Abstract
An in-house quantitative real-time PCR (qPCR) assay using TaqMan chemistry has been developed to detect NDM-1 carbapenemase genes from bacterial isolates and directly from stool samples. The qPCR amplification of blaNDM-1 DNA was linear over 10 log dilutions (r2 = 0.99), and the amplification efficiency was 1.03. The qPCR detection limit was reproducibly 1 CFU, or 10 plasmid molecules, and there was no cross-reaction with DNA extracted from several multidrug-resistant bacteria harboring other β-lactam resistance genes. Feces spiked with decreasing amounts of enterobacterial isolates producing NDM-1 were spread on ChromID ESBL and on CHROMagar KPC media and were subjected to the qPCR. The limits of carbapenem-resistant bacterial detection from stools was reproducibly 1 × 101 to 3 × 101 CFU/100 mg feces with ChromID ESBL medium. The CHROMagar KPC culture medium had higher limits of detection (1 × 101 to 4 × 103 CFU/ml), especially with bacterial isolates having low carbapenem MICs. The limits of detection with the qPCR assay were reproducibly below 1 × 101 CFU/100 mg of feces by qPCR assay. Samples spiked with NDM-1-negative bacteria were negative by qPCR. The sensitivity and specificity of the blaNDM-1 qPCR assay on spiked samples were 100% in both cases. Using an automated DNA extraction system (QIAcube system), the qPCR assay was reproducible. The use of qPCR is likely to shorten the time for blaNDM-1 detection from 48 h to 4 h and will be a valuable tool for outbreak follow-up in order to rapidly isolate colonized patients and assign them to cohorts.
INTRODUCTION
Carbapenems are the drugs of choice to treat serious infections caused by extended-spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae. This is due to their efficacy in clinical trials and to the fact that, until recently, carbapenem resistance in K. pneumoniae was rare (3, 11, 16, 20). Although several mechanisms of carbapenem resistance have been reported, most of these mechanisms, including the presence of ESBLs or AmpC enzymes in combination with porin mutations (24) and the production of Ambler class A enzymes (such as KPC and GES), class D enzymes (such as OXA-48), and metalloenzymes (such as IMP, VIM, or NDM-1), have now widely emerged (3, 10, 13, 17, 24, 27, 30, 33). The latest, NDM-1 is probably the most worrisome, in part due to the location of its gene, which is largely encountered in Escherichia coli and which is widespread in the community (10). Recently, a derivative with a point mutation, NDM-2, has also been detected in Acinetobacter baumannii (9).
blaNDM-1-producing Enterobacteriaceae are usually resistant to all classes of antibiotics, including β-lactams, which comprise penicillins, cephalosporins, monobactams, and carbapenems (9, 30, 33), leaving physicians with limited antibiotic choices for treating infected patients. Detection of carbapenemases primarily on the basis of phenotypic testing may sometimes be difficult since the MIC may remain in the susceptibility range (18). Using automated susceptibility testing, identification of these carbapenem-resistant isolates may be improved significantly (32). The presence of metalloenzymes such as IMP, VIM, and NDM-1 in clinical isolates may be confirmed using imipenem and imipenem/EDTA Etests (13). The “gold standard” for their detection is based on carbapenem hydrolysis assays, which are not easy to implement in the routine laboratory, or on molecular techniques, such as PCR (18). Identification of carriers of carbapenemase producers in the stools is mostly based on usage of screening culture media, such as ChromID ESBL and CHROMagar media (4). However, even though these media are easy to use, they require an additional 48 h before the carrier status of a patient can be established (18, 29). In order to control the spread of carbapenemase producers in hospitalized patients, effective infection control measures and controlled antibiotic usage must be complemented by the use of rapid and sensitive molecular diagnostic assays, as suggested for KPC control (29). These diagnostic tools will help to rapidly isolate colonized individuals and assign them to cohorts.
