Abstract
The prevailing resistance mechanism against glycopeptides in Gram-positive pathogens involves reprogramming the biosynthesis of peptidoglycan precursors, resulting in d-alanyl-d-lactate depsipeptide termini. Amycolatopsis balhimycina produces the vancomycin-like glycopeptide balhimycin and therefore has to protect itself from the action of the glycopeptide. We studied the roles of the accessory resistance gene orthologs vanYb, vnlRb, and vnlSb, which are part of the balhimycin biosynthetic gene cluster (represented by the subscript “b”). The VanYb carboxypeptidase cleaved the terminal d-Ala from peptidoglycan precursors, and its heterologous expression enhanced glycopeptide resistance in Streptomyces coelicolor. The VanRS-like two component system VnlRSb was not involved in glycopeptide resistance or in the expression of the vanHAX glycopeptide resistance genes. Mature A. balhimycina peptidoglycan contained mainly tri- and tetrapeptides, with only traces of the d-Ala-d-Ala-ending pentapeptides that are binding sites for the antibiotic produced. The structure of the peptidoglycan precursor is consistent with the presence of vanHAX genes, which were identified outside the balhimycin synthesis cluster. Both wild-type and non-antibiotic-producing mutant strains synthesized peptidoglycan precursors ending mainly with d-Lac, indicating constitutive synthesis of a resistant cell wall. A. balhimycina could provide a model for an ancestral glycopeptide producer with constitutively expressed resistance genes.
INTRODUCTION
Amycolatopsis balhimycina DSM 5908 (formerly described as Amycolatopsis mediterranei) was first isolated from a soil sample originating from the Himalayas (20). It belongs to the order Actinomycetales and synthesizes the glycopeptide antibiotic balhimycin. Balhimycin differs only in its glycosylation pattern from vancomycin, which is used as an antibiotic of last resort against multidrug-resistant Gram-positive bacteria. Since the activities of balhimycin are comparable to those of vancomycin both in vitro and in vivo and A. balhimycina is accessible to genetic manipulation, this strain has become a model organism for analyzing glycopeptide synthesis and resistance (32). Glycopeptide antibiotics inhibit cell wall biosynthesis in Gram-positive bacteria by binding the d-alanyl-d-alanine (d-Ala-d-Ala) terminus of peptidoglycan precursors on the outside surface of the cytoplasmic membrane (24). However, when the usage of vancomycin steadily increased in the 1980s (14), the first vancomycin-resistant enterococci (VRE) were isolated in hospitals (17). In these pathogens, cell wall biosynthesis was reprogrammed in such a way that the pentapeptide of the peptidoglycan precursor terminated in d-alanyl-d-lactate (d-Ala-d-Lac) rather than d-Ala-d-Ala, thereby causing an ∼1,000-fold lower binding affinity of the glycopeptide (5). This alteration of the cell wall precursor requires the following three genes: vanH, coding for a dehydrogenase which converts pyruvate to d-Lac (1); vanA, which codes for a d-Ala-d-Lac ligase (4); and vanX, which codes for a dd-dipeptidase that cleaves the d-Ala-d-Ala dipeptide to ensure that only altered peptidoglycan precursors terminating in d-Ala-d-Lac are built up (25). van-like genes which have similarity to the resistance genes from VRE have been found in several glycopeptide producers, such as various Amycolatopsis spp. (16).
We report here the rare case of an antibiotic producer carrying a biosynthetic gene cluster without essential resistance genes. The essential vanHAX resistance genes, located elsewhere in the chromosome, are expressed constitutively and are not controlled by the vanRS-like two component system vnlRS, resulting in constitutive production of peptidoglycan precursors terminating in d-Ala-d-Lac.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used for this study are listed in Table 1.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant feature(s) | Source or reference(s) |
|---|---|---|
| Strains | ||
| Escherichia coli strains | ||
| XL1 Blue | General cloning host | 6 |
| BL21 | Heterologous expression host | 7 |
| ET12567 | Methylation-deficient strain | 18 |
| ET12567/pUZ8002 | Transformed with plasmid pUZ8002; Kanr; donor for conjugation | 13, 18 |
| ET12567/pUB307 | Transformed with plasmid pUB307; Kanr; donor for conjugation | 18 |
| Amycolatopsis balhimycina strains | ||
| DSM5908 | Wild type | 20 |
| OP696 | Balhimycin null mutant; defective in precursor biosynthesis | 23 |
| ΔvnlR | Wild-type strain with deletion of vnlRb | This work |
| Δorf7 | Wild-type strain with deletion of orf7 | This work |
| Other strains | ||
| Streptomyces coelicolor A3(2) | Plasmid-carrying wild type; balhimycin resistant | 2 |
| Streptomyces coelicolor M145 | Plasmid-free derivative of S. coelicolor A3(2); balhimycin resistant | 13 |
| Streptomyces coelicolor vanYb | Carries vanYb under the control of its own promoter | This work |
| Streptomyces coelicolor J3201 | vanRS deletion mutant | 12 |
| Streptomyces coelicolor J3201 vanYb | vanRS deletion mutant; carries vanYb | This work |
| Streptomyces lividans TK23 | 11 | |
| Plasmids | ||
| Escherichia coli plasmids | ||
| pDrive | bla aphII lacZ′α | Qiagen |
| pGEM-T Easy | bla lacZ′α | Promega |
| pSP1 | bla ermE; nonreplicative gene disruption vector | 22 |
| pRSetB | bla; T7 expression system with His tag | Invitrogen |
| pRSetBvanY | pRSetB with integrated vanYb | This work |
| Actinomycetes plasmids | ||
| pSET152 | Actinomycetes-E. coli shuttle vector; attP int aac(IV)3 | 3 |
| pSET152vanY | vanYb with its own promoter region | This work |
| pSETermE*ΔHindIII | attP int aac(IV)3; ermE* promoter; only one HindIII site | 29 |
Media and culture conditions.
