Abstract
Among patients with tuberculosis, rifampin plasma concentrations and sputum conversion rates have been reported to be lower in Africans. Rifampin is a substrate of P-glycoprotein (coded for by the ABCB1 gene) and organic anion-transporting polypeptide 1B1 (coded for by SLCO1B1). The objectives were to identify genetic polymorphisms of drug transporters and the transcriptional regulators pregnane X receptor (PXR) and constitutive androstane receptor (CAR) with an impact on rifampin pharmacokinetics in South Africans. Fifty-seven patients with tuberculosis from Cape Town underwent pharmacokinetic sampling during treatment with rifampin, pyrazinamide, isoniazid, and ethambutol. DNA was genotyped for ABCB1, SLCO1B1, PXR, and CAR polymorphisms by using real-time PCR. NONMEM was used for data analysis. The allele frequency of the SLCO1B1 rs4149032 polymorphism was 0.70. Patients heterozygous and homozygous for this polymorphism had reductions in the bioavailability (and, thus, the area under the curve [AUC]) of rifampin of 18% and 28%, respectively. Simulations showed that increasing the daily rifampin dose by 150 mg in patients with the polymorphism would result in plasma concentrations similar to those of wild-type individuals and reduce the percentage of patients with peak plasma concentrations (Cmax) below 8 mg/liter from 63% to 31%. ABCB1, PXR, and CAR polymorphisms were not associated with differences in rifampin pharmacokinetics. SLCO1B1 rs4149032 was present in most patients and was associated with substantially reduced rifampin exposure. These data suggest that the standard recommended dose of rifampin should be reconsidered for South Africans.
INTRODUCTION
The development of resistance to rifamycins and their bactericidal effects are related to rifamycin concentrations (6, 10, 27). There is a high intersubject variability of rifampin plasma concentrations in patients with tuberculosis, some of which is explained by HIV infection, sex, formulation, and weight (8, 15, 18, 29). Rifampin concentrations are lower in patients from sub-Saharan Africa (28), which may contribute to the lower sputum culture conversion rates at 2 months in African patients reported by two recent multicenter phase 2 trials of moxifloxacin in rifampin-based antitubercular regimens (5, 7). Population differences in drug concentrations may be partly the result of differences in genes encoding drug-metabolizing enzymes or transporters. Little is known about the pharmacogenetic determinants of rifampin exposure. Since about 50% of rifampin is excreted via the bile unchanged (20), polymorphisms of drug transporters and/or their transcriptional regulators may influence rifampin pharmacokinetics. Rifampin is a substrate of the drug efflux pump P-glycoprotein, coded for by the ABCB1 gene (21). The biliary excretion of rifampin occurs after hepatocellular uptake, which is mediated primarily by organic anion-transporting polypeptide 1B1 (OATP1B1), coded for by the gene SLCO1B1 (11, 24). The SLCO1B1 single-nucleotide polymorphism (SNP) C463A has recently been associated with reduced rifampin exposure (28).
We sought to determine the effect of polymorphisms of ABCB1, SLCO1B1, and the transcriptional regulators pregnane X receptor (PXR) and constitutive androstane receptor (CAR) on rifampin pharmacokinetics. We also investigated the potential utility of dose adjustment based upon the SLCO1B1 genotype.
MATERIALS AND METHODS
Study participants.
The study received ethical approval from the University of Cape Town research ethics committee. Sixty patients attending the Delft Community Health Centre in Cape Town, South Africa, who participated in a randomized controlled trial of a micronutrient intervention (vitamin A and zinc) were studied after written informed consent was obtained. The micronutrient intervention had no effect on clinical or microbiological outcomes and was reported elsewhere previously (26). The study participants had sputum smear-positive pulmonary tuberculosis and were administered daily doses of the same fixed-dose combination tablets (Rifafour), each containing 150 mg rifampin, 75 mg isoniazid, 400 mg pyrazinamide, and 275 mg ethambutol, with directly observed administration. Patients weighing 38 to 54 kg received 3 tablets daily, those weighing 55 to 70 kg received 4 tablets daily, and one patient who was over 70 kg received 5 tablets once daily, in accordance with standard South African tuberculosis treatment guidelines (24). Participants underwent pharmacokinetic sampling after steady state was attained (at least 1 month after the start of treatment), and 4 to 8 samples per patient were collected randomly over a 7-h period. Twenty-four patients underwent pharmacokinetic sampling on a second occasion, about 1 month after the first pharmacokinetic sampling. An additional blood sample was collected for genetic analyses after separate written consent was obtained from the 60 patients.
