Abstract
Due to emerging resistance to traditional antimicrobial agents, such as ampicillin, trimethoprim-sulfamethoxazole, and chloramphenicol, azithromycin is increasingly used for the treatment of invasive Salmonella infections. In the present study, 696 isolates of non-Typhi Salmonella collected from humans, food animals, and retail meats in the United States were investigated for antimicrobial susceptibility to azithromycin. Seventy-two Salmonella enterica serotype Typhi isolates from humans were also tested. For each isolate, MICs of azithromycin and 15 other antimicrobial agents were determined by broth microdilution. Among the non-Typhi Salmonella isolates, azithromycin MICs among human isolates ranged from 1 to 32 μg/ml, whereas the MICs among the animal and retail meat isolates ranged from 2 to 16 μg/ml and 4 to 16 μg/ml, respectively. Among Salmonella serotype Typhi isolates, the azithromycin MICs ranged from 4 to 16 μg/ml. The highest MIC observed in the present study was 32 μg/ml, and it was detected in three human isolates belonging to serotypes Kentucky, Montevideo, and Paratyphi A. Based on our findings, we propose an epidemiological cutoff value (ECOFF) for wild-type Salmonella of ≤16 μg/ml of azithromycin. The susceptibility data provided could be used in combination with clinical outcome data to determine tentative clinical breakpoints for azithromycin and Salmonella enterica.
INTRODUCTION
Non-Typhi Salmonella is the second leading cause of food-borne illness in the United States (34). Each year, approximately 1.0 million people are infected with Salmonella, resulting in 19,000 hospitalizations and almost 400 deaths (34). Although antimicrobial treatment is not indicated for uncomplicated infections, antimicrobial agents are potentially life-saving for extraintestinal infections. Likewise, antimicrobial therapy is essential for treating infections of Salmonella enterica serotypes Typhi and Paratyphi, the causative agents of enteric fever (31).
Due to widespread resistance to traditional first-line drugs, such as ampicillin, trimethoprim-sulfamethoxazole, and chloramphenicol, current recommendations suggest using a fluoroquinolone (e.g., ciprofloxacin) or an extended-spectrum cephalosporin (e.g., ceftriaxone) for treating invasive and severe Salmonella infections (16, 18). However, Salmonella serotype Typhi and paratyphoidal serotypes with decreased susceptibilities to fluoroquinolones, which may be associated with inadequate responses to treatment, have become common, and reports of fluoroquinolone-resistant Salmonella strains are increasing (8, 22, 24, 25). Furthermore, extended-spectrum β-lactamase-producing Salmonella serotype Typhi and Salmonella serotype Paratyphi A have been reported (1, 32). Of even greater concern, some Salmonella isolates display concurrent resistance to quinolones and extended-spectrum cephalosporins, which may require use of an alternative antimicrobial class for management of invasive infections (27, 36).
Azithromycin is an azalide antimicrobial agent that has been demonstrated in clinical trials to be equivalent or superior to chloramphenicol, fluoroquinolones, and extended-spectrum cephalosporins for the management of uncomplicated typhoid fever (3, 4, 13–15, 30). In addition, azithromycin is increasingly considered for the management of bacillary dysentery and invasive nontyphoidal Salmonella infections (9, 10, 37, 38). Azithromycin shows excellent penetration into most tissues, and it achieves concentrations in macrophages and neutrophils that are >100-fold higher than concentrations in serum (23, 29). These properties, along with azithromycin's long half-life of 2 to 3 days, have increased azithromycin's role in the therapeutic management of infections of the reticuloendothelial system.
Prior to initiation of any antimicrobial therapy, the in vitro susceptibility of the disease-causing bacteria to potentially effective antimicrobial agents should be examined. The in vitro susceptibilities of bacteria are classified as either “susceptible,” “intermediate,” or “resistant” and are defined by clinical breakpoints. In the United States, the Food and Drug Administration (FDA) and the Clinical and Laboratory Standards Institute (CLSI) define breakpoints. In Europe, the European Committee on Antimicrobial Susceptibility Testing (EUCAST) is responsible for defining clinical breakpoints for new and existing drugs (20, 21).