Escherichia coli strains were grown in LB (26) at 37°C. Actinomycetes strains were grown in R5 medium (13) at 30°C. Media were supplemented with antibiotics when necessary to maintain plasmids.
DNA preparation and manipulation.
The methods used for the isolation and manipulation of DNA for E. coli and actinomycetes were described by Kieser et al. (13). PCR fragments were isolated from agarose gels by use of a QIAquick kit (Qiagen). Restriction endonucleases were obtained from various suppliers and were used according to their specifications.
Preparation of A. balhimycina RNA.
A. balhimycina cells were cultivated for different times. The cells were then harvested and shock frozen at −70°C. An aliquot was resuspended in 100 μl of buffer P (31) that contained 10 mg/ml of lysozyme and was then incubated for 7 min at 37°C. The RNA was extracted by use of an RNeasy minikit (Qiagen) according to the manufacturer's instructions.
Reverse transcription (RT) analysis.
From the isolated RNA, cDNA was synthesized and used as a template for amplification of a fragment containing parts of vanH and vanA by use of primers vanH1 and vanA2 and of a fragment containing parts of vanA and vanX by use of primers vanA3 and vanX4. To test for the vanYb transcript (“b” represents the balhimycin biosynthetic gene cluster), the primer pair vanY_RT_rev-vanY_RT_for was used (Table 2). Chromosomal DNA from A. balhimycina was used as a positive control, and total RNA was used to test for contamination with chromosomal DNA.
Table 2.
Primers used in this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| p2_rev | GWAGTGCCACCAYTY |
| p3_forw | GGNGAGGACGGNWSNMTNCAGGG |
| HAXges_for | GCGCTGCCTATACTGCCGCATATGACCTAC |
| HAXges_rev | CGAGCCCAGCACGGCCGAATTCGGTTATTC |
| vanH1 | GGGACAAGCCCATCAAGAAC |
| vanA2 | GAGCGGACTTGACGGAGATG |
| vanA3 | GGGTGGACCTGTTCCTCAAG |
| vanX4 | TTGGCGTCCCAGCGAATACC |
| vanY-BamHI | GGATCCGGCGACCCTGGTCAC |
| vanY-XbaI | TCTAGACGATCAGCACGCGCATGCC |
| vanY-HindIII | AAGCTTCGTCTGCGGAGCAC |
| vanY-NdeI | ATATCATATGACCTACCGCGAGTCGG |
| vanY-EcoRI | GAATTCCGTCTGCGGAGCAC |
| vanY Expressdown | TTAAGCTTCGGGCGACCCTG |
| vanY Expressup | AAGGATCCAACCTACCGCGAGTC |
| vanRinakt 1.1 | TTTATAGAGCTCATGCTGCCGGCTCGTATGTG |
| vanRinakt 1.2 | AATAATTCTAGAGGAGCAGGCGGTGGACAGAG |
| vanRinakt 2.1 | TTTATATCTAGAGCCCCGATGGTACGAGAGGCCAC |
| vanRinakt 2.2 | AATAATCTGCAGGGATGTCCTCGACGCTGATG |
| prΔvnlR.1prove | AGGAAGTTGCGCACCAGGAC |
| prΔvnlR.2prove | GACTCGCGGTAGGTCATGTG |
| vanY_RT_rev | TTCACGCACAGTTCG |
| vanY_RT_for | TCGGCACGAGGATTG |
| prΔorf7F1.1 | ATATGAATTCGTCCCACGGTGGCGTTCTAC |
| prΔorf7F1.2 | ATATTCTAGACGCGGCAAAGGTGTGCAGC |
| prΔorf7F2.1 | ATATTCTAGACAGGAGCAAAGTCTCTAGG |
| prΔorf7F2.2 | ATATGCATGCGGTGACCGTCTTCATGTCAG |
| prΔorf7.1prove | GTTGCCGGAATCGGTGACAG |
| prΔorf7.2prove | CGGTGATCGACCGGAAC |
| attBI-fwd | TTCTGGAAATCCTCGAAGGC |
| attint-rev | GTAAGCACCCGCGTACGTGT |
| ermEleft | GCACGCCTGGTCGATGTC |
| ermEright | CTTCTCCCGCAACGACTT |
Amplification of an internal vanHAX fragment.
To identify cosmids carrying vanHAX-like genes, the primer pair p2-p3 (Table 2) was used to generate a labeled probe. With this probe, the A. balhimycina cosmid library was screened by Southern hybridization.
Construction and analysis of Δorf7 and ΔvnlR mutant strains.