A variety of molecular tools have been developed and have shown their usefulness in detecting resistance genes (1, 2, 6, 12, 14, 15, 21, 25, 31). Only a few of these techniques have been designed for direct identification of resistance genes from clinical samples (8, 19). Here, we describe the development of a real-time quantitative PCR (qPCR) assay for the detection of blaNDM-1 genes directly from stools. The assay was validated by comparing qPCR with culture of diluted bacterial suspensions or of spiked stool samples with several species of NDM-1-producing bacteria on ChromID ESBL and CHROMagar KPC media. Moreover, a bacterial DNA extraction methodology that has the advantage of being able to be either used totally manually or adapted to a fully automated extraction system was used.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Ten well-characterized enterobacterial NDM-1 producers, along with 22 non-NDM-1 producers used as negative-control strains, were used for direct detection in culture or for spiking experiments, in order to validate the novel detection assay (Table 1). Recombinant plasmid pNDM-1, made of a pBKCMV backbone carrying ca. 4-kb insert expressing the NDM-1 carbapenemase, was used for qPCR optimization (22). The concentration of the plasmid solution was 1011 plasmid molecules/μl (28). MICs were determined using Etest (bioMérieux, Marcy-l'-Etoile, France) and interpreted according to CLSI (5).
Table 1.
Bacterial isolates used in this study
| Isolate no. | Bacterium | Genotype | MIC (μg/ml)a |
Reference | NDM-1 qPCR result | ||
|---|---|---|---|---|---|---|---|
| IMP | MER | ETP | |||||
| 1 | Providencia stuartii PS | Wild type | 1 | 1 | 0.12 | 13 | − |
| 2 | E. cloacae CIP 7928 | Wild type | 0.12 | 0.016 | 0.12 | 13 | − |
| 3 | Serratia marcescens SM | Wild type | 0.25 | 0.125 | 0.064 | 13 | − |
| 4 | E coli Ec1 | Wild type | 0.06 | 0.016 | 0.006 | 13 | − |
| 5 | K. pneumoniae KPS1 | OKP-A | 0.12 | 0.12 | 0.032 | 13 | − |
| 6 | E. coli | ACC-1 | 0.12 | 0.06 | 0.5 | 13 | − |
| 7 | E coli | OXA-1 | 0.06 | 0.016 | 0.006 | 13 | − |
| 8 | Proteus mirabilis | OXA-23 | 0.25 | 2 | 0.06 | 13 | − |
| 9 | C. freundii 14827984 | TEM-1, GES-1 | 0.25 | 0.03 | 0.125 | 13 | − |
| 10 | K. pneumoniae ILT-3 | CTX-M-19 | 0.064 | 0.064 | 0.064 | 13 | − |
| 11 | K. pneumoniae HPA-1 | OXA-48, CTX-M-15, SHV-1 | 1.5 | 12 | 32 | 7 | − |
| 12 | C. freundii | OXA-48,VEB-1 | >32 | >32 | >32 | 13 | − |
| 13 | E. coli | OXA-48, TEM-150 | 24 | 32 | 24 | 13 | − |
| 14 | K. pneumoniae A28006 | KPC, CTX-M2, TEM-1, SHV-1 | 16 | 32 | 24 | 13 | − |
| 15 | E. cloacae HGM | KPC | 12 | 12 | 12 | 13 | − |
| 16 | E. coli | KPC, CTX-M-9, TEM-1 | 24 | 16 | 32 | 13 | − |
| 17 | E. coli DIH-1 | VIM-19, CTX-M-15, TEM-1 | 8 | 4 | 16 | 26 | − |
| 18 | K. pneumoniae DIH-2 | VIM-19, CTX-M-15, TEM-1, SHV-1 | 8 | 4 | 16 | 26 | − |
| 19 | P. aeruginosa | VIM-4, TEM-1 | >32 | >32 | >32 | 13 | − |
| 20 | A. baumannii | IMP-4 | >32 | >32 | >32 | 13 | − |
| 21 | E. coli | IMP-1, TEM-1 | 4 | 6 | 12 | 13 | − |
| 22 | Pseudomonas aeruginosa | IMP-13 | >32 | >32 | >32 | 13 | − |
| 23 | K. pneumoniae A | NDM-1, CTX-M15, TEM-1, SHV-1, CMY-2 | >32 | >32 | >32 | 17 | + |
| 24 | K. pneumoniae B | NDM-1, CTX-M15, TEM-1, SHV-1, CMY-2 | >32 | >32 | >32 | 17 | + |
| 25 | K. pneumoniae D | NDM-1, CTX-M15, TEM-1, SHV-1, CMY-2 | >32 | >32 | >32 | 17 | + |
| 26 | K. pneumoniae F | NDM-1, CTX-M15, TEM-1, SHV-1, CMY-2 | >32 | >32 | >32 | 17 | + |
| 27 | K. pneumoniae G | NDM-1, CTX-M15, SHV-1 | 2 | 6 | 2 | 17 | + |
| 28 | K. pneumoniae I | NDM-1, CTX-M15, TEM-1, SHV-1, CMY-2 | >32 | >32 | >32 | 17 | + |
| 29 | C. freundii A | NDM-1, CTX-M15 | >32 | >32 | >32 | 22 | + |
| 30 | E. coli A | NDM-1, CTX-M15, TEM-1 | 6 | 4 | 4 | 21 | + |
| 31 | E. coli B | NDM-1, TEM-1 | 3 | 3 | 2 | 21 | + |
| 32 | E. cloacae A | NDM-1 | 8 | 6 | 6 | 17 | + |
IMP, imipenem; MER, meropenem; ETP, ertapenem.
