Abstract
Hepatitis C virus (HCV) inhibitors include direct-acting antivirals (DAAs) such as NS3 serine protease inhibitors, nucleoside and nonnucleoside polymerase inhibitors, and host-targeting antivirals (HTAs) such as cyclophilin inhibitors that have been developed in recent years. Drug-resistant HCV variants have been reported both in vitro and in the clinical setting for most classes of drugs. We report a comparative study in which the genetic barrier to drug resistance of a representative selection of these inhibitors is evaluated employing a number of resistance selection protocols. The NS3 protease inhibitors VX-950 and BILN 2061, the nucleoside polymerase inhibitor 2′-C-methylcytidine, three nonnucleoside polymerase inhibitors (thiophene carboxylic acid, benzimidazole, and benzothiadiazine), and DEB025 were included. For each drug and passage in the selection process, the phenotype and genotype of the drug-resistant replicon were determined. For a number of molecules (BILN 2061 and nonnucleoside inhibitors), drug-resistant variants were readily selected when wild-type replicon-containing cells were directly cultured in the presence of high concentrations of the inhibitor. Resistance to DEB025 could be selected only following a lengthy stepwise selection procedure. For some DAAs, the signature mutations that emerged under inhibitor pressure differed depending on the selection protocol that was employed. Replication fitness of resistant mutants revealed that the C445F mutation in the RNA-dependent RNA polymerase can restore loss of fitness caused by a number of unfit resistance mutations. These data provide important insights into the various pathways leading to drug resistance and allow a direct comparison of the genetic barriers of various HCV drugs.
INTRODUCTION
Hepatitis C virus (HCV) is a positive single-stranded RNA virus and the only member of the Hepacivirus genus within the Flaviviridae family. An estimated 170 million people are chronically infected worldwide. Three million to four million people become newly infected each year (57). Chronically infected patients are at increased risk of developing liver cirrhosis and hepatocellular carcinoma. In Western countries, infection with HCV is the most common reason for liver transplantation. The current standard of care for the management of chronic hepatitis C virus infection consists of the combination of pegylated alpha interferon (pegIFN-α) and ribavirin. This therapy is effective in only 50 to 60% of infected patients and is associated with serious side effects (44). Therefore, more tolerable, highly potent inhibitors of HCV replication are urgently needed and are currently also being developed. Antivirals that specifically target viral proteins are referred to as “direct-acting antivirals” (DAAs) for HCV. A number of NS3/NS4A protease inhibitors are currently in clinical development. The first HCV NS3/4A serine protease inhibitor to enter clinical trials was ciluprevir (BILN 2061) (54), but clinical development was halted because of cardiotoxicity. Other protease inhibitors in clinical development include danoprevir (ITMN-191), narlaprevir (SCH 900518), and vaniprevir (MK-7009); telaprevir (VX-950), boceprevir (SCH-503034), and TMC435 progressed into phase III clinical trials. Both nucleoside and nonnucleoside inhibitors of the HCV RNA-dependent RNA polymerase (RdRp) have been identified. Nucleoside analogues mimic natural polymerase substrates and cause chain termination following phosphorylation to their corresponding 5′ triphosphate. Valopicitabine (2′-C-methylcytidine [2′-CMC]) was the first nucleoside analogue to enter clinical trials. Development has been discontinued because of modest antiviral efficacy along with significant gastrointestinal side effects (2). RG7128, a prodrug of nucleoside analogue PSI-6130 (β-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine), and PSI-7977, a liver-targeted prodrug of the uridine nucleotide analogue PSI-6206 monophosphate, are in phase II clinical trials. A number of structurally unrelated nonnucleoside polymerase inhibitors have been reported; these include, but are not limited to, benzimidazoles, benzothiadiazines, thiophene derivates, benzofuranes, and imidazopyridines (14). Recently, inhibitors of other targets, such as (i) the entry process, (ii) NS4A (74), (iii) NS4B (7, 17), and (iv) NS5A (31), have also been identified (14). Not surprisingly, monotherapy with most DAAs has been associated with the rapid emergence of resistant variants (63).
On the other hand, host factors that are essential for efficient viral replication may also be good antiviral targets. Host-targeting antivirals (HTAs) may have a higher barrier to resistance than (most) DAA inhibitors. A number of cyclophilin-binding molecules such as alisporivir (DEB025), NIM811, and SCY-635 have proven to be potent inhibitors of HCV replication and have shown clinical efficacy (30, 47).
The rapid emergence of drug-resistant variants of HCV is, as is also the case with HIV, of major concern and results from several factors. These include the poor fidelity and lack of exonucleolytic proofreading capacity of the reverse transcriptase (RT) enzyme in the case of HIV (error rate, 10−5 mutations per nucleotide per genomic replication) (4) or the RdRp in the case of HCV (error rate, 10−3 to 10−5 mutations per nucleotide per genomic replication) (9) and the very high magnitude of replication (HCV, 1012 virions/day [48]; HIV, 1010 virions/day [51]). As a result, multiple viral variants known as quasispecies (4, 59) are generated. In various clinical studies for both HIV and HCV in which antiretroviral drugs or DAA inhibitors were given as monotherapy, escape mutants were shown to develop very rapidly. For example, the administration of a single dose of the nonnucleoside RT inhibitor (NNRTI) nevirapine to prevent mother-to-child HIV transmission rapidly and routinely selects for NNRTI-resistant mutants (23). Also, for HIV protease inhibitors, more than 80 mutations have been reported (56). When HCV-infected patients were treated with telaprevir for 14 days, viral breakthrough, associated with a number of mutations that confer low-level and high-level resistance to telaprevir, was noted in a significant number of patients (55). Drug-resistant variants also rapidly emerged in clinical trials with other protease inhibitors such as boceprevir (61) and nonnucleoside polymerase inhibitors (49, 69). These drug-resistant HCV variants may be present at frequencies of <1% in the quasispecies population in treatment-naïve patients (27, 35), which may result in a rapid selection during DAA treatment. These findings are in agreement with those of in vitro resistance studies in which HCV is shown to develop (often rapidly) resistance against polymerase and protease inhibitors.
HCV subgenomic replicons have been widely used in the discovery and the development of DAA inhibitors. Drug-resistant HCV replicons have been obtained for most classes of drugs. However, since different resistance selection protocols are used in different studies, it is not possible to directly compare the genetic barrier to antiviral drug resistance of various (classes of) HCV drugs. We here report a comparative study in which the genetic barrier to drug resistance of a selection of reference compounds is evaluated employing a number of resistance selection protocols. The NS3 protease inhibitors (VX-950, BILN 2061), a nucleoside polymerase inhibitor (2′-C-methylcytidine), three nonnucleoside polymerase inhibitors (thiophene carboxylic acid, benzimidazole JT-16, and benzothiadiazine A-782759) as well as the cyclophilin-binding molecule DEB025 were used for this purpose.
MATERIALS AND METHODS
Compounds.
The following molecules were included in this study: (i) the nucleoside polymerase inhibitor 2′-CMC (the active moiety of NM283) (8), (ii) the nonnucleoside polymerase inhibitors 2-[4-[[4-(acetylamino)-4′-chloro-[1,1′-biphenyl]-2-yl]methoxy]phenyl]-1-cyclohexyl-1H-benzimidazole-5-carboxylic acid (JT-16) (21), 3-{isopropyl[(trans-4-methylcyclohexyl)carbonyl]amino}-5-phenylthiophene-2-carboxylic acid (TCA) (33), and 2-[3-(1-cyclobutylamino-4-hydroxy-2-oxo-1,2-dihydro-quinolin-3-yl)-1,1-dioxo-1,4-dihydro-1l6-benzo(1,2,4)thiadiazin-7-yloxy]-acetamide (A-782759) (53), (iv) the protease inhibitors (2R,6S,12Z,13aS,14aR,16aS)-6-[[(cyclopentyloxy)carbonyl]amino]-1,2,3,6,7,8,9,10,11,13a,14,15,16,16a,-tetradecahydro-2-[[7-methoxy-8-methyl-2-[2-[(1-methylethyl)amino]-4-thiazolyl]-4-quinolinyl]oxy]-5,16-dioxocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecine-14a(5H)-carboxylic acid (BILN 2061) (28), and (1S,3aR,6aS)-2-[2S)-2-[[(2S)-2-cyclohexyl-2-(pyrazine-2-carbonylamino)acetyl]amino]-3,3-dimethylbutanoyl]-N-[(3S)-1-(cyclopropylamino)-1,2-dioxohexan-3-yl]-3,3a,4,5,6,6a-hexahydro-1H-cyclopenta[c]pyrrole-1-carboxamide (VX-950) (60), and (v) the cyclophilin-binding compound DEB025 (13). All molecules were synthesized in-house by medicinal chemists.
Selection of drug-resistant replicon.
