Abstract
We describe a novel translation inhibitor that has anti-dengue virus (DENV) activity in vitro and in vivo. The inhibitor was identified through a high-throughput screening using a DENV infection assay. The compound contains a benzomorphan core structure. Mode-of-action analysis indicated that the compound inhibits protein translation in a viral RNA sequence-independent manner. Analysis of the stereochemistry demonstrated that only one enantiomer of the racemic compound inhibits viral RNA translation. Medicinal chemistry was performed to eliminate a metabolically labile glucuronidation site of the compound to improve its in vivo stability. Pharmacokinetic analysis showed that upon a single subcutaneous dosing of 25 mg/kg of body weight in mice, plasma levels of the compound reached a Cmax (maximum plasma drug concentration) above the protein-binding-adjusted 90% effective concentration (EC90) value of 0.96 μM. In agreement with the in vivo pharmacokinetic results, treatment of DENV-infected mice with 25 mg/kg of compound once per day reduced peak viremia by about 40-fold. However, mice treated with 75 mg/kg of compound per day exhibited adverse effects. Collectively, our results demonstrate that the benzomorphan compounds inhibit DENV through suppression of RNA translation. The therapeutic window of the current compounds needs to be improved for further development.
INTRODUCTION
Dengue virus (DENV), a member of the Flavivirus genus from the Flaviviridae family, causes approximately 50 million to 100 million human infections annually (12). Besides DENV, many other flaviviruses are important human pathogens, including West Nile virus (WNV), yellow fever virus (YFV), Japanese encephalitis virus (JEV), and tick-borne encephalitis virus (TBEV). There is no clinically approved antiviral treatment for any flavivirus infection. The current treatment for flavivirus infection is only supportive. Human vaccines for flaviviruses are available only for YFV, JEV, and TBEV (12). Development of a dengue vaccine is challenging because a successful vaccine requires a balanced immune response to all four serotypes of DENV; an unbalanced vaccine could lead to enhanced virus replication mediated by serotype-cross-reactive antibodies (21). Therefore, antiviral therapy is urgently needed for treatment of flavivirus infections.
The flaviviral genome is a plus-sense RNA about 11 kb in length. The genomic RNA is composed of a 5′ untranslated region (UTR), a single open reading frame, and a 3′ UTR. The single open reading frame encodes a long polyprotein that is cleaved by a combination of viral and host proteases into 10 viral proteins, three structural proteins (capsid [C], premembrane [prM], and envelope [E]) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). The structural proteins form virus particles. The nonstructural proteins, together with host factors, form a replication complex to replicate viral RNA (17). Only two of the flavivirus proteins have enzymatic activities. NS3 functions as a viral serine protease, with NS2B as a cofactor (3, 8), an NTPase, an RNA triphosphatase (38, 39), and an RNA helicase (16). NS5 acts as a methyltransferase (7, 31) and an RNA-dependent RNA polymerase (RdRp) (1, 35). Besides functioning in viral replication, the nonstructural proteins also play important roles in evasion of the innate immune response (6, 13, 18, 22, 23) and in virus assembly (14, 19). Both viral and host factors could be targeted to block the viral infection cycle for antiviral development.
In principle, antiviral drug discovery strategies include target-based approaches targeting specific viral proteins or host proteins and cell-based approaches targeting multiple steps and proteins involved in a viral life cycle (26). The latter type of approach has successfully led to the development of BMS-790052, a potent inhibitor of NS5A that has achieved proof-of-concept in clinics for treatment of hepatitis C virus (HCV) infection (11). We have used various cell-based assays to identify anti-DENV agents. Our assays include the traditional cytopathic effect (CPE) inhibition assay (36) and modern reporter-based viral assays, including a luciferase expression replicon (24, 29), virus-like particles (30), and full-length reporter virus (43). Using the above cell-based assays, we have performed multiple high-throughput screening (HTS) campaigns. After screening libraries with diverse compound structures, we have identified many inhibitors that have antiviral activities in cell culture; however, none of the compounds identified so far has shown any in vivo efficacy (36; unpublished results).
Here we report the identification and characterization of a novel translation inhibitor that has anti-DENV activity. Besides DENV, the compound also inhibits other flaviviruses in cell culture at noncytotoxic concentrations. Chemical refinement of the original hit was carried out to improve the metabolic stability of the compound. Detailed stereochemistry analyses showed only the S,R,S enantiomer of the compound has antiviral activity. Mode-of-action analyses indicated that the active enantiomer inhibits DENV infection through suppression of viral RNA translation. Importantly, when tested in a dengue mouse viremia model, this compound significantly reduced peak viremia, demonstrating the in vivo efficacy of the inhibitor.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
BHK-21 cells (baby hamster kidney cells) and Vero cells (kidney epithelial cells from an African green monkey) were cultured in Dulbecco's modified Eagle medium (DMEM) plus 10% fetal bovine serum (FBS) and infected with virus in the presence of DMEM plus 2% FBS. A549 cells (human alveolar basal epithelial cells) were maintained in F12 medium plus 10% FBS, and the infections were carried out in F12 medium plus 2% FBS. C6/36 cells were cultured in RPMI 1640 medium with 10% FBS, and the virus infections were carried out in RPMI 1640 medium with 5% FBS.
We used the following viruses: WNV (strain 3356), YFV (17D vaccine strain), DENV-2 (strains New Guinea C and TSV01), Western equine encephalitis virus (WEEV) (strain Cova 746), and vesicular stomatitis virus (VSV) (New Jersey serotype). The sources of these viruses were reported previously (41). DENV-specific mouse monoclonal antibody 4G2 against envelope protein was prepared from a hybridoma cell line purchased from American Type Culture Collection (ATCC).
CPE-based HTS assay.
The details of the HTS assay were reported previously (36). Briefly, Huh-7 cells were infected with DENV-2 (New Guinea C strain) at a multiplicity of infection (MOI) of 1 in the presence of 5 μM test compounds. After incubation for 3 days, compound-mediated inhibition of virus-induced cytopathic effect (CPE) was determined by measuring the intracellular level of ATP using a CellTiter-Glo luminescent-cell viability assay (Promega).
Compound synthesis and enantiomer purification.
The synthetic scheme is described in the supplemental material. The isomeric mixture of compound was purified by chiral high-performance liquid chromatography (HPLC) using a Chiralcel OJ column (Chiral Technologies Europe). The purified enantiomers were dissolved at a concentration of 5 μg/ml in methanol, and their optical rotations were determined in an ADP440+ polarimeter (Bellingham and Stanley Ltd.) at room temperature.
CFI assay.
