Abstract
Selection of human cytomegalovirus variants in the presence of ganciclovir or foscarnet led to 18 DNA polymerase mutations, 14 of which had not been previously studied. Using bacterial artificial chromosome technology, each of these mutations was individually transferred into the genome of a reference strain. Following reconstitution of infectious viral stocks, each mutant was assessed for its drug susceptibility and growth kinetics in cell culture. Computer-assisted three-dimensional (3D) modeling of the polymerase was also used to position each of the mutations in one of four proposed structural domains and to predict their influence on structural stability of the protein. Among the 10 DNA polymerase mutations selected with ganciclovir, 7 (P488R, C539R, L545S, V787L, V812L, P829S, and L862F) were associated with ganciclovir resistance, whereas 2 (F595I and V946L) conferred only foscarnet resistance. Among the eight mutations selected with foscarnet, only two (T552N and S585A) conferred foscarnet resistance, whereas four (N408D, K500N, L802V, and L957F) had an impact on ganciclovir susceptibility. Surprisingly, the combination of mutations, some of which were not associated with resistance for a specific antiviral, resulted in increasing resistance effects. 3D modeling suggested that none of the mutated residues were directly involved in the polymerase catalytic site but rather had an influence on drug susceptibility by modifying the structural flexibility of the protein. Our study significantly adds to the number of DNA polymerase mutations conferring in vitro drug resistance and emphasizes the point that evaluation of individual mutations may not accurately reflect the phenotype conferred by multiple mutations.
INTRODUCTION
Human cytomegalovirus (HCMV) is an important opportunistic pathogen that causes serious morbidity and mortality in immunocompromised patients (28). Despite recent efforts to develop new anti-HCMV agents with different viral targets, ganciclovir (GCV) and its prodrug, valganciclovir (VGCV), remain the first-line agents for the prevention and treatment of HCMV systemic infections. Cidofovir (CDV) and foscarnet (FOS) are used as second-line agents due to significant toxicities and lack of oral formulations. Ultimately, all three antivirals target the HCMV DNA polymerase (Pol; pUL54) (28, 38). Because HCMV results in lifelong infections, immunosuppressed patients often require extended courses of antiviral agents. This clinical context could lead to the emergence of drug-resistant HCMV mutants that can be associated with treatment failures (8, 18, 27, 31, 34, 35, 50, 51, 54). To exert its antiviral activity, GCV must first be activated by the sequential addition of three phosphate groups to the molecule (3). In HCMV-infected cells, addition of the first phosphate group is carried out by the viral kinase UL97 protein (36, 52). In the clinic, GCV resistance usually arises first from UL97 mutations, resulting in decreased accumulation of the activated form of the antiviral, whereas subsequent UL54 mutations can confer high levels of GCV resistance and variable degrees of cross-resistance (38). CDV activation relies exclusively on cellular proteins (21), whereas FOS requires no activation steps. Thus, only mutations in the UL54 gene are expected to account for resistance to the latter antivirals.
Due to the time required to grow clinical isolates and perform standard susceptibility testing, phenotypic assessment of these isolates is often impractical and cannot be used to guide clinical decisions (38). To circumvent this problem, genotypic analyses and virtual phenotyping have become the standard in clinical practice (4, 7, 8, 11, 13, 14, 17, 40, 42). However, the use of virtual phenotypes requires a good knowledge of HCMV mutations leading to drug resistance and those occurring as a consequence of natural variations (11, 13, 14, 16, 17, 22, 38, 40, 41). In this view, bacterial artificial chromosome (BAC) technology has become the method of choice for the generation of recombinant HCMVs in order to study antiviral drug resistance (11, 12, 14, 17, 40–42). In addition, an effect between UL54 mutations and specific genetic backgrounds on antiviral resistance has been reported by a few groups (11, 15, 17, 43, 48).
Here we report on the impact of individual or multiple UL54 mutations that emerged during serial in vitro passages of a laboratory strain (29) and a clinical HCMV strain. Also, to provide insight into the mechanisms leading to drug resistance, computer-assisted three-dimensional (3D) modeling studies and structural enzyme flexibility analyses of the HCMV DNA polymerase were conducted.
MATERIALS AND METHODS
Cells and viruses.
Human foreskin fibroblasts (HFFs) were grown and maintained in Eagle's minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS). The reporter cell line U373/proUL54, derived from U373MG astrocytoma cells (29), was grown and maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS. Drug-resistant viruses selected from laboratory strain AD169 have been previously described (29). In addition, a set of sequential mutant viruses was obtained by serial passage of a pretherapy susceptible clinical isolate (5) in the presence of GCV as previously described (29). In brief, an aliquot of the pretherapy clinical isolate was passaged in the presence of increasing concentrations of GCV (1.5 to 1,000 μM). The GCV concentration was kept at the same level until good viral growth was noticed (usually one to three passages). Two plaque-purified viruses were isolated and characterized at 10, 25, 50, 300, and 1,000 μM of GCV. Plaque purification was accomplished in the presence of the corresponding concentration of GCV. Of note, further exposure to GCV was performed using viruses that had not been plaque purified.
DNA constructs.
The AD169 genome cloned as an infectious BAC (BAC-pHB5) was obtained from Martin Messerle (Max von Pettenkofer-Institut, Munich, Germany) (9). The Escherichia coli K-12 derivative BW25141 (recA1), along with plasmids pKD13, pKD46, and pCp20, was provided by Barry Wanner (Purdue University, West Lafayette, IN) (25). The EcoRI-StuI fragment from pGPol (22), corresponding to nucleotides (nt) −818 to 4949 relative to the start codon of AD169 UL54, was cloned into the SspI-EcoRI sites of pSP72 (Promega). The resulting plasmid, pSPolL, contained the entire HCMV UL54 coding sequence (nt 1 to 3726) with unique SspI (nt −109)-RsrII (nt −4) sites in the 5′ untranslated region. Plasmid pSPoldel was obtained by cloning the tetracycline resistance cassette (Tetr) from pBR322 (New England BioLabs) into the RsrII (nt −4)-BspEI (nt 3321) sites of pSPolL, therefore deleting the 5′ end along with the catalytic subdomain (nt 903 to 2964) of UL54. The transfer plasmid pSPolL-Kan was obtained by subcloning the BstBI fragment from pKD13 (25) and filling in the extremities to preserve the integrity of the FLP recombination target (FRT) sites into the RsrII site of pSPolL. The resulting transfer plasmid then contained the entire coding sequence of UL54 preceded (at position −4) by a selectable marker (kanamycin resistance) flanked by two FRT sites, allowing subsequent excision of the marker (25). HCMV UL54 mutations were first introduced into pSPolL using the QuikChange PCR-based site-directed mutagenesis kit (Stratagene). DNA fragments containing the desired mutations were then subcloned into the transfer plasmid pSPolL-Kan using different combinations of the various unique restriction sites found within the UL54 coding sequence. Double mutant plasmids were constructed by subcloning different mutated DNA fragments into pSPolL-Kan. The chloramphenicol resistance (Cmr) gene found in pCp20 (10, 25) was disrupted by collapsing of the NcoI and XhoI sites. The resulting plasmid, pCp20Ap, therefore harbored only ampicillin resistance (Ampr). Finally, the expression of the HCMV pp71 (pUL82) protein in eukaryotic cells has been shown to enhance the infectivity of HCMV DNA (2). Such a plasmid, pBK-CMV82, was obtained by cloning the PCR-amplified coding sequence of UL82 into the HindIII-BamHI sites of pBK-CMV (Stratagene).