At present, no clinical azithromycin breakpoints have been defined for Enterobacteriaceae, including Salmonella, by either the CLSI or EUCAST (5, 6, 11). By performing antimicrobial susceptibility testing on 696 isolates of non-Typhi Salmonella isolated from humans, food animals, and retail meats in the United States, we provide data on the range of azithromycin MICs observed among Salmonella enterica isolates from the United States. We further present azithromycin MICs for 72 isolates of Salmonella serotype Typhi. This information could be combined with clinical outcome data to facilitate establishment of clinical azithromycin breakpoints for Salmonella.
MATERIALS AND METHODS
In 2008, 54 state and local public health laboratories participating in the National Antimicrobial Resistance Monitoring System (NARMS) forwarded every isolate of Salmonella serotype Typhi and every 20th non-Typhi Salmonella isolate from human clinical infections to the Centers for Disease Control and Prevention (CDC). Similarly, non-Typhi Salmonella isolates from retail meats (chicken breasts, ground turkey, ground beef, and pork chops) were submitted by the states participating in the Foodborne Diseases Active Surveillance Network (FoodNet) for analysis at the U.S. Food and Drug Administration Center for Veterinary Medicine (FDA-CVM). Non-Typhi Salmonella isolates from food animals were obtained from carcass rinsates (chicken), carcass swabs (turkey, cattle, and swine), and ground products (chicken, turkey, and beef). Animal samples were collected by the U.S. Department of Agriculture's (USDA) Food Safety Inspection Service (FSIS) from federally inspected slaughter and processing plants throughout the United States and forwarded to the USDA in Athens, GA, for further analysis. In addition, non-Typhi Salmonella isolates from animal products were collected through special studies performed at the USDA. At each agency, 232 non-Typhi Salmonella isolates were randomly chosen for azithromycin susceptibility determination. Moreover, 60 isolates of Salmonella serotype Typhi submitted in 2008 and 12 additional isolates from previous years were tested for azithromycin susceptibility at the CDC. All Typhi isolates were clinical isolates collected from humans.
MICs were determined by broth microdilution using a frozen azithromycin reference panel with concentrations ranging from 0.25 to 256 μg/ml (Sensititre; Trek Diagnostics, Westlake, OH). Antimicrobial susceptibility testing was performed according to the manufacturer's instructions, with Staphylococcus aureus ATCC 29213 used as a quality control strain (5). For each isolate, a final inoculum of 5 × 105 CFU/ml was targeted. The panels were read after 18 h of incubation at 35°C.
In addition, all isolates were tested for susceptibility to 15 antimicrobial agents included on the NARMS Gram-negative panel (amikacin, ampicillin, amoxicillin-clavulanic acid, ceftiofur, ceftriaxone, cefoxitin, chloramphenicol, ciprofloxacin, gentamicin, kanamycin, nalidixic acid, sulfamethoxazole, streptomycin, trimethoprim-sulfamethoxazole, and tetracycline) and interpreted according to CLSI standards, where available (5). Additional susceptibility testing with azithromycin Etest (bioMérieux, Inc., NC) strips was performed according to the manufacturer's instructions on Mueller-Hinton II agar plates incubated at 37°C for 16 to 20 h.
An MIC histogram was constructed, and the MIC50 value, representing the MIC at which the growth of 50% of the population is inhibited, was calculated for each of the four sample groups (Salmonella serotype Typhi from humans and non-Typhi Salmonella from humans, retail meats, and animals). The histograms were further inspected to identify the wild-type MIC distribution.
Genomic DNA was prepared by lysing the bacteria at 95°C and collecting the supernatant following centrifugation. A PCR assay using previously described primers was used to screen for the following macrolide resistance genes: ereA, ereB, ermB, mefA, mphA, mphB, and mphD (28). Plasmids were extracted using the Qiagen plasmid midi prep kit (Qiagen, Valencia, CA) and electroporated into electrocompetent Escherichia coli DH10B (Invitrogen, Carlsbad, CA).
RESULTS
In 2008, 2,379 isolates of non-Typhi Salmonella were submitted to the CDC, 1,326 to the USDA, and 495 to the FDA-CVM as part of the NARMS program for enteric bacteria. At each agency, a subset of 232 isolates was randomly chosen to be tested for antimicrobial susceptibility to azithromycin. In addition, 72 isolates of Salmonella serotype Typhi were included. Among the Salmonella serotype Typhi isolates, 49 (68.1%) were isolated from blood cultures and 16 (22.2%) from stool. The sources of the remaining 7 isolates were not provided by the submitting laboratory.