A cell-based flavivirus immunodetection (CFI) assay was performed as described previously (37). A549 cells were infected with DENV-2 (MOI of 0.3) in the presence of 2-fold serial dilutions of test compounds. After incubation at 37°C for 48 h, viral antigen production was quantified by immune detection using the 4G2 antibody and goat anti-mouse IgG conjugated with horseradish peroxidase as primary and secondary antibodies, respectively. The concentration of compounds that decreased viral envelope protein production by 50% (EC50) was calculated by nonlinear regression analysis.
Viral-titer inhibition assay.
Vero cells (4 × 105 cells/well) were seeded in a 12-well plate. At 24 h postseeding, the cells were infected with the appropriate viruses at an MOI of 0.1. For DENV, WNV, YFV, and WEEV infections, samples were collected at 42 h postinfection (p.i.). For VSV, samples of culture medium were collected at 16 h postinfection. A plaque assay was carried out as reported previously (29).
Replicon assay.
The replicon assay was performed as described previously (24). A549 cells containing a luciferase-reporting replicon of DENV-2 were treated with 2-fold serial dilutions of test compounds. After incubation at 37°C for 48 h, a luciferase substrate (EnduRen [Promega]) was added according to the manufacturer's protocol. Luminescence was measured in a Clarity luminometer (Biotek). The concentration of compounds that decreased the luciferase expression by 50% (IC50) was calculated by nonlinear regression analysis.
Cell viability assay.
Cell viability was measured using the CellTiter 96 Aqueous One solution cell proliferation assay (Promega) according to the manufacturer's protocol. Approximately 1 × 104 Vero or A549 cells in 100 μl medium were seeded in a 96 well-plate. After incubation for 16 h, the cells were treated with test compound. After another incubation for 48 h, 20 μl of CellTiter 96 solution was added to each well of the 96-well plate. The cells were further incubated at 37°C for 2 h and measured for absorbance at 490 nm in a SaffireII microplate reader (Tecan, Austria).
Time-of-addition assay.
Approximately 2 × 104 Vero cells per well were seeded in a 96-well plate. At 24 h postseeding, the cells were infected with a luciferase-expressing dengue reporter virus (MOI = 4). The construction and preparation of luciferase DENV-2 were described elsewhere (43). At 0, 2, 4, 6, 8, 10, 13, 16, 20, and 24 h p.i., 2 μM NITD-2636 was added to the infected cells. Dimethyl sulfoxide (DMSO) (0.9%) was used as a negative control. At 24 h p.i., luciferase signal was measured using a Renilla luciferase assay system (Promega).
Transient replicon assay.
Ten micrograms of luciferase replicon (Rluc2A-Rep) RNA of DENV-1 (30) was electroporated into 8 × 106 BHK-21 cells. The cells were seeded in a 12-well plate (4 × 105 per well) and immediately treated with 0.2, 0.6, or 1.8 μM NITD-451 or with 0.9% DMSO as a control. At various time points, the cells were washed once with cold PBS, and 200 μl of 1× lysis buffer (Promega) was added. The plates containing the lysis buffer were sealed with Parafilm and stored at −80°C. Once samples for all time points had been collected, the cell lysates were subjected to luciferase according to the manufacturer's protocol (Promega). Specifically, 20 μl of cell lysates was transferred to a 96-well plate containing 50 μl of assay buffer plus luciferase substrate and assayed for luciferase signals in a Clarity luminescence microplate reader (BioTek).
In vitro translation inhibition assay.
The Pierce human in vitro protein expression kit for mRNA templates (Thermo Scientific) was used to assay the inhibitory effects of compounds (0.04 to 20 μM cycloheximide, NITD-451, or NITD-452) on RNA translation. Three different RNAs were used in the in vitro translation inhibition assay. (i) A DENV-1 luciferase replicon RNA contained a Renilla luciferase-FMDV 2A (foot-and-mouth disease virus 2A) fragment replacing viral C-prM-E genes. (ii) A DENV-2-UTR-Luc reporter RNA contained the DENV-2 5′UTR, the coding sequence of the first 38 amino acids of the viral capsid gene, the Renilla luciferase gene, and the DENV-2 3′UTR. (iii) A nonviral β-globin UTR-Luc reporter RNA contained the 5′ and 3′ UTRs of cellular β-globin RNA flanking the Renilla luciferase gene. All RNAs were transcribed in vitro from plasmid templates using the T7 mMessage mMachine high-yield capped RNA transcription kit (Ambion). The cDNA plasmid for DENV-1 luciferase replicon RNA was reported previously (29). The cDNA plasmids for DENV-2–UTR-Luc RNA and β-globin UTR-Luc RNA were constructed by using overlapping PCR followed by cloning the PCR products into the plasmid vector pGL4.7 (Promega) at the NheI and BamHI sites and at the NheI and NruI sites, respectively.
In vitro translation was performed following the manufacturer's protocol with modifications. Briefly, the translation reaction was assembled with preincubation of lysate (4 μl) and accessory proteins (0.8 μl) on ice for 5 min, followed by addition of the kit components H2O (16.76 μl), salt solution B (0.32 μl), amino acid (0.32 μl), RNase inhibitor (0.32 μl), and Energy Mix (0.48 μl). The amount of in vitro translation cell lysates used in the reaction was predetermined to ensure that the assay was performed within a linear range (data not shown). Next, RNA (200 ng of DENV-1–Luc replicon RNA, 4 ng of DENV-2–UTR-Luc reporter RNA, or 4 ng of β-globin UTR-Luc reporter RNA) and compound were added to the reaction to a final volume of 25 μl. The assembled reactions were mixed in microplates and incubated at 30°C for 3.5 h. Translation inhibition efficiency was determined by the luciferase bioluminescence readout measured with the luciferase assay system kit (Promega) and Clarity luminescence microplate reader. Readout values were normalized to those for the respective no-compound (with 0.45% DMSO) control. The compound concentration that decreased the luciferase expression by 50% (IC50) was calculated by nonlinear regression analysis.
In vitro rat liver microsomal stability assay.
Rat liver microsomes were obtained from BD Biosciences. Test compounds (1 μM) were incubated with rat liver microsomes in 0.1 M KH2PO4 buffer (pH 7.4) at 37°C in the presence of a cofactor solution containing 1.0 mM NADPH, 1.0 mM uridine diphosphate glucuronic acid (UDPGA), and 2 mM MgCl2. At 0, 10, 20, and 30 min, the reaction was quenched and extracted with acetonitrile. The samples were centrifuged, and the supernatants were analyzed by high-performance liquid chromatography–tandem mass spectrometry (LC/MS/MS). The in vitro half-life and intrinsic clearance were calculated according to the method in reference 27, and the hepatic extraction ratio was calculated according to the method in reference 15.