Construction of recombinant pHB5 and reconstitution of recombinant viruses.
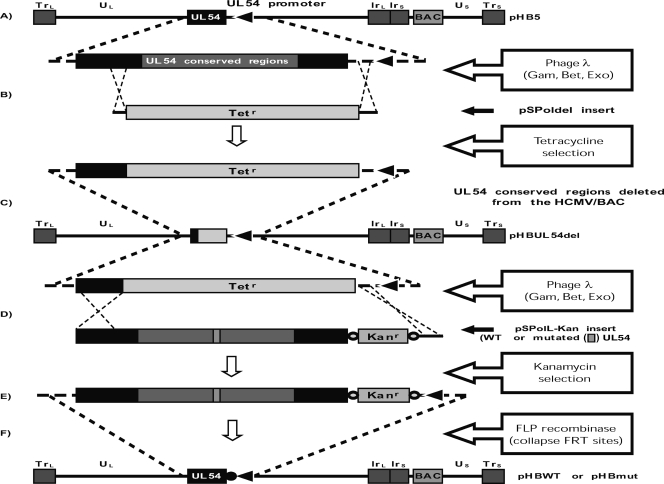
The detailed approach used to substitute UL54 alleles in pHB5 is illustrated in Fig. 1. In brief, BW25141 bacteria containing both pKD46 and the HCMV/BAC pHB5 were made electrocompetent in the presence of 1 mM l-arabinose to induce the expression of the phage λ Red recombinase elements (Gam, Bet, and Exo) from pKD46 (25). To ensure that subsequent recombination steps would lead to the introduction of the desired mutation into the HCMV/BAC sequence, a preliminary step consisting of the deletion of all UL54 conserved regions from pHB5 was performed. This was done by electroporation of the SspI-ScaI fragment of pSPoldel, consisting of Tetr flanked by regions homologous to the HCMV UL54 (at −109 to −5 and 3321 to 3565 nt) (Fig. 1B). Tetr clones were then identified, and proper integration of the Tetr gene and the absence of rearrangement within the HCMV/BAC sequence were verified by PCR and EcoRI digestion, respectively. The recombinant pHBUL54del BAC was then used in all subsequent recombination steps. SspI-ScaI (nt −109 to 3565 of the original UL54 plus the excisable Kanr inserted at position −4) DNA fragments excised from wild-type (WT) or mutated pSPolL-Kan transfer plasmids were used to reintroduce full-length UL54 genes into the sequence of pHBUL54del using the same approach (with Kanr selection instead of Tetr) (Fig. 1D). The Kanr gene was then excised from the resulting WT (pHBWT-Kan) or mutated (pHBmut-Kan) BACs by electroporation of pCp20Ap expressing the FLP recombinase that mediates site-specific homologous recombination between FRT sites flanking Kanr (Fig. 1F) (25). The resulting recombinant BACs then contained full-length WT (pHBWT) or mutated (pHBmut) UL54 genes preceded at position −4 by a short 31-nt residual FRT motif. Phenotypic characterization of bacterial clones was performed at every step of the process to identify clones in which the desired sequence of events had occurred. Mutations associated with natural polymorphisms (655L, 685S, 885T, and 898D) present in all viruses selected from the clinical strain were not included in recombinant viruses. For the purposes of PCR, sequencing, and restriction digestion analyses, BAC DNA was prepared by alkaline lysis, followed by acetate-isopropanol precipitation.
Fig. 1.
UL54 allele exchange within HCMV/BAC pHB5. (A) The AD169 genome cloned as a bacterial artificial chromosome is represented with a magnified view of the DNA polymerase (UL54) locus. (B) The pSPoldel insert was used to delete UL54 conserved regions from the HCMV/BAC sequence using phage λ Red recombinase-enhanced homologous recombination. The result is an HCMV/BAC (pHBUL54del) (C) in which nucleotides −4 to 3321 of the UL54 gene have been replaced by the Tetr gene. (D) Full-length UL54 genes, either WT or mutated (mut), are reintroduced into the HCMV/BAC sequence using pSPolL-Kan inserts. The latter consist of nucleotides −109 to 3565 from WT or mutated UL54 sequences with the insertion of a kanamycin resistance (Kanr) gene flanked by FRT sites (○). (E) These clones (pHBWT-Kan or pHBmut-Kan) contain full-length UL54 genes preceded at position −4 by Kanr and the two FRT sites. (F) Expression of the FLP recombinase catalyzes the site-specific recombination between FRT sites, therefore resulting in the excision of Kanr and leaving a short 31-nt residual FRT motif (●) at position −4. All steps were carried out in BW25141 (recA1) bacteria (25).
For the purpose of electroporation into mammalian cells, BAC DNA was prepared using Qiagen's plasmid maxikit (Qiagen, Toronto, Ontario, Canada) following the manufacturer's recommendations for very-low-copy-number plasmid/cosmid purification. HCMV/BAC DNAs were electroporated into subconfluent HFFs along with plasmid pBK-CMV82 as previously described (39). Viruses reconstituted from recombinant HCMV/BACs (RvHBWT or RvHBmut) were then allowed to grow until sufficient stocks could be frozen, usually after 14 to 21 days. Given the clonal nature of the DNA used for reconstitution of recombinant viruses, no additional plaque purification steps were performed. Viral DNA was extracted from infected HFFs. PCR primers BW1 and BW4 (1) were used to amplify a 4.28-kb (nt −343 to 3937) DNA fragment encompassing both regions of homology used in the recombination steps, the residual FRT motif, and the entire UL54 coding sequence. Both PCR primers, along with six other internal primers, were then used to sequence the entire DNA fragment. In addition, the catalytic domain of the HCMV UL97 gene, also involved in GCV resistance, was sequenced in selected recombinant viruses as previously described (4).
Susceptibility testing.