Among the 232 non-Typhi Salmonella isolates randomly selected from human isolate submissions, Salmonella enterica serotypes Enteritidis (19.0%) and Typhimurium (13.4%) were most common. Of these, 84.1% were isolated from stool cultures and 9.1% from blood cultures. The remaining 16 isolates were isolated from urine, other sources, or nonspecified sources. Among the isolates randomly selected for analysis from food animal submissions, Salmonella enterica serotypes Kentucky (16.4%), Heidelberg (9.5%), and Montevideo (7.3%) were most common, and among the isolates randomly selected from retail meats, Salmonella enterica serotypes Heidelberg (19.6%), Hadar (16.8%), and Typhimurium variant O:5− (10.8%) predominated. Among the animal isolates, 227 were obtained from the following sample sources: chicken (38.8%), cattle (37.9%), turkey (15.4%), and swine (7.9%). The remaining five isolates from the USDA were special study isolates obtained from ready-to-eat products and eggs. Among the retail meat isolates, 116 (50.0%) originated from ground turkey, 95 (40.9%) from chicken breast, 12 (5.2%) from pork chops, and 9 (3.9%) from ground beef.
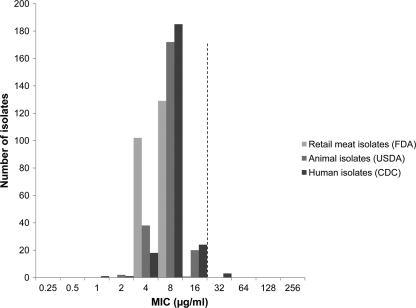
Among the non-Typhi Salmonella isolates from all sources, 64 (9.2%) displayed resistance to ceftriaxone (MIC ≥ 4 μg/ml) and 15 (2.2%) showed decreased susceptibility to ciprofloxacin (MIC ≥ 0.125 μg/ml). The azithromycin MICs among the isolates collected from humans ranged from 1 to 32 μg/ml, whereas the MICs among the animal and retail meat isolates ranged from 2 to 16 μg/ml and 4 to 16 μg/ml, respectively (Fig. 1). All three distributions peaked at 8 μg/ml, and the MIC50 value for each distribution was 8 μg/ml. Three isolates collected from humans displayed an MIC of 32 μg/ml. These isolates comprised Salmonella serotypes Kentucky, Montevideo, and Paratyphi A. The serotype Paratyphi A isolate was obtained from blood, whereas the serotype Kentucky and Montevideo isolates were obtained from stool. By PCR, these three isolates were negative for genes associated with macrolide resistance (ereA, ereB, ermB, mefA, mphA, mphB, and mphD) (28). Moreover, plasmid extractions and electroporation of plasmids into E. coli DH10B did not yield any transformants. Retesting of these isolates by Etest yielded MICs of 16 μg/ml.
Fig. 1.
Azithromycin MIC distributions for 696 isolates of non-Typhi Salmonella enterica collected from humans, food animals and retail meats through the National Antimicrobial Resistance Monitoring System (NARMS), 2008. The dashed line denotes a proposed epidemiological cutoff value (ECOFF) for WT isolates of ≤16 μg/ml.
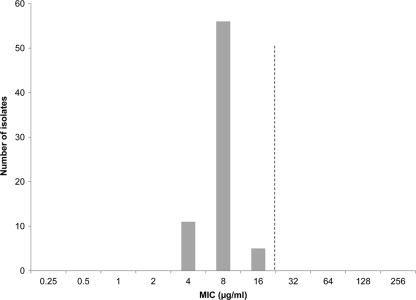
Among the 72 isolates of Salmonella serotype Typhi, 5 (7.0%) showed resistance to ampicillin (MIC ≥ 32 μg/ml), 3 (4.2%) to chloramphenicol (MIC ≥ 32 μg/ml), and 7 (9.7%) to trimethoprim-sulfamethoxazole (MIC ≥ 4 μg/ml). Fifty-eight (81.0%) isolates showed decreased susceptibility to ciprofloxacin (MIC ≥ 0.125 μg/ml). Of these, 12 (16.7%) showed clinical resistance (ciprofloxacin MIC ≥ 4 μg/ml), according to current CLSI guidelines (6). The azithromycin MICs among the Salmonella serotype Typhi isolates ranged from 4 to 16 μg/ml (Fig. 2).
Fig. 2.
Azithromycin MIC distribution for 72 isolates of S. enterica serotype Typhi collected by the National Antimicrobial Resistance Monitoring System (NARMS). The dashed line denotes a proposed epidemiological cutoff value (ECOFF) for WT isolates of ≤16 μg/ml.