In vivo study.
The Animal Care and Use Committee (IACUC) of the Novartis Institute for Tropical Diseases is registered with the Agri-Food and Veterinary Authority (AVA) of Singapore. All animal experimental protocols were approved by the institute IACUC. The pharmacokinetic profile of NITD-451 was determined in CD-1 female mice following subcutaneous (s.c.) administration of 25 mg/kg of body weight. Blood samples were collected at 0.08, 0.25, 0.75, 3, 8, and 16 h after dosing. There were three animals per time point. Plasma concentrations of NITD-451 and its metabolite were measured using an Agilent Technologies HPLC (Santa Clara, CA) and an Applied Biosystems (Foster City, CA) triple quadrupole linear ion trap mass spectrometer. Metabolite identification was performed using the software programs LightSight (Applied Biosystems, Foster City, CA) and ACD (ACDlabs, Toronto, Canada) MS Fragmenter and Manager. Pharmacokinetic parameters were calculated by a noncompartmental approach using the WinNonLin software program (Pharsight).
The in vivo efficacy of NITD-451 was evaluated in a dengue viremia model in mice (34). AG129 mice (with a knockout of the alpha/beta interferon receptor and the gamma interferon receptor), purchased from B & K Universal, were injected intraperitoneally with 0.4 ml of RPMI-1640 medium containing 5 × 106 PFU/ml of DENV-2 (strain TSV01). The infected mice were then dosed with NITD-451 or vehicle (0.1 N HCl and 10% of 10% Solutol HS15, with top up with 50 mM citrate buffer, pH 4.75) by s.c. injection. The mice (n = 6 per group) were monitored twice a day, and blood samples were collected on day 3 p.i. for viral titer determination by plaque assay. Statistical analysis was performed by using Student's t test using the SigmaPlot/SigmaStat software program (Systat Software Inc.).
RESULTS
Identification of NITD-2636.
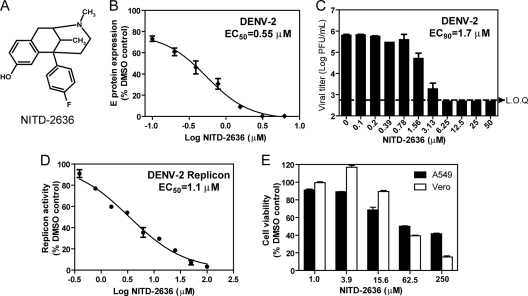
DENV infection of many mammalian cells causes a cytopathic effect (CPE). Compounds that inhibit DENV replication without cytotoxicity reduce or eliminate the CPE of infected cells. A CPE-based HTS assay was used to screen the Novartis compound library at a final compound concentration of 5 μM. The extent of CPE was quantified by measurement of the intracellular ATP level. The details of the HTS assay and screen results were recently reported (36). The HTS led to the identification of a class of compounds with a benzomorphan core as inhibitors of DENV. One such compound, NITD-2636 (Fig. 1A), exhibited 76% cell protection at 5 μM (data not shown). To confirm the antiviral activity, we analyzed the compound using a cell-based flavivirus immunodetection (CFI) assay. The CFI assay is an enzyme-linked immunosorbent assay (ELISA)-based assay that measures the amount of viral E protein in cells infected with DENV (37). The compound reduced viral E protein production in a dose-responsive manner, with an EC50 of 0.55 μM (Fig. 1B). Next, we validated the antiviral activity using a viral titer reduction assay. The compound inhibited virus production with an estimated EC90 of 1.7 μM (Fig. 1C). Treatment with 6.25 μM compound suppressed viral titers by >103-fold, to an undetectable level (limit of quantification [LOQ] = 500 PFU/ml). Furthermore, NITD-2636 inhibited a DENV-2 luciferase-reporting replicon in A549 cells, with an IC50 of 1.1 μM (Fig. 1D). Since the replicon does not reconstitute viral entry and virion assembly, these results suggest that the compound inhibits virus mainly through suppression of viral translation and/or RNA synthesis. A 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS)-based cell viability assay (with a 48-h incubation time) showed that the compound had 50% cytotoxic concentration (CC50) values of 63 and 52 μM in A549 and Vero cells, respectively (Fig. 1E), suggesting that the observed antiviral activity is not due to compound-mediated cytotoxicity.
Fig. 1.
(A) Chemical structure of NITD-2636. (B) Effect of NITD-2636 on the expression of viral envelope protein. A549 cells were infected with DENV-2 (MOI = 0.3; strain TSV01) in the presence of 2-fold serial dilutions of NITD-2636. After incubation at 37°C for 48 h, the expression of viral envelope protein was quantified by CFI assay. (C) Effect of NITD-2636 on the growth of DENV-2. Vero cells were infected with DENV-2 (MOI = 0.1). After 48 h p.t., cell culture supernatants were harvested for plaque assay using BHK-21 cells. (D) Effect of NITD-2636 on dengue virus replicon. A549 cells containing a luciferase replicon of DENV-2 were treated with NITD-2636 at the indicated concentrations for 48 h. The inhibition of viral replication by the compound was measured by the luciferase activity. (E) Cytotoxicity of NITD-2636 in Vero and A549 cells. Cytotoxicity was examined by incubation of Vero and A549 cells for 2 days with the indicated concentrations of NITD-2636. Cell viability was measured by an MTS assay. Average results and standard deviations (n = 3) are presented. LOQ, limit of quantification.
Antiviral spectrum of NITD-2636.
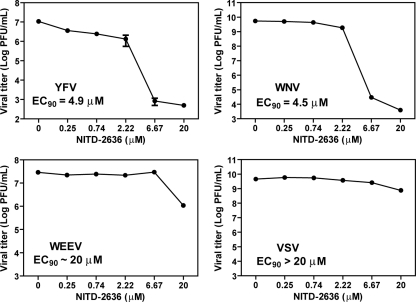
We examined the antiviral spectrum of NITD-2636 against other flaviviruses (YFV and WNV), a plus-stranded RNA alphavirus (Western equine encephalitis virus [WEEV]), and a negative-strand RNA rhabdovirus (vesicular stomatitis virus [VSV]). Vero cells were infected with YFV, WNV, WEEV, or VSV at an MOI of 0.1. The infected cells were immediately treated with various concentrations of NITD-2636. Culture medium was collected for plaque assays at 42 h p.i. (YFV, WNV, and WEEV) or 16 h p.i. (VSV). As shown in Fig. 2, NITD-2636 inhibited YFV and WNV with EC90s of 4.9 μM and 4.5 μM, respectively. In contrast, the EC90 values were around 20 μM and >20 μM against WEEV and VSV, respectively. These results indicate that compared with VSV and WEEV, flaviviruses are more sensitive to NITD-2636 inhibition.
