Abstract
Since the work done by R.A. Emerson in the 1930s, Inhibitor of striate (Isr) has been recognized as a dose-dependent genetic modifier of variegation in chlorotic leaf striping mutants of maize such as striate2 (sr2). We have shown that Isr specifically inhibits proliferation and differentiation of plastid defective cells in sr2 mutants. Leaf narrowing is due to loss of intermediate veins and ground tissue located at leaf margins, and the few remaining plastid defective cells are of irregular size and aberrant organization. The Isr gene has been cloned by targeted transposon tagging. Isr mRNA is expressed throughout young leaves, but Isr chimeras indicate that the expression of Isr at leaf margins is sufficient to suppress both the lateral expansion of sr2 leaves and the extent of striping. Isr protein appears to encode a chloroplast protein with sequence similarity to a family of bacterial phosphatases involved in carbon catabolite repression or in carbon metabolism. We propose that the action of Isr in nuclear and plastid communication could be triggered by carbon stress.
Keywords: Isr, cell proliferation, variegation, plastids, sr2, leaf development
Chloroplasts and mitochondria evolved from symbiotic prokaryotes that provided photosynthetic and respiratory functions to their primitive eukaryotic hosts. As a result, organellar transcription, translation, and protein translocation utilize prokaryotic components and mechanisms rather than eukaryotic ones (for examples in chloroplasts see Danon 1997; Settles and Martienssen 1998; Fisk et al. 1999; Hess and Borner 1999). Mechanisms that integrate the function of organelles with the physiology of the host have been the subject of considerable study since cytoplasmic effects and nuclear-cytoplasmic incompatibilities were first noted in plants (Rhoades 1943; Surpin and Chory 1997). Some mitochondrial mutants affect plastid development, suggesting that cytoplasmic organelles might respond to common regulatory cues (Surpin and Chory 1997; Fisk et al. 1999). Given their prokaryotic origin, mechanisms of nuclear-cytoplasmic interaction could be prokaryotic in nature, a notion that has received some support recently with the realization that nuclearly encoded phytochrome photoreceptors are related to prokaryotic receptor kinases (Fankhauser and Chory 1999). However, in most cases these mechanisms have eluded molecular characterization. One possible source of mutations in nuclear–cytoplasmic signaling pathways are enhancers and suppressors of mutants with defective cytoplasm. We describe the molecular characterization of such a genetic modifier in plants.
In maize, a number of mutations effect chloroplast biogenesis in only a portion of the leaf. In iojap 1 (ij1) and striate 2 (sr2), for example, plastid-defective tissues (white or yellow stripes) are mostly localized at the margins of successive leaves (blade and sheath) (Jenkins 1924; Joachim and Burnham 1953). As leaf primordia arise surrounding the shoot apex, the central portion of a leaf is clonally related to the margin of the next leaf above or below (Sharman 1942; Steffensen 1968; Coe and Neuffer 1978; McDaniel and Poethig 1988). Thus, the white or yellow stripes of these mutants do not arise from meristematic lineages in the shoot apex. Rather, stripes are restricted to single leaves, which indicates that they arise from one or more cells shortly after initiation of leaf primordia. Such striping patterns can arise from apparently null mutations (Han et al. 1992). Two mechanisms have been proposed to account for these striping patterns. One mechanism is related to sorting of mixed plastids (Kirk and Tilney-Basset 1978; for review, see Birky 1983). After repeated cell divisions, defective organelles sort into separate cell lineages. As ground tissue at the leaf margins is derived from a lineage (the L1) separate from the middle portion of the leaf (the L2), sorting could account for the pattern of marginal stripes (Coe et al. 1988b). The other mechanism concerns overpopulation of defective plastids relative to healthy ones in cells that undergo rapid cell division (Walbot and Coe 1979; Borner and Sears 1986). In this case, the biogenesis of green plastids in mutant cells cannot keep up with rapid cell division. In maize, the marginal lateral domains of lamina arise directly from the shoot apex by recruiting cells opposite to the site of leaf initiation (Sharman 1942). Consequently, the rate of cell division is significantly higher and the orientation of cell division is less polarized near the basal margins of the leaf than in other parts of the lamina (Stein and Steffensen 1959; Poethig 1984; Sylvester et al. 1990; Poethig and Szymkowiak 1995). This could account for the pattern of stripes.
Genetic modifiers of the striping mutants ij and sr2 have been characterized extensively (Coe et al. 1982, 1988b; Kermicle and Axtell 1981) and alter not only striping patterns but also leaf morphology. In the presence of these modifiers, striped mutants produce narrow leaves with few stripes, if any. For example, ij mutants in a K55 inbred background have severely narrow leaves with few stripes, whereas heterozygous sibs are normal (Coe et al. 1988b). Isr (Inhibitor of striate) is perhaps the best known genetic factor that suppresses the expression of striping mutants [for a general description, see Coe et al. (1988a) and Neuffer et al. (1997)]. The phenotypic action of Isr has been best characterized in its interaction with sr2 (Kermicle and Axtell 1981), although anecdotally, Isr has also been shown to influence the phenotype of ij and other striping factors (Emerson et al. 1935; Mazoti 1978). However, plastids affected by ij are maternally transmitted and it was this aspect of ij that was shown to be influenced by Isr (Mazoti 1978). In contrast, sr2 plants do not produce maternal albinos of heterozygous genotype (Rhoades 1943; Kermicle and Axtell 1981). Also, defective plastids show different internal ultrastructures between ij and sr2 (Thompson et al. 1983; Williams and Kermicle 1974). Therefore, the primary lesions of those two mutations might not be directly responsible for modifier-mediated phenotypic alterations in their leaves.
Isr is located on chromosome 10L, 0.025 cM distal to the R gene that encodes a transcription factor necessary for the expression of structural genes of the anthocyanin biosynthesis pathway (Walker et al. 1995). The strength of Isr action is dosage- and allele-dependent (Kermicle and Axtell 1981). As the copy number of Isr is increased, the width of leaves and the extent of stripes are proportionately reduced in sr2 mutants. There are no effects in normal maize. We have characterized the cellular phenotype of sr2 leaves in various Isr constitutions, and determined the molecular nature of Isr. Here, we show that Isr suppresses proliferation of plastid-defective cells during lateral expansion of leaf primordia, and that the Isr gene encodes a protein with sequence similarity to a family of bacterial phosphatases. We propose that Isr might mediate a carbon signaling mechanism to exert inhibitory action on cellular activities in cytoplasmically defective cells. Isr and other components of this pathway are thus strong candidates for mediating the plastid signal between nucleus and cytoplasm.
Results
Isr reduces the spacing of lateral veins at the margins of sr2 leaves
Isr behaves as a second-site modifier in that it has no discernible morphological phenotype in the absence of mutations affecting chlorophyll variegation. Therefore, all of the following studies were carried out in plants homozygous for sr2. Three different Isr alleles were examined in this study, each linked to complex alleles at the R locus, namely r-g, R-nj, and R-st. The Isr alleles are named isr, Isr-nj, and Isr-st, respectively. In a classic study of the inheritance of complex loci, R.A. Brink (1973) proposed that each of these R alleles has multiple copies of a duplicated segment, which was later proposed to carry both the R and Isr genes (Kermicle 1996). Isr-st inhibits striping more strongly than Isr-nj does, but Isr-st/isr is weaker than Isr-nj/Isr-nj. Following the notion of Kermicle and Axtell (1981), the inhibition strength of the isr, Isr-nj, and Isr-st haplotypes is equivalent to zero (0), one (1), and one and one-half (1½) copies of Isr, respectively. All of the genetic stocks were maintained in a single genetic background, W22.
In a dosage- and allele-dependent manner, Isr inhibits the extent of striping and reduces leaf width in homozygous sr2 plants (Fig. 1A; Kermicle and Axtell 1981). However, Isr does not alter leaf length (data not shown). To characterize this effect at the cellular level, leaf blades of different Isr genetic constitutions were inspected by light microscopy (see Materials and Methods). The maize leaf is comprised of three primary tissues, the epidermis, the vasculature, and the ground tissue. The epidermis is a single cell layer derived from the embryonic protoderm lineage. The vasculature and the ground tissue are derived from multiple lineages, and comprise mesophyll and vascular bundle sheath cells, as well as the phloem, xylem, and other vascular cell types that make up the regularly spaced lateral and intermediate veins within each bundle (Sharman 1942). Mesophyll and bundle sheath cells accumulate large numbers of green photosynthetic chloroplasts.
Figure 1.