The plasmid pSPΔorf7 was constructed to inactivate orf7 by an in-frame deletion of 1,314 bp. A 1,764-bp upstream fragment (frΔorf7-left) and a 1,711-bp downstream fragment (frΔorf7-right) of orf7 were amplified from cosmid DNA by PCRs using the primer pairs prΔorf7F1.1-prΔorf7F1.2 and prΔorf7F2.1-prΔorf7F2.2 (Table 2), respectively. The primers contained restriction sites at the 3′ and 5′ ends (EcoRI/XbaI and XbaI/SphI sites, respectively). Both fragments were cloned into the vector pSP1 to generate pSPΔorf7.
For the in-frame deletion (678 bp) of vnlRb, plasmid pSPΔvnlR was constructed. A 1,369-bp downstream fragment (frΔvnlR-left) and a 1,467-bp upstream fragment (frΔvnlR-right) of vnlRb were amplified from cosmid DNA by PCRs using the primer pairs vnlRinakt1.1-vnlRinakt1.2 and vnlRinakt2.1-vnlRinakt2.2 (Table 2), respectively. The primers contained restriction sites at the 5′ and 3′ ends (XbaI/PstI and XbaI/SacI sites, respectively). Both fragments were cloned into the vector pSP1 to generate pSPΔvnlR.
Wild-type A. balhimycina was transformed with pSPΔorf7 or pSPΔvnlR by use of a direct transformation method as described previously (22). The integration of the plasmids into the chromosome via homologous recombination was confirmed by PCR screening for the erythromycin resistance cassette, using primers ermEleft and ermEright (Table 2). To obtain deletion mutants, a second homologous recombination event was provoked by stressing plasmid-carrying colonies as described elsewhere (23). Colonies were examined for sensitivity to erythromycin, and the deletions were verified by PCR analysis, using primers prΔorf7.1prove and prΔorf7.2prove (Table 2) to generate a 2,585-bp fragment from the A. balhimycina Δorf7 mutant and the primers prΔvnlR.1prove and prΔvnlR.2prove (Table 2) to generate a 310-bp fragment from the A. balhimycina ΔvnlR mutant.
Bioassay for detection of active balhimycin.
Balhimycin production was determined through the use of bioassays using Bacillus subtilis ATCC 6633 as a test organism and cell supernatants from Amycolatopsis strains grown in R5 medium.
Bioassay for determination of balhimycin resistance.
Amycolatopsis and Streptomyces strains were grown for 3 days at 30°C on R5/HA plates containing balhimycin at different concentrations.
Isolation of the cell wall.
Actinomycetes cells were grown at 30°C with shaking and were then cooled rapidly and harvested by centrifugation at 10,000 × g for 10 min. The pellet was resuspended in 50 mM Tris-HCl (pH 7.0), and this solution was added dropwise to a boiling SDS solution (5%). The solution was stirred the entire time, and boiling was continued for 15 min before cooling of the mixture to room temperature. The crude cell wall material was collected, and the pellet was washed free of SDS by a continuous resuspending and washing procedure consisting of two washes with 20 ml of 1 M NaCl and then one wash with water. The washing procedure was continued until no SDS was detectable by use of a published assay (8). Glass beads (diameter, 0.1 to 0.5 mm) and 1 ml of the cell wall suspension were mixed, and the walls were broken mechanically in a FastPrep instrument (Precellys; 6 passages for 20 to 30 s at 6,500 × g) on ice. After removal of the glass beads and the unbroken cells (by centrifugation at 2,000 × g for 5 min), the broken cell walls were sedimented by centrifugation at 25,000 × g for 15 min at room temperature. The pellet was then resuspended in 3 ml of 100 mM Tris-HCl (pH 7.5) with 20 mM MgSO4. DNase (10 μg/ml) and RNase (50 μg/ml) were added, and the sample was incubated for 2 h with stirring. Afterwards, 10 mM CaCl2 and 100 μg/ml trypsin were added, and the sample was incubated for 18 h with stirring. SDS was added to a final concentration of 1%, and the sample was incubated at 80°C for 15 min. The volume was adjusted with water to 20 ml, and the cell wall was sedimented by centrifugation at 25,000 × g for 30 min. The pellet was resuspended in 10 ml 8 M LiCl and incubated at 37°C for 15 min. After harvesting, the pellet was resuspended in 10 ml of 100 mM EDTA (pH 7.0) and incubated at 37°C for 15 min. The pellet was then washed successively with water, acetone, and water. After the last step, the pellet was resuspended in a small amount of water, and the material obtained was lyophilized.
Isolation of peptidoglycan from cell walls (peptidoglycan-teichoic acid complexes).
For cell wall composition analyses, it was essential to cleave the peptidoglycan into smaller fragments (muropeptides). Our initial attempt to release muropeptides from the cell wall, i.e., the peptidoglycan-teichoic acid complex, by use of cellosyl failed, presumably because the teichoic acids inhibited the enzyme. Therefore, teichoic acid was removed with hydrofluoric acid (HF) to isolate the peptidoglycan. The purified cell wall (5 mg) was resuspended in ice-cold 48% HF. The sample was stirred for 48 h at 4°C. The sample was centrifuged at 100,000 × g for 45 min at 4°C and then washed twice with ice-cold 100 mM Tris-HCl (pH 7) and twice with ice-cold water. It was then resuspended in 750 μl of water, and potassium azide was added (final concentration, 0.05%).
Preparation and reduction of muropeptides for high-performance liquid chromatography (HPLC) analysis.
