Abstract
GS-9190 (Tegobuvir) is a novel imidazopyridine inhibitor of hepatitis C virus (HCV) RNA replication in vitro and has demonstrated potent antiviral activity in patients chronically infected with genotype 1 (GT1) HCV. GS-9190 exhibits reduced activity against GT2a (JFH1) subgenomic replicons and GT2a (J6/JFH1) infectious virus, suggesting that the compound's mechanism of action involves a genotype-specific viral component. To further investigate the GS-9190 mechanism of action, we utilized the susceptibility differences between GT1b and GT2a by constructing a series of replicon chimeras where combinations of 1b and 2a nonstructural proteins were encoded within the same replicon. The antiviral activities of GS-9190 against the chimeric replicons were reduced to levels comparable to that of the wild-type GT2a replicon in chimeras expressing GT2a NS5B. GT1b replicons in which the β-hairpin region (amino acids 435 to 455) was replaced by the corresponding sequence of GT2a were markedly less susceptible to GS-9190, indicating the importance of the thumb subdomain of the polymerase in this effect. Resistance selection in GT1b replicon cells identified several mutations in NS5B (C316Y, Y448H, Y452H, and C445F) that contributed to the drug resistance phenotype. Reintroduction of these mutations into wild-type replicons conferred resistance to GS-9190, with the number of NS5B mutations correlating with the degree of resistance. Analysis of GS-9190 cross-resistance against previously reported NS5B drug-selected mutations showed that the resistance pattern of GS-9190 is different from other nonnucleoside inhibitors. Collectively, these data demonstrate that GS-9190 represents a novel class of nonnucleoside polymerase inhibitors that interact with NS5B likely through involvement of the β-hairpin in the thumb subdomain.
INTRODUCTION
Hepatitis C virus (HCV) is a major cause of morbidity, affecting approximately 170 million people worldwide with an estimated 3 to 4 million additional new infections occurring each year (36). HCV is a positive-strand RNA virus with six major genotypes that are further divided into multiple subtypes. Due to the error-prone nature of its replication enzyme, a myriad of different viral quasispecies exists within an infected individual (32). With this high degree of viral variability, the current treatment regimen, which consists of weekly injections of pegylated alpha interferon (PEG-IFN) and twice-daily oral doses of ribavirin (RBV), is of limited efficacy and, in addition, carries significant side effects (8, 23). Although the HCV NS3/4A protease inhibitors telaprevir and boceprevir for treatment of chronic HCV infection will soon be available, these compounds will still need to be combined with the current standard of care (PEG-IFN/RBV) to be efficacious and will not cure all infected individuals (10, 14, 30). Therefore, the development of additional direct antiviral agents with diverse resistance profiles is necessary, with the ultimate goal of developing all-oral antiviral combinations that can achieve superior sustained virologic response (SVR) without the use of IFN or RBV. Thus, major efforts are under way to identify additional novel inhibitors of HCV. In particular, much emphasis has been placed on the viral polymerase NS5B as a target.
Viral polymerases are attractive targets for drug discovery and have yielded approved drugs for HIV, HBV, herpes simplex virus, and cytomegalovirus. The HCV NS5B polymerase is an RNA-dependent RNA polymerase containing canonical thumb, finger, and palm subdomains (2, 3, 19, 37, 40). Both nucleoside inhibitors (NIs) and nonnucleoside inhibitors (NNIs) of NS5B have been reported in the literature and are currently in clinical trials (4, 9, 16, 18, 26, 31, 34). NIs act as chain terminators and tend to show pan-genotypic activity compared to NNIs. However, efficacies of some nucleoside inhibitors in the clinic have been marred by significant adverse events (7). NNIs in clinical development target one of the several allosteric binding sites in the NS5B polymerase with compounds that bind in a similar manner and that demonstrate overlapping resistance profiles. Novel NNIs with resistance traits diverse from those already in clinical trials will be essential in the development of effective combination therapy and in overcoming viral resistance.
Recently, a novel class of substituted imidazopyridine compounds showing selective inhibition of HCV was reported (35). Here we report on the molecular target of the most promising member of this class, GS-9190 (Tegobuvir), which has demonstrated antiviral activity in HCV-infected patients (1, 39). By using chimeric replicons, kinetic comparison, and resistance selection, we demonstrate that GS-9190 inhibits viral replication by targeting the NS5B polymerase. Furthermore, by using data gleaned from reverse genetics and molecular modeling, we propose that GS-9190 exploits a unique pocket on NS5B and utilizes a novel binding mechanism to inhibit HCV replication.
MATERIALS AND METHODS
Replicon cell lines.
Huh-luc and Huh7-Lunet cells were obtained from ReBlikon GmbH (Mainz, Germany). All Huh7-Lunet-based replicon cell lines were grown in Dulbeccos's modified Eagle's medium (DMEM) with GlutaMAX-I (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT), 1 U/ml penicillin (Invitrogen), 1 μg/ml streptomycin (Invitrogen), and 0.1 mM nonessential amino acids (Invitrogen). Stable replicon cell lines were selected and maintained in medium containing 0.5 mg/ml G418 (Geneticin; Invitrogen). Creation of Huh7-Lunet stable genotype 1b (Con-1) and 1a (1a H77-51) subgenomic replicon cells has been reported previously (21, 29). Analogous stable subgenomic genotype 2a (JFH1) replicon cells were created by electroporating Huh7-Lunet cells with RNA transcribed from a linearized bicistronic construct encoding an HCV JFH1 internal ribosome entry site (IRES)-driven neomycin cassette and encephalomyocarditis virus-controlled coding region containing the nonstructural NS3, NS4A, NS4B, NS5A, and NS5B polyprotein sequences. Twenty-four hours after electroporation, replicon-containing cell clones were selected using 0.5 mg/ml G418. Following selection and clonal expansion, a single 2a replicon clone was chosen for further experimentation based upon sequence fidelity, replicon levels as assessed in an NS3 protease assay (38), and 50% effective concentrations (EC50s) when tested against a panel of antiviral control compounds.
Compounds.
GS-9190 was synthesized at Gilead Sciences, Inc. (Foster City, CA). BILN-2061 and 2′-C-methyl adenosine (2′CMeA) were purchased from Acme Bioscience (Belmont, CA). The ViroPharma/Wyeth HCV NS5B site IV inhibitor HCV-796 (13) was synthesized by Curragh Chemistries (Cleveland, OH). An Abbott benzothiadiazine NS5B polymerase inhibitor (A-782759) (24) was synthesized by ChemALong Laboratories (Lemont, IL).