Fig. 2.
Antiviral spectrum of NITD-2636. Vero cells were infected with indicated viruses at an MOI of 0.1. The infected cells were immediately treated with NITD-2636. Virus production was quantified using plaque assays. Average results and standard deviations (n = 3) are presented.
Time-of-addition analysis of NITD-2636.
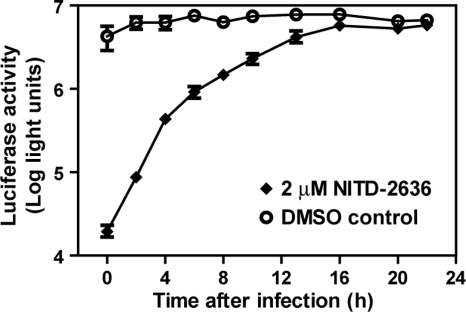
A time-of-addition experiment was performed to determine the stage of inhibition during a viral infection cycle. Vero cells were synchronously infected with a luciferase-expressing dengue reporter virus (MOI of 1). NITD-2636 (2 μM) or DMSO (0.9%) was added to the infected cells at various time points after infection (Fig. 3). Viral replication was quantified at 24 h postinfection by measuring the luciferase activity. The inhibition of luciferase expression by NITD-2636 gradually diminished when the compound was added at later time points; no suppression of luciferase activity was observed when the compound was added later than 16 h postinfection. During a single cycle of flavivirus infection, viral RNA translation occurs in the first 1 to 5 h p.i., viral RNA synthesis occurs at ≥5 h p.i., and progeny virions are released at ≥12 h p.i. (4). Our time-of-addition results suggest that NITD-2636 could block an early and a late step in the virus infection cycle; alternatively, the compound blocks only a late step in the viral infection cycle.
Fig. 3.
Time-of-addition analysis. Vero cells were infected with DENV-2 (strain New Guinea C) containing a Renilla luciferase reporter. Approximately 2 × 104 Vero cells were seeded per well of a 96-well plate. After incubation for 24 h, the cells were infected with the reporter virus at an MOI of 1. NITD-2636 was added to the infected cells at a final concentration of 2 μM at indicated time points. At 24 h p.i., the cells were assayed for luciferase activities. As controls, DMSO (0.9%) was added at various time points. Average results and standard deviations (n = 4) are presented.
Activity of the enantiomers.
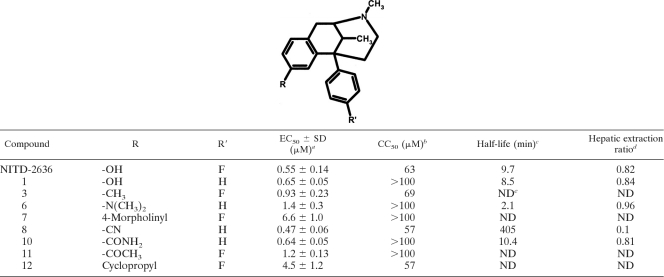
Synthetic chemistry efforts were carried out to explore the structure-activity relationship (SAR) of the benzomorphan scaffold using NITD-2636 as a starting point. Our initial attempt focused on identifying an equipotent replacement for the pharmaceutically undesirable phenolic-OH group, which underwent glucuronidation after intravenous administration of the compound in mice (data not shown). Several derivatives were synthesized, and their antiviral activities were tested in a CFI assay (Table 1). para fluorine substitution on the 6-phenyl ring of NITD-2636 had no negative impact on potency (compound 1). Substitution of the phenolic-OH group with an electron-donating methyl group (compound 3) and an electron-withdrawing cyano (compounds 8) or acetyl group (compound 11) retained antiviral activity. Replacement with a dimethyl amine (compound 6) or a polar amide group (compound 10) was well tolerated as well. In contrast, replacement with a bulky morpholine group (compound 7) or a cyclopropyl group (compound 12) decreased the potency by 12- and 8-fold, respectively.
Table 1.
Structure-activity relationship of the benzomorphan chemotype
EC50s were derived from the CFI assay (n ≥ 3).
CC50 values were derived from the cell viability assay using A549 cells.
Half-life (time required to metabolize 50% of input compound) was determined in a rat liver microsomal stability assay.
Hepatic extraction ratios (fraction of input compound that has been metabolized) were determined in a rat liver microsomal stability assay.
ND, not determined.
Active compounds were subjected to an in vitro rat liver microsomal stability test (see Materials and Methods) to predict their in vivo metabolic clearance. The assay measures the depletion of test compounds after incubation with rat liver microsomes to calculate in vitro metabolic parameters including the half-life (time required to metabolize 50% of input compound) and hepatic extraction ratio (fraction of input compound that has been metabolized). NITD-2636 exhibited a half-life of 9.7 min and a hepatic extraction ratio of 0.82 (Table 1). Among the active derivatives, compound 8 showed an improved metabolic stability, with a half-life of 405 min and a hepatic extraction ratio of 0.10 (Table 1). Compound 8 was therefore chosen for further studies.
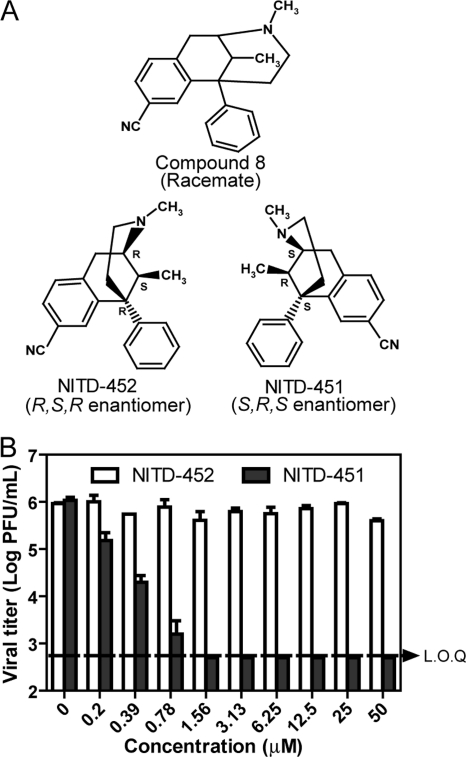
Since the synthesis of compound 8 yields a racemic mixture (42) (see the supplemental material), we sought to separate the enantiomers and to determine which stereoisomer is active against DENV. The racemic mixture was purified by chiral HPLC, resolving into two enantiomers (NITD-451 and NITD-452) with equal abundances (Fig. 4A). The S,R,S and R,S,R enantiomers were readily identified and differentiated by optical rotation. The measured optical rotation for the two enantiomers were +96.4° and −98.9°, respectively. Their configurations were further confirmed by X-ray crystallography (data not shown). Remarkably, when tested using a viral-titer inhibition assay, only the S,R,S enantiomer (NITD-451) showed antiviral activity; treatment with 1.56 μM NITD-451 suppressed viral titers by >103-fold, to an undetectable level (Fig. 4B). In contrast, the R,S,R enantiomer (NITD-452) was inactive; no significant antiviral activity was observed for NITD-452, up to a 50 μM concentration. Similar results were obtained for compounds within the same chemical class (data not shown). These results demonstrate that the S,R,S configuration is the required stereochemistry for activity against DENV.