Genotyping.
Genomic DNA was extracted from the blood samples by using the QIAamp DNA blood minikit (Qiagen Inc., Hilden, Germany). Real-time PCR using fluorescent probes for allelic discrimination was used for genotyping. Primers and probes were sourced from Applied Biosystems Inc. (Warrington, United Kingdom). Absolute quantitative PCR mix was obtained from ThermoFisherScientific (Loughborough, United Kingdom). The PCR conditions for most SNPs were an initial denaturation step at 95°C for 15 min, followed 40 to 50 cycles of denaturation at 95°C for 15 s and then annealing and extension at 60°C for 60 s, with a plate read after each cycle. For SLCO1B1 T521C, the denaturation temperature was 92°C. Each PCR mixture contained 2 μl of genomic DNA, 12.5 μl of absolute quantitative PCR mix (2×), 1.25 μl of primer mix (20×), and 1.25 μl of probe mix (20×), made up to 25 μl with water. The assays were run on a Chromo4 real-time PCR detection system (Bio-Rad Life Sciences, United Kingdom). The following SNPs were genotyped for, based on previous reports of allele frequency and functional significance: ABCB1 (C3435T, G2677T, C1236T, and rs3842), SLCO1B1 (T521C, C463A [rs11045819], and rs4149032), PXR (C63396T and T44477C), and CAR (rs2307424). All genotyping assays were performed in duplicate, and a genotype was assigned only when the two separate assays were in agreement. Haplotype analysis of the SLCO1B1 gene was performed by using Haploview software (4).
Drug plasma concentration determination.
Blood samples were centrifuged to obtain plasma, which was stored at −80°C until analysis. Rifampin concentrations were measured by using high-performance liquid chromatography with tandem mass spectroscopy (14). The lower limit of quantitation (LLOQ) of this assay was 0.08 mg/liter.
Pharmacokinetic analyses.
Pharmacokinetic data from 57 patients were available. Population nonlinear mixed-effects modeling with NONMEM software, version 7.1.2 (Icon Inc., Verona, PA), was used for pharmacokinetic analyses. Population pharmacokinetic parameter estimates, between-subject variability (BSV), within-subject variability (WSV), covariances between random effects, and residual variability were obtained by using the Laplacian estimation method with epsilon-eta interactions. Twelve percent of the data were below the LLOQ and were entirely in the absorption phase (i.e., less than 2 h after the dose had been taken). Beal's M3 method (2) was used to handle BLQ data, whereby measurements below 0.08 mg/liter were used to calculate the likelihood that the predicted concentration at that time was less than the LLOQ, while those greater than or equal to the LLOQ were used to calculate the likelihood that the predicted concentration was equal to the measured value. A transit absorption compartment model was used to account for the variability in the absorption delay (29). First-order elimination from a one-compartment model describes the rest of the structural model. The objective function value (OFV), a goodness-of-fit criterion, and visual predictive checks (VPCs) were used for model building and evaluation. A decrease in the OFV of more than 3.84 points after the addition of one model parameter was regarded as being statistically significant.
The effects of several covariates on model parameters were investigated one at a time in a stepwise fashion. Covariates investigated included study arm (vitamin A and zinc versus placebo), body weight, age, HIV status, sex, and rifampin dose. The covariates for the pharmacogenetic analyses included ABCB1 (C3435T, G2677T, C1236T, and rs3842), SLCO1B1 (T521C, C463A, and rs4149032), PXR (C63396T and T44477C), and CAR (rs2307424) polymorphisms. A nonparametric bootstrap of 200 replications was run on the final model to obtain the precision of the final parameter estimates.