Construction of mutant and chimeric replicons.
NS5B point mutations were engineered into the pFK I341 PI-Luc/NS3-3′/ET replicon construct (ReBlikon GmbH) using the Stratagene QuikChange XL mutagenesis kit (Stratagene) and appropriate primers. Mutagenesis was confirmed via sequencing of the NS5B region of the plasmid (Elim Biopharmaceuticals, Inc., Hayward, CA). The NS5B region was then subcloned into a new pFK I341 PI-Luc/NS3-3′/ET replicon backbone by using the BclI and SpeI sites, and the subcloned NS5B region was resequenced.
A plasmid carrying the chimeric 1b NS4A4B/2a replicon DNA was constructed by replacing NS4A and NS4B in the 2a JFH strain with the corresponding region derived from the 1b Con1 strain. NsiI and BamHI restriction sites were introduced into the PCR primer set 1b NS4A PCR NsiI F (5′-ACATCATGGCATGCATGTCGGCTGACCTGGAGGTCG-3′; NsiI site underlined) and 1b NS5A PCR BamHI R (5′-CAAACATCTCTTAGCCAGGATCCGGAGCATGGCGTGGAGC-3′; BamHI site underlined), which was used to amplify the HCV-1b NS4A-4B fragment. After digestion of the 1b PCR fragment and pJFH1 DNA with NsiI and BamHI, the 1b NS4A-4B fragment was ligated into pJFH1 (Toray, Inc.). The junctions of the resulting chimeric replicon (1b NS4A4B/2a) were confirmed by DNA sequencing. Overlapping PCR was employed to obtain the fragment containing 1b protease and 2a helicase by using the PCR product of 1b protease and 2a helicase as templates and primer sets 1b Protease PCR MluI F (5′-TTTCCTTTGAAAAACACGATACGCGTATACCATGGCGCCTATTAC-3′; MluI site underlined) and 2a Hel PCR NsiI R (5′-CCTCAAGGTCAGCTTGCATGCATGTGGCGATG-3′; NsiI site underlined) to incorporate the restriction sites MluI and NsiI. After digestion of the PCR fragment and 1b NS4A4B/2a DNA with MluI and NsiI, the 1b protease plus 2a helicase fragment was ligated into 1b NS4A4B/2a.
A 2a NS5B/1b chimeric replicon was created by amplifying the NS5B-3′-untranslated region (UTR) via PCR from the pJFH plasmid using the primers 2a5′NS5BBclIfw (5′-GATCGACTGATCACTCCCTGTAGCCCCGAAGAGG-3′) and 2a3′UTRSpeIrev2 (5′-AGCTTAGACTAGTACATGATCTGCAGAGAGACCAG-3′). PCR fragments were gel purified (Qiagen), digested with SpeI and BclI (New England BioLabs [NEB]), and ligated into an analogously digested and purified pFK I341 PI-Luc/NS3-3′/ET vector fragment. A β-hairpin chimera was created using multiple rounds of site-directed mutagenesis (QuikChange XL; Stratagene) to change amino acids 434 to 455 of the NS5B region in pFK I341 PI-Luc/NS3-3′/ET to that of genotype 2a (JFH1). The NS5B region was then subcloned into a fresh vector backbone by using BclI and SpeI restriction sites. All constructs were sequenced to confirm sequence fidelity.
Transient transfections.
DNA plasmids carrying replicon sequences were linearized in vitro using SpeI restriction endonuclease (NEB) followed by electrophoresis and gel purification of the linearized fragment (QIAquick gel extraction kit; Qiagen). Replicon RNA was derived from the purified template by using T7 runoff transcription (MEGAscript T7 kit; Ambion). For transfection of RNA into Huh7-Lunet cells, cells were trypsinized and washed three times with phosphate-buffered saline (PBS). A suspension of 4 × 106 cells in 400 μl PBS was mixed with 10 μg RNA and subjected to electroporation at settings of 270 V and 950 μF capacitance. Cells were then transferred into 20 ml of prewarmed culture medium and seeded into appropriate plates for further analyses.
Antiviral EC50 and CC50 determinations.
Replicon-containing cells were trypsinized and seeded in cell culture medium without G418 in white or black 96-well plates for EC50 or 50% cytotoxic concentration (CC50) analyses, respectively. Stable replicon cell lines were seeded at a density of 5,000 cells per well, while Huh7-Lunet cells transiently tranfected with replicon RNA were seeded at 20,000 cells per well postelectroporation. Ten-point dilutions of compounds were performed in dimethyl sulfoxide (DMSO) followed by further dilution in cell culture medium and subsequent addition to cell plates. Compound-treated cells were incubated for 72 h at 37C in a 5% CO2 incubator. Following incubation, CC50 values were determined by using a commercial reagent according to the manufacturer's specifications (Cell Titer Glo; Promega, Madison, WI). For luciferase-containing replicons, luciferase expression following compound treatment was quantified using a commercially available assay system (Luciferase assay system; Promega). For replicon cell lines not containing a luciferase reporter, antireplicon efficacy was determined using an NS3 protease assay described previously (38). Antiviral effects against a cell culture-adapted genotype 2a (J6/JFH) infectious virus were determined as previously reported (27). Curve fitting and EC50/CC50 values were derived using nonlinear regression analyses (GraphPad Prism). Curves were extrapolated from duplicate points, and all experiments were individually performed at least twice.
NS5B enzymatic assays.
GS-9190 inhibition of NS5B-dependent RNA polymerization activity was determined using two assay formats in vitro. The first format utilized a heteropolymeric RNA template described previously (12). The reaction was assembled in 50 mM Tris-HCl (pH 7.5), 10 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol (DTT), 1 mM EDTA, 40 ng/μl RNA template, 0.5 μCi of [α-33P]nucleoside triphosphate, and 500 μM remaining nucleoside triphosphates (NTPs). Purified NS5B enzyme was preincubated with compounds for 20 min at 34°C, after which the reaction was started by the addition of RNA template and NTPs. The radiolabeled NTP was used at the Km concentration. The reaction was allowed to proceed for 90 min and then the mixture was transferred onto 96-well DE81 filter membranes, washed three times with 100 mM Na2HPO4 and once with ethanol, and dried. Scintillation fluid was added to the wells, and counts per minute (cpm) were measured in a TopCount apparatus.