All plaque-purified variants derived from the pretherapy clinical isolate were assessed for their susceptibility profiles using a standardized plaque reduction assay (PRA) (33). The luciferase reduction assay (LRA) previously reported by our group (29) was used to assess drug susceptibilities of all recombinant viruses reported in this study. In brief, recombinant HCMVs were grown on confluent HFF monolayers in 96-well plates in the presence of serial dilutions of the antivirals. On day 5, aliquots of cell-free viruses found in culture supernatants were transferred onto freshly confluent U373/proUL54 cell monolayers that were further incubated for 48 h. In U373/proUL54 cells, HCMV replication triggers the expression of the firefly luciferase gene under the control of a specific HCMV promoter. After the cells were lysed using a luciferase-compatible lysis buffer, addition of luciferin resulted in photon emission that was measured using a standard luminometer. Emission of light (expressed in relative light units [RLU]) was then plotted against antiviral concentrations, and the concentrations that inhibit 50% of viral growth (50% effective concentrations [EC50s]) were calculated using best-fit curves (29). In its current format (i.e., transfer of cell-free viruses found in supernatants), the LRA is of limited use with clinical isolates that are typically associated with very low levels of production of cell-free viruses. We considered an EC50 ratio (concentration of the antiviral that inhibits expression of luciferase by 50% compared to that by the WT recombinant virus) of ≥1.7, with confirmation by statistical analysis (i.e., no overlap between EC50 95% confidence intervals of mutant and WT viruses), to represent a significant change in drug susceptibility.
Viral growth kinetics analyses.
Viral growth kinetics of each recombinant virus were evaluated using real-time PCR quantification of cell-free virus collected at different time points following infection with a standardized inoculum. In brief, cell-free viruses were inoculated onto HFFs in 12-well plates to obtain 120 to 150 PFU per well. Supernatants were collected on a daily basis, centrifuged at low speed to clear cellular debris (850 × g, 5 min at 4°C), and then frozen until DNA was extracted using the MagNA Pure LC total nucleic acid isolation kit (Roche Applied Science). A portion of the glycoprotein B gene (gpB) was amplified using PCR primers gpBup (TCACCCTATCGCGTGTGTTC) and gpBlo (ATAGGAGGCGCCACGTATTC). Internal probes used for melting curve analyses were CMVgB-fluo (TTAAAGGTGTGCGCCACGATGTTGCG-fluorescein) and CMVgB-red (LCRed640-TTGTAGACCACCATGATGCCCT-phosphate). Each 20-μl reaction mixture contained 5 μl of sample DNA, 5% dimethyl sulfoxide (DMSO), 1 mM MgCl2, 400 nM each PCR primer, 300 nM CMVgB-fluo, 600 nM CMVgB-red, and 2 μl of LC FastStart DNA master hybridization probes mix (Roche Applied Science). Cycling conditions were as follows: initial denaturation (95°C, 10 min), followed by 50 cycles of DNA amplification (95°C, 15 s; 56°C, 10 s; 72°C, 15 s) and then final melting curve (95°C, 0 s; 48°C, 30 s; 0.10°C/s up to 95°C with continuous fluorescence assessment). Each PCR run included the appropriate negative controls as well as an internal standard control (1,000 copies of plasmid HCMV DNA) in order to fit the results into a preestablished standard curve (101 to 109 copies of plasmid DNA). Using these conditions, 10 copies of the plasmid CMV DNA could be systematically detected, and the intra- and interassay variabilities were 15.8% and 17.7%, respectively, for 102 copies as well as 5.6% and 13.9%, respectively, for 105 copies of plasmid DNA. Results were reported as the number of HCMV genome copies per ml of supernatant. To minimize interassay variability, all viruses were evaluated in a single experiment. To confirm the initial inoculum, parallel 12-well plates were inoculated with the same virus dilution and overlaid with a semisolid medium consisting of MEM supplemented with 5% FBS and 0.5% agarose (all final concentrations). After 10 days of incubation, the plates were stained with crystal violet and the number of viral plaques was determined under a light microscope.
3D modeling and conformation analyses.
Our previous 3D model of the HCMV DNA polymerase in editing mode (49) was refined using the crystal structure of the herpes simplex virus type 1 (HSV-1) DNA polymerase (37). Briefly, conserved amino acid side chains in our model were placed in the same orientation as their analogues in the HSV-1 DNA polymerase structure to preserve nonbonded interactions. To evaluate the flexibility of the WT enzyme and the predicted influence of the various mutations, a translation-libration-screw (TLS) analysis was performed using the TLSMD program (44). The atomic temperature factors from the HSV-1 DNA polymerase structure (37) were included in our homology model for homologue residues to generate our working HCMV DNA polymerase model for TLS simulation. The proposed impact of various mutations on protein stability was evaluated using the program CUPSAT, which predicts the difference in the free energy of unfolding (ΔΔG) between a WT protein and its mutant (45). A change in protein stability measured from computed ΔΔG indicates a stabilizing mutation in case of a positive value and a destabilizing mutation for a negative value.
RESULTS
Sequential emergence of UL54 mutations following antiviral selection.
The emergence of UL54 mutations L545S, V812L, P829S, and D879G after selection with GCV and of UL54 mutations N408D, K500N, T552N, S585A, N757K, L802V, L926V, and L957F following exposure to FOS has been previously described (29). The sequential emergence of UL54 and UL97 mutations from a pretherapy clinical strain following exposure to increasing concentrations of GCV is presented in Table 1, along with drug susceptibilities of selected plaque-purified viruses. Initially, two different UL54 mutants (with P488R and C539R mutations) were detected. Eventually, however, one of the mutants (the one with the C539R mutation) overgrew the other one and was detected in all subsequent plaque-purified viruses. A total of two UL97 mutations (H520Q and G598D) and six UL54 mutations (P488R, C539R, F595I, V787L, L862F, and V946L) were observed in different plaque-purified viruses.
Table 1.
Genotypic characterization of plaque-purified variants selected with GCV from a pretherapy clinical isolate
| Varianta | Genotype |
EC50, in μM (ratio)c |
|||
|---|---|---|---|---|---|
| UL97 | UL54b | GCV | CDV | FOS | |
| Clinical isolate | WT | Baseline | 1.9 | 0.53 | 21.8 |
| 10 GCV-1 A | WT | P488R | 14.3 (7.5) | 3.9 (7.4) | 21.3 (1.0) |
| 10 GCV-1 B | WT | C539R | 12.0 (6.3) | NAd | NA |
| 25 GCV-1 A | H520Q | C539R, F595I | 113 (59) | 3.8 (7.4) | NA |
| 25 GCV-1 B | G598D | C539R, F595I | 52.5 (28) | NA | NA |
| 50 GCV-1 A, Be | H520Q | C539R, F595I | 150 (79) | 6.9 (13) | 137 (6.3) |
| 300 GCV-1 A | H520Q | C539R, F595I | NA | NA | NA |
| 300 GCV-1 B | H520Q | C539R | NA | NA | NA |
| 1,000 GCV-1 A | H520Q | C539R, V787L, L862F, V946L | NA | NA | NA |
| 1,000 GCV-1 B | H520Q | C539R, V787L, V946L | NA | NA | NA |
Numbers indicate GCV concentration at which at least two different clones (A and B) were plaque purified.
Only variations from the baseline UL54 sequence are shown.
EC50 values were obtained using a standardized PRA (33). Values in parentheses are EC50 ratios, which express the EC50 for the plaque-purified variant/EC50 for the pretherapy clinical isolate.
NA, not available. The unpurified virus obtained at 1,000 μM had a GCV EC50 value of 512 μM.
The two different clones analyzed (A and B) had identical genotypes.
Reconstitution of recombinant HCMVs based on UL54 allele exchange in pHB5.