DISCUSSION
Currently, azithromycin is recommended for the treatment of both shigellosis and invasive salmonellosis by the World Health Organization and the American Academy of Pediatrics (2, 37, 38) and is increasingly used for the management of uncomplicated enteric fever (3, 4, 13–15, 30). However, clinical breakpoints for azithromycin and Salmonella have yet to be defined. Clinical breakpoints are necessary to detect emerging and changing patterns of resistance and to guide clinicians in the selection of effective antimicrobial therapy. The first step toward defining clinical breakpoints is to collect relevant data, including (i) pharmacodynamic data of the drug, (ii) pharmacological properties of the drug, (iii) clinical outcome data, and (iv) microbiological data, i.e., MIC data for the specific pathogen in question (7, 35). In this paper, we provide MIC data for 696 isolates of non-Typhi Salmonella and 72 isolates of Salmonella serotype Typhi that could contribute to establishing susceptibility breakpoints for Salmonella and azithromycin.
The 696 non-Typhi Salmonella isolates included in the present study were collected from various sources, including humans, animals, and retail meats in the United States. The MIC distributions for the different sample sources were similar, and the majority of the isolates in each distribution displayed MIC values of 4 to 8 μg/ml. Thus, overall, azithromycin showed similar activities against non-Typhi Salmonella isolates obtained from various sources, including isolates displaying resistance to ceftriaxone and decreased susceptibility to ciprofloxacin. Our findings are consistent with a study reporting azithromycin susceptibility among non-Typhi Salmonella isolates collected in Finland (17) and are also consistent with MIC distribution data presented for Salmonella serotype Typhi and Shigella (15, 19).
The highest MIC value observed in the present study was 32 μg/ml, and it was detected in three human isolates belonging to serotypes Kentucky, Montevideo, and Paratyphi A. These isolates were all negative for ereA, ereB, ermB, mefA, mphA, mphB, and mphD, genes associated with macrolide resistance (28). In addition, attempts to transfer the resistance failed in all three isolates, indicating a lack of plasmid-mediated mechanisms. Other possible mechanisms include mutations in the 23S rRNA genes or the rlpD and rlpV genes, the last two of which encode ribosomal proteins L4 and L22, respectively (33). Further investigations will be necessary to determine whether these isolates acquired a resistance mechanism or whether they belong to the wild-type distribution and their slightly elevated MICs are due to normal variation in the testing methodologies. The latter possibility is supported by the fact that these three isolates displayed MICs of 16 μg/ml upon retesting with Etest.
Studies investigating azithromycin resistance mechanisms in Salmonella are scarce. Gunell and colleagues reported an rlpD mutation among isolates with azithromycin MICs of 64 to 128 μg/ml (17). Moreover, an isolate of Salmonella enterica serotype Stanley collected through NARMS in the United States in 1999 displayed an azithromycin MIC of 128 μg/ml and harbored the mphA gene (12; J. P. Folster et al., unpublished data).
Although clinical breakpoints are essential for defining clinical resistance, they might not always be the most sensitive tool for detecting isolates with an acquired resistance mechanism. The detection of resistance mechanisms is becoming increasingly important as surveillance programs recognize their role in the global control of antimicrobial resistance. In this context, the use of microbiological breakpoints or epidemiological cutoff values (ECOFFs) is useful. The concept of ECOFFs was introduced by EUCAST as a way of distinguishing bacteria without resistance (wild type [WT]) from those with acquired resistance (21). The WT ECOFF is expressed as ≤x μg/ml and serves to divide the bacterial population into two groups: those that are wild type (WT) and those that acquired resistance and are non-wild type (NWT) (21). Based on this study and the work of EUCAST and Gunell and coauthors, we propose an ECOFF for WT Salmonella isolates of ≤16 μg/ml of azithromycin (11, 17). This means that isolates with an MIC of ≥32 μg/ml could be considered NWT for surveillance purposes. Additional studies would be required to confirm the accuracy of the proposed ECOFF and determine whether isolates displaying an MIC value of 32 μg/ml belong to the wild-type distribution.
Whether NWT isolates should be classified as clinically resistant remains to be determined following the accumulation of clinical endpoint data. Recently, the first case of azithromycin treatment failure in a patient with invasive Salmonella infection was reported (26). The Salmonella serotype Paratyphi A isolate that initially displayed an azithromycin MIC of 64 μg/ml later displayed an MIC of 256 μg/ml when it was isolated from a second blood culture (26).