The final model was used to obtain the area under the curve (AUC) for each individual by creating an additional compartment where drug accumulated from 0 to 24 h after the dose. A Mann-Whitney U test was used to investigate the differences in AUCs between carriers and noncarriers of the rs4149032 polymorphism. Stochastic simulations to obtain drug concentration-time profiles for each individual based on the final model were then carried out to investigate the applicability of dose adjustment based on genotype.
RESULTS
Of the 60 patients, 40% were women and 16% were HIV infected. The median (2.5 percentile, 97.5 percentile) for weight, age, and body mass index were 52 kg (41 kg, 69 kg), 29 years (18 years, 55 years), and 19 kg/m2 (15 kg/m2, 25 kg/m2), respectively. Forty percent of the study participants were of black African ethnicity (mainly Xhosa), while the rest were of mixed ancestry (mainly Caucasian and African origins). Of the 60 study participants, 57 had both pharmacokinetic and genetic data, providing a total of 437 pharmacokinetic observations, which were then used for the pharmacokinetic modeling. Three of the 60 patients had no measurable rifampin concentrations over the entire sampling interval and hence could not be included in the pharmacokinetic analysis. Of these 3 patients, 2 were homozygous for the variant allele, while one was homozygous for the common allele.
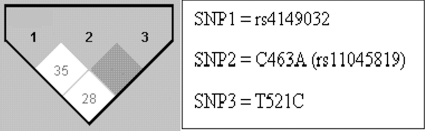
The SLCO1B1 rs4149032 polymorphism existed at an overall frequency of 0.70 in our study population. The frequency in men was 0.69, while in women it was 0.76. The frequency in black Africans was 0.93, while in the mixed-race group it was 0.59. Thirty-one individuals were homozygous (52%) for the reported variant allele, while 22 (37%) were heterozygous, and 7 (12%) were homozygous for the common allele. The remaining allele frequencies for the various SNPs that were genotyped are shown in Table 1. There were at most three individuals homozygous for any of these polymorphisms in the study population; therefore, carrier/noncarrier assignments were used for subsequent analyses. Only 3 individuals that were carriers of the SLCO1B1 521C allele were observed, and this SNP was therefore not included in any statistical analysis. Similarly, only 4 individuals were carriers of the SLCO1B1 463A allele. All polymorphisms were in Hardy-Weinberg equilibrium. The linkage disequilibrium (LD) plot of the 3 SLCO1B1 SNPs from Haploview is shown in Fig. 1. The inferred haplotypes were then tested to determine their effect on rifampin pharmacokinetics. However, results were obtained that were similar to those obtained for rs4149032 alone; hence, we proceeded with rs4149032 in our analyses.
Table 1.
Allele frequencies
| Allele | Frequency |
|---|---|
| ABCB1 C3435T | 0.26 |
| ABCB1 G2677T | 0.19 |
| ABCB1 C1236T | 0.26 |
| ABCB1 rs3842 | 0.21 |
| PXR C63396T | 0.27 |
| PXR T44477C | 0.27 |
| CAR rs2307424 | 0.11 |
| SLCO1B1 T521C | 0.05 |
| SLCO1B1 rs4149032 | 0.70 |
| SLCO1B1 C463A | 0.04 |
Fig. 1.
Linkage disequilibrium (LD) plots for the assessed SNPs in SLCO1B1. The shading within the boxes indicates the extent of LD between respective single-nucleotide polymorphisms, with white indicating a weaker LD than gray.