All reported UL54 mutations (29; present study) were individually introduced into the genome of recombinant viruses. Mutation L802M, previously studied by other groups (18, 22), was also introduced as a comparison for mutation L802V. Five double mutants were also generated. In all cases, the structural integrity of the recombinant BACs was confirmed by EcoRI digestion prior to reconstitution of live viral stocks in HFFs (data not shown). Sequence analyses of the BW1-BW4 PCR-amplified DNA fragment (nt −343 to 3937 relative to the start codon of UL54, including regions of homology used in homologous recombination steps) from all recombinant viruses confirmed the presence of the 31-nt residual FRT motif at position −4, the presence of the expected UL54 mutations, and the absence of unintended UL54 mutations. In addition, the catalytic portion of UL97 was sequenced for all recombinant viruses harboring an EC50 ratio of ≥1.5 for GCV. No UL97 mutations could be detected from our recombinant viruses.
Susceptibility profiles of recombinant HCMVs.
All recombinant viruses generated in the present study were assessed for their susceptibility to GCV, CDV, and FOS using the LRA (Table 2). Of the 10 UL54 mutations selected with GCV, 7 were found to induce a significant decrease in GCV susceptibility, whereas 2 other mutations were associated with in vitro FOS resistance only (F595I and V946L). Among the eight UL54 mutations selected with FOS, only two (T552N and S585A) were associated with in vitro FOS resistance. Four of the other six mutations were either associated with GCV and CDV in vitro resistance (N408D and K500N) or GCV in vitro resistance alone (L802V and L957F). The combination of mutations that we studied showed various effects on drug susceptibility profiles. Most notably, the combination of mutations not individually affecting FOS susceptibility (L545S/P829S or N408D/L957F) resulted in some level of in vitro resistance to this antiviral. Mutation L957F was also apparently able to modulate the level of FOS susceptibility conferred by mutation T552N alone (Table 2).
Table 2.
Phenotypic characterization of recombinant HCMV pUL54 mutants and predicted enzyme structural changes
| Strain or recombinant mutant (selectiona) | UL54 conserved region | GCV |
CDV |
FOS |
Protein stability analyses |
||||
|---|---|---|---|---|---|---|---|---|---|
| EC50b (μM) | Ratioc | EC50 (μM) | Ratio | EC50 (μM) | Ratio | Predicted ΔΔGd (kcal/mol) | Domain involved | ||
| AD169 | 0.91 (0.72–1.10) | 1.1 | 0.099 (0.079–0.119) | 1.1 | 21.6 (15.1–28.1) | 0.9 | |||
| RvHB5e | 0.81 (0.56–1.07) | 1.0 | 0.085 (0.081–0.088) | 0.9 | 17.8 (10.9–24.6) | 0.7 | |||
| RvHBWTf | 0.82 (0.60–1.04) | 0.094 (0.076–0.112) | 24.3 (16.6–32.1) | ||||||
| Mutations selected with GCV-1 (from a clinical strain) | |||||||||
| 488R (10) | IV-δC | 2.84 (2.45–3.23) | 3.5 | 0.739 (0.622–0.857) | 7.9 | 15.2 (9.8–20.5) | 0.6 | 3.24 (S) | Exo |
| 539R (10) | δC (Exo III) | 2.60 (2.08–3.12) | 3.2 | 1.247 (1.037–1.456) | 13.3 | 15.9 (10.1–21.7) | 0.7 | −3.75 (D) | Exo |
| 595I (25) | δC-II | 1.10 (0.73–1.47) | 1.3 | 0.109 (0.082–0.137) | 1.2 | 47.8 (34.2–61.4) | 2.0 | 5.05 (S) | N terminal |
| 787L (1,000) | VI | 1.50 (1.14–1.86) | 1.8 | 0.082 (0.049–0.115) | 0.9 | 58.3 (35.3–81.3) | 2.4 | 2.06 (S) | Pol |
| 862F (1,000) | III-I | 1.40 (1.14–1.65) | 1.7 | 0.080 (0.057–0.104) | 0.9 | 27.2 (23.5–30.9) | 1.1 | NIg | NI |
| 946L (1,000) | I-VII | 0.93 (0.60–1.26) | 1.1 | 0.086 (0.053–0.118) | 0.9 | 57.9 (34.1–81.7) | 2.4 | 2.49 (S) | Pol |
| Mutations selected with GCV-2 (from laboratory strain AD169) | |||||||||
| 545S (10) | δC (Exo III) | 4.13 (3.02–5.24) | 5.0 | 1.113 (0.917–1.308) | 11.9 | 24.3 (16.6–32.0) | 1.0 | 1.5 (S) | Exo |
| 812L (1,000) | III | 2.14 (1.39–2.89) | 2.6 | 0.190 (0.162–0.217) | 2.0 | 20.6 (19.6–21.5) | 0.8 | 0.51 (S) | Pol |
| 829S (300) | III | 1.68 (1.20–2.16) | 2.0 | 0.148 (0.118–0.177) | 1.6 | 27.2 (21.4–33.0) | 1.1 | 1.25 (S) | Pol |
| 879G (1,000) | III-I | 1.02 (0.64–1.39) | 1.2 | 0.086 (0.061–0.110) | 0.9 | 21.7 (16.9–26.5) | 0.9 | NI | NI |
| 545S + 829S | 5.98 (2.82–9.14) | 7.3 | 0.424 (0.350–0.498) | 4.5 | 44.7 (33.4–56.0) | 1.8 | |||
| Mutations selected with FOS-1 (from laboratory strain AD169) | |||||||||
| 408D (800) | IV (Exo II) | 3.33 (2.46–4.19) | 4.0 | 0.392 (0.342–0.442) | 4.2 | 31.5 (18.7–44.2) | 1.3 | 2.31 (S) | Exo |
| 552N (300) | δC | 1.53 (1.14–1.92) | 1.9 | 0.110 (0.072–0.149) | 1.2 | 63.0 (35.2–90.9) | 2.6 | −0.17 (D) | Exo |
| 957F (3,000) | I-VII | 2.20 (1.33–3.06) | 2.7 | 0.128 (0.089–0.166) | 1.4 | 31.8 (20.5–43.1) | 1.3 | −0.2 (D) | Pol |
| 408D + 552N | 3.45 (2.55–4.36) | 4.2 | 0.181 (0.120–0.243) | 1.9 | 97.3 (60.4–134.2) | 4.0 | |||
| 408D + 957F | 4.05 (2.47–5.62) | 4.9 | 0.420 (0.318–0.522) | 4.5 | 81.5 (79.2–83.7) | 3.3 | |||
| 552N + 957F | 1.30 (1.04–1.56) | 1.6 | 0.088 (0.050–0.125) | 0.9 | 134.1 (97.8–170) | 5.5 | |||
| Mutations selected with FOS-2 (from laboratory strain AD169) | |||||||||
| 500N (300) | δC | 2.61 (2.32–2.90) | 3.2 | 0.284 (0.227–0.341) | 3.0 | 29.3 (24.1–34.4) | 1.2 | −0.5 (D) | Exo |
| 585A (300) | δC | 1.24 (0.83–1.66) | 1.5 | 0.133 (0.115–0.150) | 1.4 | 65.1 (50.0–80.3) | 2.7 | −1.03 (D) | Exo |
| 757K (3,000) | II-VI | 0.92 (0.57–1.26) | 1.3 | 0.076 (0.048–0.103) | 0.8 | 32.5 (18.9–46.0) | 1.3 | 1.11 (S) | Pol |
| 802 Mh | VI-III | 1.39 (1.14–1.64) | 1.7 | 0.090 (0.070–0.110) | 1.0 | 65.7 (45.2–86.2) | 2.7 | 0.02 (S) | Pol |
| 802V (800) | VI-III | 1.46 (1.15–1.76) | 1.8 | 0.105 (0.087–0.124) | 1.1 | 22.8 (15.4–30.2) | 0.9 | −0.7 (D) | Pol |
| 926V (3,000) | I-VII | 1.11 (0.94–1.28) | 1.3 | 0.107 (0.066–0.147) | 1.1 | 22.7 (13.9–31.5) | 0.9 | 1.49 (S) | Pol |
| 500N + 585A | 1.95 (1.34–2.56) | 2.4 | 0.227 (0.165–0.288) | 2.4 | 66.5 (44.2–88.8) | 2.7 | |||
Selection refers to the concentration (in μM) of the selection agent at which the mutation was first detected.