Huh-7 cells containing subgenomic HCV replicons I377/NS3-3′/wild type (WT) (Huh 9-13) (39) were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco, Merelbeke, Belgium) supplemented with 10% heat-inactivated fetal calf serum (Integro, Zaandam, The Netherlands), 1× minimal essential medium nonessential amino acid solution without l-glutamine (Gibco), 100 IU/ml of penicillin and 100 μg/ml of streptomycin (Gibco), and 1 mg/ml G418 (Geneticin-selective antibiotic; Gibco). Huh-Lunet cells, which are derived from a cell clone that was generated by curing Huh-7 replicon cells with a selective drug, were cultured without G418 (19).
Various drug-resistant replicons were selected following culturing of Huh 9-13 replicon-containing cells for 11 passages (5.5 weeks) under constant antiviral pressure (1×, 2×, 5×, 25×, and 125× 50% effective concentration [EC50]). When replicon-containing cells suffered (massive cell death) from compound pressure (because of replicon disappearance), G418 and compound were removed until cells recovered. This particular observation during cell culture was designated “critical phase,” and all these events are listed in Tables 1 to 7. Thereafter, cells were recultured in the presence of the antiviral drug and G418 pressure until the predetermined 5.5 culture weeks were completed. Subsequently, Huh 9-13 cells were challenged with gradually increasing doses of antiviral pressure; e.g., cells surviving doses of 5× EC50 were subsequently challenged with 25× or 125× EC50. Since certain compounds are, at particular concentrations, toxic for the host cell, drug selection protocols were in this case not carried out with such concentrations. Following selection of drug-resistant cultures, the genotypes and phenotypes of all replicons obtained during the different selection protocols were determined. When the replicon proved ≥4-fold less susceptible to the inhibitor than the wild-type replicon in the case of resistance culture or ≥3-fold less susceptible to the inhibitor than the wild-type replicon in the case of transient transfections, the replicon/mutant was considered resistant. These thresholds were set according to Table SA1 in the supplemental material, in which the variability of the antiviral assays is calculated.
Table 1.
Mutations and antiviral phenotype of replicons obtained following culturing in the presence of BILN 2061a using various selection protocols
| Antiviral pressure (fold EC50) | Fold increaseb compared to WT | EC50 ± SDb (μM) | NS3 mutation(s) (aa 22-336)c | Critical phased |
|---|---|---|---|---|
| 1 | 2 | 0.033 ± 0.008 | S174S/C | N |
| 2 | 16 | 0.24 ± 0.08 | D168D/E/V, S174S/C | N |
| 5 | 13 | 0.19 ± 0.09 | D168D/E/V, S174S/C | N |
| 25 | 67 | 1 ± 0.4 | D168V, E176E/G | Y |
| 125 | 200 | 3 ± 0.5 | D168V, S174S/C, E176E/G | Y |
| 2-5 | 17 | 0.26 ± 0.007 | D168D/E/V, S174S/C | N |
| 2-25 | 61 | 0.92 ± 0.3 | D168V, S174S/C, E176E/G | N |
| 2-125 | 133 | 2 ± 0.2 | D168V, S174S/C, E176E/G | N |
| 5-25 | 56 | 0.84 ± 0.005 | D168D/V, S174S/C, E176E/G | N |
| 5-125 | 133 | 2 ± 0.6 | D168V, S174S/C, E176E/G | N |
| 25-125 | 133 | 2 ± 0.3 | D168V | N |
| 2-5-25 | 133 | 2 ± 0.6 | D168V, S174S/C | N |
| 2-5-125 | 133 | 2 ± 0.6 | D168V, S174S/C | N |
| 2-25-125 | 133 | 2 ± 0.5 | D168V, S174S/C, S280S/A | N |
| 5-25-125 | 200 | 3 ± 0.04 | D168V, S174S/C, E176E/G, S280S/A | N |
| 2-5-25-125 | 267 | 4 ± 0.7 | D168V, S174S/C | N |
EC50, 0.015 ± 0.01 μM; CC50, 22.6 ± 0.1 μM.
Data are mean values for 2 independent determinations of the antiviral phenotype.
Major resistance mutations are indicated in boldface. aa, amino acids.
Y, yes; N, no.
Table 7.
Mutations and antiviral phenotype of replicons obtained following culturing in the presence of DEB025a using various selection protocols
| Antiviral pressure (fold EC50) | Fold increaseb compared to WT | EC50 ± SDb (μM) | NS5A mutations (aa 26-447)c | Critical phased |
|---|---|---|---|---|
| 1 | 2 | 0.094 ± 0.01 | NP | N |
| 2 | 4 | 0.15 ± 0.05 | NP | Y |
| 5 | — | NA | ND | Y |
| 25 | — | NA | ND | Y |
| 125 | — | NA | ND | Y |
| 2-5 | 10 | 0.41 ± 0.05 | L287L/P, D320E, D409D/V | Y |
| 2-25 | — | NA | ND | Y |
| 2-125 | — | NA | ND | Y |
| 2-5-25 | 21 | 0.88 ± 0.007 | L287L/P, D320E, D409D/V | Y |
| 2-5-125 | — | NA | ND | Y |
| 2-5-25-125 | — | NA | ND | Y |
EC50, 0.041 ± 0.03 μM; CC50, 12 ± 6 μM.
Data are mean values for 2 independent determinations of the antiviral phenotype. —, cell cultures died; NA, not applicable.
The major resistance mutation is indicated in boldface. aa, amino acids; NP, none present; ND, not determined.
Y, yes; N, no.
Phenotyping of wild-type or drug-resistant Huh 9-13 replicon cells.
Antiviral assays were performed as described before (15). Briefly, cells were seeded at a density of 5 × 103 cells per well in a 96-well cell culture plate in complete DMEM. Following incubation for 24 h at 37°C (5% CO2), serial dilutions of the test compounds in complete DMEM were added in a total volume of 100 μl and cells were cultured for an additional 3 days. Replicon RNA levels were determined by a reverse transcription-quantitative PCR. Primers used for detection of HCV replicon RNA were 5′-CCG GCT ACC TGC CCA TTC-3′ (forward primer), 5′-CCA GAT CAT CCT GAT CGA CAA G-3′ (reverse primer), and 5′-FAM-ACA TCG CAT CGA GCG AGC ACG TAC-TAMRA-3′ (probe; where FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine). The EC50 was defined as the concentration of compound that reduced HCV RNA replication by 50%. The fold resistance value was calculated as the ratio of the EC50 in the resistant replicon to the EC50 in the wild-type replicon. Maximum fold resistance values were calculated as the ratio of the highest EC50 of the inhibitor tested to the mean EC50 in the wild-type replicon.
Cytostatic assays were performed as described before (15). Briefly, cells were seeded at a density of 5 × 103 cells per well in a 96-well cell culture plate in complete DMEM. After 24 h of incubation at 37°C, serial dilutions of the test compounds in complete DMEM were added. After 3 days of incubation at 37°C, cell viability was determined using the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium/phenazine methosulfate (MTS/PMS) method (Promega). The 50% cytotoxic concentration (CC50) value was defined as the concentration that inhibited the proliferation of exponentially growing cells by 50%.
Population sequencing of resistant replicon RNA.
Total RNA of the wild-type or resistant replicon population was extracted from 5 × 104 replicon cells using an RNeasy minikit (Qiagen, Venlo, The Netherlands), according to the manufacturer's instructions. cDNA fragments encompassing selected fragments of the HCV genome were amplified with 0.6 μM HCV-specific primers (see Table SA2 in the supplemental material) using a Qiagen OneStep reverse transcription-PCR kit. The reverse transcription-PCR program was as follows: 30 min at 50°C for reverse transcription and 15 min at 95°C to activate the HotStar Taq enzyme, followed by 30 cycles of 30 s at 94°C, 30 s at 60°C, 55°C, or 50°C, and 60 s at 72°C. A final elongation step of 10 min at 72°C was performed after cycling. Amplification products were purified using a Wizard SV Gel and PCR cleanup system (Promega Benelux, Leiden, The Netherlands), and nucleotide sequences were determined using the same primers (final concentration, 0.5 μM) used for reverse transcription-PCR and the BigDye Terminator (version 3.1) sequencing system (Applied Biosystems, Nieuwerkerk Ad IJssel, The Netherlands). Mutations that are detected in both wild-type and resistant Huh 9-13 replicon-containing cells were not included in the mutational analysis. Furthermore, no linkage between mutations was implied.
Clonal sequencing of wild-type replicon.
HCV RNA was isolated from Huh 9-13 cells using the RNeasy minikit (Qiagen Benelux), according to the manufacturer's instructions. cDNA fragments were synthesized using the Transcriptor high-fidelity cDNA synthesis kit (Roche Diagnostics, Vilvoorde, Belgium). The cDNAs were subjected to amplification by PCR using the 9F/9R primers (also used for population sequencing) and an AccuPrime Pfx DNA polymerase kit (Invitrogen, Merelbeke, Belgium) according to the manufacturer's instructions. This polymerase was chosen, as it possesses a proofreading 3′ to 5′ exonuclease activity. The appropriately sized product was than purified by the Wizard SV Gel and PCR cleanup system (Promega) and cloned using a TOPO TA cloning kit for sequencing (Invitrogen). Transformed TOP10 Escherichia coli cells were plated on ampicillin-LB agar plates. Colonies were randomly picked, and 96 clones were sent for sequencing using the M13F/M13R primers at Beckman Coulter Genomics (formerly Agencourt Bioscience and Cogenics; Takeley, United Kingdom).