One hundred microliters of 25 mM potassium phosphate (pH 5.5) was added to 100 μl of the peptidoglycan suspension (10 to 15 mg/ml). Ten microliters of 10 mg/ml mutanolysin (Sigma-Aldrich) was added, and the sample was incubated at 37°C for 18 h. The sample was then boiled for 10 min and centrifuged. The supernatant was mixed with the same volume of potassium borate (0.5 M). Sodium borohydride (1 to 2 mg) was added, and the sample was incubated at room temperature for 30 min. Excess borohydride was destroyed by the successive addition of phosphoric acid until a pH of 1 to 2 was reached.
Peptidoglycan precursor extraction.
Peptidoglycan precursors were extracted by a method previously described for Bacillus cereus (15), with some modifications. Briefly, mycelium was grown in the appropriate medium for 2 days before bacitracin (Sigma) was added and growth was continued for 1 h. Bacitracin binds the C55-isoprenyl pyrophosphate and prevents the transport of cell wall precursors outside the cell, resulting in the accumulation of cytoplasmic UDP-linked precursors (30). Cells were harvested, and the pellet was resuspended in 25 ml water. The solution was boiled for 20 min and then centrifuged. The supernatant was collected and lyophilized. The lyophilized supernatant was resuspended in 1 ml water and extracted with 200 μl chloroform. After vortexing and centrifugation (13,000 rpm for 2 min), the suspension was used for HPLC and HPLC-mass spectrometry (HPLC-MS).
HPLC-MS analysis of peptidoglycan precursors.
Five microliters of the sample was injected by use of an HPLC-MS instrument (XCT 6330 LC/MSD Ultra Trap system; Agilent Technologies) and a Nucleosil 100 C18 column (3 μm by 100 mm by 2 mm internal diameter). The HPLC parameters were as follows: the gradient was linear from 0% eluent A (H2O, 0.1% HCOOH) to 10% eluent B (acetonitrile, 0.06% HCOOH), at a flow rate of 400 μl/min, and took 25 min. The MS parameters were as follows: ionization alternated between positive and negative, the capillary voltage was 3.5 kV, and the temperature was 350°C. Tandem MS (MS/MS) was performed in the negative mode with the corresponding target mass.
VanY assays. (i) Fluorometric OPTA method.
The VanY activity was determined using a previously described method (33). Briefly, a calibration curve was initially made. The concentration of released d-Ala was determined by using a calibration curve in which the relative fluorescence was plotted over the free d-Ala concentration. Several concentrations of free d-Ala were measured thrice. From a stock solution of 50 mM d-Ala, different concentrations were set up in a zinc acetate (ZnAc)-NaHCO3 solution (5 mM ZnAc and 50 mM NaHCO3) in a final volume of 20 μl. The reaction was quenched by the addition of 5 μl HCl (250 mM) followed by the addition of 75 μl H2O. d-Ala was monitored by the addition of 100 μl fluoraldehyde (o-phthaldialdehyde [OPTA]) solution followed by incubation at room temperature for 5 min. After the addition of 800 μl H2O, the solution was diluted 100-fold and measured in a fluorescence reader (Infinite 200; Tecan) with a λex value of 340 nm and a λem value of 455 nm. The background value for a probe without d-Ala was subtracted.
To test the activity with the artificial substrate Nα,Nε-diacetyl-Lys-d-Ala-d-Ala, the reaction mixture was set up with 5 mM ZnAc, 50 mM NaHCO3, and 10 mM substrate. To 10 μl of this mixture, crude cell extract with VanYb (from the induced E. coli strain BL21 at a final protein concentration of 50 μg/ml) was added in a final volume of 20 μl and incubated at 37°C for 1 h. The sample was further prepared in a similar manner to that for the calibration curve before the measurement was taken in a fluorescence reader. As a blank value, crude cell extract lacking VanYb was used.
(ii) HPLC-DAD analysis.
Using HPLC, we measured the carboxypeptidase substrate Nα,Nε-diacetyl-Lys-d-Ala-d-Ala and the carboxyesterase substrate Nα,Nε-diacetyl-Lys-d-Ala-d-Lac and traced the differences in the substrate concentrations. The samples were prepared as follows. Five microliters of a 10 mM substrate solution was added to 20 μl of crude cell extract and incubated at 37°C for 30 min. Crude cell extract lacking VanYb was used as a blank. To stop the reaction, the sample was boiled. Twenty-five microliters of H2O was added, and after centrifugation, the sample was injected by use of an HPLC-DAD (diode array detector) instrument (HP 1090M chromatograph with autosampler, DAD, and HP Kayak XM 600 Chem station; Agilent Technologies) and a Nucleosil 100 C18 column. The gradient was linear from 0% eluent A (H2O, 0.1% H3PO4) to 100% eluent B (acetonitrile) at a flow rate of 2 ml/min. The substrate peaks were monitored at 210 nm and quantified by integration. The threshold of this method was 0.03 mM substrate. Therefore, values below this threshold were not detectable.
RESULTS AND DISCUSSION
Putative resistance determinants in the balhimycin biosynthetic gene cluster.