EC50s were determined using the LRA previously described (29). Results represent mean values of at least 3 independent experiments. Values in parentheses represent 95% confidence intervals.
Ratios express the EC50 for the recombinant mutant/EC50 for the wild-type recombinant. Ratios indicated in boldface type indicate a significant change (i.e., ratio of at least 1.7 combined with statistical significance).
The predicted effect on domain stability was evaluated using the program CUPSAT (45). Effects on structure are indicated in parentheses, with mutations classified as having either a stabilizing (S) or a destabilizing (D) effect on the domain structure (see Materials and Methods).
RvHB5 is the virus reconstituted from the original (unmodified) HCMV/BAC pHB5 (9).
RvHBWT is the wild-type virus reconstituted using our system (i.e., with the residual FRT motif).
NI, not included in the 3D model.
Mutation L802M was included as a comparison for mutation L802V identified in our studies.
Viral growth kinetics.
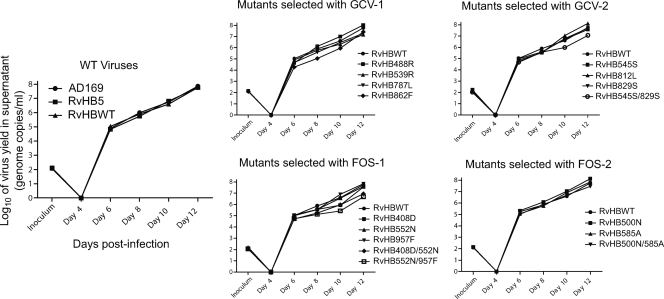
All of our recombinant viruses were assessed for their growth properties over multiple cycles of replication using a low multiplicity of infection (Fig. 2). In the first panel, the growth of the reference laboratory strain AD169 could not be distinguished from the growth curves generated using viruses reconstituted from either the original HCMV/BAC pHB5 strain (9) or our recombinant WT BAC containing the residual FRT motif (pHBWT). Among single mutations selected with either GCV or FOS, mutation L862F was associated with the greatest decrease in virus yield (between 0.3 and 0.9 log10 from day 6 to 12 compared to WT), whereas other mutations had no or only a minor impact on virus yield production. However, some of the double mutants (L545S/P829S, N408D/T552N, and T552N/L957F) consistently showed a decreased virus yield (0.3 to 1.2 log10 from day 6 to 12 compared to WT), whereas others (N408D/L957F and K500N/S585A) remained relatively unaffected.
Fig. 2.
Growth curves of selected recombinant mutants. Values presented for the inoculum were determined using a standard plaque assay. Values presented are the means of two PCR-determined titers of virus yield in supernatant. Mutants not represented, including the double mutant N408D/L957F, had growth curves similar to the one presented for the WT recombinant virus (RvHBWT).
3D modeling and conformation analyses.
Our previous models of the HCMV DNA polymerase contained 602 residues (open conformation) and 604 residues (closed conformation) and were based primarily on the bacteriophage RB69 DNA polymerase (another member of the polymerase α family to which the HCMV DNA polymerase also belongs) crystal structure, with which pUL54 shares 26% residue identity (49). Refinement of our model on the basis of the HSV-1 DNA polymerase crystal structure (37) led to a 826-residue model (residues 27 to 339, 362 to 442, 451 to 456, 474 to 599, 699 to 857, 860 to 861, 881 to 898, 903 to 1061, 1091 to 1104, and 1170 to 1228) in the open conformation (the model is available upon request). The residues' identity between the HSV-1 and HCMV DNA polymerases is 36.4% (46.1% similarity), whereas it is 42.5% (53.1% similarity) when only conserved regions (residues 295 to 988 of the HCMV DNA polymerase) are considered. The TLS determination generated a backbone displacement model with four TLS groups, including the following residues: group 1, 27 to 163; group 2, 164 to 312; group 3, 313 to 568; and group 4, 569 to 1092 (Fig. 3A). These groups correspond to the 4 regions of the DNA polymerase, namely, group 1 corresponds to the N-terminal region, group 2 to the NH2-domain, group 3 to the polymerase domains, and group 4 to the exonuclease domain. This suggests that these domains have different states of flexibility moving independently one from another, as illustrated in Fig. 3A.
Fig. 3.
Structural analysis of HCMV DNA polymerase. (A) The proposed backbone displacement of the homology 3D structure of the HCMV DNA polymerase is presented along with the DNA template. The four colors represent the four TLS groups moving independently from one another during TLS simulation (blue, residues 27 to 163; green, residues 164 to 312; red, residues 313 to 568; magenta, residues 569 to 1227). The width of the strand indicates the predicted amplitude of movement. The proposed functional assignments to these groups are indicated. (B) Model of the HCMV polymerase in editing mode with GCV, primer, and template DNA. The predicted catalytic site is also indicated. The positions of the mutations selected with GCV from the clinical isolate are indicated in blue, whereas those selected from AD169 are in yellow. The backbone color pattern is the same as for panel A. (C) Replicative model of HCMV polymerase with template and primer DNA and dTTPs. Locations of the mutations selected with FOS are indicated in either red or blue to discriminate between the two patterns of emergence that were observed. The backbone color pattern is the same as for panel A.
Predicted impact of HCMV DNA polymerase mutations on structure stability.