The second assay format utilized a poly(C) RNA template and GTP as the initiation nucleotide (20). The reaction mixture was assembled with 20 mM HEPES (pH 7.3), 12 mM KCl, 50 mM NaCl, 1 mM DTT, and 10 mM MgCl2–5 mM MnCl2, 40 ng/μl RNA template, 1 μCi of [α-33P]GTP, and 50 μM GTP. Purified enzyme was preincubated with compounds for 15 min at 34°C, after which the reaction was started by the addition of RNA template and GTP. The reaction was allowed to proceed for 60 min, and then the reaction mixture was transferred to 96-well DE81 filter membranes, washed three times with 100 mM Na2HPO4 and once with ethanol, and dried. Scintillation fluid was added to the wells, and radioactivity was measured on a TopCount analyzer (Perkin-Elmer).
Resistance selection.
Huh-Luc cells were seeded in 10-cm tissue culture dishes, grown to subconfluency and then treated with Geneticin and GS-9190 at various concentrations (G418, 0.1 to 1 mg/ml; GS-9190, 0.05 to 10 μM). Medium was changed twice weekly, and cells were passaged when necessary to maintain subconfluent levels. After 2 to 3 weeks, cells began to die and small colonies began to form. Colonies were further cultured in the presence of drugs for an additional 8 to 10 weeks. Expanded colonies of resistant cells were characterized by phenotypic evaluation to confirm reduced susceptibility to GS-9190. RNA was extracted from clones using the RNeasy midikit (Qiagen). Amplification of replicon DNA was achieved using Qiagen's one-step reverse transcription-PCR (RT-PCR) kit and combinations of HCV-specific primer pairs. Product formation was confirmed by agarose gel electrophoresis, and DNA products were submitted for sequence analysis (ELIM Biopharmaceuticals, Hayward, CA).
Quantitative RT-PCR.
Huh-9-13, a Huh7 cell line carrying the persistent I377/NS3-3′/wild-type replicon (genotype 1b), was obtained from ReBLikon GmbH (Mainz, Germany). The Huh-9-13 cells were plated at 1 × 105 cells/well in 6-well plates in DMEM (GIBCO, Carlsbad, CA) supplemented with 10% FBS (HyClone, Logan, UT), 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.1 mM nonessential amino acids (Invitrogen, Carlsbad, CA) including 1 mg/ml G-418. At 24 h after plating, the medium was replaced with G418-free medium and the cells were treated with 50 nM BILN-2061, 150 nM HCV-796, 30 nM GS-9190, or the equivalent concentration of DMSO as a negative control. Cell samples were collected at 1, 3, 6, 9, 12, 16, and 24 h posttreatment. Total RNA was extracted with the RiboPure kit (AM1924; Life Technologies Corporation, Carlsbad, CA) following the manufacturer's protocol. Extracted RNA samples were stored at −80°C until use. For the quantitative RT-PCR (QRT-PCR) assay, the Qiagen one-step QRT-PCR kit was used according to the manufacturer's protocol (Qiagen, Valencia, CA). The genotype 1b HCV NS3 gene-specific primers forward primer NS3_180FL (5′–CGGCGGACTGTCTATCATGGTGCG–6-carboxyfluorescein [FAM]–3′) and reverse NS3_180 (5′–GGTCCTGGTCCACATTGGTGT-3′) and 18S rRNA LUX (FAM) endogenous control primer set (115HM-01) were produced by Invitrogen Corporation (Carlsbad, CA). For the reverse transcriptase step, the reaction mixtures were incubated at 44°C for 30 min, and the reverse transcriptase enzyme was then degraded by heating the sample to 94°C for 10 min. The QPCR step included 38 cycles at 94°C for 15 s and 58°C for 30 s.
Modeling.
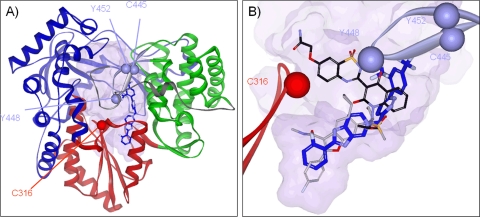
Docking studies were carried out with Accelrys' DiscoveryStudio 2.55 and 3.0 using the default settings of the CDOCKER and LigandFit algorithms. Best results were obtained when the NS5B central cavity was partitioned into 10 subsites. Figure 6, below, was prepared by aligning the backbone atoms of crystal structures 3FKQ and 3HKK. All ligands are shown within the 3FKQ protein.
Fig. 6.
Illustration of the NS5B cavity encompassing allosteric sites III and IV and the putative region targeted by GS-9190. (A) The right-hand model of the NS5B structure (PDB 3FKQ) (color coding: the palm is red, thumb is green, fingers are blue, tail is gray, and β-hairpin is mauve) with GS-9190 modeled in a potential pocket. (B) Close-up of GS-9190 (blue) in the putative binding region along with site IV inhibitor HCV-796 (gray) and site III inhibitor A-782759 (black). Positions of mutations moderating GS-9190 antiviral activity are indicated by their α carbon.
RESULTS
Activity of GS-9190 in HCV replicon and infectious virus assays.
GS-9190 is a novel imidazopyridine analogue derived from a previously described series of pestivirus replication inhibitors and optimized for selective inhibition of HCV replication (28, 35) (Fig. 1). To assess the anti-HCV activity of GS-9190, the compound was evaluated for inhibitory effects on the replication of genotype 1a and 1b subgenomic replicons along with NS3 protease inhibitor BILN-2061 (17), the nucleoside inhibitor 2′CMeA (6), and the nonnucleoside inhibitor HCV-796 (13) (Table 1). GS-9190 proved to be a highly potent and selective inhibitor of genotype 1 replicons, displaying subnanomolar to nanomolar potencies as measured by the EC50, with minimal cytotoxicity in all cell lines tested (50% cytotoxic concentration of >50 μM). Similar to BILN-2061 and HCV-796, although GS-9190 was highly active against both the 1a and 1b genotypes, potency against 1b was consistently 5- to 10-fold higher than that against genotype 1a, suggesting that genotypic differences may influence GS-9190 antiviral activity. To further gauge cross-genotype antiviral activity, GS-9190 was also evaluated against a genotype 2a (JFH-1) replicon. Interestingly, GS-9190 lost more than 1,500-fold activity over genotype 1a and over 20,000-fold over genotype 1b in the genotype 2a replicon assay. To further confirm the reduced effectiveness of GS-9190 against genotype 2a, the compound was also tested against a cell culture-adapted infectious virus (J6/JFH-1). As was observed in the 2a replicon assay, GS-9190 displayed a loss in potency against the genotype 2a virus with an EC50 of 2.9 μM. Taken together, these results show that GS-9190 is a potent inhibitor of HCV genotype 1 and suggest that differences in the nonstructural protein region between genotypes may influence compound potency.