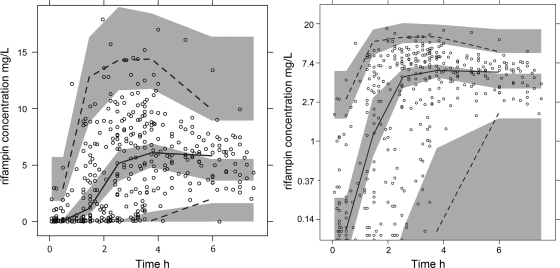
Based upon a VPC (Fig. 2), the model was found to predict the data acceptably, particularly in the absorption phase, which contained many observations. Only a fraction of the dosing interval had been captured, which may explain why the model predictions are higher than those observations at later times. Population parameter estimates from the final model bootstrap and the 95% confidence intervals (CIs) are shown in Table 2. Oral clearance (CL/F) and the apparent volume of distribution (V/F) were allometrically scaled for weight (3), and this resulted in a drop in the objective function value of 6 points compared to a model without weight. The association of SLCO1B1 rs4149032 with differences in oral bioavailability relative to the nominal population value of 1 was incorporated in a categorical manner. Heterozygotes had 18% lower bioavailability, and homozygotes had 28% lower bioavailability than homozygotes for the SLCO1B1 rs4149032 allele. The inclusion of this effect also resulted in a 21% decrease in the between-subject variability for the CL/F. The SLCO1B1 rs41490932 polymorphism was associated with apparent increases in the mean transit time of 9% for heterozygotes and 18% for homozygotes. However, as this effect was not statistically significant, it was not included in the final model. A mixture model with three subpopulations was investigated as an alternative way of identifying phenotypic polymorphisms but did not improve the OFV compared with the genetic polymorphism-based method.
Fig. 2.
Visual predictive check of the final model. The open circles indicate the observations. The upper dotted line represents the 95th percentile of the observation. The continuous line represents the median of the observations. The lower dotted line represents the 5th percentile of the observations. The shaded areas are the simulated confidence intervals for the corresponding percentiles. For a clearer picture of the low concentrations, a log-transformed visual predictive check of the same model is shown on the right. The VPC is truncated at the LLOQ for both predictions and observations.
Table 2.
Population pharmacokinetic parameter estimatesa
| Parameter | Estimated value (95% CI) |
|---|---|
| CL/F (liters/h/70 kg) | 11 (10, 13) |
| V/F (liters/70 kg) | 50 (41, 53) |
| ka (1/h) | 1.1 (0.9, 1.4) |
| MTT (h) | 1.6 (1.3, 1.8) |
| NN | 19 (fixed) |
| Additive error (mg/liter) | 0.03 (0.02, 0.04) |
| Proportional error | 0.30 (0.24, 0.33) |
| Effect of female sex on V/F (%) | −30 (−21, −35) |
| Effect of female sex on MTT (%) | −30 (−21, −35) |
| Effect of SLCO1B1 rs41490932 on F in | |
| heterozygotes (%) | −18 (−15, −25) |
| Effect of SLCO1B1 rs41490932 on F in | |
| variant homozygotes (%) | −28 (−19, −34) |
| Effect of dose on MTT (%) | −27 (22, 36) |
| BSV of F | 0.15 (0.10, 0.16) |
| BSV of CL | 0.20 (0.17, 0.32) |
| BSV of MTT | 0.52 (0.45, 0.84) |
| Correlation between BSV of CL and MTT | 0.86 (0.81, 0.96) |
| WSV of F | 0.21 (0.12, 0.25) |
| WSV of CL | 0.32 (0.24, 0.56) |
| WSV of V | 0.29 (0.20, 0.39) |
| WSV of MTT | 0.59 (0.45, 0.70) |
| Correlation between WSV of V and MTT | −0.40 (−0.29, −0.765) |
ka, first-order absorption rate constant; NN, number of transit compartments; MTT, mean transit time; BSV, between-subject variability; WSV, within-subject variability.
ABCB1 C3435T, C1236T, CAR rs23047424, PXR T44477C, and C63396T polymorphisms did not have a statistically significant association with CL/F, V/F, or relative bioavailability. The ABCB1 G2677T polymorphism resulted in a 19% increase in the CL/F and a 19% increase in the mean transit time. Although the inclusion of the ABCB1 G2677T polymorphism in the model decreased the OFV by 4 points (P < 0.05), as the VPC was unchanged, it was not included in the final model for reasons of parsimony.
Women had a V/F that was 30% lower than that of men. At the same time, women had a 30% longer mean transit time, showing that women have a longer absorption delay than men. In contrast, both men and women given higher doses (600 or 750 mg of rifampin daily) were found to have a 27% shorter absorption delay than those given 450 mg daily. Individuals with a higher dose also had CL/F reduced by 18%, but this was not statistically significant and was not included in the final model. The assignment of the micronutrient study arm (vitamin A and zinc versus placebo) had no significant effect on rifampin pharmacokinetics.