Serial dilutions for spiking experiments.
For the spiking experiments, 10 NDM-1 producers, along with K. pneumoniae HPA-1 producing OXA-48 and CTX-M-15 and E. coli 16 producing KPC and CTX-M-9, were used (Table 1). Bacterial suspensions of these strains with an optical density at 600 nm of 0.5 were serially diluted in phosphate-buffered saline (PBS); nine 10-fold dilutions were made. To quantify the viable bacteria in each dilution step, a blood agar plate (bioMérieux), a ChromID ESBL plate (bioMérieux), and a CHROMagar KPC plate (CHROMagar, Paris, France) were inoculated with 100 μl of a suspension and incubated overnight at 37°C; the number of colonies that grew was counted on the following day.
E. coli Ec1 (Table 1, isolate 4) was grown overnight. Nine-hundred-microliter aliquots of this bacterial suspension were subsequently spiked with 100 μl of serially diluted NDM-1 producers of K. pneumoniae A, Enterobacter cloacae A, and E. coli B. Two-hundred-microliter spiked bacterial suspensions were subjected to DNA extraction.
Spiked fecal samples were made by adding 100 μl of each dilution in PBS to 900 μl of a fecal suspension that was obtained by suspending 6 g of freshly pooled feces from 3 healthy volunteers in 60 ml of distilled water. A fecal suspension without the addition of a bacterial strain was used as a negative control. Two hundred microliters of spiked stool sample was subjected to DNA extraction. Each fecal suspension was plated onto sheep blood-containing tryptic soy agar, ChromID ESBL medium, and CHROMagar KPC medium. Viable bacteria were counted after 24 h at 37°C, and growth on selective media was compared to qPCR results. All experiments were performed in triplicate.
DNA extraction from cultured bacterial colonies.
Fresh overnight bacterial cultures or spiked E. coli Ec1 cultures were used for DNA extraction with a QIAamp DNA minikit (Qiagen, Les Ulis, France) according to the protocol suggested by the manufacturer. Extracted bacterial DNA was eluted from the columns in 200 μl elution buffer and stored at −20°C.
DNA extraction from stool samples.
The QIAamp DNA stool minikit (Qiagen) was used to manually extract bacterial DNA from resuspended stool samples according to the manufacturer's suggestions. The combined action of InhibitEX, a unique adsorption resin, and an optimized buffer leads to removal of PCR inhibitors. Extracted DNA was eluted in 200 μl elution buffer and stored at −20°C. The convenient QIAamp spin-column procedure provides rapid purification of nucleic acids. Purification of DNA using the QIAamp DNA stool minikit was subsequently automated on the QIAcube apparatus (Qiagen), which enables simultaneous DNA extractions from 12 samples in less than 2 h with the well-established Qiagen spin-column kits and eliminates the need for tedious manual steps. The manually and automated extracted DNAs were compared in the qPCR assay.
NDM-1 real-time PCR assay.