The A. balhimycina balhimycin biosynthetic gene cluster does not include the resistance-mediating vanHAX genes (27). By screening a cosmid library and by the ongoing genome sequencing project (T. Weber, personal communication), these genes could be identified elsewhere in the chromosome, at least 2 Mb away from the cluster. Within the balhimycin biosynthesis gene cluster, we have identified several genes potentially involved in glycopeptide resistance, including orf7 (showing >50% homology to a Na+/H+ antiporter gene) and homologues of the known enterococcal accessory resistance genes vanR, vanS, and vanY (named vnlR, vnlS, and vanYb, respectively, from here on). The in-frame deletion mutant lacking orf7 exhibited no detectable difference in balhimycin production and resistance compared to the wild-type strain, indicating that orf7 is not involved in the resistance mechanism.
Characterization of VanYb.
The presence of the vanY dd-carboxypeptidase gene increases the level of glycopeptide resistance in enterococci harboring the vanHAX genes (33). In contrast to the enterococcal enzyme, VanYb encoded by the balhimycin biosynthetic gene cluster lacks a predicted membrane association site. VanYb was overexpressed as a His-tagged protein in E. coli to analyze its biochemical activity by the OPTA method (see Materials and Methods for details). dd-Carboxypeptidase activity was assessed by measuring the release of d-Ala from the diacetyl-Lys-d-Ala-d-Ala substrate. Cell extract from the VanYb-overproducing strain but not from a control strain showed considerable dd-carboxypeptidase activity, resulting in hydrolysis of 45.6% of the substrate. Heat inactivation of the sample abolished the dd-carboxypeptidase activity. Using a newly established HPLC-based assay, we could not detect a possible dd-carboxyesterase activity against diacetyl-Lys-d-Ala-d-Lac, although in this assay, 70% of diacetyl-Lys-d-Ala-d-Ala was digested by the cell extract from the VanYb overproduction strain. These results demonstrate that VanYb has dd-carboxypeptidase activity and not the dd-carboxyesterase activity as described for other carboxypeptidases (33).
The functionality of VanYb was tested by heterologous expression of the corresponding gene in Streptomyces coelicolor, which harbors the inducible vanHAX genes but lacks a vanY gene (2). The first approach to introduce vanYb, placing the gene under the control of the strong constitutive promoter PermE* by using the integrative plasmid pSET152ermE*ΔHindIII (29), did not produce transformants, possibly because the constitutive expression of dd-carboxypeptidase degraded the cell wall precursors and was thus lethal. In contrast, it was possible to introduce the plasmid pSET152 (3), carrying vanYb under the control of its own promoter, resulting in the recombinant strain S. coelicolor vanYb, for which the expression of vanYb was proven by RT-PCR (data not shown). This vanYb expression strain showed a higher level of balhimycin resistance than the wild-type strain (50 μg/ml versus 10 μg/ml) in an agar diffusion test.
The vanYb gene under the control of its own promoter was also transferred into the glycopeptide-sensitive mutant S. coelicolor J3201, in which the vanHAX genes cannot be induced due to the absence of the regulatory two-component system (TCS) VanRS (10, 12). The presence of vanYb did not confer balhimycin resistance in this background, indicating that VanYb alone is unable to confer resistance and requires functional vanHAX genes to increase the resistance level in S. coelicolor. These results indicate that the vanYb gene is functional in S. coelicolor, depending on the presence of the vanHAX genes, and are in accordance with the observation that a Nonomuraea vanY mutant is resistant to glycopeptide, albeit at a reduced level (19, 28).
Analysis of vnlR function in A. balhimycina.
In enterococci and S. coelicolor, the transcription of vanHAX genes is controlled by a TCS encoded by vanR and vanS (10). Our RT-PCR experiments detected a vanHAXb transcript at all time points measured, including time points long before the onset of balhimycin production (data not shown). Thus, in A. balhimycina, the vanHAX genes are expressed constitutively.
We then constructed an A. balhimycina in-frame vnlRb deletion mutant and confirmed the absence of vnlRb by PCR analyses and Southern hybridization (data not shown). The ΔvnlRb mutant was able to grow normally in the presence of up to 100 μg/ml balhimycin and produced balhimycin at a level comparable to that of the wild type (∼100 mg/liter), indicating that vnlRb is not required for the expression of glycopeptide resistance. RT-PCR analysis of the ΔvnlRb mutant confirmed the same vanHAX and vanY expression level as that in the wild type, showing that vnlRb does not regulate the vanHAX and vanY genes in A. balhimycina.
Comparison of cell wall compositions of S. coelicolor A3(2) and A. balhimycina.
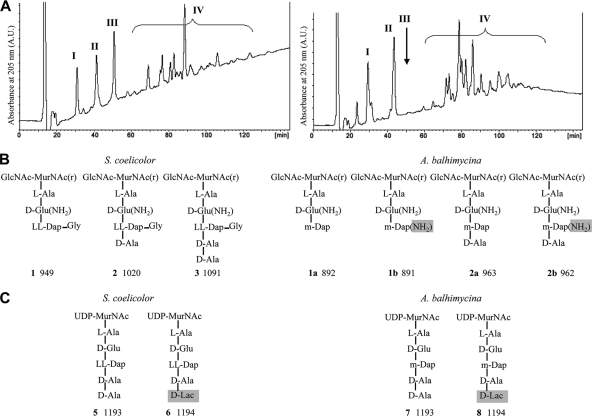
In order to investigate possible differences in the peptidoglycan structures of a glycopeptide producer and nonproducer, the muropeptides were released from cell wall peptidoglycan by use of mutanolysin, reduced with sodium borohydride, and analyzed by HPLC-MS. Monomeric muropeptides containing tri-, tetra-, and pentapeptides with a glycine branch were detected in the peptidoglycan from S. coelicolor grown in the absence of vancomycin (Fig. 1A) and were confirmed by MS analysis (Table 3). The basic structure of the mature S. coelicolor peptidoglycan is in accordance with the findings of Hong et al. (10), who analyzed the cytoplasmic peptidoglycan precursor. Unlike the precursor, the mature peptidoglycan carries a glutamine residue at position 2 of the peptide (Fig. 1B and C), showing that like the case in other Gram-positive bacteria, an amidation step occurs during the synthesis and/or maturation of the cell wall. Interestingly, pentapeptides were virtually absent in the mature peptidoglycan isolated from S. coelicolor grown in the presence of vancomycin (data not shown) when the vanHAX resistance genes were expressed. Thus, S. coelicolor alters its cell wall peptidoglycan in response to exogenous glycopeptide.