Site-directed mutagenesis.
Various published and drug-selected resistance mutations were introduced in pFK I389 Lucibineo EI NS3-3′ET (71), including D168V in NS3 (BILN 2061), S282T in NS5B (2′-CMC), C316Y in NS5B (A-782759), T389A in NS5B (JT-16), M414T in NS5B (A-782759), M423T in NS5B (TCA), C445F in NS5B (A-782759, JT-16, TCA), P495L in NS5B (JT-16), Y452H in NS5B (A-782759), C316Y and C445F in NS5B (A-782759), and C445F and Y452H in NS5B (A-782759).
Initially, the NS5B or NS3 gene sequences were excised from the pFK I389 Lucubineo EI NS3-3′ET construct by SpeI-XhoI or NotI-MluI restriction digestion and subcloned to construct pCRII-HCV5B or pCRII-NS3. Mutations (single or combinations) were introduced into pCRII-HCV5B or pCRII-NS3 (see Table SA3 in the supplemental material). For the double mutants, the C316Y and Y452H mutations were constructed in pCRII-HCV5B containing the C445F mutation. The construction of the T389A mutant will be described elsewhere. Site-directed mutagenesis was performed using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The 50-μl reaction mixture contained 1× reaction buffer, 50 ng plasmid solution, 125 ng forward primer, 125 ng reverse primer, 1 μl deoxynucleoside triphosphate mix, and 2.5 U PfuTurbo DNA polymerase. Thermal cycling was performed as follows: denaturation at 95°C 30 s, followed by 12 cycles (for 1 mutation) or 16 cycles (for 2 mutations) of 30 s at 95°C, 1 min at 55°C, and 7 min at 68°C. Following temperature cycling, the reaction mixture was cooled down to 4°C and digested with 10 U DpnI to remove the methylated, nonmutated parental supercoiled double-stranded DNA template. The mutated pCRII-HCV5B or pCRII-NS3 plasmid was then transformed in One Shot TOP10 chemically competent E. coli cells (Invitrogen), and positive colonies were subcultured the next day. Plasmid DNA was collected by a Wizard Plus SV miniprep kit (Promega Benelux) and digested with SpeI-XhoI for NS5B or NotI-MluI for NS3 to collect the mutated NS5B/NS3 fragments. The mutated fragments were ligated into digested pFK I389 Lucibineo EI NS3-3′ET and subsequently transformed into One Shot TOP10 chemically competent E. coli cells. Plasmid DNA was collected by the Wizard Plus SV miniprep kit. To control for the correct plasmid insert, a restriction digest was performed. In addition, to confirm the presence of the desired mutations, the entire NS5B and NS3 inserts were sequenced. The M423T mutant (a kind gift from W. Zhong, Gilead Sciences) was obtained in a similar way as the other NS3 and NS5B mutants. In short, the genotype 1b PI-Rluc replicon plasmid was created from the pFK I341 PI-Luc/NS3-3′ET construct (38) by replacing the firefly luciferase reporter gene with a Renilla luciferase reporter sequence (Promega Benelux). The 1b PI-Rluc construct encoding an M423T mutation in NS5B was produced by replacing the wild-type NS5B BclI-SpeI fragment with an analogous fragment in which the M423T mutant sequence had previously been introduced via site-directed mutagenesis using the QuikChange site-directed mutagenesis kit of Stratagene.
Transient transfection. (i) In vitro transcription.
Total DNA isolated with the Wizard Plus SV miniprep kit from an overnight E. coli culture transformed with mutant HCV replicon was linearized using AseI (New England BioLabs, Germany) and ScaI (Promega Benelux) or only SpeI (Promega Benelux) in the case of M423T. The linearized plasmid was phenol-chloroform extracted, ethanol precipitated, and dissolved in RNase-free water. In vitro transcription was performed on 5 μg DNA by a RiboMAX large-scale RNA production system-T7 enzyme mix (Promega Benelux) according to the manufacturer's instructions. After 2 h at 37°C, an additional 5 μl of enzyme mix (T7) was added and the reaction mixture was incubated for an extra 2 h at 37°C. Transcription was ended by adding RQ1 RNase-free DNase (Promega Benelux). RNA was purified and collected by using the RNeasy miniprotocol for RNA cleanup (RNeasy minikit; Qiagen Benelux). The concentration and purity of RNA were spectrophotometrically measured.
(ii) Transfection.
Twenty micrograms of in vitro-transcribed RNA was mixed with 400 μl of a suspension of 4 × 106 Huh-Lunet cells in a cuvette with a gap width of 0.4 cm (VWR International, Leuven, Belgium). After one pulse at 1,600 V with an ECM 830 Electro Square Porator (BTX Harvard Apparatus), cells were immediately transferred into 20 ml of complete Dulbecco's modified Eagle medium without G418. One hundred-microliter aliquots of the cell suspension were seeded in a 96-well plate (Iwaki, Asahi Techno Glass, Japan) previously filled with serial dilutions of the test compounds in complete Dulbecco's modified Eagle medium without G418. Cells were allowed to proliferate for 4 days at 37°C, after which the luciferase activity was determined using a Steady-Glo luciferase assay system (Promega Benelux); the luciferase signal was measured using a Luminoskan Ascent apparatus (Thermo, Vantaa, Finland). The EC50 was defined as the concentration of compound that reduced the luciferase signal by 50% compared to the signal for nontreated transfected cells.
Replication fitness.
Transfections were performed in Huh-Lunet cells as described above, with the exception that cells were transfected with 5 μg RNA. To stabilize the input mutant RNA, an additional 5 μg tRNA (Sigma-Aldrich, Bornem, Belgium) was added. Transfected cells were immediately transferred to 24 ml of complete Dulbecco's modified Eagle medium without G418, and a 2-ml aliquot of the cell suspension was added to a 6-well plate (Iwaki). Cells were collected at 4 h (normalization point) and 1, 2, 3, and 4 days after transfection to compare luciferase values with wild-type values, after extraction of the tRNA background signal. Cells transfected with the GND replicon (a replication-deficient subgenomic replicon encoding a GDD-to-GND mutation in NS5B [62]) were used as a negative control reflecting background activity from the residual input RNA.
Three-dimensional structures of HCV inhibitor binding to NS5B.
Different HCV NS5B RdRp structures, 1YVF (52), 1OS5 (40), and 2BRK (16), containing A-782759, thiophene carboxylic acid, and JT-16, respectively, were superimposed onto a reference structure, 1GX6 (6), using the Dalilite server (Holmer). Structures were loaded in the Chimera program to create three-dimensional figures. Colors used are blue for the fingers domain (residues 1 to 188 and 228 to 286), yellow for the palm domain (residues 188 to 227 and 287 to 370), and pink for the thumb domain (residues 371 to 563). The different inhibitors are highlighted in green carbons and a transparent surface. Mutations are presented as colored balls.
RESULTS
Huh 9-13 replicon-containing cells were cultured for 11 consecutive passages (5.5 weeks) under continuous antiviral pressure (either 1×, 2×, 5×, 25×, or 125× EC50) with a selection of inhibitors. Massive cell death was documented in those cultures where it appeared as a critical phase. Surviving Huh 9-13 cells were subsequently challenged with gradually increasing concentrations of compound for 11 passages per concentration. At the end of the selection protocol, the genotype (amino acid changes are presented in the tables; codon changes are presented in Table SA4 in the supplemental material) and antiviral phenotype of the replicons (see Tables 1 to 7) thus obtained were determined.
Protease inhibitors (BILN 2061 and VX-950).
BILN 2061-resistant variants were readily selected at all steps of the resistance selection procedure, even when wild-type replicon-containing cells were directly cultured in the presence of high drug concentrations (25× or 125× the EC50) (Table 1). Replicons with a reduced susceptibility (∼20-fold) to BILN 2061 were obtained following selection at concentrations as low as 2× the EC50. Selecting resistance in the presence of concentrations of 25× and 125× the EC50 (either directly or gradually) resulted in pronounced resistance (>50-fold increase in EC50). VX-950-resistant variants were also readily selected in all protocols when wild-type replicon-containing cells were directly cultured in the presence of high drug concentrations (5× the EC50) (Table 2).
Table 2.
Mutations and antiviral phenotype of replicons obtained following culturing in the presence of VX-950a using various selection protocols
| Antiviral pressure (fold EC50) | Fold increaseb compared to WT | EC50 ± SDb (μM) | NS3 mutations (aa 1-336)c | Critical phased |
|---|---|---|---|---|
| 1 | 1 | 0.78 ± 0.4 | T22T/I, E176E/G | Y |
| 2 | 3 | 2.0 ± 0.1 | T54T/S, E176E/G, I248I/V, T254T/A, S280S/A | Y |
| 5 | 5 | 3.6 ± 1.4 | T54T/A, A156A/S/T | Y |
| 2-5 | 7 | 5.8 ± 1.0 | T54T/S, V170V/A, E176G, T177T/A, T254A, S280A | Y |
EC50, 0.74 ± 0.8 μM; CC50, 24.1 ± 4.4 μM.