The real-time PCR Rotor gene 6000 amplification/detection system (Qiagen) was used for the amplification and detection of the blaNDM-1 amplicon (154 bp) by use of the TaqMan technology. The forward primer sequence (primer NDM-1-For; 5′-ATT AGC CGC TGC ATT GAT-3′) and the reverse primer sequence (primer NDM-1-Rev; 5′-CAT GTC GAG ATA GGA AGT G-3′) specific for the detection of blaNDM-1 were designed in-house. In this study, the blaNDM-1-specific probe (NDM-1-Probe; 5′-FAM-CTG [+C]CA [+G]AC [+A]TT [+C]GG TGC-TAMRA-3′ (where the bracketed nucleotides with plus signs are locked nucleic acid [LNA] nucleotides) was designed in-house and was labeled with 6-carboxyfluorescein (FAM) at the 5′ end and the 6-carboxytetramethylrhodamine (TAMRA) quencher at the 3′ end. The sensitivity of the TaqMan assay was optimized by evaluating different concentrations of the primers (200, 300, 600, and 900 nM) and probe (100, 200, and 300 nM). The concentrations of the primers and probe used in this study that gave the best detection limits were 400 nM for blaNDM-1 primers and 200 nM for the blaNDM-1 probe. The Quantitect virus kit (Qiagen) was used as recommended by the manufacturer: The 25-μl qPCR mixture contained 5 μl of 5× qPCR master mix, 1.25 μl of 20× probe mix (8 μM NDM-1-For, 8 μM NDM-1-Rev, 4 μM NDM-1-Probe), 7.75 μl of water, and 10 μl of DNA sample (Qiagen). qPCR was performed under the following conditions: 5 min at 95°C and 50 cycles of 15 s at 95°C and 1 min at 60°C. One PCR experiment (including mix preparation and amplification) takes less than 2 h.
Interpretation of results, standard curve, and sensitivity test on plasmids.
A sample was considered positive by qPCR if it crossed the fixed threshold limit of 0.05. To determine the efficiency of the qPCR, threshold cycle (CT) values obtained from a series of template DNA dilutions were graphed on the y axis versus the log of the dilution on the x axis.
RESULTS
Analytical sensitivity and precision of blaNDM-1 qPCR assay.
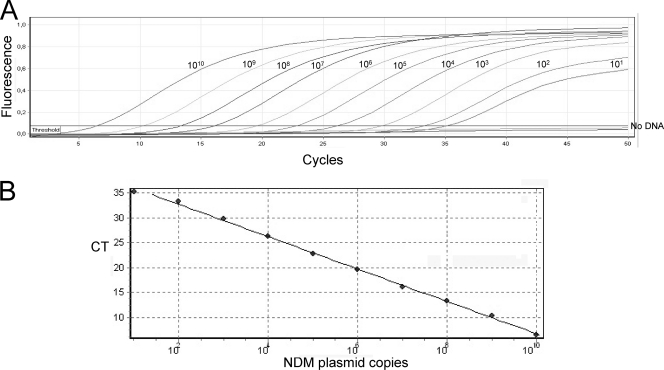
The analytical sensitivity of the blaNDM-1 qPCR assay was determined after serially diluting known concentrations of plasmid DNA. The amplification was linear over 10 log dilutions (r2 = 0.998; slope, −3.24), and the amplification efficiency was 1.03 on purified plasmid DNA (Fig. 1A and B).
Fig. 1.
Amplification curve (A) and standard curve (B) of blaNDM-1 qPCR assay using serial (10-fold) dilutions of recombinant plasmid DNA harboring blaNDM-1. CT values were obtained for each dilution and plotted against the number of plasmid copies per reaction. The numbers above each amplification curve correspond to the plasmid copy number present in the PCR mixture.
The detection limit of the real-time blaNDM-1 assay was reproducibly 10 plasmid molecules/PCR. Beyond 10 copies, results were not reproducible, most likely due to the well-recognized stochastic properties of qPCR on highly diluted nucleic acids. The analytical sensitivity of the assay did not change upon its utilization.
The performance of the assay on purified plasmid DNA (105 NDM-1-carrying plasmid) over 10 runs indicated that the qPCR assay was highly stable and precise. Overall, a mean CT of 21.1 and a standard deviation of 0.5 were obtained.
Cross-reactivity with other β-lactamase producers.
The specificities of the primers and probe for the detection of blaNDM-1 genes were evaluated by the BLAST search program, available at www.ncbi.nlm.nih.gov. No matches to the primers and probe sequences other than those for the blaNDM-1 genes were found. In addition, the blaNDM-1 qPCR assay was negative (absence of any amplification signal) with DNA extracted from the 22 NDM-1-negative bacterial pathogens listed in Table 1. Of these 22 isolates negative for the blaNDM-1 gene, 10 were imipenem susceptible and 12 were intermediate or resistant by means of another carbapenemase gene (4).