Fig. 1.
Separation and proposed structures of cell wall muropeptides. (A) Separation of cell wall muropeptides by HPLC. Fraction I, disaccharide tripeptide (Tri); fraction II, disaccharide tetrapeptide (Tetra); fraction III, disaccharide pentapeptide (Penta); fraction IV, multimers. The disaccharide pentapeptide is present in S. coelicolor (left) and virtually absent in A. balhimycina (right). (B) Proposed muropeptide structures of S. coelicolor, including Tri(Gln)Gly (1), Tetra(Gln)Gly (2), and Penta(Gln)Gly (3), and of A. balhimycina, including Tri(Gln) (1a), Tri(Gln)(NH2) (1b), Tetra(Gln) (2a), and Tetra(Gln)(NH2) (2b). The amidation of Dap is highlighted in gray. The letter “r” indicates that MurNAc was reduced during sample preparation. The measured molecular masses (Da) of the reduced muropeptides (Na+ form) are given below the structures. (C) Proposed structures of precursors from S. coelicolor, including the pentapeptide precursor (5) and pentadepsipeptide precursor (6), and from A. balhimycina, including the pentapeptide precursor (7) and pentadepsipeptide precursor (8). The neutral masses calculated from the measured molecular masses of the H+ forms are given below the structures.
Table 3.
MS analysis of reduced monomeric muropeptides
| Organism | Proposed muropeptidea | Molecular mass of compound (Da) |
Molecular mass after chemical methylation (Da) |
No. of methyl groupsb | ||
|---|---|---|---|---|---|---|
| Measured | Calculated | Measured | Calculated | |||
| E. coli | Tetra(Glu) Na+ | 964.31 | 964.40 | 1,006.29 | 1,006.45 | 3 |
| S. aureus | Tetra(Gln)Gly3 Na+ | 1,090.50 | 1,090.49 | 1,104.40 | 1,104.50 | 1 |
| Tetra(Gln)Gly4 Na+ | 1,147.52 | 1,147.51 | 1,161.42 | 1,161.53 | 1 | |
| Tetra(Gln)Gly5 Na+ | 1,204.53 | 1,204.37 | 1,218.44 | 1,218.55 | 1 | |
| S. coelicolor | Tri(Gln)Gly Na+ | 949.39 | 949.379 | 977.48 | 977.41 | 2 |
| Tetra(Gln)Gly Na+ | 1,020.40 | 1,020.41 | 1,048.52 | 1,048.44 | 2 | |
| Penta(Gln)Gly Na+ | 1,091.41 | 1,091.45 | 1,119.51 | 1,119.48 | 2 | |
| A. balhimycina | Tri(Gln)(NH2) Na+ | 891.28 | 891.34 | 905.31 | 905.36 | 1 |
| Tri(Gln) Na+ | 892.31 | 892.35 | 920.34 | 920.38 | 2 | |
| Tetra(Gln)(NH2) Na+ | 962.37 | 962.38 | 976.48 | 976.40 | 1 | |
| Tetra(Gln) Na+ | 963.37 | 963.39 | 991.52 | 991.42 | 2 | |
Muropeptides consist of two sugar residues (MurNAc and GlcNAc) and three to five stem amino acids. E. coli muropeptides contain a glutamate (Glu) residue at the second position, and S. aureus muropeptides contain a glutamine (Gln) residue. NH2 marks the amidation of a carboxylic group within the muropeptide. The number of glycine (Gly) residues linked to the ε-amino group of the third amino acid is given.
The number of methyl groups was calculated from the increase in molecular mass after chemical methylation and is indicative of the number of free (unamidated) carboxylic groups.
The basic structure of the stem peptides in the peptidoglycan of A. balhimycina was established and differed from that of S. coelicolor, Staphylococcus aureus, or E. coli (Fig. 1; Table 3). MS analysis of the monomeric muropeptides showed that A. balhimycina peptidoglycan does not contain the glycine branch at meso-Dap (22) which is present in S. coelicolor and that the peptide is mostly monoamidated and sometimes diamidated at positions 2 and 3, respectively.