In order to evaluate if differences in the combined-genotype-associated effects on bioavailability were also associated with differences in overall drug exposures, the AUC from 0 to 24 h after dose administration (AUC0–24) was generated from the final model for each individual. After stratification by SLCO1B1 rs4149032 genotype, there was no statistically significant difference in the AUC0–24 between heterozygotes and homozygotes. However, a Mann-Whitney U test showed the median AUC0–24 of 43 mg · h/liter in individuals with the polymorphism to be significantly lower than the median of 56 mg · h/liter found for those without the polymorphism (P < 0.05).
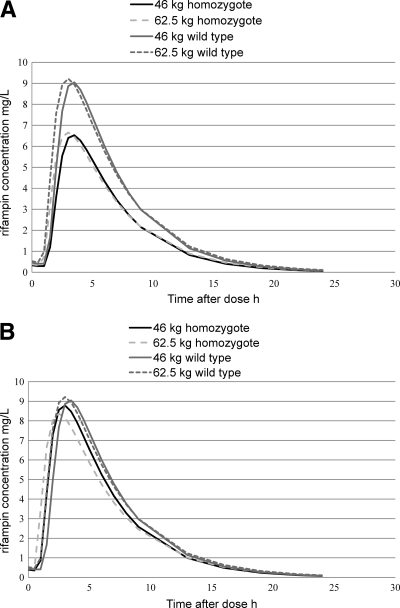
Simulations were carried out to obtain typical values for men weighing 46 kg and 62.5 kg, which represent the weights in the middle of the standard dosing weight bands (9) for patients receiving 450-mg and 600-mg rifampin doses once daily (Fig. 3A). Using a reference minimum Cmax of 8 mg/liter (18), a typical individual without the polymorphism was predicted to achieve adequate plasma concentrations, while a typical individual with the polymorphism was underdosed (Fig. 3A). Since the bioavailability was shown to be 28% lower in homozygous individuals with the polymorphism, we simulated the effect of a 150-mg dose increase for both homo- and heterozygous SLCO1B1 rs4149032 individuals, as we were unable to establish a statistically significant difference in the AUCs between them. Thus, 46-kg carriers of the variant allele would receive 600 mg instead of the currently recommended 450 mg, and 62.5-kg carriers of the variant allele would receive 750 mg once daily (Fig. 3B).
Fig. 3.
Predicted rifampin plasma concentration-time profiles for typical individuals weighing 46 and 62.5 kg homozygous for the common allele and minor alleles, respectively, with standard weight-based dosing (A) and after an additional 150 mg for those homozygous for the common allele (B).
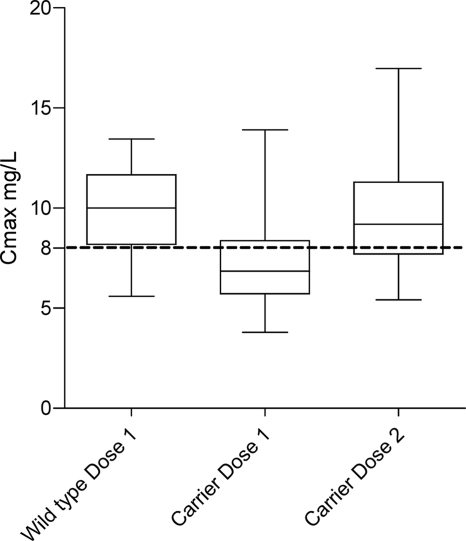
Simulations of a single typical individual (Fig. 3) do not take into account population variability or residual variability. We carried out stochastic simulations based on our actual data set using either the currently recommended doses or a genotype-based dose whereby SLCO1B1 rs41490932 carriers received an additional 150 mg of rifampin daily. The genotype-based doses resulted in a predicted reduction in the proportion of carriers with a Cmax of <8 mg/liter from 69% to 34%. The percentage of patients with a Cmax of <8 mg/liter in the entire study population was reduced from 63% to 31% (Fig. 4).
Fig. 4.