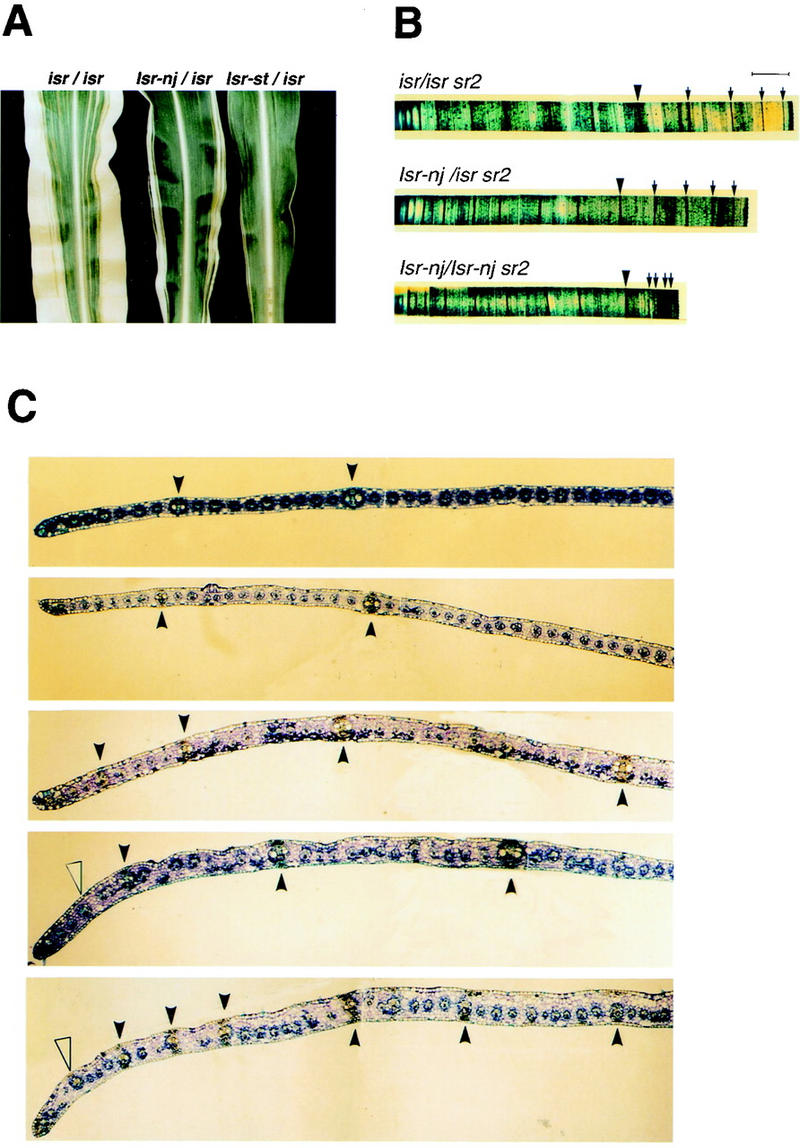
Leaf phenotypes and cross sections of leaf margins of various Isr constitutions. (A) Leaf phenotypes of sr2 in the presence of isr, Isr-nj, and Isr-st. Leaves are from sr2 plants of isr/isr, Isr-nj/isr, and Isr-st/isr (from left to right). Isr reduces both leaf width and the extent of striping of sr2. (B) Half blades of sr2 leaves of three different genotypes of Isr. Half leaf blades from midribs (left) to margins (right) were stained in Toluene blue dye solution. Leaves are from sr2 plants of isr/isr, Isr-nj/isr, and Isr-nj/Isr-nj. The eighth lateral veins from the midribs are marked as large arrowheads. Laterals after the eighth one are shown as small arrows. Bar, 0.5 cm. (C) Cross sections of leaf margins of various Isr constitutions. From top to bottom, the genetic constitutions of Sr2 and Isr are isr/isr Sr2, isr/isr sr2, Isr-nj/isr sr2, Isr-st/isr sr2, and Isr-st/Isr-st sr2. (Closed arrowheads) Large bundle sheaths bordered with hypodermal sclerenchyma cells on both abaxial and adaxial surfaces. (Open arrowheads) Large bundle sheaths that are associated with sclerenchyma cells on only one side of the leaf.
Whole mounts of half leaves carrying zero, one, and two copies of Isr (isr/isr, Isr-nj/isr, and Isr-nj/Isr-nj, respectively) were aligned for comparison (Fig. 1B). Samples were stained in Toluidine Blue, which stains lateral veins much darker than intermediate veins. Because there were slight variations in leaf width even in the same genetic background, the samples that showed similar lateral spacing near midribs were aligned and compared. The eighth lateral vein from the midrib is indicated by a large arrowhead in each case (Fig. 1B). Leaf width between the midrib and the eighth lateral vein was similar among the samples, as was the total number of lateral veins from the margin to midrib. Also, the intervals between laterals were comparable up to the eighth vein. However, the spacing of lateral veins near leaf margins became narrower as the copy number of Isr increased (small arrows in Fig. 1B). In sr2 plants with the equivalent of three copies of Isr (Isr-st/Isr-st), laterals at the leaf margins were frequently so close that individual veins were not easily identifiable (data not shown).
To examine the cellular organization of leaf margins, cross sections were examined. Along with samples from Sr2 controls, sections of sr2 blades with various Isr constitutions were aligned as shown in Figure 1C. In normal maize leaf blades, lateral and intermediate veins can be distinguished by their size, and by the neighboring sclerenchyma tissue (top). Lateral veins are surrounded both abaxially and adaxially with hypodermal sclerenchyma cells (solid arrowheads), whereas primary intermediate veins are only associated with sclerenchyma on one side (Russell and Evert 1985). At the margins of Isr-st/isr sr2/sr2 and Isr-st/Isr-st sr2/sr2 leaves (bottom two samples of Fig. 1C), unusually large bundles associated with sclerenchyma cells on only one side were observed (open arrowheads). These probably result from the failure of lateral veins to be fully developed.
Control Sr2 and sr2 leaves in isr plants showed similar spacing of lateral veins (top two sections of Fig. 1C). As the copy number of Isr was increased in sr2, intervals between laterals near leaf margins became narrower, and many fewer intermediate veins developed. Figure 2A highlights the third and fourth laterals from the marginal edges of Sr2 and sr2 leaves (the uppermost and the lowermost sections of Fig. 1C, respectively). In the Sr2 sample, 26 intermediate veins developed between the laterals. The sr2 leaf was of Isr-st/Isr-st that was equivalent to three copies of Isr. Only one intermediate between the poorly developed laterals was found in this leaf (Fig. 2B). A similar reduction in mesophyll cells was observed in these leaves. Interestingly, blade margins in high-copy Isr leaves (Isr-st/isr and Isr-st/Isr-st sr2) were much thicker than in isr/isr leaves (Figs. 1C and 2A). Aberrations in the size and number of cell layers were observed. Mesophyll cells were irregularly placed and their sizes were highly variable. In contrast, bundle sheath and epidermal cells were relatively uniform and unaffected. In wild-type leaf blades, two layers of mesophyll cells are formed between adjacent bundles with one mesophyll cell layer between bundle sheath and epidermis (Russell and Evert 1985). In defective tissues, three or four mesophyll cell layers were found either side of each vein, and often two layers of mesophyll cells were placed between bundle sheath and epidermis (Fig. 2B and C, b). The number of intermediate veins between laterals was highly variable, as was the size and number of mesophyll cells. From this study, it was concluded that reduction of leaf width was largely due to a drastic reduction in intermediate veins and the surrounding mesophyll cells at leaf margins.
Figure 2.
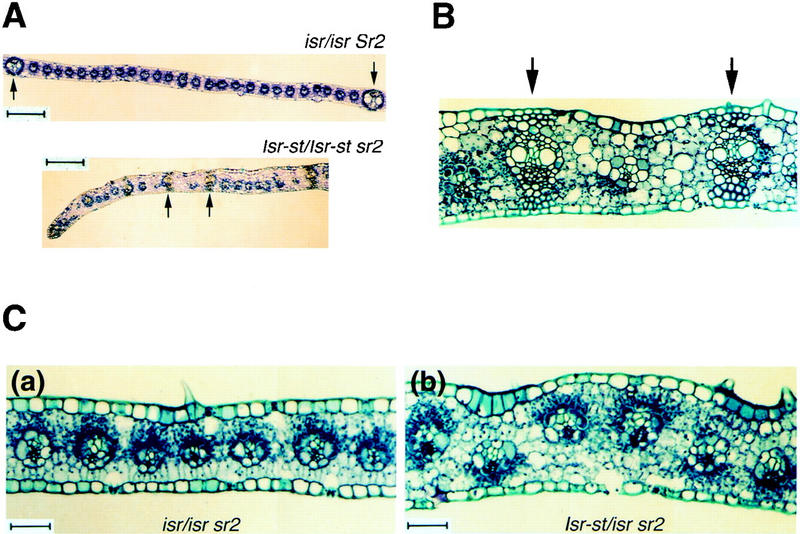
Comparison of the intervals of lateral veins in blade margins (A,B) and cross sections of leaf margins with green sectors (C). (A) The third and fourth laterals from the marginal edges are indicated by arrows. The samples are from isr/isr Sr2 and Isr-st/Isr-st sr2 leaves shown in Fig. 1C. The normal leaf carries 26 intermediate veins between the third and fourth laterals. There is only one intermediate vein between the laterals in the sample of Isr-st/Isr-st sr2. Bars, 200 μm. (B) Isr-st/Isr-st sr2 of Fig. 2A was magnified to show cellular organization between the lateral veins (arrows). (C) From cross sections of sectored leaf margins, green sectors were identified by dark-stained plastids. (a) isr/isr sr2; (b) Isr-st/isr sr2. Dark-stained organelles are either green plastids or nuclei. Defective albino plastids are not visible in those sections. Bar, 50 μm.