In summary, the azithromycin MIC distributions for Salmonella serotype Typhi and non-Typhi Salmonella isolates were concordant, and the majority of the isolates showed MICs of 4 to 8 μg/ml. Based on data from this study and the work of others (11, 17), we suggest an ECOFF for WT Salmonella of ≤16 μg/ml of azithromycin. Moreover, as azithromycin use increases for the management of invasive salmonellosis, we encourage national and international antimicrobial susceptibility testing consensus groups to consider the MIC distribution data presented here to contribute to the establishment of clinical breakpoints for azithromycin and Salmonella enterica.
ACKNOWLEDGMENTS
We thank the public health laboratories that participate in the NARMS, the Retail Foods Survey Working Group, and the FSIS laboratories for submitting the isolates.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
This work was supported by interagency agreements that the CDC and USDA have with the FDA Center for Veterinary Medicine (FDA-CVM).
Footnotes
Published ahead of print on 20 June 2011.
REFERENCES
- 1. Al Naiemi N., et al. 2008. Extended-spectrum-beta-lactamase production in a Salmonella enterica serotype Typhi strain from the Philippines. J. Clin. Microbiol. 46:2794–2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. American Academy of Pediatrics 2009. Shigella infections, p. 584–589, 593–596 In Pickering L. K. (ed.), Red Book: 2009 report of the Committee on Infectious Diseases, 28th ed American Academy of Pediatrics, Elk Grove Village, IL [Google Scholar]
- 3. Butler T., et al. 1999. Treatment of typhoid fever with azithromycin versus chloramphenicol in a randomized multicentre trial in India. J. Antimicrob. Chemother. 44:243–250 [DOI] [PubMed] [Google Scholar]
- 4. Chinh N. T., et al. 2000. A randomized controlled comparison of azithromycin and ofloxacin for treatment of multidrug-resistant or nalidixic acid-resistant enteric fever. Antimicrob. Agents Chemother. 44:1855–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. CLSI M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. CLSI M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7. Clinical and Laboratory Standards Institute 2008. Development of in vitro susceptibility testing criteria and quality control parameters, 3rd ed Approved standard. CLSI M23-A3. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. Crump J. A., et al. 2008. Clinical response and outcome of infection with Salmonella enterica serotype Typhi with decreased susceptibility to fluoroquinolones: a United States FoodNet multicenter retrospective cohort study. Antimicrob. Agents Chemother. 52:1278–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crump J. A., Mintz E. D. 2010. Global trends in typhoid and paratyphoid fever. Clin. Infect. Dis. 50:241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DuPont H. L. 2009. Clinical practice. Bacterial diarrhea. N. Engl. J. Med. 361:1560–1569 [DOI] [PubMed] [Google Scholar]
- 11. European Committee on Antimicrobial Susceptibility Testing 2011. Breakpoint tables for interpretation of MICs and zone diameters, version 1.3. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/EUCAST_breakpoints_v1.3_pdf.pdf
- 12. Folster J. P., Rickert R., Barzilay E. J., Whichard J. M. 2009. Identification of the aminoglycoside resistance determinants armA and rmtC among non-Typhi Salmonella isolates from humans in the United States. Antimicrob. Agents Chemother. 53:4563–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frenck R. W., Jr., et al. 2004. Short-course azithromycin for the treatment of uncomplicated typhoid fever in children and adolescents. Clin. Infect. Dis. 38:951–957 [DOI] [PubMed] [Google Scholar]
- 14. Frenck R. W., Jr., et al. 2000. Azithromycin versus ceftriaxone for the treatment of uncomplicated typhoid fever in children. Clin. Infect. Dis. 31:1134–1138 [DOI] [PubMed] [Google Scholar]
- 15. Girgis N. I., et al. 1999. Azithromycin versus ciprofloxacin for treatment of uncomplicated typhoid fever in a randomized trial in Egypt that included patients with multidrug resistance. Antimicrob. Agents Chemother. 43:1441–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guerrant R. L., et al. 2001. Practice guidelines for the management of infectious diarrhea. Clin. Infect. Dis. 32:331–351 [DOI] [PubMed] [Google Scholar]