Box (interquartile range)-and-whisker (95th percentiles) plot of simulated variation in the Cmax based on the final model, with weight, sex, and genotype distributions in our data set. Simulations were performed by using the currently recommended doses (dose 1), and a genotype-based dose whereby SLCO1B1 rs41490932 carriers received an additional 150 mg of rifampin daily (dose 2).
DISCUSSION
Our study is the first to show that the SLCO1B1 rs4149032 polymorphism is associated with low-level rifampin exposure. We found low rifampin concentrations in a high proportion of patients with tuberculosis, which is consistent with other reports of African patients (15, 23, 28). The lower rifampin concentrations observed for Africans could be accounted for by the SLCO1B1 rs4149032 polymorphism, which has been reported to occur at a frequency of 75% in the Yoruba from Nigeria (and 70% in our study population), compared with 29% in Caucasians and 56% in Asians (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=4149032). The high allele frequency of the SLCO1B1 rs4149032 polymorphism among South Africans suggests that higher doses of rifampin may be appropriate for South African populations. Increasing the rifampin dose could improve sputum culture conversion rates, which were reported to be lower in Africans than in non-Africans in two previously reported multicenter studies (5, 7). There is in vitro animal and human evidence that increasing the rifampin dose improves its antituberculosis activity, and higher doses are unlikely to result in a significant increase in adverse events (16). Currently, genotyping prior to treatment would be difficult to implement, particularly in our resource-limited setting. Since the majority of the South Africans that we studied are carriers of the rs4149032 polymorphism, increasing the dose of rifampin across the board will benefit not only those with the polymorphism but also, possibly, those without it, as higher doses are likely to be more effective and appear to be safe.
We investigated the effects of several polymorphisms of drug transporters and transcriptional regulators in an attempt to explain the high variability that was previously reported for rifampin (29) pharmacokinetics. For PXR, it is possible that we did not observe a pharmacogenetic effect because the induction of PXR by rifampin may have overridden any potential genetic effects. Indeed, the amount of PXR induction that occurs due to the administration of rifampin was shown to be inversely proportional to the baseline PXR activity (12). The trend that we observed of increased CL/F values for PXR C63396T carriers is, however, consistent with the decrease in atazanavir plasma concentrations found for individuals with this polymorphism (22). High levels of P-glycoprotein activity may limit the distribution of rifampin in the body, leading to lower V/F values and higher plasma concentrations, as was previously suggested for digoxin (19). However, we did not find women to have higher CL/F values than men, which indicates that the role of P-glycoprotein may be more important for determining the distribution volume than the extent of oral absorption. The increase in the CL/F and the mean transit time combined with the decrease in the V/F seen with the ABCB1 G2677T/A polymorphism, though not statistically significant, is suggestive of this SNP resulting in higher levels of P-glycoprotein activity and/or expression, consistent with previous reports (17).
An important finding is the effect of the SLCO1B1 rs4149032 polymorphism on rifampin exposure. This SNP significantly lowered bioavailability in our study population. This intronic SNP was recently shown (in Caucasians) to be in LD with the functional variant C463A (rs11045819), termed SLCO1B1*4 (13). LD data of the SLCO1B1 gene from the HapMap project show that there is stronger LD in Caucasians than in Africans (http://hapmap.ncbi.nlm.nih.gov/cgi-perl/gbrowse/hapmap28_B36/). Different patterns of LD in Africans have been attributed to greater genetic diversity as a result of migration, admixture with other African and non-African populations, as well as the longer lineage of the African population (25). This could explain why C463A may not be correlated with rs4149032 in our African population, as suggested by haplotype analyses. Recently, C463A was shown to decrease the rifampin AUC by 36% in a mixed cohort of Africans and Caucasians (28). However, this allele was uncommon in our small study population.
The shorter mean transit time in individuals receiving higher doses suggests a saturation of the transporter-mediated mechanisms involved in absorption (P-glycoprotein), first-pass effects (P-glycoprotein and OATP1B1), and elimination (P-glycoprotein and OATP1B1) and is in agreement with the dose-dependent rifampin pharmacokinetics reported in previous studies (1). A mixed-order elimination model was investigated, but this failed to significantly improve the model.