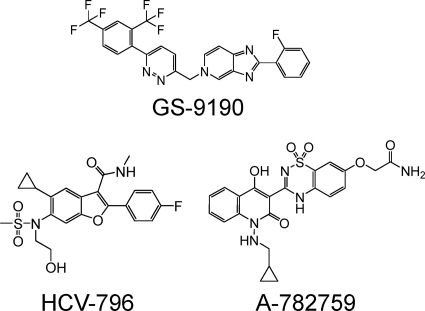
Fig. 1.
Structures of GS-9190, HCV-796, and A-782759.
Table 1.
Antiviral activities of GS-9190 and other known HCV inhibitors against HCV genotypes 1 and 2a
| Genotype(s) | EC50 (nM) |
|||
|---|---|---|---|---|
| GS-9190 | BILN-2061 | 2′-CMeA | HCV-796 | |
| Subgenomic replicons | ||||
| 1a (H77) | 13.8 ± 9.6 | 7.8 ± 5.0 | 420.5 ± 306.5 | 15.0 ± 7.1 |
| 1b (Con-1) | 0.8 ± 0.7 | 0.8 ± 0.7 | 217.5 ± 90.8 | 4.0 ± 2.7 |
| 2a (JFH-1) | 21,900 ± 19,000 | 73.0 ± 43.3 | 420.3 ± 94.2 | 144.0 ± 71.1 |
| 1a/1b/2a repliconsa | >50,000 | >50,000 | >50,000 | >50,000 |
| HCV | ||||
| 2a (J6/JFH-1) | 2,900 ± 1250 | 86.9 ± 34.0 | 527.5 ± 324.7 | 104.1 ± 15.1 |
| 2a (J6/JFH-1)a | >50,000 | >50,000 | >50,000 | >50,000 |
Data shown are the CC50 (in nM) rather than the EC50.
The NS5B genotype determines susceptibility to GS-9190.
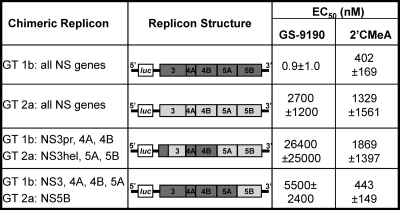
We next attempted to identify which HCV nonstructural protein(s) may be involved in the GS-9190 mechanism of action. In biochemical assays, no or poor activity was observed when GS-9190 was tested against the NS3/4A serine protease, NS3 RNA helicase, NS5B polymerase (using both heterologous and homopolymeric RNA as template), and HCV IRES-mediated translation (Table 2 and data not shown). Although GS-9190 lacked activity in these in vitro assays, it is possible that GS-9190 may possess an inhibitory function requiring the context of viral replication complexes and/or cellular factors. We took advantage of the apparent activity differences between genotypes 1b and 2a replicons for GS-9190 and constructed a series of chimeric replicons incorporating mixtures of viral proteins derived from the 1b and 2a genotypes (Fig. 2). GS-9190 was tested against the chimeric and wild-type replicons in transient assays along with appropriate control compounds. Interestingly, the predictive determinant of GS-9190 activity in these chimeric replicons appeared to be associated with the NS5B polymerase gene. Introduction of the GT2a NS5B gene alone into the 1b replicon drastically reduced the susceptibility of GS-9190 (>6,000-fold) to a level similar to that of the GT2a replicon. Hence, these findings imply that the NS5B genotype is a main contributing determinant of GS-9190 susceptibility and also that the compound may target the NS5B polymerase to inhibit HCV replication.
Table 2.
IC50 values of GS-9190 in NS5B-dependent polymerase assays in vitro
| Compound | IC50 (μM) |
|
|---|---|---|
| Heterologous RNA template | Poly(C) template | |
| GS-9190 | 71 | >100 |
| 2′CMeA | 2 | NDa |
| 3′dGTP | 1 | 0.2 |
ND, not done.
Fig. 2.
The NS5B genotype determines susceptibility to GS-9190. Chimeric replicons containing mixed combinations of genotype 1b and 2a nonstructural gene sequences were tested transiently in Huh7-Lunet cells. Chimeras were analyzed in a minimum of two independent experiments.
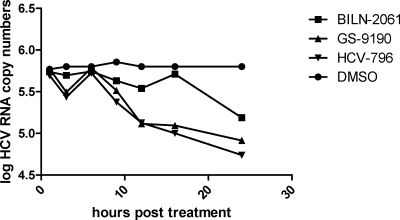
To further explore the inhibitory nature of GS-9190, we analyzed the kinetics of replicon RNA inhibition by using a real-time RT-PCR detection method (33). The drop in replicon RNA copy number following GS-9190 treatment was monitored over time compared to the protease inhibitor BILN-2061 and NNI HCV-796 (Fig. 3). BILN-2061 is expected to have slower inhibition kinetics, since it prevents assembly of new replication complexes (RC) but has no effect on preexisting RC. In contrast, a direct inhibitor of the NS5B polymerase is expected to have faster kinetics since it can inhibit the activity of existing RC. Using seven time points in a 24-hour period, we found that the kinetics of replicon inhibition following GS-9190 treatment was nearly identical to that observed following treatment with HCV-796, further suggesting that GS-9190 does not function as a protease inhibitor and, rather, may directly act on the RNA replication machinery.
Fig. 3.
The kinetics of RNA reduction following GS-9190 treatment is similar to that of the NNI HCV-796. 1b replicon cells were treated with inhibitors at 30 times the EC50, and total RNA was extracted and subjected to real-time analyses at various time points following treatment. Data shown are averages of samples run in triplicate.
NS5B mutations confer resistance to GS-9190.