The effects of Isr are limited to cells with defective plastids
In white margins of sr2 plants, small green sectors frequently ran through mesophyll layers and/or veins, which allowed the contribution of normal and plastid-defective cells to the aberrant tissues to be assessed. In these sections, green chloroplasts and nuclei are stained deep dark blue, whereas defective white plastids are not so conspicuous. In the absence of Isr, or in low copy (Isr-nj/isr), white and green cells had relatively uniform sizes and were found in single mesophyll layers between bundle sheath and epidermis (Fig. 2C, a). However, in the presence of high copy Isr (Isr-st or Isr-st/Isr-st), white and green sectors had differently organized mesophyll cell layer. A majority of plastid-defective mesophyll cells were tiered in more than one cell layer between bundle sheath and epidermis (Fig. 2C, b). In contrast, green cells had relatively uniform sizes, and were mostly in single cell layers and rarely in double layers. In sectored tissues, green mesophyll cell layers were never thicker than albino cell layers. The data suggested that aberrant cellular organization in Isr leaves is restricted to plastid-defective cells.
Tagging and cloning Isr using transposon Ac
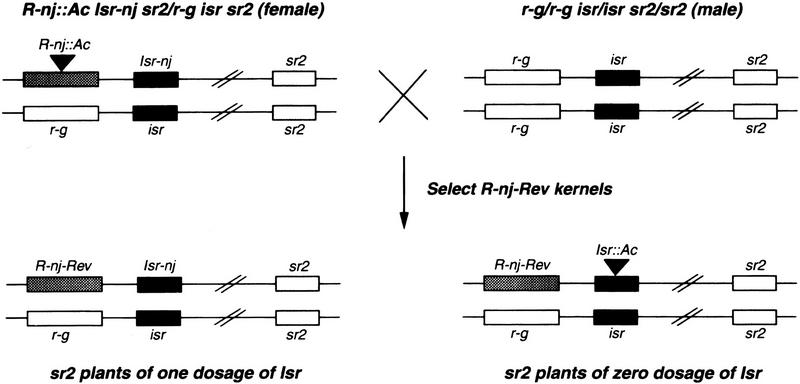
To molecularly tag Isr, we took advantage of an Ac element closely linked at the R locus (Dellaporta et al. 1988). A parental line of genotype R-nj::Ac/r-g Isr-nj/isr sr2/sr2 was constructed in which Ac was located at R-nj. As shown in Figure 3, R-nj::Ac/r-g sr2/sr2 plants as female parent were crossed with homozygous r-g/r-g isr/isr sr2/sr2 plants. F1 kernels of R-nj-Rev phenotype in which Ac had excised from R-nj, were planted in the field to inspect the expression of sr2 phenotypes. If Ac at a R-nj locus had transposed into Isr and subsequently interfered with the expression of Isr, the sr2 phenotype of R-nj-Rev/r-g Isr::Ac/isr should resemble ones of r-g/r-g isr/isr. Among ∼5000 F1 R-nj-Rev/r-g progeny, two sr2 plants were phenotypically indistinguishable from r-g/r-g sr2. Both plants produced germinal revertants in subsequent generations, indicating they carried new unstable alleles of isr. Only one putative Ac-tagged Isr plant was subjected to further molecular analysis. Southern blot hybridization with Ac probes was performed to identify a specific Ac element responsible for the mutation of Isr. A 6.8-kb BamHI fragment carrying part of Ac cosegregated with the new allele of isr, and was absent in Isr germinal revertants (data not shown).
Figure 3.
Genetic strategy of tagging Isr by Ac. R-nj::Ac Isr-nj sr2/r-g isr sr2 as a female parent were crossed with r-g/r-g isr/isr sr2/sr2. R-nj-Rev kernels were selected and planted. Plants that showed similar sr2 phenotype to isr/isr, were chosen as putative Isr::Ac/isr plants, and propagated to obtain germinal revertants in subsequent generations.
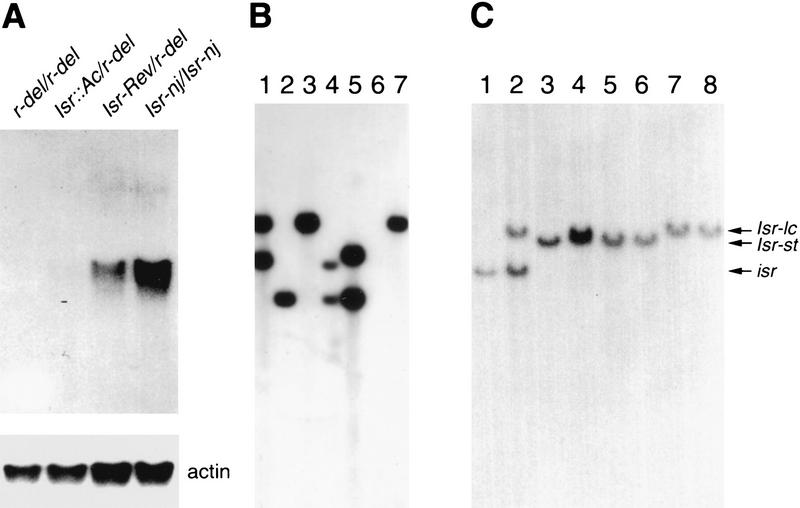
The 6.8-kb BamHI fragment was cloned into a bacteriophage λ vector. Figure 4D shows a restriction map of the 6.8-kb genomic DNA clone. Somatic and germinal revertants were analyzed by Southern blots using a 1.8-kb EcoRI fragment from this clone as a probe. Figure 4A shows a leaf with a somatic revertant sector from an Isr::Ac plant. The leaf appeared to be a genetic chimera of Isr. The left half of the leaf showed the phenotype of Isr-Rev, whereas the other half was of Isr::Ac. The chimeric phenotype was restricted to sheath and blade tissue from a single leaf (data not shown). To examine which part of the leaf contained the Isr-Rev allele, the leaf was longitudinally divided into six parts (LI, LII, LIII, RIII, RII, and RI), and DNA was isolated from each part. Figure 4B and C show Southern analysis of six parts of the leaf, along with Isr-nj, isr, germinal revertant (Isr-Rev/isr), and Isr::Ac (Isr::Ac/isr). A 6.8-kb Ac-hybridizing DNA fragment was detected in lane LII, LIII, RIII, RII, RI, and Isr::Ac, whereas lane Isr-nj, isr, LI, and Isr-Rev did not carry the Ac fragment. A genomic DNA fragment that comigrated with the Ac fragment weakly hybridized with the probe and was detected in all of the lanes. Southern analysis revealed that the Ac element was no longer present in the farthest left part (LI) of the chimerical leaf and a germinal revertant (Isr-Rev). The same blot was reprobed with a 1.8-kb EcoRI-flanking DNA (Fig. 4C). Only the farthest left part of the leaf (LI) and a germinal revertant (Isr-Rev) carried the same size of DNA fragment as Isr-nj did. The rest of the chimerical leaf (LII, LIII, RIII, RII, and RI) had the same DNA fragment found in Isr::Ac plants. Isr::Ac and its derivatives were heterozygous with isr, and carried isr-specific polymorphic DNA fragments. Southern analysis thus demonstrated that the phenotypic chimera arose from functional recovery of Isr at marginal tissue. This data strongly argue that the expression of Isr only at leaf margins is sufficient to suppress lateral expansion of leaves. To examine whether the cloned genomic fragment was from the transcribed region of Isr, Northern analysis was performed with total cellular RNA from seedlings of the following genotypes; r-del/r-del, r-del/Isr::Ac, r-del/Isr-Rev, and Isr-nj/Isr-nj. Previous genetic data showed that the deficiency in r-del included Isr (Kermicle and Axtell 1981). As expected, r-del/r-del and Isr::Ac/r-del failed to produce Isr mRNA (Fig. 5A). A 1.5-kb mRNA was detected from plants of Isr-Rev/r-del and Isr-nj/Isr-nj. It should be noticed that Isr-nj/Isr-nj accumulated much more mRNA than Isr-Rev/r-del. Thus, the level of Isr mRNA was positively correlated with the copy number of Isr.