In conclusion, the SLCO1B1 rs4149032 polymorphism exists at a high frequency in the African population and is associated with low rifampin concentrations (<8 mg/liter). Because of the high frequency of the carrier state in Cape Town (70%), a nonselective increase of the daily dose of 150 mg would double the number of patients (31% to 63%) reaching this concentration target. The public health implication of our data is that an increased dose of rifampin should be considered for South Africans.
ACKNOWLEDGMENTS
This work was made possible by grants from South African TB/HIV Training (SATBAT) (NIH/FIC 5U2RTW007373 and 5U2RTW007370); the National Research Foundation (NRF) South Africa (2067444 and RCN 180353/S50); the Norwegian Programme for Development, Research, and Higher Education (NUFUPRO-2007/10183); the Research Council of Norway (RCN) (183694/S50); the United Kingdom Medical Research Council (G0800247); and the South African Medical Research Council. E.C. is a recipient of a scholarship from the Clinical Infectious Diseases Research Initiative (CIDRI) Wellcome Trust Fund 412164. H.M. and G.M. received partial support from SATBAT through the Fogarty International Center (5U2RTW007370/3 and 5U2RTW007373).
We are grateful to Wynand Smythe and Jan-Stefan Van Der Walt for assistance with model building and evaluation. We thank the Delft study team and participants.
Footnotes
Published ahead of print on 27 June 2011.
REFERENCES
- 1. Agrawal S., et al. 2004. Bioequivalence trials of rifampicin containing formulations: extrinsic and intrinsic factors in the absorption of rifampicin. Pharmacol. Res. 50:317–327 [DOI] [PubMed] [Google Scholar]
- 2. Ahn J. E., Karlsson M. O., Dunne A., Ludden T. M. 2008. Likelihood based approaches to handling data below the quantification limit using NONMEM VI. J. Pharmacokinet. Pharmacodyn. 35:401–421 [DOI] [PubMed] [Google Scholar]
- 3. Anderson B. J., Holford N. H. 2008. Mechanism-based concepts of size and maturity in pharmacokinetics. Annu. Rev. Pharmacol. Toxicol. 48:303–332 [DOI] [PubMed] [Google Scholar]
- 4. Barrett J. C., Fry B., Maller J., Daly M. J. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 [DOI] [PubMed] [Google Scholar]
- 5. Burman W. J., et al. 2006. Moxifloxacin versus ethambutol in the first 2 months of treatment for pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 174:331–338 [DOI] [PubMed] [Google Scholar]
- 6. Diacon A. H., et al. 2007. Early bactericidal activity of high-dose rifampin in patients with pulmonary tuberculosis evidenced by positive sputum smears. Antimicrob. Agents Chemother. 51:2994–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dorman S. E., et al. 2009. Substitution of moxifloxacin for isoniazid during intensive phase treatment of pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 180:273–280 [DOI] [PubMed] [Google Scholar]
- 8. Gurumurthy P., et al. 2004. Decreased bioavailability of rifampin and other antituberculosis drugs in patients with advanced human immunodeficiency virus disease. Antimicrob. Agents Chemother. 48:4473–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Health and Medical Publishing Group 2010. South African medicines formulary, 9th ed Health and Medical Publishing Group, Cape Town, South Africa [Google Scholar]
- 10. Jayaram R., et al. 2003. Pharmacokinetics-pharmacodynamics of rifampin in an aerosol infection model of tuberculosis. Antimicrob. Agents Chemother. 47:2118–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim R. B. 2003. Organic anion-transporting polypeptide (OATP) transporter family and drug disposition. Eur. J. Clin. Invest. 33(Suppl. 2):1–5 [DOI] [PubMed] [Google Scholar]
- 12. Lamba J., Lamba V., Strom S., Venkataramanan R., Schuetz E. 2008. Novel single nucleotide polymorphisms in the promoter and intron 1 of human pregnane X receptor/NR1I2 and their association with CYP3A4 expression. Drug Metab. Dispos. 36:169–181 [DOI] [PubMed] [Google Scholar]