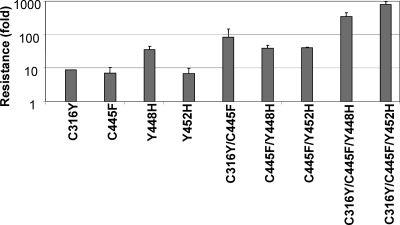
To further confirm that NS5B polymerase is the target of inhibition by GS-9190, we performed drug resistance selection using stable genotype 1b replicon cells treated with various concentrations of GS-9190. Individual colonies resulting from the selection were expanded, followed by a phenotypic assay to assess resistance to GS-9190. The nonstructural coding regions from resistant replicons were then reverse transcribed and sequenced to identify deviations from the wild-type sequence that may confer compound resistance. Interestingly, although several mutations were found in the NS3, NS4B, and NS5A genes, the majority of mutations uncovered were clustered in the NS5B region (Table 3). All found mutations were then individually reengineered into the wild-type replicon, and the susceptibility to GS-9190 was determined. None of the mutations detected in NS3, NS4B, or NS5A individually conferred resistance to GS-9190. However, several mutations in NS5B, C316Y, C445F, and Y452H, significantly reduced susceptibility to GS-9190, each resulting in a 7- to 10-fold shift over the wild-type genotype 1b replicon. Moreover, an independent resistance selection using a closely related GS-9190 analogue identified yet another NS5B mutation, Y448H (denoted by the * in Table 3). When Y448H was engineered into the wild-type 1b replicon, susceptibility to GS-9190 was reduced 36-fold, the most pronounced effect imparted by any single mutation tested. In addition to analyzing the effect of individual NS5B mutations on GS-9190 resistance, mutations were also tested in combinations (Fig. 4). Combining NS5B mutant residues led to an even greater degree of resistance. Most strikingly, although less-than-10-fold resistance was observed when C316Y, C445F, and Y452H were individually engineered into the replicon, combining these mutations led to a nearly 1,000-fold change in EC50. Taken together, these results indicate that mutations in the NS5B gene alone are sufficient to confer resistance to GS-9190, and they further corroborate that the NS5B polymerase is the target by which GS-9190 exhibits its antiviral effects.
Table 3.
Mutations in NS5B genes confer resistance to GS-9190a
| Mutation in GS-9190r replicon | Gene | Fold shift in GS-9190 EC50 vs. WT |
|---|---|---|
| Wild type | 1 | |
| A391V | NS3 | 0.2 |
| C432S | NS3 | 0.3 |
| Q93R | NS4B | 0.5 |
| V362A | NS5A | 0.6 |
| C316Y | NS5B | 8.8 |
| C445F | NS5B | 7.1 |
| Y452H | NS5B | 6.9 |
| R465G | NS5B | 1.1 |
| W571R | NS5B | 1.2 |
| Y448H* | NS5B | 36 |
WT, wild type.
, Y488H was the result of independent resistance selection using a closely related GS-9190 analogue.
Fig. 4.
The degree of GS-9190 resistance correlates with the number of NS5B mutations. NS5B mutations were engineered either alone or in combination via site-directed mutagenesis into wild-type 1b replicons. The antiviral phenotype of the constructs was evaluated in Huh7-Lunet cells transiently transfected with the constructs to determine the effect on the GS-9190 EC50. Resistance was characterized as the average fold change in the GS-9190 EC50 versus the wild-type genotype 1b replicon from multiple experiments. Data are mean values ± standard deviations for at least two independent experiments.
The β-loop region in the NS5B thumb subdomain is involved in the GS-9190 interaction.
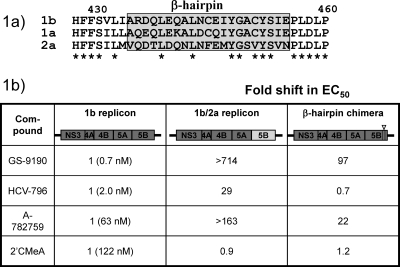
In order to more precisely define which region of NS5B might be involved in interacting with GS-9190, we compared the amino acid sequences of genotypes 1b, 1a, and 2a NS5B in the regions of resistance mutations identified from resistance selection experiments. The β-hairpin region between amino acids 435 and 455 was of particular interest for further study since it is characterized by sequence similarity between genotypes 1a and 1b yet a high degree of divergence between genotypes 1b and 2a (Fig. 5A). To test whether the β-hairpin region played a role in the differential susceptibility of genotype 1 and genotype 2a replicons to GS-9190, we constructed and tested a chimeric replicon in which all viral nonstructural protein sequences were derived from genotype 1b except for the β-hairpin region, which was changed to that of genotype 2a (Fig. 5B). The β-hairpin chimeric replicon was 97-fold less sensitive to GS-9190 than the wild-type 1b replicon. In contrast, no change in susceptibility to the nucleoside polymerase inhibitor 2′CMeA was observed. With this intriguing finding, we then went on to test two known NNIs that bind to the NS5B palm subdomain: HCV-796, which binds to NNI site IV, and the benzothiadiazine inhibitor A-782759, which binds to NNI site III (Fig. 1). HCV-796, known to bind near the catalytic active site of NS5B, was 29-fold more potent against the wild-type 1b replicon than against the 1b/2a NS5B chimeric replicon. However, consistent with the reported binding mechanism, HCV-796 remained equipotent against the β-hairpin chimera. The benzothiadiazine A-782759 also displayed a significant decrease in potency against the 1b/2a NS5B chimera compared to the wild-type 1b replicon. However, like GS-9190, the potency of A-782759 was significantly decreased against the β-hairpin chimera. This result is in agreement with the resistance mutation data for A-782759, showing that a mutation in the β-hairpin (Y448H) confers resistance (24). The notable parallel activities between GS-9190 and A-782759 against the chimeric replicons suggest that the antiviral activity of GS-9190 likely results from inhibition of NS5B via interaction with a pocket that overlaps with the binding site of A-782759 and involves the β-hairpin.
Fig. 5.
Replacement of the genotype 1b β-hairpin sequence in NS5B with that of genotype 2a (amino acids 435 to 455) significantly reduces replicon susceptibility to GS-9190. The antiviral phenotype of chimeric replicons was assessed in transiently transfected Huh7-Lunet cells and analyzed 72 h post-inhibitor addition. EC50s shown are averages of at least two independent experiments, each carried out in duplicate.
Cross-resistance studies demonstrated a unique resistance profile for GS-9190.