Figure 4.
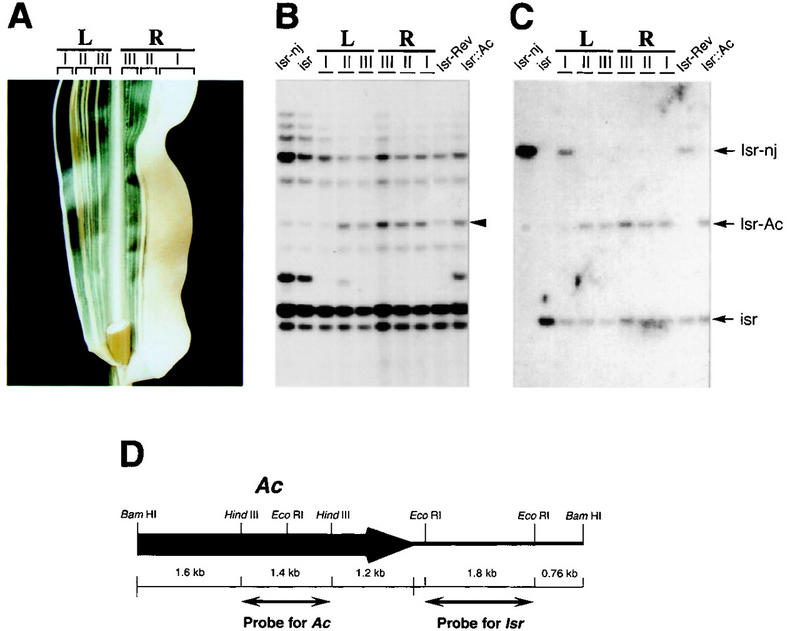
Analysis of somatic reversion of Isr::Ac sr2 and a restriction map of the 6.8-kb BamHI genomic fragment carrying Ac. (A) A chimerical leaf was from an Isr::Ac sr2 plant. The left half (L) of the leaf appears to be of Isr-Rev; the right half (R) is Isr::Ac. The leaf was divided into six parts (LI, LII, LIII, RIII, RII, and RI) for DNA extraction. (B) Along with Isr-nj, isr, a germinal revertant (Isr-Rev), and an Isr::Ac plant, the six parts of the leaf were analyzed by Southern hybridization using an Ac internal DNA fragment as a probe. (Arrowhead) A 6.8-kb genomic DNA that cosegregates with the phenotype of Isr. (C) The same blot was reprobed with a 1.8-kb EcoRI DNA fragment flanking the Ac element (for probe, see Fig. 4D). Isr::Ac and Isr-Rev including the chimerical leaf are heterozygous with isr. (D) A cloned 6.8-kb BamHI genomic DNA carried a part of Ac (bold line with arrowhead) and genomic DNA (thin line). An 1.8-kb EcoRI DNA fragment flanked by Ac was used as a probe for Isr detection. A 1.6-kb HindIII fragment was used to detect Ac elements.
Figure 5.
Northern analysis of Isr and Southern analysis of Isr alleles and unequal cross-overs between R-st and Lc. (A) Total cellular RNA was extracted from seedlings of r-del/r-del, r-del/Isr::Ac, r-del/Isr-Rev, and Isr-nj/Isr-nj. Northern hybridization was performed with an EcoRI 1.8-kb-flanking DNA probe (see Fig. 4D) (top). To evaluate equal loading of RNA samples, the same blot was reprobed with a maize actin cDNA (bottom). (B) HindIII cut genomic DNA was analyzed with Southern hybridization using the 1.8-kb-flanking DNA as a probe. R genotypes from lane 1 to lane 7 are the following; R-r/R-r, r-g/r-g, R-nj/R-nj, R-st/r-g, r-g Lc/r-g Lc, r-del/r-del, and B73 inbred. (C) HindIII cut genomic Southern analysis was performed on unequal cross-overs between R-st and Lc (lanes 5–8) along with r-g (lane 1), r-g Lc (lane 2), R-st (lane 3), and R-st Lc (lane 4). Lanes 5 and 6 are from independent recombinants of genotype R-st Isr-st (ΔLc ΔIsr-lc). Lanes 7 and 8 are from independent recombinants of genotype R-st (ΔIsr-st ΔLc) Isr-lc.
Southern analysis of Isr alleles
Genetic data have shown that Isr is present in one or two copies, depending on R genetic constitution (Kermicle and Axtell 1981). R-r carries two copies of Isr, whereas R-nj, R-st, and r-g are flanked by a single copy of Isr. Also, Isr is present next to Lc, a displaced R gene that is located two map units distal to R. To molecularly confirm the copy number of Isr in various R constitutions, Southern analysis was performed with the 1.8-kb-flanking DNA as a probe. The following genetic stocks were used for this study; R-r/R-r, r-g/r-g, R-nj/R-nj, R-st/r-g, r-g Lc/r-g Lc, and r-del/r-del. All of the R stocks used in this study are in a W22 genetic background. Also, the Isr genetic constitution of the inbred line B73 was examined. DNA from seedlings was digested with HindIII. Figure 5B is an autoradiograph of a Southern blot probed with the 1.8-kb Isr DNA. R-r and r-g Lc carried two copies of Isr, whereas R-nj, r-g, and R-st carried one copy of Isr. This was consistent with genetic data. Isr has been extensively utilized as a genetic marker to study the molecular organization of the R locus. Therefore, intragenic recombinants at R should be excellent genetic materials to map the cloned Isr genomic DNA. R-st and Lc are located 2 map units apart on the same chromosome and are flanked by Isr-st and Isr-lc, respectively. R-st Lc stocks and their intragenic recombinant derivatives were examined for this study (Eggelston et al. 1995).
Plants carrying the following recombinant haplotypes were kindly supplied by Dr. Kermicle (University of Wisconsin, WI), and were utilized to map the cloned DNA; R-st Isr-st (ΔLc ΔIsr-Lc), and R-st (ΔIsr-st ΔLc) Isr-lc. These haplotypes have lost Lc Isr-lc and Isr-st Lc, respectively. Allele-specific RFLPs (restriction fragment length polymorphism) were examined by Southern hybridization (Fig. 5C). As reference alleles, r-g, r-g Lc, R-st, and R-st Lc were used for this study (Fig. 5C, lanes 1,2,3,4, respectively). Lanes 5 and 6 are from independent recombinants (g333 and g326) of haplotype R-st Isr-st (ΔLc ΔIsr-lc), and lanes 7 and 8 are from independent recombinants (g315 and g301) of haplotype R-st (ΔIsr-st ΔLc) Isr-lc. An Isr-st-specific RFLP was detected in R-st, R-st Lc, and two R-st Isr-st (ΔLc ΔIsr-lc) lines, respectively (Fig. 5C, lanes 3,4,5,6). An Isr-lc-specific RFLP was detected in r-g Lc, R-st Lc, and two R-st (ΔIsr-st ΔLc) Isr-lc lines respectively (Fig. 5C, lanes 2,4,7,8). Thus, Southern analysis with the cloned genomic DNA perfectly fulfilled what was expected from previous genetic data on Isr.
Isr encodes proteins homologous to a class of bacterial phosphatases involved in carbon catabolite repression or in carbon metabolisms
To obtain sequence information of an Isr locus, a genomic library (from Dr. Steve Briggs at Pioneer Hi-Bred International, Johnston, IA, USA) and a cDNA library (from Dr. Alice Barkan at University of Oregon, OR) were screened with the 1.8-kb EcoRI fragment as a probe. Both libraries were constructed using seedlings of a maize inbred line, B73. On the basis of restriction enzyme analyses and Northern hybridization, it was found that a 3.8-kb genomic DNA fragment contained all of the coding region of Isr. Transcription sites were determined by primer extension (data not shown). A cDNA of a 1.5-kb insert and the 3.8-kb genomic DNA were sequenced and aligned to define the protein-coding region and exon/intron boundaries. The genomic structure and the nucleotide sequence of Isr are shown in Figure 6A and B, respectively.
Figure 6.