Single-residue mutations can cause a reduction in hydrophobic area, overpacking, or loss of electrostatic interactions and thus lead to changes in protein stability. The proposed impact of individual mutations on the stability of the UL54 protein domains is presented in Table 2. The positions of the residues involved relative to the putative active site in either the editing (mutations selected with GCV) or replicative (mutations selected with FOS) mode are presented in Fig. 3B and C, respectively. None of the mutations are located directly in the putative enzyme catalytic site. However, some of the interactions, including local modifications in hydrophobicity, steric clashes, and the formation of new hydrogen bonds, could be proposed using our model. The mutations appear to have various effects on the structure, either stabilizing or destabilizing it. Interestingly, there is a correlation between the location of mutations conferring drug resistance and its proposed effect on the protein structure. In fact, most (7 out of 9) variations appearing in the polymerase domain (residues 569 to 1227) stabilize the protein structure, whereas mutations appearing in other domains have either stabilizing or destabilizing effects.
DISCUSSION
In the present report, we describe recombinant phenotyping for UL54 mutations that emerged following in vitro exposure to either GCV or FOS. Eleven of these mutations represent new additions to the growing number of UL54 mutations with confirmed involvement in in vitro drug resistance (38). To provide an insight into the mechanisms by which those mutations could affect drug susceptibility, computer-assisted 3D modeling of the HCMV pUL54 and structural effects analyses were also performed.
In addition to the 12 UL54 mutations described in our previous work (29), 6 additional UL54 mutations were selected from a clinical strain following serial passage in the presence of GCV. In contrast to what was observed in mutants derived from the laboratory strain AD169, multiple variants were selected from the clinical strain for a given drug concentration. First, two distinct UL54 mutants were initially selected. Both harbored comparable levels of GCV resistance, but only one of those could be detected after further exposure to GCV (Table 1). Once again, the sequential emergence of multiple UL54 mutations led not only to high-level GCV resistance but also to cross-resistance to CDV and FOS.
The BAC technology has recently emerged as a powerful tool to study the involvement of individual HCMV mutations in drug resistance (11, 12, 14, 17, 40–42). Due to the high number of UL54 mutations to be tested, we opted for a highly efficient selectable marker that can be excised from the viral genome, leaving a short 31-nt residual FRT motif in the 5′ untranslated region of UL54. The presence of this short residual FRT motif did not have any apparent effects on viral drug susceptibility profiles (Table 2) or viral growth kinetics (Fig. 2). The lack of interference of such a short residual FRT motif has also been documented in other studies for UL97 (14, 40) and UL54 (17). Of the 19 individual UL54 mutations studied using our system, 5 (N408D [22], L545S [22], V787L [54], V812L [23], and L802M [18, 22]) were previously studied using marker transfer experiments. In those cases, our results show an excellent correlation with previous results. The only significant difference was seen with mutation V812L, which did not confer FOS resistance, as reported by others (23). Overall, we have confirmed the role of seven new mutations associated with GCV resistance (P488R, K500N, C539R, L802V, P829S, L862F, and L957F) and four new mutations associated with FOS resistance (T552N, S585A, F595I, and V946L).
Surprisingly, however, some of the mutations selected during our study did not confer resistance to the corresponding antiviral but, rather, seemed to confer in vitro resistance to another antiviral (N408D, K500N, F595I, L802V, V946L, and L957F) or had no apparent effects on drug resistance (N757K, D879G, and L926V). Any of these variations could represent UL54 modifications that favor the subsequent emergence of mutations conferring resistance to the corresponding antiviral or might act in synergy with other UL54 mutations. Similarly, the stepwise emergence of mutations in the HIV-1 protease has been well documented (24, 53). The importance of the genetic background has recently been outlined in a few studies in which three mutations identified in clinical specimens (G698D, Y818C, and L845P) could not be transferred in a reference strain or conferred a severe replication defect (17, 20). In the case of mutations G698D and Y818C, other UL54 mutations, not themselves associated with drug resistance, were also present in the clinical specimens (17). In another study, a set of six UL54 mutations not proposed to be involved in drug resistance not only was associated with partial compensation of the fitness loss conferred by mutation D301N but also significantly influenced its susceptibility to all three antivirals (15). Finally, even though the combination of UL54 mutations was initially reported to have rather additive effects (22), synergism between known resistance mutations was also reported to impact CDV (11, 23) and FOS (11) resistance in vitro.
Some of the double mutants that we have generated also support those findings. Indeed, two of our double mutants (with L545S/P829S and N408D/L957F mutations) exhibited resistance to FOS, while none of the single mutations were able to confer such a phenotype (Table 2). In addition, mutation L957F was also apparently able to modulate the FOS resistance level conferred by mutation T552N alone. CDV susceptibility was also modulated in some of our double mutants. Due to the large number of mutations presented here, not all possible double mutants were generated. We therefore cannot exclude the possibility that some other combinations might have similar modulating effects that would explain their selection with either GCV or FOS in cell culture.
Results from our past (49) and present studies strongly suggest that structural and flexibility changes in the DNA polymerase, rather than direct involvement of the catalytic sites, are responsible for the drug resistance phenotypes conferred by specific UL54 mutations. It is therefore plausible that the combined effects of UL54 mutations on the protein may provide structural changes that are compatible with drug resistance. 3D modeling and predicted structural changes have proven to be precious tools in understanding HIV-1 resistance to antiretroviral agents (46). Structural flexibility analyses of our HCMV model suggested four TLS groups with different states of flexibility, corresponding to an exonuclease domain, a polymerase/C-terminal domain, an NH2-domain, and the N-terminal region. In the present study, four of the six mutations conferring FOS resistance were located in either the polymerase or N-terminal domain and had a stabilizing effect on structure flexibility, whereas five of the six mutations associated with both GCV and CDV resistance were located in the exonuclease domain and exhibited either stabilizing or destabilizing effects on structure stability (Table 2).
Another important observation from our model is that, whether GCV or FOS was used in the selection process, initial UL54 mutations to emerge were located within the proposed exonuclease domain of the protein (Table 2). In one recent study, a recombinant virus with UL54 mutation D413A located in motif Exo II exhibited GCV and CDV resistance but remained susceptible to FOS (19). However, the presence of the mutation was associated with the accelerated development of resistance to maribavir, which is not a polymerase inhibitor. Moreover, after a limited number of passages in the absence of selective pressure, the plating efficiency of the unpurified D413A recombinant virus under 400 μM FOS was about 30-fold that of the parental strain passaged under the same conditions (19). Even though the putative FOS-resistant plaque-purified clones were not further characterized, those results strongly suggest that at least some mutations in the exonuclease domain could facilitate the subsequent development of other mutations associated with in vitro drug resistance. This phenomenon has been well studied in HSV-1 DNA polymerase mutants with mutations involving critical Exo motif residues (30, 32). In our study, three mutations (N408D, C539R, and L545S) at strictly conserved residues within motifs Exo II and III (29) emerged early in the selection process and might have facilitated the subsequent emergence of other UL54 mutations. Alternatively, these mutations could have also influenced the protein structure, allowing the emergence of additional mutations.