We next wanted to test the cross-resistance of GS-9190 against previously reported NS5B mutations selected by other NNIs. Noting the possibility of an overlapping binding site, insinuated by the β-hairpin chimeric data, we also tested the site III NNI A-782759 and the site IV NNI HCV-796 (Fig. 1). The antiviral effects of these three compounds were analyzed against replicons containing either the NS5B site I mutation P495L, the site II mutation M423T, the site III mutation M414T, the site IV mutation Y448H, or the palm site mutation C316Y (Table 4). As previously reported, resistance to HCV-796 only occurred in replicons carrying the C316Y mutation, consistent with the binding region of the compound. The site III inhibitor A-782759 also demonstrated a resistance profile consistent with the literature in which the primary site of resistance is at M414, with Y448H and C316Y also conferring lower levels of resistance as well. Like HCV-796 and A-782759, GS-9190 showed some degree of EC50 shift in response to C316Y, albeit significantly less than observed with the other NNIs. Like A-782759, significant resistance to GS-9190 occurred in response to the Y448H mutant. However, unlike A-782759, replicons bearing the M414T mutation were still susceptible to GS-9190, suggesting that GS-9190 does not interact with NS5B in a manner identical to A-782759. Taken together, the resistance profile of GS-9190 appears to be unique among NNIs, sharing only partial resistance features with the two NNIs interacting with this region of NS5B.
Table 4.
Cross-resistance of GS-9190 with common NS5B drug-selected mutations
| Mutation in NS5B | Fold shift in EC50 vs. WT |
||
|---|---|---|---|
| GS-9190 | HCV-796 | A-782759 | |
| Wild type | 1 | 1 | 1 |
| P495L | 0.83 | 0.3 | 6.4 |
| M423T | 0.75 | 0.98 | 0.5 |
| M414T | 0.83 | 1.3 | >57 |
| Y448H | 36 | 0.92 | 35 |
| C316Y | 8.8 | 61 | >16 |
Model of the GS-9190 interaction site in NS5B.
Based on the above biological data and the apo-enzyme crystal structure of NS5B, we conducted modeling to explore the possible interactions between GS-9190 and NS5B. Despite the absence of viral polymerase inhibition in biochemical assays, the overlap in the resistant mutations with site III and IV inhibitors led us to probe for potential GS-9190 interaction sites in the NS5B cavity. Using various publicly available crystal structures (PDB 1C2P, 3FKQ, 3HKK, and 1GX5), we employed docking methods to identify a potential high-affinity site for GS-9190. The only structure to produce an orientation with significant docking scores was 3FKQ (Fig. 6A). Here, GS-9190 is partially inserted into the site IV inhibitor pocket and extends across the site III inhibitor pocket along the β-hairpin. This pose is most consistent with the mutations observed and the general structure-activity relationships exhibited by this compound series (unpublished data). In the absence of evidence of a direct interaction of GS-9190 with NS5B, Fig. 6B illustrates how the putative GS-9190 binding site would encompass parts of the space occupied by both site III and site IV inhibitors. However, we must give serious caution that the hypothetical interactions between NS5B and GS-9190 might not reflect the actual interaction mode based on the fact that the NS5B crystal structure was derived from bacterially expressed recombinant NS5B proteins for which GS-9190 failed to demonstrate inhibitory activity in biochemical assays. Nonetheless, these interaction models may shed some light into the possible mode of interaction between NS5B polymerase and GS-9190.
DISCUSSION
The combining of multiple direct-acting antiviral agents with diverse resistance profiles will be the key to a truly effective direct antiviral therapy. Here we have reported on a novel HCV-specific inhibitor, GS-9190 (Tegobuvir), that is highly potent against genotype 1 HCV, the most prevalent genotype in the Western world and one that is significantly refractory to the current standard of care. Although clearly possessing antiviral activity as demonstrated in vitro and in HCV-infected patients (1, 39), little was known about the exact mechanism of action of GS-9190 and other imidazopyridine analogues. Like most other anti-HCV compounds that have been optimized specifically for activity against genotype 1, GS-9190 exhibits reduced potency against genotype 2a. Here we were able to take advantage of this genotype-dependent difference in potency to explore the mechanism by which GS-9190 inhibits HCV replication.
As mentioned, previous analyses of GS-9190 in in vitro assays for effects on the NS3 serine protease, NS3 RNA helicase, IRES-directed translation, and NS5B polymerase showed no inhibition, suggesting that GS-9190 may be dependent on the cellular context of HCV replication for activity. We therefore created a series of chimeric replicons carrying various combinations of nonstructural proteins derived from genotypes 1b and 2a and assessed their antiviral phenotypes in a transient cell-based replicon assay. Using the combined data from the chimeras, we found that the NS5B genotype defined sensitivity to GS-9190, identifying the NS5B polymerase as a likely target of activity. This conclusion was further corroborated by a kinetics experiment using real-time quantitative PCR to measure replicon RNA inhibition, in which GS-9190 behaved in nearly an identical manner to the NS5B NNI HCV-796 and faster than the NS3 protease inhibitor BILN-2061, suggesting GS-9190 inhibits replication.
To further identify the mechanism of action of GS-9190, we performed resistance selections in genotype 1b replicon cells. Replicon sequences were obtained from phenotypically resistant colonies, and individual mutations were reengineered into wild-type 1b replicons. Using this method, we uncovered that the only mutations conferring GS-9190 resistance were those found in NS5B. Specifically, C316Y, C445F, and Y452H individually induced 7- to 10-fold reductions in susceptibility to GS-9190. When a closely related analogue of GS-9190 was used to select for drug resistance, an additional mutation, Y448H, was identified. This particular mutation resulted in 36-fold resistance to GS-9190. Furthermore, combining these mutations had an even greater effect on GS-9190 potency. Taken together, these data pinpoint NS5B as the target by which GS-9190 enacts antiviral activity. In addition, by using a β-hairpin chimeric replicon, we found specifically that the 20-amino-acid of the β-hairpin region of the polymerase thumb subdomain played a significant role in establishing GS-9190 potency. The involvement of this region as a site for inhibition of HCV replication is not unprecedented, as a secondary resistance mutation for the benzothiadiazine NNI A-782759 has previously been reported to reside in the β-hairpin (Y448H) (24). We confirmed this finding in our study, as A-782759 was markedly less active against the β-hairpin chimeric replicon. Interestingly, the amino acid Y448 was not changed in this chimera, suggesting that resistance arising from sequence modification within the β-hairpin may be due to gross conformational change versus alterations of compound interactions with specific residues. Cross-resistance analyses of the site III inhibitor A-782759 and site IV inhibitor HCV-796 with GS-9190 revealed that GS-9190 has a different drug resistance profile from that of the known site III and site IV inhibitors, further implying that GS-9190 has an NS5B interaction mechanism distinct from other NNIs.