Fig. 4.
Antiviral activities of the enantiomers. (A) Chemical structures of compound 8 and its stereoisomers. (B) Antiviral activities of the enantiomers. The effects of NITD-451 and NITD-452 on the growth of DENV-2 were determined in the viral titer reduction assay as described in Materials and Methods. LOQ, limit of quantification.
NITD-451 inhibits DENV RNA translation.
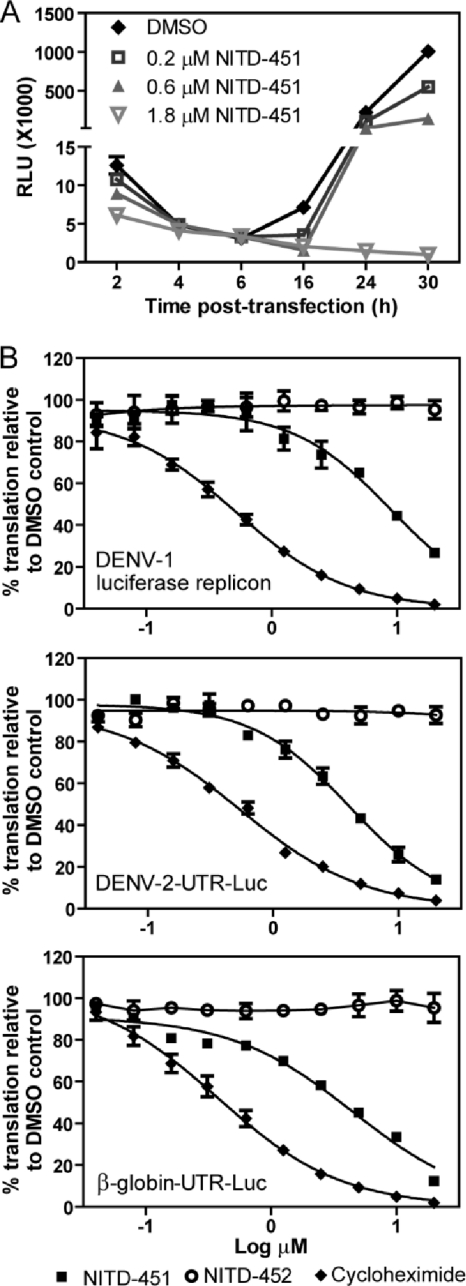
We assessed the mechanism of action of NITD-451. Since the original hit compound, NITD-2636, was active in the replicon assay and there was no change in the pharmacophore of the chemical structure, we hypothesized that, similar to NITD-2636, NITD-451 inhibits viral translation and/or replication. To test this hypothesis, we performed a transient-transfection assay using a DENV-1 luciferase-reporting replicon. The replicon was in vitro transcribed and transfected into BHK-21 cells, followed by treatment with NITD-451. Luciferase activities were monitored at various time points posttransfection (p.t.) (Fig. 5A). In the DMSO-treated cells (as a control), luciferase activity peaked within 2 h p.t. and gradually diminished afterwards. At ≥16 h p.t., the luciferase activity increased again and peaked at 30 h posttransfection. The 2-h peak of the luciferase signal represented translation of the input replicon RNA, whereas the 30-h peak represented RNA synthesis (20). In the NITD-451-treated cells, the compound inhibited the luciferase signals in a dose-responsive manner at both early and late times after RNA transfection. At 2 h p.t., treatment with 1.8 μM NITD-451 inhibited translation of the input replicon RNA by 52%; at 30 h p.t., the compound suppressed luciferase activity by >99% (Fig. 5A). The results suggest that NITD-451 inhibits viral RNA translation, leading to suppression of viral RNA synthesis.
Fig. 5.
Inhibition of RNA translation. (A) Transient replicon assay. A luciferase-reporting replicon (10 μg) was electroporated into BHK-21 cells. The transfected cells were immediately incubated with 0.9% DMSO or NITD-451 at the indicated concentrations. Luciferase activities were measured at 2, 4, 6, 16, 24, and 30 h posttransfection. (B) In vitro translation inhibition assay. A luciferase reporter replicon RNA of DENV-1 (strain Western Pacific) or reporter RNAs (DENV-2–UTR-Luc and β-globin UTR-Luc) were incubated with the human cell lysates (from the Pierce human in vitro protein expression kit) in the presence of NITD-451, NITD-452, cycloheximide, or DMSO (0.45%). After incubation at 30°C for 3.5 h, luciferase activity was measured by a luciferase assay system (Promega). Average results and standard deviations (n = 3) are presented for all experiments.
To confirm that NITD-451 inhibits translation, we established an in vitro translation system using the Pierce human in vitro protein expression kit. In vitro-transcribed DENV-1 luciferase-reporting replicon RNA was incubated with the human cell lysates in the presence of NITD-451, NITD-452, cycloheximide (a known translation inhibitor), or DMSO (a negative control). After incubating the translation reactions at 30°C for 3.5 h, luciferase activity was measured. As expected, cycloheximide strongly inhibited translation of the replicon RNA, with an estimated IC50 of 0.52 μM. NITD-451 also reduced the luciferase signal in a dose-dependent manner, with an IC50 of 8.9 μM. In contrast, NITD-452, the enantiomer that is inactive in the antiviral assays, did not inhibit the translation of replicon RNA (Fig. 5B, top panel). These results demonstrate that the antiviral activity of NITD-451 is in part due to its inhibition of viral translation.