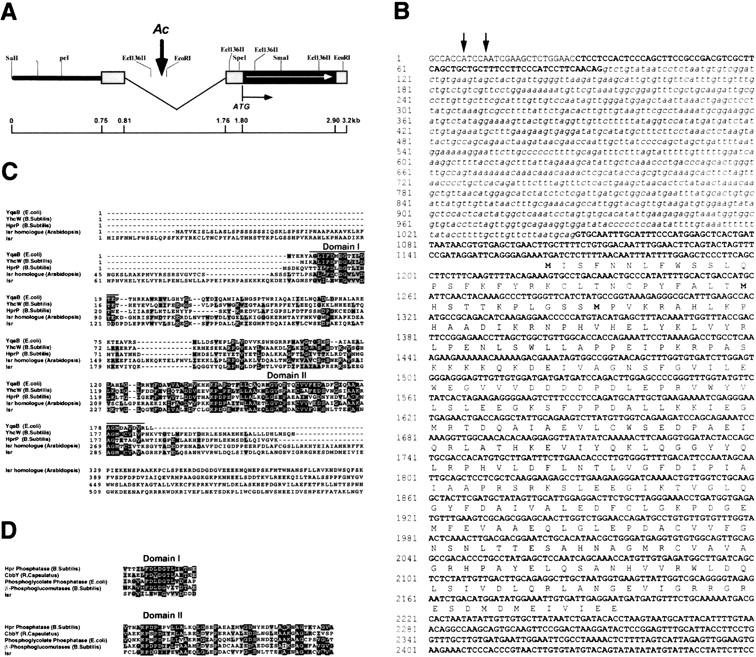
The genomic organization and amino acid sequence comparison of Isr. (A) Bold line indicates the sequence upstream of the transcription region of Isr. Untranslated regions are shown as gray boxes. The insertion site of Ac is shown as a vertical arrow in an intron. The protein-coding region is shown as a black box with horizontal arrows. (B) cDNA sequences are in bold characters and an intron sequence is written in italics. Arrows indicate transcription initiation sites. Deduced amino acids are listed under the corresponding codons. (C) The total length of yhcW (B. subtilis), yqaB (E. coli), Isr homolog (Arabidopsis), Isr, and hprP (B. subtilis) proteins are aligned. Conserved amino acids are in black boxes, whereas similar amino acids are in gray boxes. The most conserved peptides are denoted as domain I and II. (D) Alignment of most conserved peptides (Domain I and II) of Isr with functionally defined proteins.
The Isr gene contains one intron of 1068-nucleotides long in the untranslated leader sequence and has one ORF of 1032 bases. The ORF encodes 344 amino acids. The deduced Isr protein has a molecular mass of ∼38.7 kD and is predicted to be acidic (pI = 5.75). The carboxy-terminal 244 amino acids of this predicted protein shows significant similarity to a group of genes from plants and bacterial genomes. A closely related homolog in Arabidopsis has emerged from genome sequencing, and has more extensive similarity in this 244 amino acid domain (Fig. 6C). Bacterial genes, yqaB of Escherichia coli and yhcW of Bacillus subtilis, have been identified by genomic DNA sequencing analyses, but their functions are not known. Because of sequence similarities that are described below, these two genes have been classified to belong to the cbbY/cbbZ family of Calvin-Benson-Bassham (CBB) cycle. This similarity extends throughout the length of the proteins, but is most pronounced in two domains (Domain I and II) of 17 amino acids and 71 amino acids (Fig. 6C). The positions and distances of identical amino acids and these peptide domains were well conserved throughout the proteins. Extensive sequence similarities across such diverse species suggested that Isr could be engaged in evolutionarily conserved cellular processes. The two peptide domains that are highly conserved among Isr homologs are also commonly found in a class of bacterial phosphatases of carbon metabolism. Figure 6D shows aligned comparison of these two peptide domains among Isr and functionally defined bacterial genes. Isr homologs include phosphoglucomutase, which is involved in sucrose metabolism and phosphoglycolate phosphatase of the carbon respiratory pathway. These phosphatases are related to the Rhodobacter capsulatus cbbY gene, as well as a group of carbohydrate phosphatases in yeast and mammalian cells (Galinier et al. 1998). The biochemical function of one Isr homolog in Bacillus, hprP (previously yvoE), has been demonstrated recently as a P-Ser-Hpr phosphatase (Galinier et al. 1998). This phosphatase dephosphorylates Hpr, a histidine-rich phosphocarrier protein. In Bacillus subtilis, Hpr is a key component of the phosphotransferase system for carbon catabolite repression (CCR) in response to glucose. The extensive similarity with a class of bacterial phosphatases suggested a possible role of Isr in carbon regulation or carbohydrate metabolism. This analogy is further discussed below.
Both maize and Arabidopsis ORFs have an amino-terminal extension that encodes a basic, serine-rich peptide that ends in maize with the sequence I-V-A. These features suggest that Isr encodes a chloroplast protein, and that the first 104 amino acids encode a transit peptide that is cleaved after import into the chloroplast (the consensus cleavage signal is C/I-X-C/A). An Isr homolog is found in photosynthetic bacterial genomes, such as those of Synechocystis and Rhodobacter spp.
Isr mRNA present in most cells of leaf tissues
Anatomical studies on sr2 leaves and a genetic chimera of Isr::Ac demonstrated that Isr inhibits proliferation of ground tissue and spacing between lateral veins located at leaf margins. To ask whether the expression pattern of Isr might show spatial specificity or could be altered in the presence of sr2, in situ anti-sense RNA hybridization was performed with young seedlings. Along with r-del as a negative control, Isr-nj Sr2 and Isr-nj sr2 were examined. Figure 7 shows in situ anti-sense RNA hybridization with transverse sections of seedling stems. In situ hybridization with r-del, in which the Isr locus was deleted, revealed no signal except for cell walls of large vascular tissues (Fig. 7D). Signals around cell walls often result from nonspecific binding of anti-sense RNA probes. Isr mRNA was detected in most cells in transverse sections of Isr-nj Sr2 leaves (Fig. 7A). In the oldest leaves, cells near the midrib exhibited less hybridization than other parts of the blade. As shown in Figure 7B, the inner cells of midribs are much bigger with larger vacuoles than neighboring cells. Therefore, differences in signal intensities between midrib and other parts could result from differences in the sizes of cells and vacuoles, rather than the abundance of Isr mRNA inside cells. From this study, we conclude that the local effect of Isr on sr2 leaves was not due to spatial specificity in the expression of Isr. To test whether sr2 mutation could alter the expression of Isr, Isr-nj sr2 were examined at the seedling stage. As shown in Figure 7C, the expression pattern of Isr-nj sr2 was almost identical to one of Isr-nj Sr2. Also, there was no difference in the level of Isr mRNA between sr2 and Sr2 (data not shown). Therefore, it is unlikely that the sr2 mutation itself has a direct influence on the action of Isr. Rather, our data indicates that Isr can only exercise inhibitory action on cell proliferation in white stripes.
Figure 7.
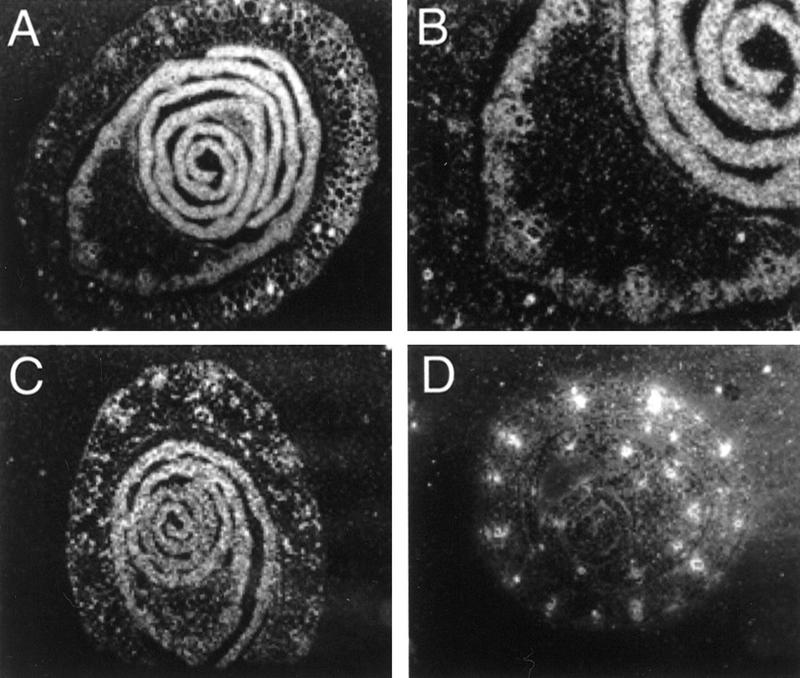
In situ hybridization. Transverse sections of seedling whorls were examined with in situ anti-sense RNA hybridization, and viewed under dark-field. r-del served as a negative control (D). Isr-nj sr2 (A) and Isr-nj Sr2 (C) showed signals in most cells. Fig. 7B is a close-up view of a part of Fig. 7A.