Using the NS5B crystal structure, we were able to map these mutations and found that their locations centered on the β-hairpin and near the catalytic active site. Interestingly, although the Y448H mutation was previously shown to be involved in resistance to benzothiadiazines (NNI site III), the primary site of resistance for that class is M414 (25). We observed no M414 mutations in GS-9190-resistant replicons. Furthermore, additional studies evaluating GS-9190 cross-resistance showed no alteration of the GS-9190 EC50 in replicons harboring the NS5B M414T mutation. These data, along with a model overlapping GS-9190, HCV-796, and A-782759 in a putative NS5B pocket, suggest that the mode by which GS-9190 interacts with NS5B is different from other palm-site NNIs.
Given the preponderance of evidence obtained in cell-based assays that NS5B is the target of GS-9190, it is bewildering that the compound is inactive in in vitro NS5B polymerase assays. There are several possible hypotheses that may explain this apparent discrepancy: (i) GS-9190 may only interact with the NS5B protein in the context of the intact replication complexes. It is conceivable that the conformation of NS5B in the context of the replication complexes is different from that of the C-terminally truncated recombinant NS5B protein used in the polymerase assays and that the precise conformation needed to allow binding with GS-9190 may not be adopted in the purified recombinant enzyme preparation. (ii) To address this possibility, we evaluated the effect of GS-9190 on the polymerization activity of endogenous replication complexes by employing membrane fractions isolated from replicon cells (15). However, GS-9190 failed to show inhibitory activity in this assay (data not shown), suggesting that it is unlikely that the lack of direct interaction between GS-9190 and purified NS5B protein can be ascribed to the absence of other viral or cellular proteins in the replication complexes or due to certain conformational differences. (iii) The interaction of GS-9190 and NS5B may occur during protein translation and before the enzyme adopts its final conformation. It is possible that the binding pocket for GS-9190 is formed transiently during the translation process. GS-9190 may require metabolic activation before it can interact with the enzyme and exert antiviral activity. To explore this hypothesis, we tested the effects of several known cytochrome P450 (CYP) inhibitors on the antiviral activity of GS-9190 in a replicon assay. These inhibitors include 1-benzylimidazole, α-naphthoflavone, and tetramethoxystilbene, which have been shown to be either nonselective CYP inhibitors or selective for certain CYP isoforms (5, 11, 22). To our surprise, GS-9190 showed significantly reduced antiviral activity (up to 50- to 90-fold) in the replicon assay when tested in combination with the CYP inhibitors (Table 5). Note that the concentrations of the CYP inhibitors used in this experiment were chosen based on their lack of cytotoxic or antireplicon effect when tested individually in the replicon assay. Furthermore, these CYP inhibitors had no effect on the antiviral activity of 2′CMeA, suggesting that this detrimental effect is specific for GS-9190. These data suggest that GS-9190 may undergo an intracellular activation step, likely through a CYP-mediated oxidative reaction, to produce the active metabolite and subsequently interact with and inhibit the NS5B polymerase function. This hypothesis could explain the lack of inhibitory activity of GS-9190 in NS5B biochemical and biophysical assays in vitro. Follow-up studies are under way to further investigate the molecular mechanism of this effect and how it relates to the GS-9190 antiviral mechanism of action. In conclusion, given the data from this current study, it is clear that GS-9190 exerts its antiviral activity through targeting the NS5B polymerase via a mechanism different from other NNIs. The locations of the primary resistance mutations further suggest that inhibition of NS5B by GS-9190, or most likely by an intracellular metabolite from GS-9190, is via a potentially new mode of binding.
Table 5.
GS-9190 replicon EC50s in the presence of cytochrome P450 inhibitors
| Assay | GS-9190 EC50 in the presence of CYP inhibitor at indicated concn (μM) |
||||||
|---|---|---|---|---|---|---|---|
| 1-benzylimidazole |
α-Naphthoflavone |
Tetramethoxystilbene |
|||||
| 0 | 44 | 400 | 0.044 | 0.44 | 0.44 | 4.4 | |
| GS-9190 EC50 | 6.5 | 18.7 | 381 | 17.9 | 563.7 | 16 | 334.1 |
| 2′CMeA EC50 | 150 | NDa | 98 | 114 | 103 | 122 | 96 |
ND, not done.
Footnotes
Published ahead of print on 11 July 2011.
REFERENCES
- 1. Bavisotto L., et al. 2007. Antiviral, pharmacokinetic and safety data for GS-9190, a non-nucleoside HCB NS5B polymerase inhibitor, in a phase-1 trial in HCB genotype 1 infected subjects. Hepatology 46(Suppl. 1):255A [Google Scholar]
- 2. Behrens S. E., Tomei L., De Francesco R. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12–22 [PMC free article] [PubMed] [Google Scholar]
- 3. Biswal B. K., et al. 2005. Crystal structures of the RNA-dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J. Biol. Chem. 280:18202–18210 [DOI] [PubMed] [Google Scholar]
- 4. Carroll S. S., et al. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979–11984 [DOI] [PubMed] [Google Scholar]
- 5. Chun Y. J., et al. 2009. Potent inhibition of human cytochrome P450 1B1 by tetramethoxystilbene. Toxicol. Lett. 189:84–89 [DOI] [PubMed] [Google Scholar]
- 6. Eldrup A. B., et al. 2004. Structure-activity relationship of purine ribonucleosides for inhibition of hepatitis C virus RNA-dependent RNA polymerase. J. Med. Chem. 47:2283–2295 [DOI] [PubMed] [Google Scholar]
- 7. Fleischer R., Boxwell D., Sherman K. E. 2004. Nucleoside analogues and mitochondrial toxicity. Clin. Infect. Dis. 38:e79–e80 [DOI] [PubMed] [Google Scholar]
- 8. Fried M. W., et al. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975–982 [DOI] [PubMed] [Google Scholar]
- 9. Gane E. J., et al. 2010. Oral combination therapy with a nucleoside polymerase inhibitor (RG7128) and danoprevir for chronic hepatitis C genotype 1 infection (INFORM-1): a randomised, double-blind, placebo-controlled, dose-escalation trial. Lancet 376:1467–1475 [DOI] [PubMed] [Google Scholar]