Next, we examined whether the compound selectively inhibits viral RNA translation. Two Renilla luciferase reporter RNAs were subjected to an in vitro translation inhibition assay. One reporter RNA contained the first 210 nucleotides (nt) of the DENV-2 genome (including the 5′ UTR and the N-terminal 38-amino-acid-coding sequence of capsid gene), a luciferase gene, and the complete 3′ UTR of DENV-2. The other RNA contained the 5′ and 3′ UTRs of cellular β-globin RNA flanking a luciferase gene. As shown in Fig. 5B (middle and bottom panels), the translation of both reporter RNAs was inhibited by NITD-451, with IC50s of 4.1 and 4.5 μM, respectively. As positive controls, cycloheximide inhibited the translation of the two RNAs with IC50s of 0.54 and 0.37 μM, respectively. The results demonstrate that both NITD-451 and cycloheximide nonselectively inhibit DENV-2 translation in vitro.
To exclude the possibility that the compound exerts its antiviral activity through inhibition of viral enzymatic activities, we analyzed the compound in DENV RdRp (25), protease (2), ATPase (40), and methyltransferase (5) assays. None of the viral enzyme activity was inhibited by the compound up to 100 μM, the highest tested concentration (data not shown).
In vivo pharmacokinetics of NITD-451.
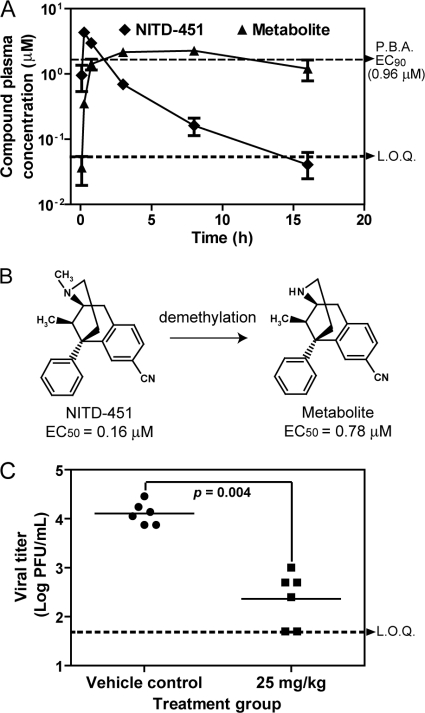
We determined the pharmacokinetic parameters of NITD-451 in mice. CD-1 mice received a single s.c. injection of 25 mg/kg of NITD-451. Blood samples were obtained at indicated time points for analysis (0.08, 0.25, 0.75, 3, 8, and 16 h) (Fig. 6A). Following the single s.c. injection, the compound was rapidly absorbed, with a Tmax (time required to reach the maximum plasma drug concentration [Cmax] after dosing) of 15 min. Plasma levels of NITD-451 reached a Cmax of 4.3 μM. To examine whether the compound concentration in plasma was above the EC90 value of NITD-451, we measured the antiviral activity of the compound in the presence of 40% human serum in cell culture using a CFI assay. The reason for addition of 40% human serum to the antiviral assay was to include the effect of the compound binding to protein in blood on antiviral efficacy. The results showed that NITD-451 had a protein-binding-adjusted EC90 value of 0.96 μM. As shown in Fig. 6A, plasma levels of NITD-451 were above the protein-binding-adjusted EC90 for at least 1 h.
Fig. 6.
In vivo pharmacokinetics and efficacy of NITD-451. (A) Plasma concentrations of NITD-451 and its metabolite after s.c. administration in CD-1 mice at a dosage of 25 mg/kg of body weight (n = 3 per time point). Protein-binding-adjusted (P.B.A.) EC90 and LOQ (limit of quantification) are indicated by dotted lines. (B) Demethylation of NITD-451. Metabolite identification and chemical resynthesis indicate that NITD-451 underwent N-demethylation in mice. The structure of demethylated metabolite of NITD-451 is shown. The EC50s for NITD-451 and its demethylated metabolite were derived from a CFI assay. (C) In vivo efficacy of NITD-451. AG129 mice were intraperitoneally inoculated with 2 × 106 PFU of DENV-2 (strain TSV01) on day 0. The mice (6 animals per group) were subcutaneously dosed with NITD-451 once a day. The peak viremia on day 3 p.i. was quantified by plaque assay. There was a significant difference in viremia (P = 0.004) between the mice treated with vehicle and the mice treated with NITD-451.
Besides the parental compound, a metabolite of NITD-451 was also observed in the plasma samples (Fig. 6A). It appeared gradually following parental compound administration, reaching its Cmax at approximately 8 h, and was readily detected in the plasma up to 16 h. Metabolite identification with the LightSight software program indicated that the parent compound underwent N-demethylation. The identity of the in vivo metabolite was further verified by LC/MS analysis using a chemically synthesized metabolite as the reference. Antiviral CFI assay of the resynthesized N-demethylated metabolite showed an EC50 of 0.78 μM (Fig. 6B). The discrepancy in metabolism of NITD-451 observed in the in vitro rat liver microsomal stability study and in vivo mouse experiment might be attributed to different animal species used.
In vivo efficacy in a DENV viremia mouse model.
A dengue viremia mouse model (34) was used to examine the in vivo efficacy of NITD-451. AG129 (lacking alpha/beta interferon and gamma interferon receptors) mice were infected with DENV-2, treated immediately with NITD-451 (s.c. dosing at 25 and 75 mg/kg of body weight in mice once per day), and measured for peak viremia on day 3 postinfection. For the 25-mg/kg treatment group, about a 40-fold reduction in viremia was achieved upon treatment; no adverse events were observed in the treated animals. Student's t test indicates that compared with the vehicle control, the viremia reduction in the 25-mg/kg-treated mice was statistically significant, with a P value of 0.004 (Fig. 6C). However, the mice treated with 75 mg/kg NITD-451 appeared to be sick, with hypersensitivity symptoms, and were euthanized on day 1 postinfection. The results indicate that the therapeutic window of NITD-451 is small.