Discussion
The action of Isr is only phenotypically detectable in striping mutants such as sr1, sr2, and j1 (japonica 1). In this study, sr2 has been chosen, because various genetic constitutions of Isr have been incorporated into sr2 mutants of a single genetic background (Kermicle and Axtell 1981). In this material, leaves exhibit bold albino stripes mainly at the leaf margins and most stripes are restricted to single leaves, indicating that they arise during leaf initiation. Isr suppresses the extent of this striping. We have determined the molecular and cellular role of Isr in this phenomenon, allowing us to speculate as to the nature of the plastid signal in regulating photosynthetic cell fate.
Isr acts early during leaf development
Three stages of leaf development (meristem founder stage, primordial stage, and postprimordial stage) are well correlated with distinct stages of vein differentiation (for review, see Freeling and Lane 1993; Nelson and Dengler 1997). A midvein pre-exists in a shoot meristem and differentiates into P0 or P1 leaf primordium. During the primordial stage, when leaf founder cells divide uniformly, lateral veins are differentiated and are complete in the P3–P4 leaf primordium which is ∼6-mm long (Sharman 1942; Sylvester et al. 1990). Sharman (1942) has shown that lateral veins are present at the end of the primordial stage of leaf growth. The postprimordial stage is characterized by ligule formation and by basipetal development of primary intermediate veins (Nelson and Dengler 1997). Although the total number of laterals of sr2 leaves was similar among different Isr genetic constitutions, intermediate veins between laterals became fewer in sr2 leaves in the presence of Isr. In an extreme case, only one intermediate vein developed between lateral veins (Fig. 2B). Therefore, Isr action should take place after the primordial stages of leaf development when lateral veins are complete.
Isr regulates the proliferation of plastid-defective tissue at the margins of sr2 leaves
The following anatomical observations argue that Isr is involved in proliferation and differentiation of plastid-defective cells at the margins of mature sr2 leaves. First of all, the total number of lateral veins in leaf blades was similar among different Isr genetic constitutions (Fig. 1B). Secondly, the spacing of laterals at the margins became narrower as the copy number of Isr was increased, and the number of intermediates between laterals in defective tissues was very low (Fig. 1C). Thirdly, the size of the remaining mesophyll cells was highly variable, especially in leaves of high Isr dosage (e.g., Isr-st/Isr-st). Many mesophyll cells were abnormally large and the layer of mesophyll cells was increased, whereas the size and shape of epidermal and bundle sheath cells were unaffected by Isr. Green-sectored tissues showed that aberrant mesophyll layers largely consisted of cells with defective plastids (see Fig. 2C). Fourthly, expression of Isr mRNA is not dependent on mutations in sr2, nor is it specific to the margins of leaves. Even though Isr mRNA is ubiquitous, phenotypic defects can be observed mostly at margins of sr2 leaves in which white stripes are located. Lastly, a somatic reversion event at the leaf margin of Isr::Ac (see Fig. 4A) reduced not only leaf width but also the extent of albino stripes. These observations confirm that the inhibitory action of Isr is restricted to cells carrying sr2-affected plastids.
As described in the Introduction, striping might arise either from sorting-out of heteroplastidic cells into separate cell lineages or by dilution of healthy plastids in successive cell divisions. In either case, the development of stripes is related to cellular proliferation. It has been well documented that partitioning of organelles in eukaryotes is tightly coupled with the cell cycle (for review, see Yaffe 1991; Warren and Wickner 1996). In yeast, several mutants in mitochondrial and vacuolar inheritance have delayed cytokinesis and accumulate multiply budded cells (McConnell and Yaffe 1990; Xu and Wickner 1996). Variation of cell sizes in Isr-affected tissues most likely results from interference with cell division. One explanation for this might be the failure of partitioning of defective plastids during rapid and prolonged cell division in the presence of Isr. In maize leaf primordia, the rate of cell division is significantly higher and the orientation of cell division is less polarized near the basal margins than in other parts of the lamina, which may account for the positioning of stripes (Poethig 1984; Poethig and Szymkowiak 1995).
Isr affects nuclear-plastid signaling in albino stripes
In higher plants, a subset of nuclear mutations that block the development of plastids also affect cellular differentiation in leaves (Kirk and Tilnew-Basset 1978). DCL (defective chloroplasts and leaves) of tomato and DAG (differentiation and greening) of Antirrhinum majus, encode plastid-located proteins (Chatterjee et al. 1996; Keddie et al. 1996). In those mutants, palisade cells failed to expand into typical columnar shapes and were small and of spherical shape in a cell-autonomous fashion. There are several differences between the action of dcl or dag and that of Isr/sr2. First of all, defective leaf tissues of dcl or dag have normal numbers of cells. Secondly, dcl- or dag- affected cells are smaller than normal cells, whereas cells of defective tissues have highly variable sizes in Isr/sr2. Finally, DCL and DAG proteins are located in plastids and might be involved in plastid biogenesis coupled with cell differentiation. In contrast, Isr has no effect on its own and acts only on proliferation of leaf cells in which plastids fail to green.
Williams and Kermicle (1974) examined the ultrastructure of sr2-affected plastids from leaves with zero, one, or two copies of Isr. They failed to detect any morphological differences in defective plastids of those Isr constitutions. Therefore, it is very unlikely that the action of Isr is additive with sr2 with respect to plastid biogenesis. That is, the inhibitory action of Isr should be exercised directly on cellular processes rather than on plastid biogenesis itself. Aberrant cellular organization is most conspicuous in mesophyll layers that consist of cells with albino plastids (Fig. 2C). Reduction of cell number and variation in cell size indicate that cell division and cell expansion are not properly maintained. It is also possible that inappropriate cell death might be provoked by Isr during lateral expansion of sr2 leaf primordia, although no evidence for this was found (Martienssen 1997). In either case, Isr interferes with cellular activities for lateral expansion at the margins, which eliminates intermediate veins and associated ground tissue that have defective plastids.
Carbon stress could be the signaling cue for the action of Isr
In striate leaves, cells of non-photosynthetic plastids should import carbohydrates from cells carrying chloroplasts, in which fixed carbon is generated photosynthetically. Accordingly, carbon fluxes should be altered in albino cells. The following observations prompt us to propose that the action mode of Isr is closely related to carbon-stresses. (1) Isr-mediated cellular defects are manifest mainly in albino cells. (2) Isr homologs belong to a class of bacterial phosphatases involved in either carbon catabolite repression or carbon respiratory metabolism. This pathway was discovered in B. subtilis by Jacques Monod during World War II (Monod 1942) and was characterized in great detail in E. coli in the 1960s. As in maize, conditional phenotypic effects have been observed in bacteria, especially under carbon challenge, in mutants of some of these Isr homologs. Although mutants in hprP have not yet been described in B. subtilis, mutants in hprK (which encodes the cognate kinase) have no phenotypic effect except in conditions of carbohydrate challenge, when they fail to repress certain glycolytic enzymes (Galinier et al. 1998). Mutants in cbbY in the photosynthetic bacteria Rhodobacter also have no phenotype, but deletion of the whole operon (cbbXYZ) results in loss of photoautotrophic growth (Gibson and Tabita 1997). It is therefore tempting to suggest that the Isr protein plays a conditional role in carbohydrate signaling for cellular activities such as cell division and expansion. Especially, the role of Hpr as a universal sensor of glucose availability in B. subtilis could be informative in speculating the possible role of Isr in higher plants. Specifically, the loss of Isr would have no effect on cellular function, unless they were challenged by external carbohydrates. Such a challenge might be apparent in the nonphotosynthetic albino stripes found in striate mutants. These cells must take up carbohydrates from neighboring green cells. Under these circumstances, Isr might act to specifically induce the equivalent of CCR in higher plants, which results in failure of cellular proliferation. Such a mechanism might even involve signaling between the plastid and the nucleus. Plastid deficiencies result in striped plants with reduced vigor, and cells that have lost functional plastids might be prohibited from proliferation by the Isr protein. Intriguingly, a plant homolog of HPr itself has also been recently reported in the monocotyledonous plant Asparagus officianalis (Saito et al. 1997). Isr should serve as an excellent probe to reveal the molecular nature of a regulatory circuit of inter-organellular communication in higher plants.
Materials and methods
Plant materials
The following R stocks carrying different Isr constitutions in a W22-inbred genetic background were obtained from Dr. Kermicle at the University of Wisconsin (Madison, WI); R-r, standard, R-st sr2, R-nj, r-g, r-g sr2, r-g Lc, r-del, and R-nj::Ac. For fine mapping of Isr, R-st Lc and unequal cross-over progeny, R-st Isr-st (ΔLc ΔIsr-lc), R-st (ΔIsr-st ΔLc) Isr-lc, (ΔR-st ΔIsr-st) Lc Isr-lc were also obtained from Dr. Kermicle. For Isr tagging, R-nj::Ac/r-g sr2 lines were developed by sequential back-crossing of R-nj::Ac to r-g sr2. Plants were bred at the Upland farm of Cold Spring Harbor Laboratory (Cold Spring Harbor, NY) and at the University Farm of Gyeongsang National University (Chinju, S. Korea).