The system was evaluated on a number of well-defined clinical isolates producing NDM-1 from different geographic origins and expressing several other β-lactamase genes. The results are summarized in Table 1. Of the 10 isolates positive for the blaNDM-1 gene, 2 were of intermediate susceptibility to imipenem (1 < MIC ≤ 4 μg/ml) and 8 were resistant (MIC > 4 μg/ml). All were identified using this assay, with CT values ranging from 19 to 20. Overall, there was 100% concordance between the genotype of the bacteria and the NDM-1 qPCR assay results (Table 1).
Spiking of bacterial culture.
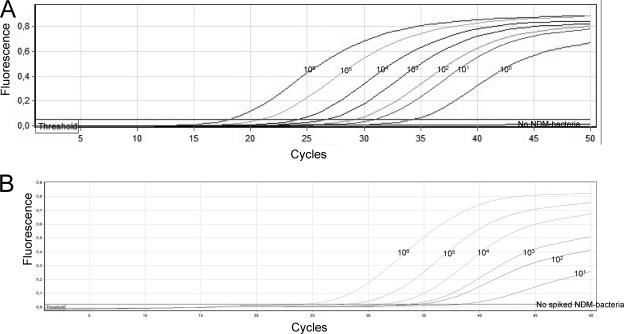
The analytical sensitivity of the blaNDM-1 qPCR assay together with the extraction method was determined after serially diluting NDM-1 producers into a fresh overnight E. coli Ec1 culture and subsequent manual and automated extraction. Both manual and automated extraction yielded similar-quality DNA, as revealed by similar amplification efficiencies and amplification standard curves (data not shown). The assay's amplification was linear over 9 log dilutions (r2 = 0.994; slope, −2.92), and the amplification efficiency was 1.19 (Fig. 2A). The limit of the linear range of the qPCR was reproducibly 1 × 101 CFU. The qPCR allowed detection of 1 CFU; however, the CT value was out of the linear range of the qPCR.
Fig. 2.
(A) Amplification curves of blaNDM-1 qPCR assay using serial (10-fold) dilutions of NDM-1-producing K. pneumoniae A in the reference E. coli Ec1 isolate. The numbers above each amplification curve correspond to the theoretical numbers of bacterial CFU present in each PCR. (B) NDM-1-producing E. coli B in fecal samples. The numbers above each amplification curve correspond to the theoretical bacterial count/ml of fecal suspension, which corresponds to about 100 mg of feces.
Detection of NDM producers by culture using chromogenic media.
Recently, two chromogenic and selective culture media have been evaluated for detecting carbapenemase-producing Enterobacteriaceae. The chromogenic ChromID ESBL culture medium (bioMérieux), which contains cefpodoxime as a selector and which is routinely used for screening ESBL producers (26), was also capable of detecting carbapenemase-producing Enterobacteriaceae (4) of the IMP, VIM, and KPC types, given their high level of resistance to cephalosporins and to carbapenems. Similarly, CHROMagar KPC (CHROMagar Company, Paris, France), which contains at least a carbapenem molecule as the selector for resistance and which was initially designed for screening for KPC producers (29), was capable of detecting carbapenemase-producing isolates with MIC for imipenem values of >4 μg/ml (4). Higher detection limits with MICs below 4 μg/ml were observed (4).
With serially diluted NDM-1 producers the lowest limit of detection of NDM-1 producers ranged from 1 × 101 to 4 × 101 CFU/ml for ChromID ESBL (Table 2). For CHROMagar KPC, the lowest limit of detection of NDM-1 producers ranged from 1 × 101 to 4 × 103 CFU/ml (Table 2). Detection limits were higher than those of the ChromID ESBL, especially for strains with low MICs for carbapenems, as shown previously (4, 18) (Table 2).
Table 2.