Alterations in the HCMV pUL54 sequence have been reported to impair the replicative capacities of some mutants (1, 11, 15, 17, 19, 20, 23, 26, 51). In our study, no severe replication impairment was noted for the single mutants. Only mutation L862F was associated with a moderate decrease in viral yield production. However, some double UL54 mutants were associated with reduced replicative capacities (≤1.2 log10) compared to the single mutants or the WT recombinant virus (Fig. 2).
The UL54 mutations described in our study were selected in vitro, so their potential clinical significance remains to be elucidated. However, some UL54 mutations described in our study, such as L545S and V812L, were also identified in the clinical setting (38). Mutations N408D and V787L were further identified in clinical specimens from patients failing antiviral therapy (38). By comparison with EC50 ratios published by other groups for previously well-studied mutations (38), EC50 ratios obtained with our LRA tend to be slightly lower. Nevertheless, UL97 mutations associated with EC50 ratios as low as 1.9 have been described in the clinical setting and can result in clinical or virological failure (38, 47). On the other hand, it must be emphasized that in vitro resistance does not always correlate with clinical failure. For example, our own studies (6, 7) showed that even well-established GCV-resistant UL97 mutants were sometimes associated with asymptomatic infections.
In conclusion, we have demonstrated the involvement of 11 new UL54 mutations associated with in vitro drug resistance. More importantly, our results indicate that virtual phenotypes based solely on the sum of EC50 values conferred by individual mutations might not always accurately reflect the susceptibility profiles of viruses with multiple UL54 mutations.
ACKNOWLEDGMENTS
This study was supported by research grants from the Canadian Institutes of Health Research (CIHR; MOP-62794) to Guy Boivin. Guy Boivin holds a Canada Research Chair on Emerging Viruses and Antiviral Resistance. Christian Gilbert received Ph.D. scholarships from Le Fond de Recherche en Santé du Québec and the CIHR.
We thank Tomas Cihlar (Gilead Science) for many useful discussions at multiple steps of this project. We also thank Martin Messerle (Max von Pettenkofer-Institut, Munich, Germany) and Barry Wanner (Purdue University, West Lafayette, IN), who kindly provided important materials for our project.
Footnotes
Published ahead of print on 27 June 2011.
REFERENCES
- 1. Baldanti F., et al. 1996. Single amino acid changes in the DNA polymerase confer foscarnet resistance and slow-growth phenotype, while mutations in the UL97-encoded phosphotransferase confer ganciclovir resistance in three double-resistant human cytomegalovirus strains recovered from patients with AIDS. J. Virol. 70:1390–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baldick C. J., Jr., Marchini A., Patterson C. E., Shenk T. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Biron K. K., et al. 1985. Metabolic activation of the nucleoside analog 9-[(2-hydroxy-1-(hydroxymethyl)ethoxy]methyl)guanine in human diploid fibroblasts infected with human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 82:2473–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boivin G., et al. 2001. Rate of emergence of cytomegalovirus (CMV) mutations in leukocytes of patients with AIDS who are receiving valganciclovir as induction and maintenance therapy for CMV retinitis. J. Infect. Dis. 184:1598–1602 [DOI] [PubMed] [Google Scholar]
- 5. Boivin G., et al. 1997. A case of ganciclovir-resistant cytomegalovirus (CMV) retinitis in a patient with AIDS: longitudinal molecular analysis of the CMV viral load and viral mutations in blood compartments. AIDS 11:867–873 [DOI] [PubMed] [Google Scholar]
- 6. Boivin G., Goyette N., Gilbert C., Humar A., Covington E. 2005. Clinical impact of ganciclovir-resistant cytomegalovirus infections in solid organ transplant patients. Transpl. Infect. Dis. 7:166–170 [DOI] [PubMed] [Google Scholar]
- 7. Boivin G., et al. 2004. Absence of cytomegalovirus-resistance mutations after valganciclovir prophylaxis, in a prospective multicenter study of solid-organ transplant recipients. J. Infect. Dis. 189:1615–1618 [DOI] [PubMed] [Google Scholar]
- 8. Boivin G., et al. 2009. Cytomegalovirus resistance in solid organ transplant recipients treated with intravenous ganciclovir or oral valganciclovir. Antivir. Ther. 14:697–704 [PubMed] [Google Scholar]
- 9. Borst E. M., Hahn G., Koszinowski U. H., Messerle M. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320–8329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cherepanov P. P., Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 11. Chevillotte M., Ersing I., Mertens T., von Einem J. 2010. Differentiation between polymorphisms and resistance-associated mutations in human cytomegalovirus DNA polymerase. Antimicrob. Agents Chemother. 54:5004–5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chevillotte M., Schubert A., Mertens T., von Einem J. 2009. Fluorescence-based assay for phenotypic characterization of human cytomegalovirus polymerase mutations regarding drug susceptibility and viral replicative fitness. Antimicrob. Agents Chemother. 53:3752–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chevillotte M., et al. 2010. A new tool linking human cytomegalovirus drug resistance mutations to resistance phenotypes. Antiviral Res. 85:318–327 [DOI] [PubMed] [Google Scholar]
- 14. Chou S. 2010. Recombinant phenotyping of cytomegalovirus UL97 kinase sequence variants for ganciclovir resistance. Antimicrob. Agents Chemother. 54:2371–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chou S., Lurain N. S., Thompson K. D., Miner R. C., Drew W. L. 2003. Viral DNA polymerase mutations associated with drug resistance in human cytomegalovirus. J. Infect. Dis. 188:32–39 [DOI] [PubMed] [Google Scholar]
- 16. Chou S., et al. 1999. Interstrain variation in the human cytomegalovirus DNA polymerase sequence and its effect on genotypic diagnosis of antiviral drug resistance. Antimicrob. Agents Chemother. 43:1500–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chou S., et al. 2010. Recombinant phenotyping of cytomegalovirus sequence variants detected after 200 or 100 days of valganciclovir prophylaxis. Transplantation 90:1409–1413 [DOI] [PubMed] [Google Scholar]
- 18. Chou S., et al. 1997. Evolution of mutations conferring multidrug resistance during prophylaxis and therapy for cytomegalovirus disease. J. Infect. Dis. 176:786–789 [DOI] [PubMed] [Google Scholar]
- 19. Chou S., Marousek G. I. 2008. Accelerated evolution of maribavir resistance in a cytomegalovirus exonuclease domain II mutant. J. Virol. 82:246–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chou S., Marousek G. I., Van Wechel L. C., Li S., Weinberg A. 2007. Growth and drug resistance phenotypes resulting from cytomegalovirus DNA polymerase region III mutations observed in clinical specimens. Antimicrob. Agents Chemother. 51:4160–4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cihlar T., Chen M. S. 1996. Identification of enzymes catalyzing two-step phosphorylation of cidofovir and the effect of cytomegalovirus infection on their activities in host cells. Mol. Pharmacol. 50:1502–1510 [PubMed] [Google Scholar]