Peptidoglycan from wild-type A. balhimycina, the glycopeptide mutant OP696 (23), or the highly glycopeptide-resistant species Amycolatopsis japonicum (21) contained no or only traces of pentapeptides, irrespective of whether the cells were grown in balhimycin production or nonproduction growth medium. Pentadepsipeptides were not detectable in either S. coelicolor or A. balhimycina due to hydrolysis of the acid-labile d-Ala-d-Lac bond during sample preparation. Since the pentapeptides, but not the tetra- and tripeptides, represent possible binding sites for the antibiotic, we hypothesize that Amycolatopsis species generally and constitutively produce a cell wall virtually without pentapeptides to minimize possible glycopeptide binding to their cell wall.
Cell wall precursor analysis of A. balhimycina strains.
UDP-linked peptidoglycan precursors were isolated from cell extracts and analyzed by HPLC-MS/MS (Fig. 1 and 2). S. coelicolor grown in the absence of balhimycin contained only the sensitive precursor, UDP-MurNAc-l-Ala-d-Glu-m-Dap-d-Ala-d-Ala (1,193 Da), and cells grown with glycopeptide produced both sensitive and d-Lac-containing (UDP-MurNAc-l-Ala-d-Glu-m-Dap-d-Ala-d-Lac) (1,194 Da) precursors, consistent with the presence of the inducible-resistance vanHAX genes (9).
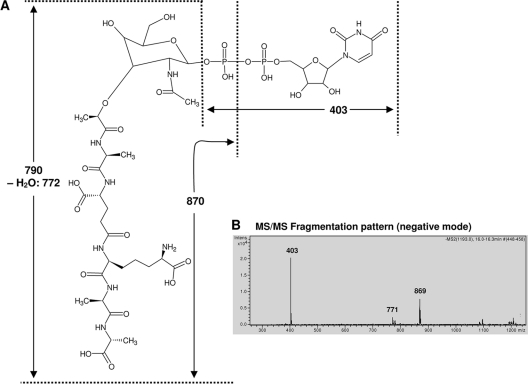
Fig. 2.
MS/MS fragmentation of cell wall precursors. (A) Structure of the UDP-linked cell wall precursor and its MS/MS fragmentation pattern. The neutral mass is given. (B) MS/MS analysis in negative ion mode of the precursor from A. balhimycina with a molecular mass of 1,193 Da. The fragmentation of the precursor with a mass of 1,193 Da resulted in a fragment with a mass of 403 Da corresponding to the identical UDP part of the molecule. Further fragments of 790 Da and 870 Da contained the peptide part of the molecule and confirmed the presence of the expected pentapeptide moiety.
Wild-type A. balhimycina or the nonproducing mutant OP696 (23) grown in production medium contained only the d-Lac-ending precursor UDP-MurNAc-l-Ala-d-Glu-m-Dap-d-Ala-d-Lac, whereas wild-type cells grown under nonproducing conditions contained both types of precursors (with d-Lac or d-Ala).
Concluding remarks.
Taken together, these data indicate that A. balhimycina carries homologues of all genes known to be involved in high-level resistance in enterococci. In contrast to the case for enterococci, the glycopeptide producer constitutively expresses the resistance genes even when it is not producing the antibiotic. This might be a safety mechanism to protect nonproducing cells living in a population with neighboring glycopeptide producer cells.
ACKNOWLEDGMENTS
The Deutsche Forschungsgemeinschaft (DFG) supported this work by the SFB 766 program “Bacterial Cell Envelope: Structure, Function and Infection Interface.”
We thank Katrin Welzel and Stefan Pelzer from Combinature Biopharm AG, Berlin, Germany, for screening the cosmid library, Mark Buttner and Hee-Jeon Hong for providing the Streptomyces coelicolor J3201 mutant, and Graeme Nicholson from the Institute of Organic Chemistry of the University of Tübingen for performing the methylation of muropeptides.
Footnotes
Published ahead of print on 20 June 2011.
REFERENCES
- 1. Arthur M., Molinas C., Dutka-Malen S., Courvalin P. 1991. Structural relationship between the vancomycin resistance protein VanH and 2-hydroxycarboxylic acid dehydrogenases. Gene 103:133–134 [DOI] [PubMed] [Google Scholar]
- 2. Bentley S. D., et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147 [DOI] [PubMed] [Google Scholar]
- 3. Bierman M., et al. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49 [DOI] [PubMed] [Google Scholar]
- 4. Bugg T. D., Dutka-Malen S., Arthur M., Courvalin P., Walsh C. T. 1991. Identification of vancomycin resistance protein VanA as a d-alanine:d-alanine ligase of altered substrate specificity. Biochemistry 30:2017–2021 [DOI] [PubMed] [Google Scholar]
- 5. Bugg T. D., et al. 1991. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry 30:10408–10415 [DOI] [PubMed] [Google Scholar]
- 6. Bullock W. O., Fernandez J. M., Short J. M. 1987. XL1-Blue, a high efficiency plasmid transforming recA Escherichia coli strain with beta galactosidase selection. Biotechniques 5:376–379 [Google Scholar]
- 7. Davanloo P., Rosenberg A. H., Dunn J. J., Studier F. W. 1984. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 81:2035–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hayashi K. 1975. A rapid determination of sodium dodecyl sulfate with methylene blue. Anal. Biochem. 67:503–506 [DOI] [PubMed] [Google Scholar]
- 9. Hong H. J., Hutchings M. I., Hill L. M., Buttner M. J. 2005. The role of the novel Fem protein VanK in vancomycin resistance in Streptomyces coelicolor. J. Biol. Chem. 280:13055–13061 [DOI] [PubMed] [Google Scholar]