Growth conditions
The phenotypes of mature sr2 plants were measured at the University farm of Gyeongsang National University. Daily high and low temperature of the growing site was typically at 30°C day and at 20°C night during the growing season. Seedlings were germinated on a commercial soil and were grown for 10 days in a growth chamber. The temperature in the growth chamber was constant at 25°C with an air circulation system attached. The light/dark cycle of the chamber was 12 hr each.
Leaf sample preparation for anatomical study and in situ hybridization
Leaves were harvested from ∼20 mature plants of each genotype that were about to shed pollen in the field. Whole mounts were prepared from 2-cm strips of leaf blade according to Evans and Poethig (1995). At 10 cm above the blade sheath, half leaves were transversely cut from midrib to margin. Samples were fixed in three parts ethanol and one part glacial acetic acid at room temperature for 24 hr, and cleared in 70% ethanol in the cold room until needed. Samples were stained overnight at room temperature in 1 part 0.05% toluidine blue (0.01 m sodium acetate at pH 4.4) to 15 parts H2O. Samples were destained at 4°C. Sequential photographs were taken of the adaxial surface from midrib to margin with an Axiophot photomicroscope (ZEISS, Germany). To obtain cross sections of leaf samples, a standard tissue-embedding method was used. Leaf samples were fixed in FAA solution for 1 day at room temperature. FAA (100 ml) consisted of 50 ml of EtOH, 10 ml of formalin (37%), and 5 ml of glacial acetic acid in 35 ml of distilled water. Samples were vacuum infiltrated in the fixative for 3–5 min until the tissue sank. For paraffin embedding, samples were subjected to EtOH dehydration. Dehydrated samples were transferred into xylene to replace EtOH in graded steps. After the final xylene step, several small pieces of solid paraffin were added to the xylene and stored at 42°C overnight. The step was repeated for 2 days until the paraffin no longer dissolved. Then, the uncapped vial was transferred to a 58°C oven. Melted paraffin was refilled into the vial twice a day for 3 days. On the hot side of the embedding tray, paraffin and samples were poured into paper boats and the specimen were arranged appropriately.
Using a standard rotary microtome, the specimen was cut at 8-μm thickness for in situ hybridization and at 12–14-μm thickness for anatomical study. Paraffin ribbons were fixed on polylysine-treated slides at 45°C overnight. After being deparaffinized and brought to water, slides were dipped into staining solution sections of 0.1% aqueous toluidine blue for 15 sec and washed with distilled water. Slides were brought to xylene after a graded EtOH series and coverslips were put on the semi-dried samples.
For in situ anti-sense RNA hybridization, 35S-labeled, single-stranded RNA probes were synthesized by in vitro transcription from a linearized Isr cDNA clone. Synthesized probes were alkali hydrolyzed for hybridization. Following published methods (Jackson 1992; Langdale 1993), leaf samples on slides were subjected to prehybridization treatments with proteinase K followed by acetic anhydride. After dehydration, samples were hybridized with 35S-labeled riboprobes. After hybridization at 45°C overnight, slides were sequentially washed four times in a solution of 5 mm DTT and 4× SSC at room temperature. Excess RNA was removed in RNase A (1 mg/ml) solution at 37°C for 30 min. Final washes were done twice at 57°C in 0.1× SSC. Dehydrated slides were coated with Kodak NTB-2 emulsion and were stored at 4°C. After development, slides were examined by dark- and light-field microscopy.
Preparation of genomic DNA and Southern blot analysis
Genomic DNA was prepared from mature leaves or seedlings using a urea extraction procedure (K. Cone, pers. comm.). Purified DNA (5 μg) was digested with appropriate restriction endonucleases, size-fractionated on a 0.8% agarose gel, and transferred onto a nylon membrane (GeneScreen Plus, Du Pont). Filters were hybridized with probes in hybridization buffer containing 0.5 m Na2HPO4, 7% SDS, 1 mm EDTA, and 1% BSA (Church and Gilbert 1984). Final washes of the filters were done in 0.2× SSC and 0.1% SDS solution for 30 min at 65°C.
Manipulation of RNA
For RNA preparation, whole seedlings above the ground were harvested and immediately ground in liquid nitrogen. Total cellular RNA was prepared by a guanidinium thiocyanate extraction procedure. RNA concentrations were measured by UV absorbance after DNase treatment. A total of 30 mg of RNA per lane were fractionated by electrophoresis on 1.3% formaldehyde agarose gels. RNA gel was transferred onto nylon membranes (GeneScreen Plus, Du Pont). Filters were incubated with probes radiolabeled with random primers in hybridization buffer containing 0.5 m Na2HPO4, 7% SDS, 1 mm EDTA, and 1% BSA. Final washes of the filters were done in 0.2× SSC and 0.1% SDS solution for 30 min at 65°C.
To determine 5′ termini of Isr mRNA, primer extensions were conducted, with slight modification, by a standard method. A 29-mer oligonucleotide that was complementary to the upstream sequence of Isr cDNA was end labeled and annealed at 58°C with 3 μg of poly(A)+ RNAs from normal seedlings of B73. As a control, 3 μg of poly(A)+ RNAs from r-del seedlings were added to the labeled primer. The annealed primers were extended with AMV reverse transcriptase (BRL) at 48°C. The primer extensions were electrophoresed on a 6% sequencing gel alongside of a sequencing reaction of an Isr genomic clone primed with the same oligonucleotide.
Cloning of Ac-tagged Isr genomic DNA and screening Isr genomic DNA and cDNA
Genomic DNA completely digested with BamHI was fractionated on 15%–35% glycerol gradient containing 10 mm Tris (pH 8.0), 0.2 m NaCl, and 2 mm EDTA. Fractions (4–10 kb) of DNA were collected and precipitated with ethanol. λEMBL4 arms precut with BamHI were ligated with purified DNA at ratios designed to obtain multiple insertions (Han et al. 1992). Plaques containing Isr genomic DNA were detected with Ac as a probe. Genomic DNA and cDNA libraries used in this study were obtained from S. Briggs (Pioneer Inc.) and A. Barkan (University of Oregon, Eugene). Both libraries were made from the maize inbred line, B73. Screening and subcloning of Isr genomic DNA and cDNA were performed by standard methods.
DNA sequencing
Sequential series of overlapping deletions from both ends were constructed using Erase-A-Base kit (Promega) and Mungbean nuclease according to the manufacture's manual. Nucleotide sequencing was performed by dideoxy chain-termination method with a Sequenase kit (US Biochemical).
Acknowledgments
We thank Lee Yeong Gee (Gyeongsang National University, South Korea) and Tim Mulligan (Cold Spring Harbor Laboratory, NY) for field assistance. C.-d. Han thanks Dong-jin Kang Han for helping with pollination and genetic records. We thank Mary Byrne for critical reading of the manuscript and for help with sequence analysis. This project is supported by the academic research fund of Ministry of Education in Republic of Korea, to C.-d. Han (971-0601-007-2 and 1998-019-D00131). Initial field work for this project were performed at Cold Spring Harbor Laboratory supported by grants from the Department of Energy (DE-FG02 91ER20047) and National Institutes of Health (R01 GM46851-02). We also acknowledge a grant from Korea Science and Engineering Foundation to PMBBRC (Plant Molecular Biology and Biotechnology Research Center).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL cdhan@nongae.gsnu.ac.kr; FAX 82 591 7599363.