- 22. Cihlar T., Fuller M. D., Cherrington J. M. 1998. Characterization of drug resistance-associated mutations in the human cytomegalovirus DNA polymerase gene by using recombinant mutant viruses generated from overlapping DNA fragments. J. Virol. 72:5927–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cihlar T., Fuller M. D., Mulato A. S., Cherrington J. M. 1998. A point mutation in the human cytomegalovirus DNA polymerase gene selected in vitro by cidofovir confers a slow replication phenotype in cell culture. Virology 248:382–393 [DOI] [PubMed] [Google Scholar]
- 24. Condra J. H. 1998. Resistance to HIV protease inhibitors. Haemophilia 4:610–615 [DOI] [PubMed] [Google Scholar]
- 25. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ducancelle A., et al. 2006. A novel mutation in the UL54 gene of human cytomegalovirus isolates that confers resistance to foscarnet. Antivir. Ther. 11:537–540 [PubMed] [Google Scholar]
- 27. Erice A., et al. 1997. Antiviral susceptibilities and analysis of UL97 and DNA polymerase sequences of clinical cytomegalovirus isolates from immunocompromised patients. J. Infect. Dis. 175:1087–1092 [DOI] [PubMed] [Google Scholar]
- 28. Gilbert C., Boivin G. 2005. Human cytomegalovirus resistance to antiviral drugs. Antimicrob. Agents Chemother. 49:873–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gilbert C., Boivin G. 2005. New reporter cell line to evaluate the sequential emergence of multiple human cytomegalovirus mutations during in vitro drug exposure. Antimicrob. Agents Chemother. 49:4860–4866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hwang Y. T., Liu B. Y., Coen D. M., Hwang C. B. 1997. Effects of mutations in the Exo III motif of the herpes simplex virus DNA polymerase gene on enzyme activities, viral replication, and replication fidelity. J. Virol. 71:7791–7798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jabs D. A., et al. 2001. Longitudinal observations on mutations conferring ganciclovir resistance in patients with AIDS and cytomegalovirus retinitis: The Cytomegalovirus and Viral Resistance Study Group Report Number 8. Am. J. Ophthalmol. 132:700–710 [DOI] [PubMed] [Google Scholar]
- 32. Kuhn F. J., Knopf C. W. 1996. Herpes simplex virus type 1 DNA polymerase. Mutational analysis of the 3′-5′-exonuclease domain. J. Biol. Chem. 271:29245–29254 [DOI] [PubMed] [Google Scholar]
- 33. Landry M. L., et al. 2000. A standardized plaque reduction assay for determination of drug susceptibilities of cytomegalovirus clinical isolates. Antimicrob. Agents Chemother. 44:688–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Limaye A. P., Corey L., Koelle D. M., Davis C. L., Boeckh M. 2000. Emergence of ganciclovir-resistant cytomegalovirus disease among recipients of solid-organ transplants. Lancet 356:645–649 [DOI] [PubMed] [Google Scholar]
- 35. Limaye A. P., et al. 2002. High incidence of ganciclovir-resistant cytomegalovirus infection among lung transplant recipients receiving preemptive therapy. J. Infect. Dis. 185:20–27 [DOI] [PubMed] [Google Scholar]
- 36. Littler E., Stuart A. D., Chee M. S. 1992. Human cytomegalovirus UL97 open reading frame encodes a protein that phosphorylates the antiviral nucleoside analogue ganciclovir. Nature 358:160–162 [DOI] [PubMed] [Google Scholar]
- 37. Liu S., et al. 2006. Crystal structure of the herpes simplex virus 1 DNA polymerase. J. Biol. Chem. 281:18193–18200 [DOI] [PubMed] [Google Scholar]
- 38. Lurain N. S., Chou S. 2010. Antiviral drug resistance of human cytomegalovirus. Clin. Microbiol. Rev. 23:689–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marchini A., Liu H., Zhu H. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martin M., Gilbert C., Covington E., Boivin G. 2006. Characterization of human cytomegalovirus (HCMV) UL97 mutations found in a valganciclovir/oral ganciclovir prophylactic trial by use of a bacterial artificial chromosome containing the HCMV genome. J. Infect. Dis. 194:579–583 [DOI] [PubMed] [Google Scholar]
- 41. Martin M., Goyette N., Boivin G. 2010. Contrasting effects on ganciclovir susceptibility and replicative capacity of two mutations at codon 466 of the human cytomegalovirus UL97 gene. J. Clin. Virol. 49:296–298 [DOI] [PubMed] [Google Scholar]
- 42. Martin M., Goyette N., Ives J., Boivin G. 2010. Incidence and characterization of cytomegalovirus resistance mutations among pediatric solid organ transplant patients who received valganciclovir prophylaxis. J. Clin. Virol. 47:321–324 [DOI] [PubMed] [Google Scholar]
- 43. Mousavi-Jazi M., et al. 2001. Variations in the cytomegalovirus DNA polymerase and phosphotransferase genes in relation to foscarnet and ganciclovir sensitivity. J. Clin. Virol. 23:1–15 [DOI] [PubMed] [Google Scholar]
- 44. Painter J., Merritt E. A. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62:439–450 [DOI] [PubMed] [Google Scholar]
- 45. Parthiban V., Gromiha M. M., Schomburg D. 2006. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 34:W239–W242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Piana S., Carloni P., Rothlisberger U. 2002. Drug resistance in HIV-1 protease: flexibility-assisted mechanism of compensatory mutations. Protein Sci. 11:2393–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schreiber A., et al. 2009. Antiviral treatment of cytomegalovirus infection and resistant strains. Expert Opin. Pharmacother. 10:191–209 [DOI] [PubMed] [Google Scholar]
- 48. Scott G. M., Weinberg A., Rawlinson W. D., Chou S. 2007. Multidrug resistance conferred by novel DNA polymerase mutations in human cytomegalovirus isolates. Antimicrob. Agents Chemother. 51:89–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shi R., Azzi A., Gilbert C., Boivin G., Lin S. X. 2006. Three-dimensional modeling of cytomegalovirus DNA polymerase and preliminary analysis of drug resistance. Proteins 64:301–307 [DOI] [PubMed] [Google Scholar]
- 50. Smith I. L., et al. 1997. High-level resistance of cytomegalovirus to ganciclovir is associated with alterations in both the UL97 and DNA polymerase genes. J. Infect. Dis. 176:69–77 [DOI] [PubMed] [Google Scholar]
- 51. Springer K. L., et al. 2005. How evolution of mutations conferring drug resistance affects viral dynamics and clinical outcomes of cytomegalovirus-infected hematopoietic cell transplant recipients. J. Clin. Microbiol. 43:208–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sullivan V., et al. 1992. A protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells. Nature 358:162–164 [DOI] [PubMed] [Google Scholar]
- 53. Turner D., Schapiro J. M., Brenner B. G., Wainberg M. A. 2004. The influence of protease inhibitor resistance profiles on selection of HIV therapy in treatment-naive patients. Antivir. Ther. 9:301–314 [PubMed] [Google Scholar]
- 54. Weinberg A., et al. 2003. Mutations conferring foscarnet resistance in a cohort of patients with AIDS and cytomegalovirus retinitis. J. Infect. Dis. 187:777–784 [DOI] [PubMed] [Google Scholar]