- 13. Lubomirov R., et al. 2010. ADME pharmacogenetics: investigation of the pharmacokinetics of the antiretroviral agent lopinavir coformulated with ritonavir. Pharmacogenet. Genomics 20:217–230 [DOI] [PubMed] [Google Scholar]
- 14. McIlleron H., et al. 2007. Elevated gatifloxacin and reduced rifampicin concentrations in a single-dose interaction study amongst healthy volunteers. J. Antimicrob. Chemother. 60:1398–1401 [DOI] [PubMed] [Google Scholar]
- 15. McIlleron H., et al. 2006. Determinants of rifampin, isoniazid, pyrazinamide, and ethambutol pharmacokinetics in a cohort of tuberculosis patients. Antimicrob. Agents Chemother. 50:1170–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mitnick C. D., McGee B., Peloquin C. A. 2009. Tuberculosis pharmacotherapy: strategies to optimize patient care. Expert Opin. Pharmacother. 10:381–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moriya Y., et al. 2002. Effects of polymorphisms of MDR1, MRP1, and MRP2 genes on their mRNA expression levels in duodenal enterocytes of healthy Japanese subjects. Biol. Pharm. Bull. 25:1356–1359 [DOI] [PubMed] [Google Scholar]
- 18. Perlman D. C., et al. 2005. The clinical pharmacokinetics of rifampin and ethambutol in HIV-infected persons with tuberculosis. Clin. Infect. Dis. 41:1638–1647 [DOI] [PubMed] [Google Scholar]
- 19. Sakaeda T., et al. 2001. MDR1 genotype-related pharmacokinetics of digoxin after single oral administration in healthy Japanese subjects. Pharm. Res. 18:1400–1404 [DOI] [PubMed] [Google Scholar]
- 20. Sanofi-Aventis 2007. Rifadin (rifampin) product monograph, version 3.0:15. Sanofi-Aventis, Quebec, Canada [Google Scholar]
- 21. Schuetz E. G., Schinkel A. H., Relling M. V., Schuetz J. D. 1996. P-glycoprotein: a major determinant of rifampicin-inducible expression of cytochrome P4503A in mice and humans. Proc. Natl. Acad. Sci. U. S. A. 93:4001–4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Siccardi M., et al. 2008. Association of a single-nucleotide polymorphism in the pregnane X receptor (PXR 63396C→T) with reduced concentrations of unboosted atazanavir. Clin. Infect. Dis. 47:1222–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tappero J. W., et al. 2005. Serum concentrations of antimycobacterial drugs in patients with pulmonary tuberculosis in Botswana. Clin. Infect. Dis. 41:461–469 [DOI] [PubMed] [Google Scholar]
- 24. Tirona R. G., Leake B. F., Wolkoff A. W., Kim R. B. 2003. Human organic anion transporting polypeptide-C (SLC21A6) is a major determinant of rifampin-mediated pregnane X receptor activation. J. Pharmacol. Exp. Ther. 304:223–228 [DOI] [PubMed] [Google Scholar]
- 25. Tishkoff S. A., Williams S. M. 2002. Genetic analysis of African populations: human evolution and complex disease. Nat. Rev. Genet. 3:611–621 [DOI] [PubMed] [Google Scholar]
- 26. Visser M. E., et al. 10 November 2010, posting date The effect of vitamin A and zinc supplementation on treatment outcomes in pulmonary tuberculosis: a randomized controlled trial. Am. J. Clin. Nutr. [Epub ahead of print.] doi:10.3945/ajcn.110.001784 [DOI] [PubMed] [Google Scholar]
- 27. Weiner M., et al. 2005. Association between acquired rifamycin resistance and the pharmacokinetics of rifabutin and isoniazid among patients with HIV and tuberculosis. Clin. Infect. Dis. 40:1481–1491 [DOI] [PubMed] [Google Scholar]
- 28. Weiner M., et al. 2010. Effects of tuberculosis, race, and human gene SLCO1B1 polymorphisms on rifampin concentrations. Antimicrob. Agents Chemother. 54:4192–4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wilkins J. J., et al. 2008. Population pharmacokinetics of rifampin in pulmonary tuberculosis patients, including a semimechanistic model to describe variable absorption. Antimicrob. Agents Chemother. 52:2138–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]