- 10. Hezode C., et al. 2009. Telaprevir and peginterferon with or without ribavirin for chronic HCV infection. N. Engl. J. Med. 360:1839–1850 [DOI] [PubMed] [Google Scholar]
- 11. Hodson P. V., Qureshi K., Noble C. A., Akhtar P., Brown R. S. 2007. Inhibition of CYP1A enzymes by alpha-naphthoflavone causes both synergism and antagonism of retene toxicity to rainbow trout (Oncorhynchus mykiss). Aquat. Toxicol. 81:275–285 [DOI] [PubMed] [Google Scholar]
- 12. Hung M., Gibbs C. S., Tsiang M. 2002. Biochemical characterization of rhinovirus RNA-dependent RNA polymerase. Antiviral Res. 56:99–114 [DOI] [PubMed] [Google Scholar]
- 13. Kneteman N. M., et al. 2009. HCV796: a selective nonstructural protein 5B polymerase inhibitor with potent anti-hepatitis C virus activity in vitro, in mice with chimeric human livers, and in humans infected with hepatitis C virus. Hepatology 49:745–752 [DOI] [PubMed] [Google Scholar]
- 14. Kwo P., et al. 2009. 44th Annu. Meet. Eur. Assoc. Study Liver, Copenhagen, Denmark, 22 to 26 April 2009, abstr. 4 European Association for the Study of the Liver, Geneva, Switzerland [Google Scholar]
- 15. Lai V. C., Dempsey S., Lau J. Y., Hong Z., Zhong W. 2003. In vitro RNA replication directed by replicase complexes isolated from the subgenomic replicon cells of hepatitis C virus. J. Virol. 77:2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lam A. M., et al. 2010. PSI-7851, a pronucleotide of β-D-2′-deoxy-2′-fluoro-2′-C-methyluridine monophosphate: a potent and pan-genotype inhibitor of hepatitis C virus replication. Antimicrob. Agents Chemother. 54:3187–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lamarre D., et al. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186–189 [DOI] [PubMed] [Google Scholar]
- 18. Le Pogam S., et al. 2010. RG7128 alone or in combination with pegylated interferon-α2a and ribavirin prevents hepatitis C virus (HCV) replication and selection of resistant variants in HCV-infected patients. J. Infect. Dis. 202:1510–1519 [DOI] [PubMed] [Google Scholar]
- 19. Lesburg C. A., et al. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937–943 [DOI] [PubMed] [Google Scholar]
- 20. Lohmann V., Korner F., Herian U., Bartenschlager R. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lohmann V., et al. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113 [DOI] [PubMed] [Google Scholar]
- 22. Madan A., Parkinson A., Faiman M. D. 1993. Role of flavin-dependent monooxygenases and cytochrome P450 enzymes in the sulfoxidation of S-methyl N,N-diethylthiolcarbamate. Biochem. Pharmacol. 46:2291–2297 [DOI] [PubMed] [Google Scholar]
- 23. Manns M. P., et al. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958–965 [DOI] [PubMed] [Google Scholar]
- 24. Mo H., et al. 2005. Mutations conferring resistance to a hepatitis C virus (HCV) RNA-dependent RNA polymerase inhibitor alone or in combination with an HCV serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 49:4305–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nguyen T. T., et al. 2003. Resistance profile of a hepatitis C virus RNA-dependent RNA polymerase benzothiadiazine inhibitor. Antimicrob. Agents Chemother. 47:3525–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Olsen D. B., et al. 2004. A 7-deaza-adenosine analog is a potent and selective inhibitor of hepatitis C virus replication with excellent pharmacokinetic properties. Antimicrob. Agents Chemother. 48:3944–3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paulson M., et al. 2008. Int. Symp. Hepatitis C Virus Rel. Vir., San Antonio, TX, 5 to 9 October 2008, abstr. 023 ICMS Pty, Ltd., Southbank, Victoria, Australia [Google Scholar]
- 28. Puerstinger G., et al. 2006. Substituted 5-benzyl-2-phenyl-5H-imidazo[4,5-c]pyridines: a new class of pestivirus inhibitors. Bioorg. Med. Chem. Lett. 16:5345–5349 [DOI] [PubMed] [Google Scholar]
- 29. Robinson M., et al. 2010. Novel hepatitis C virus reporter replicon cell lines enable efficient antiviral screening against genotype 1a. Antimicrob. Agents Chemother. 54:3099–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sarrazin C., Zeuzem S. 2010. Resistance to direct antiviral agents in patients with hepatitis C virus infection. Gastroenterology 138:447–462 [DOI] [PubMed] [Google Scholar]
- 31. Shi S. T., et al. 2009. Preclinical characterization of PF-00868554, a potent nonnucleoside inhibitor of the hepatitis C virus RNA-dependent RNA polymerase. Antimicrob. Agents Chemother. 53:2544–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Simmonds P. 2004. Genetic diversity and evolution of hepatitis C virus: 15 years on. J. Gen. Virol. 85:3173–3188 [DOI] [PubMed] [Google Scholar]
- 33. Targett-Adams P., et al. 2011. Small molecules targeting hepatitis C virus-encoded NS5A cause subcellular redistribution of their target: insights into compound mode of action. J. Virol. 85:6353–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Villano S. A., et al. 2007. Antiviral activity of the non-nucleoside polymerase inhibitor, HCV-796, in combination with pegylated interferon alfa-2b in treatment-naive patients with chronic HCV. J. Hepatol. 46:S24 [Google Scholar]
- 35. Vliegen I., et al. 2009. Substituted imidazopyridines as potent inhibitors of HCV replication. J. Hepatol. 50:999–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wasley A., Alter M. J. 2000. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 20:1–16 [DOI] [PubMed] [Google Scholar]
- 37. Yamashita T., et al. 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273:15479–15486 [DOI] [PubMed] [Google Scholar]
- 38. Yang H., Delaney W. E., IV 2006. A novel fluorescence-based protease assay using the endogenous NS3/4A protease activity present in the total cell lysates of HCV replicon cells. J. Clin. Virol. 36:S109–S110 [Google Scholar]
- 39. Zeuzem S., et al. 2010. 60th Annu. Meet. Am. Assoc. Study Liver Dis., abstr. LB-1 Boston, MA, 29 October to 2 November 2010 American Association for the Study of Liver Diseases, Alexandria, VA [Google Scholar]
- 40. Zhong W., et al. 2000. Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:9134–9143 [DOI] [PMC free article] [PubMed] [Google Scholar]