Data are mean values for ≥2 independent determinations of the antiviral phenotype.
Major resistance mutations are indicated in boldface. aa, amino acids.
Y, yes; N, no.
BILN 2061 resistance is associated with mutations R155Q/K, A156T/V, and D168V/A (41, 68). Mutations at amino acid positions R155 and A156 of the HCV NS3 confer resistance to all NS3 protease inhibitors to date. The main resistance mutation for BILN 2061 identified in the present study is D168V. Whenever this substitution is present in the replicon, at least a 60-fold reduced susceptibility is noted. Mutations S174C and S280A were not reported earlier in the context of BILN 2061 resistance. However, it is unlikely that these mutations contribute to the reduced susceptibility for BILN 2061. E176G was described previously as a cell-culture-adaptive mutation (68).
The dominant resistance mutation reported in the literature for VX-950 in vitro is A156S/V/T (36). However, this mutation could be selected only as a multispecies by culturing replicon-containing cells directly in the presence of 5× the EC50. This can be explained by the low concentrations of VX-950 that were used in this study (3 μM), while in previous studies, relatively high concentrations were used (14 and 28 μM) (36). However, substitutions at positions T54 and V170, which were previously reported to be associated with low-level resistance, were observed (63, 73). Like in the BILN 2061-resistant replicons, a previously reported adaptive mutation (E176G) was identified (25). Mutation T254A is most likely an adaptive mutation, as was earlier reported for T254I (25). Mutations at positions T22, T177, I248, and S280 have not been described before. Remarkably, mutation S280A was also observed in some BILN 2061-resistant replicons. These mutations are likely adaptive mutations or may have emerged randomly.
Polymerase inhibitors. (i) Nucleoside analogue (2′-CMC).
A concentration of 25× EC50 (11 μM) cleared replicons from Huh 9-13 cells (Table 3). No resistance against 2′-CMC could be obtained following 5.5 weeks of culturing under all compound concentrations studied. This is in accordance with previously described observations (3, 29). The mutation A281T/A that was detected in the replicon when Huh 9-13 cells were cultured under the pressure of 1× EC50 of 2′-CMC and the T390I/T and A421T/A mutations detected when they were cultured under 2× and then 5× EC50 (the 2-5× EC50 protocol) of 2′-CMC are most likely adaptive mutations that do not contribute to resistance (Table 3).
Table 3.
Mutations and antiviral phenotype of replicons obtained following culturing in the presence of 2′-CMCa using various selection protocols
| Antiviral pressure (fold EC50) | Fold increaseb compared to WT | EC50 ± SDb (μM) | NS5B mutation(s) (aa 246-536)c | Critical phased |
|---|---|---|---|---|
| 1 | 1 | 0.40 ± 0.04 | A281A/T | N |
| 2 | 1 | 0.41 ± 0.07 | NP | N |
| 5 | 2 | 0.99 ± 0.1 | NP | Y |
| 25 | — | NA | ND | Y |
| 2-5 | 2 | 1 ± 0.4 | T390T/I, A421A/T | Y |
| 2-25 | — | NA | ND | Y |
| 5-25 | — | NA | ND | Y |
| 2-5-25 | — | NA | ND | Y |
EC50, 0.43 ± 0.1 μM; CC50, 26 ± 10 μM.
Data are mean values for ≥2 independent determinations of the antiviral phenotype. —, cell cultures died; NA, not applicable.
Major resistance mutations are indicated in boldface. aa, amino acids; NP, none present; ND, not determined.
Y, yes; N, no.
(ii) Nonnucleoside analogues. (a) Benzothiadiazine (A-782759).
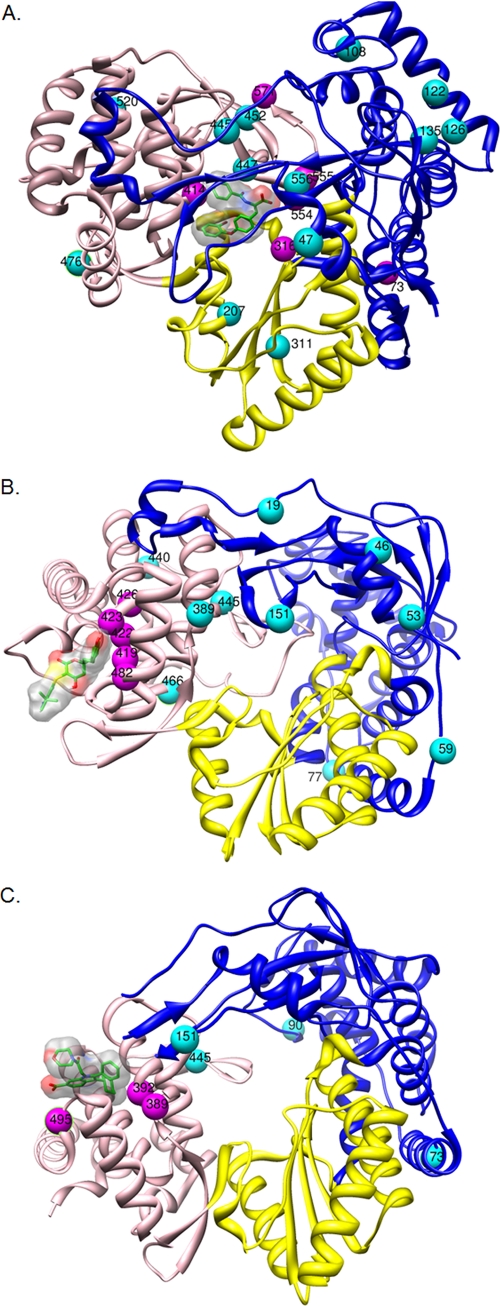
Under none of the conditions studied was A-782759 able to clear the cells of their replicons. Low-level resistance (4- to 5-fold) against A-782759 was observed when replicon-containing cells were cultured in the presence of 1× EC50 (0.075 μM) or 2× EC50 (0.15 μM) of the molecule (Table 4). Mutation S556G was present as a quasispecies in the cultures of replicons cultured in the presence of 2× EC50 or higher of the compound. Replicons cultured in the presence of 5× EC50 (0.38 μM) or using the 2-5× protocol exerted low-level resistance (11- or 13-fold). The mutation C445C/F was identified in both cases. C445C/F was also identified in the replicons obtained in some of the stepwise-culture protocols. C445R has previously been reported in A-782759-resistant replicons (43). M414T, described as being the dominant resistance mutation for A-782759 (46), was observed in the viral genome selected in the presence of the 2-125×, 5-125×, 2-5-25×, 2-5-125×, and 2-25-125× protocols. Mutation W571P was observed in replicons of the 5×, 25×, 2-25-125×, and 5-25-125× cultures and has not been reported earlier to be a benzothiadiazine resistance mutation. Nonetheless, the mutation appears to contribute to resistance (compare 2× and 25× EC50s in Table 4). Mutation G554D and/or Y555C was identified in the 125×, 2-5×, 2-125×, 2-5-25×, 2-5-125×, 2-25-125×, and 2-5-25-125× cultures and has previously been reported for another benzothiadiazine (i.e., A-837093) (42). Of note, the A73S mutation was selected in replicons of all stepwise-culture protocols. Although this mutation is located in the finger domain outside the benzothiadiazine binding site (Fig. 1A), it may contribute to resistance (compare 2× and 2-25× EC50s). However, a mutation at the nearby residue M71 was previously reported to emerge during resistance selection with a benzothiadiazine derivative; this mutation did not confer resistance to the benzothiadiazine. The C316Y mutation, identified in the replicons that were selected using the 5-125×, 2-5-125×, 5-25-125×, and 2-5-25-125× protocols, has previously been reported for A-837093 (10). Mutations at positions L47, V108, V122, L126, D135, A207, C311, I447, S476, Y452, and T520 have not been described before to contribute to benzothiadiazine resistance. These residues are not located in close proximity to the binding site of A-782759 (Fig. 1A).
Table 4.