DISCUSSION
A number of approaches to identifying flavivirus inhibitors have been established (26). Using a viral infection cell-based HTS approach, we have identified inhibitors of DENV with distinct mechanisms (36). In this study, we report the identification and characterization of a novel class of compounds that has both in vitro and in vivo anti-DENV activities. This class of inhibitors contains a benzomorphan core structure and exerts antiviral activity through suppression of viral RNA translation. The translation inhibition as a mechanism-of-action of the compound is supported by five lines of evidence. (i) Time-of-addition experiments showed that when added at ≥16 h p.t., the compound lost its antiviral activity, indicating that the compound inhibits an early step of viral infection. (ii) The compound inhibited a DENV replicon as efficiently as it inhibited a complete virus infection, suggesting that it does not suppress viral entry and virion assembly. (iii) The compound did not have any activity against in vitro assays of viral protease, ATPase, methyltransferase, and RdRp. (iv) Results from transient transfection of a luciferase replicon showed that the compound inhibited input RNA translation inside cells. (v) The compound directly suppressed in vitro translation of a reporter RNA. The molecular details of how NITD-451 inhibits RNA translation remain to be determined. Although our results have demonstrated that the compound suppresses viral translation, we could not exclude the possibility that the compound may inhibit DENV through another mechanism(s). This notion is indicated by the discrepancy between the EC50 (0.16 μM) (Fig. 6B) for NITD-451 in cell culture and the IC50 (8.9 μM) (Fig. 5B) determined by the in vitro translation assay.
The in vivo potency of NITD-451 in inhibiting host translation remains to be determined. In vitro translation analysis showed that the compound nonselectively inhibited viral and host RNA translation (Fig. 5B). This result raised the concern of toxicity due to the compound-mediated inhibition of host translation. On the other side, the compound showed selectivity in inhibiting flaviviruses. For example, treatment with 6.67 μM NITD-2636 suppressed the viral titers of DENV (Fig. 1C), YFV, and WNV (Fig. 2) by greater than 103-, 104-, and 105-fold, respectively, whereas the same treatment did not reduce the viral titers of nonflaviviruses (WEEV and VSV) (Fig. 2). The latter result argues that the observed antiflavivirus activity is not due to compound-mediated cytotoxicity. Our cell culture results showed that both the initial hit NITD-2636 and its derivative lead NITD-451 had selectivity indices (SI) (= CC50/EC50) of >50. In agreement with the cell culture results, in vivo efficacy experiment showed that treatment of DENV-infected mice with 25 mg/kg of NITD-451 reduced peak viremia by about 40-fold without any adverse effect. These results demonstrate that NITD-451 has a therapeutic window both in vitro and in vivo. We speculate that under these conditions, although the compounds inhibited both viral and host translation, the effect of suppression of viral translation on DENV replication is more dramatic than the effect of suppression of host translation on toxicity, leading to the observed therapeutic window. The differential effects of translation inhibition on viral replication and on cytotoxicity could be different among evolutionarily distant viruses (e.g., positive-strand RNA DENV versus negative-strand RNA VSV), resulting in a distinct antiviral spectrum. Alternatively, since VSV and WEEV replicate much faster than DENV and other flaviviruses, the faster-replicating VSV and WEEV may be less sensitive to translation inhibition or low cytotoxicity than the slow-replicating DENV and other flaviviruses. Nevertheless, it should be noted that when dosed at 75 mg/kg of NITD-451, the AG129 mice did exhibit adverse symptoms, suggesting that the therapeutic window of the current compound should be improved for further development. The key to achieving a better therapeutic window relies on the feasibility of modifying the compound to preferentially inhibit viral translation. In a recent HCV study, benzimidazole compounds were shown to selectively bind and induce a conformational change of viral internal ribosome entry site (IRES) RNA, which weakens the ribosome docking and ultimately leads to inhibition of IRES-driven translation in HCV (28).
Although the target of NITD-451 remains to be determined, the interaction between the target and the inhibitor is specific. This is strongly suggested by the facts that only the S,R,S enantiomer NITD-451 was active in inhibiting DENV replication and RNA translation, whereas the R,S,R enantiomer NITD-452 was not active in either assay. A chemoproteomics approach could be used for target deconvolution (32). This approach requires synthesis of an inhibitor derivative containing a cross-linking chemical group; such a cross-linking inhibitor could be used to pull down the target, which could then be decoded by mass spectrometry analysis. For the chemoproteomics approach, an SAR of the compound should be established before the cross-linking inhibitor could be synthesized.
Targeting a host factor for antiviral development has the advantage of posing a high hurdle for the emergence of resistance. In line with this notion, we were not able to raise resistant DENV after multiple attempts using either complete virus infection or a replicon system (data not shown). In HCV antiviral development, alisporivir (Debio-025, an inhibitor of host cyclophilin required for HCV replication) showed a much lower rate of emergence of resistance than inhibitors of viral protease and polymerase did in patients and cell culture (9, 10, 33). However, targeting of a host factor for antiviral development is confronted with two major challenges. The first challenge is potential toxicity. As discussed above, the compound should selectively inhibit the role of the host protein in viral replication without affecting its normal cellular function. The second challenge is whether targeting a host factor or pathway could achieve in vivo efficacy. We recently reported an inhibitor of a host pyrimidine synthesis enzyme, dihydroorotate dehydrogenase (DHODH) (36). Although this inhibitor potently suppressed DENV in cell culture (EC50, 2.4 nM), the compound did not show any in vivo efficacy in the DENV AG129 mouse model. The lack of in vivo efficacy is most likely due to the exogenous uptake of pyrimidine from diet. The external source of pyrimidine could maintain a high concentration of pyrimidine in plasma, masking the compound-mediated antiviral activity.
Finally, the current study points out the importance of understanding the targets of “hits” identified from the cell-based antiviral screening. “Hits” identified from the phenotypic cell-based screening could potentially inhibit host or viral targets. The number of host factors required for a productive viral infection cycle is much higher than the number of virally encoded proteins (10 viral proteins in the case of flavivirus). Consequently, the probability of identifying inhibitors of host targets is much higher than the probability of identifying inhibitors of viral targets. It is therefore critical to biologically profile the “hits” to understand their antiviral mechanisms and targets before major chemistry resources are committed.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sebastian Sonntag for critical reading of the manuscript. We also thank colleagues at the Novartis Institute for Tropical Diseases and from the Novartis Institutes for BioMedical Research for helpful discussions and support during the course of this study.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 5 July 2011.