- 17. Gunell M., et al. 2010. In vitro activity of azithromycin against nontyphoidal Salmonella enterica. Antimicrob. Agents Chemother. 54:3498–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hohmann E. L. 2001. Nontyphoidal salmonellosis. Clin. Infect. Dis. 32:263–269 [DOI] [PubMed] [Google Scholar]
- 19. Howie R. L., Folster J. P., Bowen A., Barzilay E. J., Whichard J. M. 2010. Reduced azithromycin susceptibility in Shigella sonnei, United States. Microb. Drug Resist. 16:245–248 [DOI] [PubMed] [Google Scholar]
- 20. Kahlmeter G., Brown D. 2004. Harmonization of antimicrobial breakpoints in Europe—can it be achieved? Clin. Microbiol. Newsl. 26:187–192 [Google Scholar]
- 21. Kahlmeter G., et al. 2003. European harmonization of MIC breakpoints for antimicrobial susceptibility testing of bacteria. J. Antimicrob. Chemother. 52:145–148 [DOI] [PubMed] [Google Scholar]
- 22. Keddy K. H., Smith A. M., Sooka A., Ismail H., Oliver S. 2010. Fluoroquinolone-resistant typhoid, South Africa. Emerg. Infect. Dis. 16:879–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lalak N. J., Morris D. L. 1993. Azithromycin clinical pharmacokinetics. Clin. Pharmacokinet. 25:370–374 [DOI] [PubMed] [Google Scholar]
- 24. Lynch M. F., et al. 2009. Typhoid fever in the United States, 1999-2006. JAMA 302:859–865 [DOI] [PubMed] [Google Scholar]
- 25. Maskey A. P., et al. 2008. Emerging trends in enteric fever in Nepal: 9124 cases confirmed by blood culture 1993-2003. Trans. R. Soc. Trop. Med. Hyg. 102:91–95 [DOI] [PubMed] [Google Scholar]
- 26. Molloy A., et al. 2010. First report of Salmonella enterica serotype Paratyphi A azithromycin resistance leading to treatment failure. J. Clin. Microbiol. 48:4655–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nordmann P., et al. 2008. Multidrug-resistant Salmonella strains expressing emerging antibiotic resistance determinants. Clin. Infect. Dis. 46:324–325 [DOI] [PubMed] [Google Scholar]
- 28. Ojo K. K., et al. 2004. The mef(A) gene predominates among seven macrolide resistance genes identified in gram-negative strains representing 13 genera, isolated from healthy Portuguese children. Antimicrob. Agents Chemother. 48:3451–3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Panteix G., et al. 1993. In-vitro concentration of azithromycin in human phagocytic cells. J. Antimicrob. Chemother. 31(Suppl. E):1–4 [DOI] [PubMed] [Google Scholar]
- 30. Parry C. M., et al. 2007. Randomized controlled comparison of ofloxacin, azithromycin, and an ofloxacin-azithromycin combination for treatment of multidrug-resistant and nalidixic acid-resistant typhoid fever. Antimicrob. Agents Chemother. 51:819–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parry C. M., Threlfall E. J. 2008. Antimicrobial resistance in typhoidal and nontyphoidal salmonellae. Curr. Opin. Infect. Dis. 21:531–538 [DOI] [PubMed] [Google Scholar]
- 32. Pokharel B. M., et al. 2006. Multidrug-resistant and extended-spectrum beta-lactamase (ESBL)-producing Salmonella enterica (serotypes Typhi and Paratyphi A) from blood isolates in Nepal: surveillance of resistance and a search for newer alternatives. Int. J. Infect. Dis. 10:434–438 [DOI] [PubMed] [Google Scholar]
- 33. Roberts M. C. 2008. Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol. Lett. 282:147–159 [DOI] [PubMed] [Google Scholar]
- 34. Scallan E., et al. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Turnidge J., Paterson D. L. 2007. Setting and revising antibacterial susceptibility breakpoints. Clin. Microbiol. Rev. 20:391–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Whichard J. M., et al. 2007. Human Salmonella and concurrent decreased susceptibility to quinolones and extended-spectrum cephalosporins. Emerg. Infect. Dis. 13:1681–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. World Health Organization 2005. Guidelines for the control of shigellosis, including epidemics due to Shigella dysenteriae type 1. http://www.searo.who.int/LinkFiles/CAH_Publications_shigella.pdf
- 38. World Health Organization 2005. The treatment of diarrhoea: a manual for physicians and other senior health workers, 4th rev. http://whqlibdoc.who.int/publications/2005/9241593180.pdf