Mutations and antiviral phenotype of replicons obtained following culturing in the presence of A-782759a using various selection protocols
| Antiviral pressure (fold EC50) | Fold increaseb compared to WT | EC50 ± SDb (μM) | NS5B mutation(s) (aa 1-591)c | Critical phased |
|---|---|---|---|---|
| 1 | 4 | 0.31 ± 0.05 | NP | N |
| 2 | 5 | 0.38 ± 0.03 | S556S/G | N |
| 5 | 11 | 0.86 ± 0.6 | C445C/F, S556S/G, W571P | N |
| 25 | 120 | 9 ± 0.7 | S556S/G, W571P | N |
| 125 | 293 | 22 ± 10 | G554G/D, Y555Y/C, S556S/G | Y |
| 2-5 | 13 | 0.99 ± 0.3 | A73A/S, C445C/F, Y555C/R/Y/H, S556S/G | N |
| 2-25 | 67 | 5 ± 1 | A73A/S, S556S/G | N |
| 2-125 | 240 | 18 ± 7 | A73A/S, M414 M/T, G554G/D, S556S/G | N |
| 5-25 | 80 | 6 ± 0.5 | A73A/S, V122V/L, L126L/V, D135D/G, C445C/F, S556S/G | N |
| 5-125 | >440 | >33 | L47L/M, A73A/S, C316C/Y, M414 M/T, C445C/F, T520T/I, S556S/G | Y |
| 25-125 | 200 | 15 ± 1 | A73A/S, C445C/F, S476S/N, S556S/G | N |
| 2-5-25 | 107 | 8 ± 0.6 | A73A/S, M414 M/T, G554G/D, Y555Y/C, S556S/G | N |
| 2-5-125 | 293 | 22 ± 2 | A73A/S, C311C/Y, C316C/Y, M414 M/T, Y555Y/C, S556S/G | Y |
| 2-25-125 | >440 | >33 | A73A/S, V108V/A, D135D/G, M414 M/T, C445C/F, Y452Y/H, G554G/D, S556S/G, W571P | N |
| 5-25-125 | >440 | >33 | L47L/M, A73A/S, A207A/G, C316C/Y, C445C/F, S556S/G, W571P | N |
| 2-5-25-125 | >440 | >33 | A73A/S, C316C/Y, C445C/F, I447I/M, G554G/D, S556S/G | N |
EC50, 0.075 ± 0.01 μM; CC50 > 33 μM.
Data are mean values for ≥2 independent determinations of the antiviral phenotype.
Major resistance mutations are indicated in boldface. aa, amino acids; NP, none present.
Y, yes; N, no.
Fig. 1.
Three-dimensional structures of NS5B with indication of the amino acid mutations identified following culture in the presence of A-782759 (A), TCA (B), and JT-16 (C). The palm, thumb, and finger domains of HCV NS5B are depicted in yellow, pink, and blue, respectively. The corresponding inhibitor is shown in green. Mutated amino acids conferring resistance are labeled in purple; other mutated amino acids are labeled in magenta.
(b) TCA.
At none of the concentrations used was TCA able to cure cells of their replicons (Table 5). No resistance mutations were observed in the 1× and 2× EC50 selection protocols. Cultures from the 5× and 2-5× conditions exhibited low-level resistance. The C445F mutation, located outside the TCA binding pocket (Fig. 1B), was also identified in the genome of replicons that had been selected using 25× EC50 and all but one stepwise-culture protocol (2-125×). The low-level resistance observed in cultures from the 5× and 2-5× conditions might be attributed to the M426T mutation, as this amino acid is positioned near the TCA binding site (Fig. 1B). E440E/G and K151K/R most likely present adaptive mutations. L419M, M423T/I/V, and I482L, previously reported to be dominant TCA resistance mutations, were detected in several but not all selection protocols (33). T389T/D likely presents an adaptive mutation (compare 5× and 2-25× EC50s). Although the R422R/K mutation, identified in replicons selected using the 2-125×, 5-125×, and 2-5-125× protocols, is located in the TCA binding pocket (5), this mutation was not earlier reported to be a TCA resistance mutation. Moreover, it was described that R422 is present in the thumb binding pocket of phenylalanine derivatives (72). Mutations at positions T19, S46, T53, V59, T77, and L466 have not previously been described to contribute to TCA resistance. These residues are not located in close proximity to the binding site of TCA (Fig. 1B) and are therefore probably not involved in resistance to TCA.
Table 5.
Mutations and antiviral phenotype of replicons obtained following culturing in the presence of TCAa using various selection protocols
| Antiviral pressure (fold EC50) | Fold increaseb compared to WT | EC50 ± SDb (μM) | NS5B mutations (aa 1-591)c | Critical phased |
|---|---|---|---|---|
| 1 | 1 | 0.21 ± 0.02 | NP | N |
| 2 | 1 | 0.32 ± 0.03 | NP | N |
| 5 | 2 | 0.65 ± 0.2 | M426T, E440E/G, C445C/F | N |
| 25 | 17 | 5 ± 1 | T389T/D, L419L/M, M423 M/T, C445C/F | N |
| 125 | 70 | 21 ± 5 | M423 M/T, E440E/G | Y |
| 2-5 | 2 | 0.57 ± 0.2 | K151K/R, M426T, E440E/G, C445C/F | N |
| 2-25 | 33 | 10 ± 4 | L419L/M, M423 M/T, C445F | N |
| 2-125 | >110 | >33 | T19T/P, T53T/I, R422R/K,M423 M/T, M426T, E440E/G, I482I/L | Y |
| 5-25 | 33 | 10 ± 2 | S46S/G, K151K/R, M423 M/T/V/A, C445C/F | N |
| 5-125 | 77 | 23 ± 3 | S46S/G, L419L/M, R422R/K, M423 M/T, E440E/G, C445C/F | N |
| 25-125 | 100 | 30 ± 2 | V59V/A, T77T/P, L419L/M, M423T, M426T, E440E/G, C445C/F | N |
| 2-5-25 | 13 | 4 ± 2 | K151K/R, L419L/M, M423 M/T/V/A, M426T, C445C/F | N |
| 2-5-125 | 67 | 20 ± 0.8 | L419L/M, R422R/K, M423 M/T, C445C/F | N |
| 2-25-125 | 33 | 10 ± 0.9 | M423T, C445F, L466L/V | Y |
| 5-25-125 | 80 | 24 ± 6 | S46S/G, M423 M/T/V/A, E440E/G, C445C/F | N |
| 2-5-25-125 | 80 | 24 ± 2 | T53T/I, L419L/M, M423 M/T, M426T, C445C/F | N |
EC50, 0.30 ± 0.2 μM; CC50 >33 μM.
Data are mean values for ≥2 independent determinations of the antiviral phenotype.
Major resistance mutations are indicated in boldface. aa, amino acids; NP, none present.
Y, yes; N, no.
(c) Benzimidazole (JT-16).
JT-16-resistant variants were readily selected in all protocols and were also selected when wild-type replicon-containing cells were directly cultured in the presence of high drug concentrations (5× the EC50) (Table 6). Concentrations of ≥10× EC50 proved cytotoxic.
Table 6.
Mutations and antiviral phenotype of replicons obtained following culturing in the presence of JT-16a using various selection protocols
| Antiviral pressure (fold EC50) | Fold increaseb compared to WT | EC50 ± SDb (μM) | NS5B mutation(s) (aa 1-591)c | Critical phased |
|---|---|---|---|---|
| 1 | 2 | 2 ± 0.5 | C445C/F | N |
| 2 | 5 | 6 ± 1 | A73A/S, T389T/A, C445C/F | Y |
| 5 | 14 | 17 ± 0.6 | K90K/R, K151K/R, P495A | Y |
| 2-5 | 11 | 14 ± 3 | T389T/A, L392L/I, C445C/F | N |
EC50, 1.2 ± 0.8 μM; CC50, 23 ± 5 μM.
Data are mean values for ≥2 independent determinations of the antiviral phenotype.
Major resistance mutations are indicated in boldface. aa, amino acids.
Y, yes; N, no.
Antiviral resistance to benzimidazole derivatives has been shown to be mainly associated with mutations at positions P495, P496, and V499 in the thumb domain (26, 64). Mutation P495A was detected under the condition 5× EC50. Substitutions at position T389 were not previously reported to be involved in benzimidazole resistance. However, experiments with mutants with the T389A and T389S mutations revealed that these mutations reduced the susceptibility to JT-16 (our unpublished results). L392I, which is located in the proximity of T389 (Fig. 1C), was earlier reported to be responsible for benzimidazole resistance (50). The C445F mutation is a known resistance mutation for benzofuran compounds such as HCV-796 (22) but is probably a compensatory mutation in benzimidazole-resistant replicons. Mutations observed outside the thumb domain (A73S, K90R, K151R) are likely adaptive mutations or may have emerged randomly in that particular population and may be of no biological relevance. These residues are not located in close proximity to the binding site of JT-16 (Fig. 1C).
Cyclophilin inhibitors.
Resistance to the cyclophilin-binding compound DEB025 could be selected only following a stepwise selection procedure (Table 7). When wild-type replicon-containing cells were directly cultured in the presence of 5×, 25×, or 125× EC50, the replicon-containing cells were cured of their replicon. Only a stepwise increase of compound concentration resulted in DEB025-resistant replicons (2-5× and 2-5-25× EC50 protocols). The highest reduction in antiviral sensitivity was obtained with replicon cultured using protocol 2-5-25× EC50. Mutation D320E in the NS5A protein, reported to be the mutation that contributes the most to resistance (12), was observed in the DEB025-resistant replicons.
Phenotypic analysis of resistant HCV variants.