REFERENCES
- 1. Ackermann M., Padmanabhan R. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926–39937 [DOI] [PubMed] [Google Scholar]
- 2. Bodenreider C., et al. 2009. A fluorescence quenching assay to discriminate between specific and nonspecific inhibitors of dengue virus protease. Anal. Biochem. 395:195–204 [DOI] [PubMed] [Google Scholar]
- 3. Chambers T. J., Grakoui A., Rice C. M. 1991. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J. Virol. 65:6042–6050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chambers T. J., Hahn C. S., Galler R., Rice C. M. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649–688 [DOI] [PubMed] [Google Scholar]
- 5. Chung K. Y., et al. 2010. Higher catalytic efficiency of N-7-methylation is responsible for processive N-7 and 2′-O methyltransferase activity in dengue virus. Virology 402:52–60 [DOI] [PubMed] [Google Scholar]
- 6. Daffis S., et al. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Egloff M. P., Benarroch D., Selisko B., Romette J. L., Canard B. 2002. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 21:2757–2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Falgout B., Miller R. H., Lai C. J. 1993. Deletion analysis of dengue virus type 4 nonstructural protein NS2B: identification of a domain required for NS2B-NS3 protease activity. J. Virol. 67:2034–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Flisiak R., et al. 2009. The cyclophilin inhibitor Debio 025 combined with PEG IFNalpha2a significantly reduces viral load in treatment-naive hepatitis C patients. Hepatology 49:1460–1468 [DOI] [PubMed] [Google Scholar]
- 10. Flisiak R., et al. 2008. The cyclophilin inhibitor Debio-025 shows potent anti-hepatitis C effect in patients coinfected with hepatitis C and human immunodeficiency virus. Hepatology 47:817–826 [DOI] [PubMed] [Google Scholar]
- 11. Gao M., et al. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gubler D., Kuno G., Markoff L. 2007. Flaviviruses, p. 1153–1253 In Knipe D. M., Howley P. M. (ed.), Fields virology, 5th ed., vol. 1 Lippincott William & Wilkins, Philadelphia, PA [Google Scholar]
- 13. Guo J., Hayashi J., Seeger C. 2005. West Nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 79:1343–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kummerer B. M., Rice C. M. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76:4773–4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lau Y. Y., et al. 2002. The use of in vitro metabolic stability for rapid selection of compounds in early discovery based on their expected hepatic extraction ratios. Pharm. Res. 19:1606–1610 [DOI] [PubMed] [Google Scholar]
- 16. Li H., Clum S., You S., Ebner K. E., Padmanabhan R. 1999. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J. Virol. 73:3108–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lindenbach B. D., Thiel H.-J., Rice C. M. 2007. Flaviviridae: the viruses and their replication, p. 1101–1152 In Knipe D. M., Howley P. M. (ed.), Fields virology, 5th ed., vol. 1 Lippincott William & Wilkins, Philadelphia, PA [Google Scholar]
- 18. Liu W., et al. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 79:1934–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu W. J., Chen H. B., Khromykh A. A. 2003. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J. Virol. 77:7804–7813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lo L., Tilgner M., Bernard K., Shi P.-Y. 2003. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus using a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 77:10004–10014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miller N. 2010. Recent progress in dengue vaccine research and development. Curr. Opin. Mol. Ther. 12:31–38 [PubMed] [Google Scholar]
- 22. Munoz-Jordan J. L., et al. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004–8013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Munoz-Jordan J. L., Sanchez-Burgos G. G., Laurent-Rolle M., Garcia-Sastre A. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. U. S. A. 100:14333–14338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ng C. Y., et al. 2007. Construction and characterization of a stable subgenomic dengue virus type 2 replicon system for antiviral compound and siRNA testing. Antiviral Res. 76:222–231 [DOI] [PubMed] [Google Scholar]
- 25. Niyomrattanakit P., et al. 2011. A fluorescence-based alkaline phosphatase-coupled polymerase assay for identification of inhibitors of dengue virus RNA-dependent RNA polymerase. J. Biomol. Screen. 16:201–210 [DOI] [PubMed] [Google Scholar]
- 26. Noble C. G., et al. 2010. Strategies for development of dengue virus inhibitors. Antiviral Res. 85:450–462 [DOI] [PubMed] [Google Scholar]
- 27. Obach R. S. 1999. Prediction of human clearance of twenty-nine drugs from hepatic microsomal intrinsic clearance data: an examination of in vitro half-life approach and nonspecific binding to microsomes. Drug Metab. Dispos. 27:1350–1359 [PubMed] [Google Scholar]
- 28. Parsons J., et al. 2009. Conformational inhibition of the hepatitis C virus internal ribosome entry site RNA. Nat. Chem. Biol. 5:823–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Puig-Basagoiti F., et al. 2006. Triaryl pyrazoline compound inhibits flavivirus RNA replication. Antimicrob. Agents Chemother. 50:1320–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qing M., Liu W., Yuan Z., Gu F., Shi P. Y. 2010. A high-throughput assay using dengue-1 virus-like particles for drug discovery. Antiviral Res. 86:163–171 [DOI] [PubMed] [Google Scholar]
- 31. Ray D., et al. 2006. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 80:8362–8370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rix U., Superti-Furga G. 2009. Target profiling of small molecules by chemical proteomics. Nat. Chem. Biol. 5:616–624 [DOI] [PubMed] [Google Scholar]
- 33. Robida J. M., Nelson H. B., Liu Z., Tang H. 2007. Characterization of hepatitis C virus subgenomic replicon resistance to cyclosporine in vitro. J. Virol. 81:5829–5840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schul W., Liu W., Xu H. Y., Flamand M., Vasudevan S. G. 2007. A dengue fever viremia model in mice shows reduction in viral replication and suppression of the inflammatory response after treatment with antiviral drugs. J. Infect. Dis. 195:665–674 [DOI] [PubMed] [Google Scholar]
- 35. Tan B. H., et al. 1996. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology 216:317–325 [DOI] [PubMed] [Google Scholar]
- 36. Wang Q., et al. 2011. Inhibition of dengue virus through suppression of host pyrimidine biosynthesis. J. Virol. 85:6548–6556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Q. Y., et al. 2009. A small-molecule dengue virus entry inhibitor. Antimicrob. Agents Chemother. 53:1823–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wengler G., Wengler G. 1991. The carboxy-terminal part of the NS 3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology 184:707–715 [DOI] [PubMed] [Google Scholar]
- 39. Wengler G., Wengler G. 1993. The NS3 nonstructural protein of flaviviruses contains an RNA triphosphatase activity. Virology 197:265–273 [DOI] [PubMed] [Google Scholar]
- 40. Xu T., et al. 2005. Structure of the dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 Å. J. Virol. 79:10278–10288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yin Z., et al. 2009. An adenosine nucleoside inhibitor of dengue virus. Proc. Natl. Acad. Sci. U. S. A. 106:20435–20439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yokoyama N., et al. 1979. Syntheses, analgetic activity, and physical dependence capacity of 5-phenyl-6,7-benzomorphan derivatives. J. Med. Chem. 22:537–553 [DOI] [PubMed] [Google Scholar]
- 43. Zou G., Xu H. Y., Qing M., Wang Q.-Y., Shi P.-Y. 2011. Development and characterization of a stable luciferase dengue virus for high-throughput screening. Antiviral Res. 91:11–19 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.