Spiking of fecal samples with NDM-1-producing enterobacterial isolate
| Isolate no. | Bacterium | IMP MIC (μg/ml) | Lowest limit of detection in water (CFU/ml) |
Lowest limit of detection in stoola (CFU/ml of stool)a |
Lowest limit od detection of NDM-1 qPCR |
|||
|---|---|---|---|---|---|---|---|---|
| ChromID ESBL | CHROMagar KPC | ChromID ESBL | CHROMagar KPC | CFU/ml of stool | No. of DNA copies/ml of stoolb | |||
| 23 | K. pneumoniae A | >32 | 1 × 101 | 1 × 101 | 2 × 101 | 1 × 102 | 101 | 1 × 102 |
| 24 | K. pneumoniae B | >32 | 2 × 101 | 1 × 101 | 2 × 101 | 5 × 101 | 2 × 101 | 2 × 102 |
| 25 | K. pneumoniae D | >32 | 3 × 101 | 2 × 101 | 2 × 101 | 2 × 101 | 101 | 4 × 102 |
| 26 | K. pneumoniae F | >32 | 1 × 101 | 2 × 101 | 1 × 101 | 2 × 101 | 2 × 101 | 2 × 102 |
| 27 | K. pneumoniae G | 2 | 2 × 101 | 3 × 102 | 1 × 102 | 4 × 103 | 101 | 2 × 102 |
| 28 | K. pneumoniae I | >32 | 1 × 101 | 3 × 101 | 2 × 101 | 6 × 101 | 101 | 6 × 102 |
| 29 | C. freundii A | >32 | 1 × 101 | 1 × 101 | 1 × 101 | 1 × 101 | 3 × 101 | 6 × 102 |
| 30 | E. coli A | 6 | 2 × 101 | 4 × 101 | 2 × 101 | 3 × 102 | 101 | 2 × 102 |
| 31 | E. coli B | 3 | 1 × 101 | 2 × 103 | 1 × 101 | 3 × 103 | 101 | 2 × 102 |
| 32 | E. cloacae A | 8 | 4 × 101 | 3 × 102 | 2 × 101 | 8 × 101 | 3 × 101 | 103 |
Fecal suspensions spiked with serially diluted NDM-1 producers showed similar detection limits with ChromID ESBL medium and CHROMagar KPC medium, with the limits being slightly higher for CHROMagar KPC medium, especially with isolates with low MICs for imipenem (Table 2).
Detection by qPCR of blaNDM-1 in spiked fecal suspensions.
The real-time blaNDM-1 assay was capable of detecting 10 to 30 CFU equivalents/100 mg of feces, which corresponds to less than 1 CFU per PCR, or 100 to 1,000 copies of the blaNDM-1 gene/100 mg of feces (which corresponds to 1 to 10 copies per PCR). Beyond 10 CFU, which means less than 1 CFU equivalent/PCR, results were not reproducible (Fig. 2B). The analytical sensitivity of the assay did not change upon the utilization of different blaNDM-1-positive strains with different carbapenem sensitivity patterns: Citrobacter freundii, E. cloacae, E. coli, or K. pneumoniae blaNDM-1-positive strains.
Automated extraction using QIAcube.
To assess whether the automation of the purification procedure has any impact on the nucleic acid quantity or quality, aliquots from the same stool samples were purified in manual and QIAcube automated procedures, and the blaNDM-1-specific qPCR was performed using the resulting end products. No difference in terms of sensitivity and specificity was observed (data not shown).
DISCUSSION
Early identification of NDM producers among both bacteria that are causing clinical infections and bacteria that are colonizers is mandatory to prevent their spread. Identification of NDM-1 producers in clinical infections should be suspected on any decreased susceptibility to carbapenems in Enterobacteriaceae, especially in E. coli, where frank resistance to carbapenems may not always be obvious (18). In the routine laboratory, detection of carbapenem resistance is mainly based on phenotypic methods, but they often need technical changes and are time-consuming (13, 17, 18). ChromID ESBL culture medium is reliable for screening for NDM-1 producers in a carriage state directly from fecal samples. The lowest limits of detection of NDM-1 producers ranged from 1 × 101 to 4 × 101 CFU/ml for ChromID ESBL medium (Table 2). The excellent ability of that medium to detect NDM-1 producers is based on the fact that those producers were also resistant to expanded-spectrum cephalosporins and is in part due to the broad-spectrum hydrolytic properties of the NDM-1 β-lactamase (23, 30, 33). Therefore, detection of this β-lactamase was possible using ChromID ESBL, even though a strain may not express an ESBL (E. coli B, for example; Table 1). The CHROMagar culture medium may be proposed for use in the follow-up of an outbreak of NDM-1 producers after identification of the first case and after it is determined that this medium is sensitive enough for detecting the specific NDM-1 producer strain responsible for the outbreak.