To confirm the impact of the mutations reported above on the actual phenotype, recombinant replicons with a single mutation or two mutations were generated. Most of these had a resistance profile that is in accordance with the profiles described in earlier studies (18, 20, 32, 37, 46, 58) (Table 8). Y452H, not earlier identified in replicons resistant to benzothiadiazines, resulted in low-level (7-fold) resistance to this molecule. Also, P495L (a benzimidazole resistance mutation) resulted in low-level (8-fold) resistance to the benzothiadiazines.
Table 8.
Effect of selected mutations on the antiviral phenotype of replicons
| Mutation | Fold increase in EC50 compared to WTa |
|||||
|---|---|---|---|---|---|---|
| Protease inhibitors |
Nonnucleoside RdRp inhibitors |
Nucleoside RdRp inhibitor 2′-CMC | ||||
| BILN 2061 | VX-950 | A-782759 | TCA | JT-16 | ||
| D168V (NS3) | >265 | 0.20 ± 0.16 | 0.90 ± 0.44 | 0.24 ± 0.19 | 0.85 ± 0.31 | 1.2 ± 0.31 |
| S282T | ND | 1.98 ± 1.0 | 1.4 ± 0.89 | 1.3 ± 0.40 | 0.96 ± 1.1 | 4 ± 0.78 |
| C316Y | 0.32 ± 0.30 | 0.21 ± 0.080 | 607 ± 177 | 0.31 ± 0.35 | 0.52 ± 0.056 | 0.83 ± 0.39 |
| 316-445 | 0.74 ± 0.45 | 1.3 ± 1.3 | >719 | 0.27 ± 0.15 | 0.72 ± 0.42 | 0.88 ± 039 |
| M414T | 1.4 ± 0.70 | 0.83 ± 0.14 | 285 ± 112 | 2 ± 1.5 | 0.84 ± 0.72 | 0.62 ± 0.33 |
| M423T | ND | 1.3 ± 0.31 | 0.56 ± 0.21 | 56 ± 11 | 0.88 ± 0.15 | ND |
| C445F | 0.93 ± 0.19 | 0.77 ± 0.66 | 3 ± 0.67 | 0.17 ± 0.14 | 1.1 ± 0.21 | 0.93 ± 0.39 |
| Y452H | 0.90 ± 0.60 | 0.37 ± 0.37 | 7 ± 1.4 | 1.6 ± 1.3 | 1.9 ± 1.8 | 1.6 ± 0.28 |
| 445-452 | 1.2 ± 0.97 | 0.65 ± 0.69 | 4 ± 1.5 | 0.35 ± 0.14 | 1.2 ± 0.47 | 0.57 ± 0.14 |
| P495L | ND | 2 ± 1.1 | 8 ± 0.63 | ND | >30 | 0.71 ± 0.33 |
Resistance was set as ≥3-fold EC50 increase compared to WT EC50. Data are mean values for ≥2 independent experiments. Boldface indicates resistance. ND, not determined.
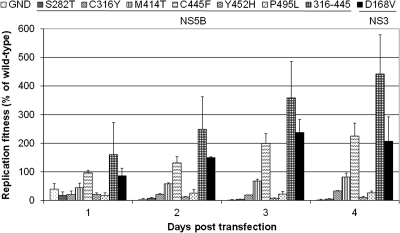
All recombinant mutations (except for C445F, D168V, and the combination C316Y-C445F) resulted in reduced replication fitness (Fig. 2). Mutations S282T, C316Y, Y452H, and P495L resulted in a >50% reduction in replication fitness.
Fig. 2.
Replication capacity of replicons carrying different resistance mutations transfected in Huh-Lunet cells. Huh-Lunet cells were transiently transfected with the mutant replicons. RNA replication was measured by means of a luciferase assay for 4 days posttransfection. Data are normalized to the value at 4 h posttransfection to normalize for transfection efficiency. Values shown are expressed as a percentage of the wild-type value at the corresponding time point posttransfection. Data are mean values ± standard deviations for at least three independent experiments.
Preexisting mutations in wild-type replicon.
The level of preexisting mutations in wild-type Huh 9-13 replicons was quantified by means of clonal sequencing. A total of 96 colonies were analyzed, and 90 sequences were obtained (see Table SA5 in the supplemental material). In 3 of 89 clones (1/90 sequences had a frameshift deletion), the C445F mutation was identified. Mutation T520I was detected in 3 of 84 clones, and D310G was detected in 1 of 89 clones. All three mutations were observed by the less sensitive population sequencing method when replicons were cultured under specific compound pressure.
DISCUSSION
Selection of in vitro drug-resistant variants of novel HCV inhibitors has been extensively used to identify and characterize mutations associated with drug-resistant phenotypes. A variety of resistance selection protocols have been used in such studies; it is hence not possible to directly compare the genetic barrier to antiviral drug resistance of various (classes of) HCV drugs. In the present study, barriers towards resistance development were compared side by side for HCV DAAs of different classes and a host-targeting antiviral (DEB025) by using different resistance selection protocols. The genetic barrier depends not only on the nature and number of resistance mutations but also on their impact on viral replication, the genetic background of the virus, and the variation in the viral population.
Protease inhibitors are associated with a low genetic barrier to resistance. They have been shown to select resistant HCV variants in vitro in less than 10 cell passages in the replicon cell culture system (37, 66, 67). Low- to medium-level (V36, T54, and R155) and high-level (A156) resistance mutations are readily identified in a significant number of patients treated with telaprevir (55). Remarkably, using the selection protocols of this study, the signature high-level resistance mutation A156S/T was observed only as a quasispecies in one of the selection protocols. This can probably be explained by the fact that replicon-containing cells could not be selected in the presence of concentrations above 5× EC50, since the next concentration in this protocol (25× EC50) proved toxic to the host cells. In an independently carried out selection experiment with VX-950 (in which cultures were directly incubated with a high concentration, i.e., 14 μM), VX-950-resistant replicons that carried (only) the resistance mutation A156T/S were selected. Thus, the particular selection protocol used may have an important impact on the selection of drug resistance mutations. In contrast to VX-950, resistance mutations against BILN 2061 (a molecule with a much larger selectivity index than VX-950) were already selected (as quasispecies) following culturing in concentrations of 2-fold the EC50.
Nucleoside HCV polymerase inhibitors have been reported to have a high barrier to resistance (45), and no preexisting mutations to nucleoside inhibitors could be identified in treatment-naïve HCV-infected patients, whereas variants naturally resistant to nonnucleoside inhibitors were observed at a low frequency (27, 35). No resistance-associated mutations were detected following RG7128 monotherapy at various doses for 2 weeks (34). For valopicitabine (NM283; the prodrug of 2′-CMC), an average of 14 to 16 weeks of treatment was needed to select for the S282T mutation (1). In agreement with these findings, we were not able to select for 2′-CMC-resistant variants using the standard protocols employed in the current study. However, we were able to obtain 2′-CMC-resistant replicons (carrying mutation S282T) when a more gradually increasing selection protocol was used (total duration, 3.5 months; data not shown).
Most nonnucleoside RdRp inhibitors have a low genetic barrier to resistance, and depending on the drug class, the target site and the mechanism of action of a variety of resistance mutations are readily selected in vitro (10, 33, 64, 65). Employing the different resistance selection protocols described in this study, variants resistant to the various nonnucleoside inhibitors studied were readily selected. Mutations that were reported earlier to be responsible for the respective drug-resistant phenotypes were identified. However, the emergence of signature mutations clearly depended on the drug resistance selection protocol that was used. For instance, when replicons were cultured in the presence of 5× the EC50 of the benzimidazole JT-16, mutation P495A emerged, whereas T398A was identified in cultures that were selected using a lower concentration or the stepwise selection protocol. Interestingly, T389A requires a transition, whereas P495A requires a transversion (see Table SA4 in the supplemental material). It was previously demonstrated for HIV that the nucleotide substitution pattern can provide important information on drug resistance (24).
Interestingly, mutation C445F emerged under JT-16 and TCA pressure, although this mutation has not been reported to be associated with resistance against these inhibitors (Table 8). Clonal sequencing revealed the preexistence at a prevalence of 3.4% of this mutation in the wild-type Huh 9-13 replicon (see Table SA5 in the supplemental material). Moreover, replicons carrying this mutation were markedly more fit than wild-type replicons (Fig. 2). Altogether, these observations suggest that C445F appears under compound pressure to compensate for the loss of replication fitness caused by unfit drug resistance mutations. For example, the replication fitness levels of mutants with the C316Y mutation and both the C316Y and C445F mutations compared to WT (at 96 h) were 33% and 440%, respectively (Fig. 2).
