S. Typhimurium isolates (phage types DT104L and DT012) of equine, cat, and avian origin associated with the presence of the extended-spectrum beta-lactamase-encoding gene blaCTX-M-1 . An integron with a variable region of 800 bp containing the aadB gene cassette was also found in another S. Typhimurium isolate, 08-02885, which additionally carried a 3′-CS-defective integron with an aadA2 gene cassette. We assume that this isolate contains a novel variant similar to SGI1-C (34) with additional acquired resistance to gentamicin, but this assumption needs further investigation. The SGI1 has historically been linked to S. Typhimurium, and we have demonstrated that, despite the clonal diversity of the S. Typhimurium isolates (shown by PFGE and MLVA analyses), there is low genetic variation in the genomic island, since among 23 isolates, 21 carry the classical SGI1, 1 the SGI1-M variant, and another, as mentioned above, a potential new variant.
Fig. 3.
SGI1 variants corresponding to the resistance gene repertoire found within the isolates (S. Agona and S. Paratyphi B dT+, SGI1; S. Typhimurium, SGI1 and SGI1-M; S. Derby, SGI1-A and SGI1-C; S. Albany, SGI1-F; S. Newport, SGI1-L; S. Kentucky, SGI1-K1). The scheme is based on information from Amar et al. (3), Boyd et al. (9), Cloeckaert et al. (13), Doublet et al. (18), and Vo et al. (39). Only antimicrobial resistance genes are shown. *, not present in all isolates.
The virulotyping results for strains carrying SGI1 indicated that only little or no variation was found for most genes incorporated in SPIs and for the fimbrial marker. Thus, the genes ssaQ , mgtC , spi4_D , sopB , and bcfC were present throughout all serovars in all 38 isolates. It was observed that some virulence patterns were serovar specific, e.g., all S. Newport and most S. Typhimurium strains shared a common virulotype, which was characterized in both by the presence of eight genes. The association of a certain serovar with a virulence-associated gene panel was also confirmed by the microarray results in this study and a previous study conducted by Huehn et al. (27) within the Med-Vet-Net project on a wide collection (77 isolates, 10 of them SGI positive) of European Salmonella isolates belonging to different serovars. The presence of genes in the SGI1 that can play a role in the virulence of the carrier isolates has been mentioned previously (6, 20). The data presented here and by Huehn et al. (27) show that, despite minor differences, SGI1-positive isolates carry more or less identical virulence gene repertoires. Mapping of the unconventional isolates did not reveal any aberrations either in the SGI1 integration sites (8) or in the flanking regions. Nevertheless, in some isolates, the left and right junctions (1 S . Agona and 3 S . Derby isolates) or only the right junction (one S. Typhimurium isolate) could not be detected by conventional PCR, although the presence of SGI1 was proved by real-time PCR targeting S004 in the precursor study carried out by Amar et al. (3). For the S. Typhimurium isolate in which only the left junction was detectable, one might assume genetic rearrangements or single-nucleotide polymorphisms (SNPs) at the ends of SGI1. For the S . Derby and the S . Agona isolates, we proved SGI1 was not integrated in a secondary chromosomal attachment site between the genes sodB and purR , as suggested by Doublet et al. (17). To elucidate the exact genetic organization of this region, further mapping and sequencing experiments will be undertaken.
In summary, we set up a panel of strains from different origins that can be used as a reference collection for molecular studies. We could observe great phenotypic and genetic hetero-geneity of SGI1-positive isolates. Apart from the resistance genes typical of SGI1 [blaPSE-1, floR, sul1, tet(G), and aadA2], other chromosomal or plasmid-located resistance determinants were detected. As shown by other authors (9, 10, 13–18, 34, 39), several genetic events led to a large number of SGI1 variants, which can be spread through vertical and horizontal transfer. Due to their virulence gene repertoires and their resistance properties, some of these variants/isolates may become clinically relevant. The data presented provide a solid basis for cooperation in resistance and virulence studies leading to a scientifically based risk characterization.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Gramatke, K. Thomas, W. Barownick, E. Junker, M. Jaber, J. Ledwolorz, and G. Berendonk (Salmonella and Antimicrobial Resistance Reference Laboratories of the Federal Institute for Risk Assessment) for their assistance. We also thank the Med-Vet-Net WP21 Project Group, which provided the isolates used in this study, and especially F. Aarestrup (DTU, Lyngby, Denmark), M. Kirchner (VLA, Weybridge, United Kingdom), S. Granier (AFSSA, Maison-Alfort, France), I. Luzzi (ISS, Rome, Italy), M. Moreno (UCM, Madrid, Spain), B. Nagy (VMRI, Budapest, Hungary), M. Torpdahl (SSI, Copenhagen, Denmark), J. Szych (PZH, Warsaw, Poland), and K. Hopkins and C. Amar (HPA, London, United Kingdom). We also thank A. Cloeckaert, R. La Ragione, and E. Liebana for their invaluable advice.