Unambiguous identification of NDM-1 by phenotypic methods is rather difficult (19). Altogether, these difficulties delay the response to an outbreak and/or its epidemiologic surveillance. Alternative strategies, including qPCR, have proven their usefulness for detecting KPC (8) and for the management of an epidemic situation. Hindiyeh et al. (8) described the development and verification of a qPCR assay for detection of blaKPC genes directly in perianal swab specimens. Carbapenem-resistant organisms, all belonging to K. pneumoniae, were isolated from 25.1% of 187 perianal swab samples, while PCR assays detected blaKPC-3 genes in 28.9% of the samples (8). Direct detection of blaKPC by PCR shortened the time required to identify patients colonized or infected with carbapenem-resistant organisms and was more sensitive than culture. Similarly, blaNDM-1 qPCR showed excellent sensitivity and specificity, with detection limits lower than those of the culture techniques and with a significant gain of time to results (4 h versus 48 h).
Routine laboratories often receive low numbers of samples that require immediate processing and that are therefore often processed manually. However, processing of small numbers of samples requires a lot of hands-on time that could be more profitably used for more valuable work. Here we evaluated the option of automating low-throughput sample processing on the highly versatile QIAcube apparatus, which enables numerous different purification procedures to be performed using a single instrument. The evaluation was facilitated by the fact that the column technologies used on the QIAcube directly correspond to those used in the manual kit procedures, providing a direct and reliable reference for each purification procedure automated on the QIAcube. qPCR results were comparable using these two extraction procedures. Taken altogether, with 2 h of automated extraction and less than 2 h of PCR, the NDM-1 qPCR yielded results in less than 4 h, compared with the 2 to 3 days required using conventional methods.
The genetics of multidrug-resistant bacteria have taught us that while it is difficult to prevent their emergence, active surveillance and early detection are mandatory for preventing further spread. Here, we developed a qPCR assay for use directly on stool samples for screening for NDM-1 carriers. This qPCR assay is also capable of detecting the newly described variant NDM-2 (9), since the nucleotide change between the two variants is located outside the primer and probe binding sequences. We found that the assay had good sensitivity and specificity and excellent agreement with culture screening techniques. Routine screening for carriage of NDM producers should be based on selective culture media, while the molecular techniques for screening directly from stools will reveal their usefulness in an outbreak situation for preventing the further spread of NDM producers.
ACKNOWLEDGMENTS
This work was funded by grants from INSERM (U914), by the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539, Université Paris XI, Paris, France), and by the European Community's Seventh Framework Programme FP7/2007-2013 under grant agreement no. 241742.
Footnotes
Published ahead of print on 20 June 2011.
REFERENCES
- 1. Birkett C. I., et al. 2007. Real-time TaqMan PCR for rapid detection and typing of genes encoding CTX-M extended-spectrum β-lactamases. J. Med. Microbiol. 56:52–55 [DOI] [PubMed] [Google Scholar]
- 2. Bisiklis A., Papageorgiou F., Frantzidou F., Alexiou-Daniel S. 2007. Specific detection of blaVIM and blaIMP metallo-beta-lactamase genes in a single real-time PCR. Clin. Microbiol. Infect. 13:1201–1203 [DOI] [PubMed] [Google Scholar]
- 3. Bush K. 2010. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr. Opin. Microbiol. 13:558–564 [DOI] [PubMed] [Google Scholar]
- 4. Carrër A., Fortineau N., Nordmann P. 2010. Use of ChromID ESBL medium for detecting carbapenemase-producing Enterobacteriaceae. J. Clin. Microbiol. 48:1913–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing. CLSI M100-S20U. Update June 2010 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. Cole J. M., Schuetz A. N., Hill C. E., Nolte F. S. 2009. Development and evaluation of a real-time PCR assay for detection of Klebsiella pneumoniae carbapenemase genes. J. Clin. Microbiol. 47:322–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cuzon G., Naas T., Lesenne A., Benhamou M., Nordmann P. 2010. Plasmid-mediated carbapenem-hydrolysing OXA-48 beta-lactamase in Klebsiella pneumoniae from Tunisia. Int. J. Antimicrob. Agents 36:91–93 [DOI] [PubMed] [Google Scholar]