Inhibitors of host factors that are crucial in viral replication may have a higher barrier to resistance than (most) DAA inhibitors. Culturing of cell-containing replicons in the presence of concentrations of 5× EC50 or higher of DEB025 resulted in complete clearance of the cells from their replicons (Table 7) and thus did not result in the selection of drug-resistant variants. We observed that resistance against the cyclophilin-binding molecule DEB025 emerged only following a lengthy stepwise selection procedure (2-5-25× EC50 protocol). Replicons that were selected to replicate in the presence of 25-fold EC50 did not survive when they were cultured in the presence of a higher concentration (125× EC50) of the inhibitor. Remarkably, the reduced susceptibility in the DEB025 resistance selection (maximum 21-fold reduction) was very moderate compared to the reduced susceptibilities to the DAAs in this study. As reported earlier, mutation D320E conferred low-level resistance (3.9-fold) to DEB025 by reducing the need of the HCV replicon for cyclophilin A-dependent isomerization of NS5A (8).
In addition to the determination of the resistant phenotype and genotype, (i) codon analysis was performed (see Table SA5 in the supplemental material), and (ii) a comparison of the critical phases following drug pressure was carried out (Tables 1 to 7). Some molecules readily resulted in a so-called critical phase in the replicon-containing cultures. This means that only a few remaining cells in the culture carry a replicon that possesses the drug resistance mutation(s). Culturing of replicon-containing cells with DEB025 or 2′-CMC resulted in critical phases in (almost) all culture protocols studied. For BILN 2061 and JT-16, the stepwise protocols did not induce massive cell death, whereas dosing the highest concentrations of monotherapy resulted in a critical phase. For A-782759 as well as for thiophene carboxylic acid treatment, critical phases were observed after dosing the highest concentrations of monotherapy and after some stepwise protocols. On the basis of (i) the fold resistance, (ii) the appearance of resistance mutations, (iii) the codon analysis, and (iv) the critical phases, it can therefore be concluded that host-targeting antivirals (e.g., DEB025) and nucleoside polymerase inhibitors have a higher in vitro barrier to resistance than other HCV DAA classes. This conclusion is largely in agreement with the findings of earlier reported in vitro resistance studies with single molecules and data from clinical studies.
A large body of work on the barrier to resistance of antiretroviral drugs has been gathered. As for HCV nonnucleoside inhibitors, first-generation NNRTIs are associated with a low barrier to resistance, limiting their clinical use. However, second-generation NNRTIs like etravirine and rilpivirine possess an increased resistance barrier, with multiple mutations being necessary to result in resistance (70). Both HCV and HIV protease inhibitors have a low barrier to resistance. The barrier to resistance of nucleos(t)ide reverse transcriptase inhibitors (NRTIs) is more complex for HIV than for HCV. For instance, lamivudine is considered an NRTI with a low barrier to resistance, since only one mutation (M184V) is required to confer high-level resistance to lamivudine. When lamivudine is used as a part of highly active antiretroviral therapy, this mutation is almost always the first to emerge. Other nucleosides, such as zidovudine, require the sequential development of multiple thymidine analogue resistance mutations (TAMs), and resistance to the nucleoside phosphonate tenofovir develops very slowly (11).
In conclusion, in this comparative study we demonstrate that different DAAs may have very different barriers toward resistance. Nucleoside analogues and a cyclophilin-binding molecule(s) have a much higher barrier to resistance than protease and nonnucleoside RdRp inhibitors. A number of factors have to be considered with regard to barriers to resistance: (i) the number of genotypic changes needed for resistance, (ii) the time and difficulty required to select resistant replicons, and (iii) the observed level of resistance of selected replicons. In addition, the development of drug resistance and acquisition of the responsible mutations may depend on the particular selection protocol used. Mutation C445F in the RdRp may compensate for the loss of replication fitness caused by unfit drug resistance mutations. Our observations highlight the importance of in-depth studies on the in vitro drug resistance genotypes and phenotypes of inhibitors when (combination) clinical studies are planned.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a fellowship to Leen Delang from the Research Foundation Flanders (FWO) and grant G.0728.09N from the FWO.
We thank Katrien Geerts and Mieke Flament for excellent technical assistance and G. Pürstinger and P. Herdewijn for the synthesis of the HCV inhibitors. We thank Bin Li, Kai Lin, and Frauke Fischer (Novartis) for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 27 June 2011.
REFERENCES
- 1. Afdhal N., et al. 2006. Valopicitabine (NM283), alone or with peginterferon, compared to peg interferon/ribavirin (pegifn/RBV) re-treatment in hepatitis C patients with prior non-response to pegifn/RBV: week 24 results. J. Hepatol. 44:S1916356583 [Google Scholar]
- 2. Afdhal N., et al. 2007. Valopicitabine (NM283), alone or with peginterferon, compared to peginterferon/ribavirin (pegIFN/RBV) retreatment in patients with HCV-1 infection and prior non-response to pegIFN/RBV: one-year results. J. Hepatol. 46:S5 [Google Scholar]
- 3. Ali S., et al. 2008. Selected replicon variants with low-level in vitro resistance to the hepatitis C virus NS5B polymerase inhibitor PSI-6130 lack cross-resistance with R1479. Antimicrob. Agents Chemother. 52:4356–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anastassopoulou C. G., Kostrikis L. G. 2006. Global genetic variation of HIV-1 infection. Curr. HIV Res. 4:365–373 [DOI] [PubMed] [Google Scholar]
- 5. Biswal B. K., et al. 2005. Crystal structures of the RNA-dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J. Biol. Chem. 280:18202–18210 [DOI] [PubMed] [Google Scholar]
- 6. Bressanelli S., Tomei L., Rey F. A., De Francesco R. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bryson P. D., et al. 2010. A small molecule inhibits HCV replication and alters NS4B's subcellular distribution. Antiviral Res. 87:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carroll S. S., et al. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979–11984 [DOI] [PubMed] [Google Scholar]
- 9. Castro C., Arnold J. J., Cameron C. E. 2005. Incorporation fidelity of the viral RNA-dependent RNA polymerase: a kinetic, thermodynamic and structural perspective. Virus Res. 107:141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen C. M., et al. 2007. Activity of a potent hepatitis C virus polymerase inhibitor in the chimpanzee model. Antimicrob. Agents Chemother. 51:4290–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clavel F., Hance A. J. 2004. HIV drug resistance. N. Engl. J. Med. 350:1023–1035 [DOI] [PubMed] [Google Scholar]
- 12. Coelmont L., et al. 2010. DEB025 (alisporivir) inhibits hepatitis C virus replication by preventing a cyclophilin A induced cis-trans isomerisation in domain II of NS5A. PLoS One 5:e13687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coelmont L., et al. 2009. Debio 025, a cyclophilin binding molecule, is highly efficient in clearing hepatitis C virus (HCV) replicon-containing cells when used alone or in combination with specifically targeted antiviral therapy for HCV (STAT-C) inhibitors. Antimicrob. Agents Chemother. 53:967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Delang L., Coelmont L., Neyts J. 2010. Antiviral therapy for hepatitis C virus: beyond the standard of care. Viruses 2:826–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delang L., et al. 2009. Statins potentiate the in vitro anti-hepatitis C virus activity of selective hepatitis C virus inhibitors and delay or prevent resistance development. Hepatology 50:6–16 [DOI] [PubMed] [Google Scholar]
- 16. Di Marco S., et al. 2005. Interdomain communication in hepatitis C virus polymerase abolished by small molecule inhibitors bound to a novel allosteric site. J. Biol. Chem. 280:29765–29770 [DOI] [PubMed] [Google Scholar]
- 17. Einav S., et al. 2008. Discovery of a hepatitis C target and its pharmacological inhibitors by microfluidic affinity analysis. Nat. Biotechnol. 26:1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Flint M., et al. 2009. Selection and characterization of hepatitis C virus replicons dually resistant to the polymerase and protease inhibitors HCV-796 and boceprevir (SCH 503034). Antimicrob. Agents Chemother. 53:401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Friebe P., Boudet J., Simorre J. P., Bartenschlager R. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hang J. Q., et al. 2009. Slow binding inhibition and mechanism of resistance of non-nucleoside polymerase inhibitors of hepatitis C virus. J. Biol. Chem. 284:15517–15529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hashimoto H. January 2003. Fused cyclic compounds and medicinal use thereof. Patent EP02743728 EP(WO2003000254 A1)
- 22. Howe A. Y., et al. 2008. Molecular mechanism of hepatitis C virus replicon variants with reduced susceptibility to a benzofuran inhibitor, HCV-796. Antimicrob. Agents Chemother. 52:3327–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jourdain G., et al. 2004. Intrapartum exposure to nevirapine and subsequent maternal responses to nevirapine-based antiretroviral therapy. N. Engl. J. Med. 351:229–240 [DOI] [PubMed] [Google Scholar]