- 10. Hong H. J., et al. 2004. Characterization of an inducible vancomycin resistance system in Streptomyces coelicolor reveals a novel gene (vanK) required for drug resistance. Mol. Microbiol. 52:1107–1121 [DOI] [PubMed] [Google Scholar]
- 11. Hopwood D. A., et al. 1983. Cloning Streptomyces genes for antibiotic production. Trends Biotechnol. 1:42–48 [Google Scholar]
- 12. Hutchings M. I., Hong H. J., Buttner M. J. 2006. The vancomycin resistance VanRS two-component signal transduction system of Streptomyces coelicolor. Mol. Microbiol. 59:923–935 [DOI] [PubMed] [Google Scholar]
- 13. Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 14. Kirst H. A., Thompson D. G., Nicas T. I. 1998. Historical yearly usage of vancomycin. Antimicrob. Agents Chemother. 42:1303–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kohlrausch U., Hoeltje J. V. 1991. One-step purification procedure for UDP-N-acetylmuramyl-peptide murein precursors from Bacillus cereus. FEMS Microbiol. Lett. 62:253–257 [DOI] [PubMed] [Google Scholar]
- 16. Lechevalier M. P., Prauser H., Labeda D. P., Ruan J. S. 1986. Two new genera of nocardioform Actinomycetes—Amycolata gen. nov. and Amycolatopsis gen. nov. Int. J. Syst. Bacteriol. 36:29–37 [Google Scholar]
- 17. Leclercq R., Derlot E., Duval J., Courvalin P. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319:157–161 [DOI] [PubMed] [Google Scholar]
- 18. MacNeil D. J., et al. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61–68 [DOI] [PubMed] [Google Scholar]
- 19. Marcone G. L., et al. 2010. Novel mechanism of glycopeptide resistance in the A40926 producer Nonomuraea sp. ATCC 39727. Antimicrob. Agents Chemother. 54:2465–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nadkarni S. R., et al. 1994. Balhimycin, a new glycopeptide antibiotic produced by Amycolatopsis sp. Y-86,21022. Taxonomy, production, isolation and biological activity. J. Antibiot. (Tokyo) 47:334–341 [DOI] [PubMed] [Google Scholar]
- 21. Nishikiori T., et al. 1984. Production by actinomycetes of (S,S)-N,N′-ethylenediamine-disuccinic acid, an inhibitor of phospholipase C. J. Antibiot. (Tokyo) 37:426–427 [DOI] [PubMed] [Google Scholar]
- 22. Pelzer S., Reichert W., Huppert M., Heckmann D., Wohlleben W. 1997. Cloning and analysis of a peptide synthetase gene of the balhimycin producer Amycolatopsis mediterranei DSM5908 and development of a gene disruption/replacement system. J. Biotechnol. 56:115–128 [DOI] [PubMed] [Google Scholar]
- 23. Puk O., et al. 2004. Biosynthesis of chloro-beta-hydroxytyrosine, a nonproteinogenic amino acid of the peptidic backbone of glycopeptide antibiotics. J. Bacteriol. 186:6093–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reynolds P. E. 1989. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 8:943–950 [DOI] [PubMed] [Google Scholar]
- 25. Reynolds P. E., Depardieu F., Dutka-Malen S., Arthur M., Courvalin P. 1994. Glycopeptide resistance mediated by enterococcal transposon Tn1546 requires production of VanX for hydrolysis of d-alanyl-d-alanine. Mol. Microbiol. 13:1065–1070 [DOI] [PubMed] [Google Scholar]
- 26. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27. Shawky R. M., et al. 2007. The border sequence of the balhimycin biosynthesis gene cluster from Amycolatopsis balhimycina contains bbr, encoding a StrR-like pathway-specific regulator. J. Mol. Microbiol. Biotechnol. 13:76–88 [DOI] [PubMed] [Google Scholar]
- 28. Sosio M., Stinchi S., Beltrametti F., Lazzarini A., Donadio S. 2003. The gene cluster for the biosynthesis of the glycopeptide antibiotic A40926 by Nonomuraea species. Chem. Biol. 10:541–549 [DOI] [PubMed] [Google Scholar]
- 29. Stegmann E., et al. 2006. Genetic analysis of the balhimycin (vancomycin-type) oxygenase genes. J. Biotechnol. 124:640–653 [DOI] [PubMed] [Google Scholar]
- 30. Stone K. J., Strominger J. L. 1971. Mechanism of action of bacitracin: complexation with metal ion and C55-isoprenyl pyrophosphate. Proc. Natl. Acad. Sci. U. S. A. 68:3223–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thompson C. J., Kieser T., Ward J. M., Hopwood D. A. 1982. Physical analysis of antibiotic-resistance genes from Streptomyces and their use in vector construction. Gene 20:51–62 [DOI] [PubMed] [Google Scholar]
- 32. Wohlleben W., Stegmann E., Süssmuth R. D. 2009. Molecular genetic approaches to analyse glycopeptides biosynthesis. Methods Enzymol. 458:459–486 [DOI] [PubMed] [Google Scholar]
- 33. Wright G. D., Molinas C., Arthur M., Courvalin P., Walsh C. T. 1992. Characterization of VanY, a dd-carboxypeptidase from vancomycin-resistant Enterococcus faecium BM4147. Antimicrob. Agents Chemother. 36:1514–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]