- 8. Hindiyeh M., et al. 2008. Rapid detection of blaKPC carbapenemase genes by real-time PCR. J. Clin. Microbiol. 46:2879–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaase M., et al. 2011. NDM-2 carbapenemase in Acinetobacter baumannii from Egypt. J. Antimicrob. Chemother. 66:1260–1262 [DOI] [PubMed] [Google Scholar]
- 10. Kumarasamy K. K., et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Livermore D. M., et al. 2007. CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59:165–174 [DOI] [PubMed] [Google Scholar]
- 12. Mendes R. E., et al. 2007. Rapid detection and identification of metallo-beta-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. J. Clin. Microbiol. 45:544–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miriagou V., et al. 2010. Acquired carbapenemases in Gram-negative bacterial pathogens: detection and surveillance issues. Clin. Microbiol. Infect. 16:112–122 [DOI] [PubMed] [Google Scholar]
- 14. Naas T., Cuzon G., Truong H., Bernabeu S., Nordmann P. 2010. Evaluation of a DNA microarray, the check-points ESBL/KPC array, for rapid detection of TEM, SHV, and CTX-M extended-spectrum beta-lactamases and KPC carbapenemases. Antimicrob. Agents Chemother. 54:3086–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Naas T., Oxacelay C., Nordmann P. 2007. Identification of CTX-M-type extended-spectrum-β-lactamase genes using real-time PCR and pyrosequencing. Antimicrob. Agents Chemother. 51:223–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Naas T., Poirel L., Nordmann P. 2008. Minor extended-spectrum β-lactamases. Clin. Microbiol. Infect. 14(Suppl. 1):42–52 [DOI] [PubMed] [Google Scholar]
- 17. Nordmann P., Cuzon G., Naas T. 2009. The real threat of KPC carbapenemase-producing bacteria. Lancet Infect. Dis. 9:321–331 [DOI] [PubMed] [Google Scholar]
- 18. Nordmann P., Poirel L., Carrër A., Toleman M. A., Walsh T. R. 2011. How to detect NDM-1 producers. J. Clin. Microbiol. 49:718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oxacelay C., Ergani A., Naas T., Nordmann P. 2007. Rapid detection of CTX-M-producing Enterobacteriaceae in urines samples. J. Antimicrob. Chemother. 64:986–989 [DOI] [PubMed] [Google Scholar]
- 20. Paterson D. L., Bonomo R. A. 2005. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pitout J. D., Hamilton N., Church D. L., Nordmann P., Poirel L. 2007. Development and clinical validation of a molecular diagnostic assay to detect CTX-M-type β-lactamases in Enterobacteriaceae. Clin. Microbiol. Infect. 13:291–297 [DOI] [PubMed] [Google Scholar]
- 22. Poirel L., Lagrutta E., Taylor P., Pham J., Nordmann P. 2010. Emergence of metallo-β-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob. Agents Chemother. 54:4914–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Poirel L., et al. 2011. Extremely drug-resistant Citrobacter freundii isolate producing NDM-1 and other carbapenemases identified in a patient returning from India. Antimicrob. Agents Chemother. 55:447–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Queenan A. M., Bush K. 2007. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20:440–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Randegger C. C., Hachler H. 2001. Real-time PCR and melting curve analysis for reliable and rapid detection of SHV extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 45:1730–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Réglier-Poupet H., et al. 2008. Performance of ChromID ESBL, a chromogenic medium for detection of Enterobacteriaceae producing extended-spectrum β-lactamases. J. Med. Microbiol. 57:310–315 [DOI] [PubMed] [Google Scholar]
- 27. Rodriguez-Martinez J. M., Nordmann P., Fortineau N., Poirel L. 2010. VIM-19, a metallo-beta-lactamase with increased carbapenemase activity from Escherichia coli and Klebsiella pneumoniae. Antimicrob. Agents Chemother. 54:471–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29. Samra Z., et al. 2008. Evaluation of CHROMagar KPC for rapid detection of carbapenem-resistant Enterobacteriaceae. J. Clin. Microbiol. 46:3110–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walsh T. R. 2010. Emerging carbapenemases: a global perspective. Int. J. Antimicrob. Agents 36:S8–S14 [DOI] [PubMed] [Google Scholar]
- 31. Woodford N., Fagan E. J., Ellington M. J. 2006. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J. Antimicrob. Chemother. 57:154–155 [DOI] [PubMed] [Google Scholar]
- 32. Woodford N., et al. 2010. Comparison of BD Phoenix, Vitek2, and Microscan automated systems for detection and inference of mechanisms responsible for carbapenem resistance in Enterobacteriaceae. J. Clin. Microbiol. 48:2999–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yong D., et al. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]