- 24. Keulen W., Boucher C., Berkhout B. 1996. Nucleotide substitution patterns can predict the requirements for drug-resistance of HIV-1 proteins. Antiviral Res. 31:45–57 [DOI] [PubMed] [Google Scholar]
- 25. Krieger N., Lohmann V., Bartenschlager R. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614–4624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kukolj G., et al. 2005. Binding site characterization and resistance to a class of non-nucleoside inhibitors of the hepatitis C virus NS5B polymerase. J. Biol. Chem. 280:39260–39267 [DOI] [PubMed] [Google Scholar]
- 27. Kuntzen T., et al. 2008. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naive patients. Hepatology 48:1769–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lamarre D., et al. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186–189 [DOI] [PubMed] [Google Scholar]
- 29. Lawitz E., et al. 2006. Valopicitabine (NM283) plus PEG-interferon in treatment-naive hepatitis C patients with HCV genotype-1 infection: HCV RNA clearance during 24 weeks of treatment. Hepatology 44:223A [Google Scholar]
- 30. Lawitz E., et al. 2009. Safety and antiviral efficacy of 14 days of the cycophilin inhibitor NIM811 in combination with pegylated interferon alpha 2A in relapsed genotype 1 HCV infected patients. J. Hepatol. 50:S379 [Google Scholar]
- 31. Lemm J. A., et al. 2010. Identification of hepatitis C virus NS5A inhibitors. J. Virol. 84:482–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Le Pogam S., et al. 2006. In vitro selected Con1 subgenomic replicons resistant to 2′-C-methyl-cytidine or to R1479 show lack of cross resistance. Virology 351:349–359 [DOI] [PubMed] [Google Scholar]
- 33. Le Pogam S., et al. 2006. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J. Virol. 80:6146–6154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Le Pogam S., et al. 2010. RG7128 alone or in combination with pegylated interferon-alpha 2a and ribavirin prevents hepatitis C virus (HCV) replication and selection of resistant variants in HCV-infected patients. J. Infect. Dis. 202:1510–1519 [DOI] [PubMed] [Google Scholar]
- 35. Le Pogam S., et al. 2008. Existence of hepatitis C virus NS5B variants naturally resistant to non-nucleoside, but not to nucleoside, polymerase inhibitors among untreated patients. J. Antimicrob. Chemother. 61:1205–1216 [DOI] [PubMed] [Google Scholar]
- 36. Lin C., et al. 2005. In vitro studies of cross-resistance mutations against two hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061. J. Biol. Chem. 280:36784–36791 [DOI] [PubMed] [Google Scholar]
- 37. Lin C., et al. 2004. In vitro resistance studies of hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061—structural analysis indicates different resistance mechanisms. J. Biol. Chem. 279:17508–17514 [DOI] [PubMed] [Google Scholar]
- 38. Lohmann V., Hoffmann S., Herian U., Penin F., Bartenschlager R. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lohmann V., et al. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113 [DOI] [PubMed] [Google Scholar]
- 40. Love R. A., et al. 2003. Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme. J. Virol. 77:7575–7581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu L., et al. 2004. Mutations conferring resistance to a potent hepatitis C virus serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 48:2260–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lu L. J., et al. 2006. Selection and characterization of hepatitis C virus replicons resistant to a potent polymerase inhibitor A-837093. Hepatology 44:351A [Google Scholar]
- 43. Lu L. J., Mo H. M., Pilot-Matias T. J., Molla A. 2007. Evolution of resistant M414T mutants among hepatitis C virus replicon cells treated with polymerase inhibitor A-782759. Antimicrob. Agents Chemother. 51:1889–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Manns M. P., Wedemeyer H., Cornberg M. 2006. Treating viral hepatitis C: efficacy, side effects, and complications. Gut 55:1350–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McCown M. F., et al. 2008. The hepatitis C virus replicon presents a higher barrier to resistance to nucleoside analogs than to nonnucleoside polymerase or protease inhibitors. Antimicrob. Agents Chemother. 52:1604–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mo H., et al. 2005. Mutations conferring resistance to a hepatitis C virus (HCV) RNA-dependent RNA polymerase inhibitor alone or in combination with an HCV serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 49:4305–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nelson D. R., et al. 2009. Efficacy and safety of the cyclophilin inhibitor Debio 025 in combination with pegylated interferon alpha-2A and ribavirin in previously null-responder genotype 1 HCV patients. J. Hepatol. 50:S40 [Google Scholar]
- 48. Neumann A. U., et al. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103–107 [DOI] [PubMed] [Google Scholar]
- 49. Nicolas O., et al. 2009. Genotypic and phenotypic analysis of HCV Ns5B variants selected from patients treated with VCH-916. J. Hepatol. 50:S349 [Google Scholar]
- 50. Penuel E., et al. 2006. Variable non-nucleoside inhibitor susceptibility among untreated HCV-infected patient samples, abstr. 18. Abstr. 1st Int. Workshop Hepatitis C: Resistance New Compounds [Google Scholar]
- 51. Perelson A. S., Neumann A. U., Markowitz M., Leonard J. M., Ho D. D. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582–1586 [DOI] [PubMed] [Google Scholar]
- 52. Pfefferkorn J. A., et al. 2005. Inhibitors of HCV NS5B polymerase. Part 1. Evaluation of the southern region of (2Z)-2-(benzoylamino)-3-(5-phenyl-2-furyl)acrylic acid. Bioorg. Med. Chem. Lett. 15:2481–2486 [DOI] [PubMed] [Google Scholar]
- 53. Pratt J. K., et al. 2005. Inhibitors of HCV NS5B polymerase: synthesis and structure-activity relationships of N-1-heteroalkyl-4-hydroxyquinolon-3-yl-benzothiadiazines. Bioorg. Med. Chem. Lett. 15:1577–1582 [DOI] [PubMed] [Google Scholar]
- 54. Reiser M., et al. 2005. Antiviral efficacy of NS3-serine protease inhibitor BILN-2061 in patients with chronic genotype 2 and 3 hepatitis C. Hepatology 41:832–835 [DOI] [PubMed] [Google Scholar]
- 55. Sarrazin C., et al. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767–1777 [DOI] [PubMed] [Google Scholar]
- 56. Shahriar R., et al. 2009. Nonpolymorphic human immunodeficiency virus type 1 protease and reverse transcriptase treatment-selected mutations. Antimicrob. Agents Chemother. 53:4869–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shepard C. W., Finelli L., Alter M. J. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558–567 [DOI] [PubMed] [Google Scholar]
- 58. Shi S. T., et al. 2008. In vitro resistance study of AG-021541, a novel nonnucleoside inhibitor of the hepatitis C virus RNA-dependent RNA polymerase. Antimicrob. Agents Chemother. 52:675–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Simmonds P. 2004. Genetic diversity and evolution of hepatitis C virus—15 years on. J. Gen. Virol. 85:3173–3188 [DOI] [PubMed] [Google Scholar]
- 60. Summa V. 2005. VX-950 (Vertex/Mitsubishi). Curr. Opin. Investig. Drugs 6:831–837 [PubMed] [Google Scholar]
- 61. Susser S., et al. 2009. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology 50:1709–1718 [DOI] [PubMed] [Google Scholar]
- 62. Targett-Adams P., McLauchlan J. 2005. Development and characterization of a transient-replication assay for the genotype 2a hepatitis C virus subgenomic replicon. J. Gen. Virol. 86:3075–3080 [DOI] [PubMed] [Google Scholar]
- 63. Thompson A. J., Mchutchison J. G. 2009. Antiviral resistance and specifically targeted therapy for HCV (STAT-C). J. Viral Hepat. 16:377–387 [DOI] [PubMed] [Google Scholar]
- 64. Tomei L., et al. 2003. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 77:13225–13231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tomei L., et al. 2004. Characterization of the inhibition of hepatitis C virus RNA replication by nonnucleosides. J. Virol. 78:938–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tong X., et al. 2008. Characterization of resistance mutations against HCV ketoamide protease inhibitors. Antiviral Res. 77:177–185 [DOI] [PubMed] [Google Scholar]
- 67. Tong X., et al. 2006. Identification and analysis of fitness of resistance mutations against the HCV protease inhibitor SCH 503034. Antiviral Res. 70:28–38 [DOI] [PubMed] [Google Scholar]
- 68. Trozzi C., et al. 2003. In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J. Virol. 77:3669–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Villano S., et al. 2006. Analysis of HCV NS5B genetic variants following monotherapy with HCV-796, a nonnucleoside polymerase inhibitor, in treatment-naïve HCV-infected patients. Hepatology 44:607A–608A [Google Scholar]
- 70. Vingerhoets J., et al. 2005. TMC125 displays a high genetic barrier to the development of resistance: evidence from in vitro selection experiments. J. Virol. 79:12773–12782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vrolijk J. M., et al. 2003. A replicon-based bioassay for the measurement of interferons in patients with chronic hepatitis C. J. Virol. Methods 110:201–209 [DOI] [PubMed] [Google Scholar]
- 72. Wang M., et al. 2003. Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase. Crystal structures and mechanism of inhibition. J. Biol. Chem. 278:9489–9495 [DOI] [PubMed] [Google Scholar]
- 73. Welsch C., et al. 2008. Molecular basis of telaprevir resistance due to V36 and T54 mutations in the NS3-4A protease of the hepatitis C virus. Genome Biol. 9:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang W., et al. 2008. Selection of replicon variants resistant to ACH-806, a novel hepatitis C virus inhibitor with no cross-resistance to NS3 protease and NS5B polymerase inhibitors. Antimicrob. Agents Chemother. 52:2043–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.