This work was funded by the Federal Institute for Risk Assessment (BfR: 46-001 and 45-004) and the EU Network of Excellence Med-Vet-Net (WP21 and -26).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 24 June 2011.
REFERENCES
- 1. Ahmed A. M., Hussein A. I., Shimamoto T. 2007. Proteus mirabilis clinical isolate harbouring a new variant of Salmonella genomic island 1 containing the multiple antibiotic resistance region. J. Antimicrob. Chemother. 59:184–190 [DOI] [PubMed] [Google Scholar]
- 2. Akiba M., et al. 2006. Detection and characterization of variant Salmonella genomic island 1s from Salmonella Derby isolates. Jpn. J. Infect. Dis. 59:341–345 [PubMed] [Google Scholar]
- 3. Amar C. F., et al. 2008. Real-time PCRs and fingerprinting assays for the detection and characterization of Salmonella Genomic Island-1 encoding multidrug resistance: application to 445 European isolates of Salmonella, Escherichia coli, Shigella, and Proteus. Microb. Drug Resist. 14:79–92 [DOI] [PubMed] [Google Scholar]
- 4. Anderson E. S., Ward L. R., de Saxe M. J., de Sa J. D. 1977. Bacteriophage-typing designations of Salmonella typhimurium. J. Hyg. 78:297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Antunes P., Machado J., Peixe L. 2007. Dissemination of sul3-containing elements linked to class 1 integrons with an unusual 3′ conserved sequence region among Salmonella isolates. Antimicrob. Agents Chemother. 51:1545–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bamforth J., et al. 2009. Salmonella Genomic Island 1 influences expression of virulence-associated genes in early stationary phase and enhances killing of Caenorhabditis elegans for Salmonella enterica serovar Typhimurium DT104, p. 104 In Proceedings of the 3rd ASM Conference on Salmonella: Biology, Pathogenesis, and Prevention. ASM Press, Washington, DC [Google Scholar]
- 7. Boyd D. A., Peters G. A., Ng L., Mulvey M. R. 2000. Partial characterization of a genomic island associated with the multidrug resistance region of Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 189:285–291 [DOI] [PubMed] [Google Scholar]
- 8. Boyd D., et al. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boyd D., Cloeckaert A., Chaslus-Dancla E., Mulvey M. R. 2002. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob. Agents Chemother. 46:1714–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boyd E. F., Almagro-Moreno S., Parent M. A. 2009. Genomic islands are dynamic, ancient integrative elements in bacterial evolution. Trends Microbiol. 17:47–53 [DOI] [PubMed] [Google Scholar]
- 11. Clinical and Laboratory Standards Institute 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 7 th ed. (M7-A7), vol. 26, no. 2. CLSI, Wayne, PA [Google Scholar]
- 12. Clinical and Laboratory Standards Institute 2006. Performance standards for antimicrobial susceptibility testing, 16th international supplement (M100-S16), vol. 26, no. 3. CLSI, Wayne, PA [Google Scholar]
- 13. Cloeckaert A., Praud K., Doublet B., Demartin M., Weill F. W. 2006. Variant Salmonella genomic island 1-L antibiotic resistance gene cluster in Salmonella enterica serovar Newport. Antimicrob. Agents Chemother. 50:3944–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Doublet B., et al. 2003. Variant Salmonella genomic island 1 antibiotic resistance gene cluster in Salmonella enterica serovar Albany. Emerg. Infect. Dis. 9:585–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doublet B., et al. 2004. Salmonella genomic island 1 multidrug resistance gene clusters in Salmonella enterica serovar Agona isolated in Belgium in 1992 to 2002. Antimicrob. Agents Chemother. 48:2510–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doublet B., Boyd D., Mulvey M. R., Cloeckaert A. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 55:1911–1924 [DOI] [PubMed] [Google Scholar]
- 17. Doublet B., Golding G. R., Mulvey M. R., Cloeckaert A. 2008. Secondary chromosomal attachment site and tandem integration of the mobilizable Salmonella genomic island 1. PLoS One 3:e2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Doublet B., et al. 2008. Novel insertion sequence- and transposon-mediated genetic rearrangements in genomic island SGI1 of Salmonella enterica serovar Kentucky. Antimicrob. Agents Chemother. 52:3745–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Emborg H.-D., Hammerum A. M. (ed.). 2007. DANMAP 2006. Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. National Food Institute, Technical University of Denmark, Søborg, Denmark: http://www.danmap.org/pdfFiles/Danmap_2006.pdf [Google Scholar]