References
- Birky CW. Relaxed cellular controls and organelle Heredity. Science. 1983;222:468–475. doi: 10.1126/science.6353578. [DOI] [PubMed] [Google Scholar]
- Brink RA. Paramutation. Annu Rev Genet. 1973;7:129–152. doi: 10.1146/annurev.ge.07.120173.001021. [DOI] [PubMed] [Google Scholar]
- Borner T, Sears BB. Plastome mutants. Plant Mol Biol Rep. 1986;4:69–92. [Google Scholar]
- Chatterjee M, Sparvoli S, Edmunds C, Garosi P, Findlay K, Martin C. DAG, a gene required for chloroplast differentiation and palisade development in Antirrhinum majus. EMBO J. 1996;15:4194–4207. [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe EH, Neuffer MG. Embryo cells and their destinies in the corn plants. In: Subtelny S, Sussex IM, editors. The clonal basis of development. New York, NY: Academic Press; 1978. pp. 113–129. [Google Scholar]
- Coe EH, Neuffer MG, Hoisington DA. The genetics of corn. In: Sprague GF, Dudley JW, editors. Corn and corn improvement. Madison, WI: ASA-CSSA-SSSA; 1988a. pp. 81–258. [Google Scholar]
- Coe EH, Thompson DG, Walbot V. Nuclear genes and chloroplast modifications in maize. Stadler Symp. 1982;14:29–46. [Google Scholar]
- Coe EH, Thompson D, Walbot V. Phenotypes mediated by the iojap genotype in maize. Am J Bot. 1988b;75:634–644. doi: 10.1002/j.1537-2197.1988.tb13486.x. [DOI] [PubMed] [Google Scholar]
- Danon A. Translational regulation in the chloroplast. Plant Physiol. 1997;115:1293–1298. doi: 10.1104/pp.115.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta S, Greenblatt I, Kermicle JL, Hicks JB, Wessler S. Molecular cloning of the maize R-nj allele by transposon-tagging with Ac. In: Gustafson JP, Appels R, editors. Chromosome structure and function: Impact of new concepts, 18th Stadler Genetics Symposium. New York, NY: Plenum Press; 1988. pp. 263–282. [Google Scholar]
- Eggleston WB, Alleman M, Kermicle JL. Molecular organization and germinal instability of R-stippled maize. Genetics. 1995;141:347–360. doi: 10.1093/genetics/141.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson RA, Beadle GW, Fraser AC. A summary of linkage studies in maize. Cornell Univ Agric Exp Sta Mem. 1935;180:3–83. [Google Scholar]
- Evans MMS, Poethig RS. Gibberellins promote vegetative phase change and reproductive maturity in maize. Plant Physiol. 1995;108:475–487. doi: 10.1104/pp.108.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. Light receptor kinases in plants. Curr Biol. 1999;9:123–126. doi: 10.1016/s0960-9822(99)80078-5. [DOI] [PubMed] [Google Scholar]
- Fisk DG, Walker MB, Barkan A. Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 1999;18:2621–2630. doi: 10.1093/emboj/18.9.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Lane B. The maize leaf. In: Freeling M, Walbot V, editors. The maize handbook. Berlin, Germany: Springer-Verlag; 1993. pp. 17–28. [Google Scholar]
- Galinier A, Kravanja M, Engelmann R, Hengstenberg W, Kilhoffer M-C, Deutscher J, Haiech J. New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc Natl Acad Sci. 1998;95:1823–1828. doi: 10.1073/pnas.95.4.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson JL, Tabita FR. Analysis of the cbbXYZ operon in Rhodobacter sphaeroides. J Bacteriol. 1997;179:663–669. doi: 10.1128/jb.179.3.663-669.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C-d, Coe EH, Martienssen RA. Molecular cloning and characterization of iojop (ij), a pattern striping gene of maize. EMBO J. 1992;11:4037–4046. doi: 10.1002/j.1460-2075.1992.tb05497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess WR, Borner T. Organellar RNA polymerases of higher plants. Int Rev Cytol. 1999;190:1–59. doi: 10.1016/s0074-7696(08)62145-2. [DOI] [PubMed] [Google Scholar]
- Jackson D. In situ hybridization in plants. In: Gurr SJ, Bowles DJ, McPherson MJ, editors. Molecular plant pathology: A practical approach. Vol. 1. Oxford, UK: IRC Press; 1992. pp. 163–174. [Google Scholar]
- Jenkins MT. Heritable characters of maize. XX. iojap-striping, a chlorophyll defect. J Hered. 1924;15:467–472. [Google Scholar]
- Joachim G, Burnham CR. Inheritance of Waseca stripe. Maize Genet Coop News Lett. 1953;27:66. [Google Scholar]
- Keddie JS, Carroll B, Jones JDG, Gruissem W. The DCL gene of tomato is required for chloroplast development and palisade cell morphogenesis in leaves. EMBO J. 1996;15:4208–4217. [PMC free article] [PubMed] [Google Scholar]
- Kermicle JL. In: Epigenetic mechanisms of gene regulation. Russo VEA, Martienssen RA, Riggs AD, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- Kermicle JL, Axtell JD. Modification of Chlorophyll striping by the R region. Maydica. 1981;XXVI:185–197. [Google Scholar]
- Kirk JTO, Tilney-Basset RAE. The plastids. Their chemistry, structure, growth and inheritance. 2nd edn. Amsterdam, The Netherlands: Elsevier North Holland; 1978. [Google Scholar]
- Langdale J. In situ hybridization. In: Freeling M, Walbot V, editors. The maize handbook. Berlin, Germany: Springer-Verlag; 1993. pp. 17–28. [Google Scholar]
- Martienssen R. Cell death: Fatal induction in plants. Curr Biol. 1997;7:534–537. doi: 10.1016/s0960-9822(06)00274-0. [DOI] [PubMed] [Google Scholar]
- Mazoti LB. Variaciones heredables de los plastidos produicas por genes nucleares. Revista Fac Agron Univ Nac La Planta. 1978;54:301–328. [Google Scholar]
- McConnell S, Stewart LC, Talin A, Yaffe MP. Temperature-sensitive yeast mutants defective in mitochondrial inheritance. J Cell Biol. 1990;111:967–976. doi: 10.1083/jcb.111.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel CN, Poethig RS. Cell lineage patterns of the shoot apical meristem in the germinating corn embryo. Planta. 1988;175:13–22. doi: 10.1007/BF00402877. [DOI] [PubMed] [Google Scholar]
- Monod J. “Recherches sur la croissance des cultures bacteriennes.” Ph.D. thesis. Paris, France: University of Paris; 1942. [Google Scholar]
- Nelson T, Dengler N. Leaf vascular pattern formation. Plant Cell. 1997;9:1121–1135. doi: 10.1105/tpc.9.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer MG, Coe EH, Wessler SR. Mutants of maize. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Poethig RS. Cellular parameters of leaf morphogenesis in maize and tobacco. In: White RA, Dickison WC, editors. Contemporary problems in plant anatomy. Orlando, FL: Academic Press; 1984. pp. 235–260. [Google Scholar]
- Poethig RS, Szymkowiak EJ. Clonal analysis of leaf development in maize. Maydica. 1995;40:67–76. [Google Scholar]
- Rhoades MM. Genic induction of an inherited cytoplasmic difference. Proc Natl Acad Sci. 1943;29:327–329. doi: 10.1073/pnas.29.11.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SH, Evert RF. Leaf vasculature in Zea mays L. Planta. 1985;164:448–458. doi: 10.1007/BF00395960. [DOI] [PubMed] [Google Scholar]
- Settles AM, Martienssen R. Old and new pathways of protein export in chloroplasts and bacterial. Trends Cell Biol. 1998;8:494–501. doi: 10.1016/s0962-8924(98)01387-7. [DOI] [PubMed] [Google Scholar]
- Sharman BC. Developmental anatomy of the shoot of Zea Mays L. Ann Bot. 1942;22:245–282. [Google Scholar]
- Steffensen DM. A reconstruction of cell development in the shoot apex of maize. Am J Bot. 1968;55:354–369. [Google Scholar]
- Stein OL, Steffensen D. Ratidation-induced genetic markers in the study of leaf growth in Zea. Am J Bot. 1959;46:485–489. [Google Scholar]
- Surpin M, Chory J. The co-ordination of nuclear and organellar genome expression in eukaryotic cells. Essays Biochem. 1997;32:113–125. [PubMed] [Google Scholar]
- Sylvester AW, Cande WZ, Freeling M. Division and differentiation during normal and liguless-1 maize leaf development. Development. 1990;110:985–1000. doi: 10.1242/dev.110.3.985. [DOI] [PubMed] [Google Scholar]
- Thompson D, Walbot V, Coe EH. Plastid development in iojap- and chloroplast mutator-affected maize plants. Am J Bot. 1983;70:940–950. [Google Scholar]
- Walbot V, Coe EH. Nuclear gene iojap conditions a programmed change to ribosome-less plastids in Zea mays. Proc Natl Acad Sci. 1979;76:2760–2764. doi: 10.1073/pnas.76.6.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EL, Robbins TP, Bureau TE, Kermicle J, Dellaporta SL. Transposon-mediated chromosomal rearrangements and gene duplications in the formation of the maize R-r complex. EMBO J. 1995;14:2350–2363. doi: 10.1002/j.1460-2075.1995.tb07230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G, Wickner W. Organelle inheritance. Cell. 1996;84:395–400. doi: 10.1016/s0092-8674(00)81284-2. [DOI] [PubMed] [Google Scholar]
- Williams E, Kermicle JL. Fine structure of plastids in maize leaves carrying the striate-2 gene. Protoplasma. 1974;79:401–408. [Google Scholar]
- Xu Z, Wickner W. Thioredoxin is required for vacuole inheritance in Saccharomyces cerevisiae. J Cell Biol. 1996;132:787–794. doi: 10.1083/jcb.132.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MP. Organelle inheritance in the cell cycle. Trends Cell Biol. 1991;1:160–164. doi: 10.1016/0962-8924(91)90017-4. [DOI] [PubMed] [Google Scholar]