- 20. Giraud E., et al. 2009. Salmonella Genomic Island 1 modulates the expression of virulence-related loci in acidic conditions in Salmonella enterica serovar Typhimurium DT104, p. 116 In Proceedings of the 3rd ASM Conference on Salmonella: Biology, Pathogenesis, and Prevention. ASM Press, Washington, DC [Google Scholar]
- 21. Glynn M. K., et al. 1998. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N. Engl. J. Med. 338:1333–1338 [DOI] [PubMed] [Google Scholar]
- 22. Grimont P. A. D., Weill F.-X. 2007. Antigenic formulae of the Salmonella serovars. WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur, Paris, France [Google Scholar]
- 23. Guerra B., Malorny B., Schroeter A., Helmuth R. 2003. Multiple resistance mechanisms in fluoroquinolone-resistant Salmonella isolates from Germany. Antimicrob. Agents Chemother. 47:2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guerra B., Junker E., Miko A., Helmuth R., Mendoza M. C. 2004. Characterization and localization of drug resistance determinants in multidrug-resistant, integron-carrying Salmonella enterica serotype Typhimurium strains. Microb. Drug Resist. 10:83–91 [DOI] [PubMed] [Google Scholar]
- 25. Hammerl J. A., et al. 2010. pSGI15, a small ColE-like qnrB19 plasmid of a Salmonella enterica serovar Typhimurium strain carrying Salmonella genomic island 1 (SGI1). J. Antimicrob. Chemother. 65:173–175 [DOI] [PubMed] [Google Scholar]
- 26. Huehn S., Bunge C., Junker E., Helmuth R., Malorny B. 2009. Poultry associated Salmonella enterica subsp. enterica serovar 4,12:d:− reveals high clonality and a distinct pathogenicity gene repertoire. Appl. Environ. Microbiol. 75:1011–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huehn S., et al. 2010. Virulotyping and antimicrobial resistance typing of Salmonella enterica serovars relevant to human health in Europe. Foodborne Pathog. Dis. 7:523–535 [DOI] [PubMed] [Google Scholar]
- 28. Kado C. I., Liu S. T. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Larsson J. T., et al. 2009. Development of a new nomenclature for Salmonella Typhimurium multilocus variable number of tandem repeats analysis (MLVA). Euro Surveill. 14:19174. [PubMed] [Google Scholar]
- 30. Levesque C., Piche L., Larose C., Roy P. H. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Levings R. S., Lightfoot D., Partridge S. R., Hall R. M., Djordjevic S. P. 2005. The genomic island SGI1, containing the multiple antibiotic resistance region of Salmonella enterica serovar Typhimurium DT104 or variants of it, is widely distributed in other S. enterica serovars. J. Bacteriol. 187:4401–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lindstedt B. A., Vardund T., Aas L., Kapperud G. 2004. Multiple-locus variable-number tandem-repeats analysis of Salmonella enterica subsp. enterica serovar Typhimurium using PCR multiplexing and multicolor capillary electrophoresis. J. Microbiol. Methods 59:163–172 [DOI] [PubMed] [Google Scholar]
- 33. McClelland M., et al. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856 [DOI] [PubMed] [Google Scholar]
- 34. Mulvey M. R., Boyd D. A., Olson A. B., Doublet B., Cloeckaert A. 2006. The genetics of Salmonella genomic island 1. Microbes Infect. 8:1915–1922 [DOI] [PubMed] [Google Scholar]
- 35. Rodríguez I., et al. 2009. Extended-spectrum β-lactamases and AmpC β-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003-07. J. Antimicrob. Chemother. 64:301–309 [DOI] [PubMed] [Google Scholar]
- 36. Rotger R., Casadesús J. 1999. The virulence plasmids of Salmonella. Int. Microbiol. 2:177–184 [PubMed] [Google Scholar]
- 37. Schroeter A., Hoog B., Helmuth R. 2004. Resistance of Salmonella isolates in Germany. J. Vet. Med. B Infect. Dis. Vet. Public Health 51:389–392 [DOI] [PubMed] [Google Scholar]
- 38. Threlfall E. J. 2002. Antimicrobial drug resistance in Salmonella: problems and perspectives in food- and water-borne infections. FEMS Microbiol. Rev. 26:141–148 [DOI] [PubMed] [Google Scholar]
- 39. Vo A. T., van Duijkeren E., Fluit A. C., Gaastra W. 2007. A novel Salmonella genomic island 1 and rare integron types in Salmonella Typhimurium isolates from horses in The Netherlands. Antimicrob. Agents Chemother. 59:594–599 [DOI] [PubMed] [Google Scholar]
- 40. Weill F. X., Fabre L., Grandry B., Grimont P. A., Casin I. 2005. Multiple-antibiotic resistance in Salmonella enterica serotype Paratyphi B isolates collected in France between 2000 and 2003 is due mainly to strains harboring Salmonella genomic islands 1, 1-B, and 1-C. Antimicrob. Agents Chemother. 49:2793–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. White P. A., McIver C. J., Rawlinson W. D. 2001. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 45:2658–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.