Abstract
A carbapenem-resistant Escherichia coli strain (DVR22) was recovered from a stool specimen from a patient with traveler's diarrhea who had traveled to India. Molecular screening led to the first identification of NDM-1 in Spain. The blaNDM-1 gene was located in a conjugative plasmid of ca. 300 kb that also contained the blaCTX-M-15, blaTEM-1, ΔblaDHA-1, and armA genes. In addition, blaNDM-1 was preceded by an ISAba125 insertion element only found in Acinetobacter spp.
TEXT
The emergence of carbapenem resistance among Enterobacteriaceae is a major cause of concern since carbapenems currently represent the treatment of choice for severe infections caused by multidrug-resistant strains producing extended-spectrum β-lactamases (ESBLs) (8).
In addition to commonly known carbapenem-hydrolyzing enzymes in Enterobacteriaceae (IMP, VIM, KPC, and OXA-48), a novel class B metallo-β-lactamase (NDM-1) has recently been described. This enzyme, first identified in Klebsiella pneumoniae and Escherichia coli clinical isolates recovered in Sweden from a traveler returning from India, confers resistance to all β-lactams except aztreonam (22). Since then, several reports have identified blaNDM genes worldwide that have typically been associated with multidrug-resistant strains (1, 5, 12, 16–18, 23).
A 40-year-old Spanish Caucasian male reported intermittent abdominal discomfort, fever, and bloody diarrhea about 5 days before returning from India. He visited a local dispensary in India where treatment with ofloxacin and ornidazole tablets (twice a day) was prescribed for 5 days. The patient reported to the Hospital Clinic of Barcelona 1 day after his return, still complaining of bloody diarrhea, although with fewer unformed stools. He was afebrile, without any sign of dehydration, and the rest of the physical examination was normal. The diarrhea resolved spontaneously over the next 9 days.
A carbapenem-resistant E. coli (DVR22) strain was recovered from the stool samples of the patient, and after isolation and identification, antimicrobial susceptibility profiling analysis performed with both BD Phoenix (Becton Dickinson, Franklin Lakes, NJ) and Etest strips (AB bioMérieux, Solna, Sweden) indicated that strain DVR22 was resistant to all the antibiotics tested except tigecycline (MIC of 0.75 μg/ml), fosfomycin (MIC of 32 μg/ml), and colistin (MIC of 0.5 μg/ml) (Table 1), presenting MICs of 8 μg/ml and 16 μg/ml for imipenem and meropenem, respectively, 24 μg/ml for ertapenem, and 6 μg/ml for doripenem (CLSI breakpoints from broth microdilution tests were used to classify the MICs obtained by Etest [7]). Screening for carbapenemase/MBL production yielded positive results when using either the cloverleaf test (modified Hodge test) or imipenem-EDTA Etest strips. PCR screening for β-lactamase genes followed by DNA sequencing using specific primers (NDM-1 F, 5′-CCAATATTATGCACCCGGTCG-3′, and NDM-1 R 5′-ATGCGGGCCGTATGAGTGATTG-3′) (2, 14, 21) identified the presence of blaNDM-1, blaCTX-M-15, blaTEM-1, and a partial sequence of the blaDHA-1 gene. In addition, screening for aminoglycoside resistance genes (21) identified the armA gene, encoding a 16S rRNA methylase conferring resistance to aminoglycosides.
Table 1.
In vitro susceptibilities of E. coli DVR22 and E. coli transconjugant expressing NDM-1 carbapenemase
| Antibiotic(s) | MIC (μg/ml) in: |
||
|---|---|---|---|
| E. coli DVR22 | E. coli J53 DVR22T | E. coli J53 | |
| Amoxicillin | >256 | >256 | 4 |
| Amoxicillin + clavulanatea | 32 | 32 | 4 |
| Piperacillin + tazobactamb | >256 | >256 | 1 |
| Cefoxitin | >256 | >256 | 2 |
| Cefotaxime | >256 | >256 | 0.094 |
| Ceftazidime | >256 | >256 | 0.125 |
| Cefepime | 256 | >256 | 0.25 |
| Imipenem | 8 | 16 | 0.19 |
| Meropenem | 16 | 12 | 0.023 |
| Doripenem | 6 | 4 | 0.032 |
| Ertapenem | 24 | 18 | 0.008 |
| Aztreonam | >256 | >256 | 64 |
| Gentamicin | >8 | >8 | <1 |
| Amikacin | >32 | >32 | <4 |
| Tobramycin | >8 | >8 | <2 |
| Ciprofloxacin | >32 | 0.008 | 0.008 |
Clavulanate was used at a fixed concentration of 2 μg/ml.
Tazobactam was used at a fixed concentration of 4 μg/ml.
In order to study the transferability of the resistance phenotype, a biparental mating between DVR22 and the E. coli strain J53AziR was conducted and transconjugants were selected on LB agar plates containing 1 μg/ml imipenem and 100 μg/ml sodium azide (Sigma Chemical Co., St. Louis, MO). PCR and susceptibility profiling showed that all transconjugants had become resistant to all the β-lactams and aminoglycosides tested (Table 1) and had also acquired the blaNDM-1, blaCTX-M-15, blaTEM-1, ΔblaDHA-1, and armA genes. Plasmid analysis by S1 nuclease–pulsed-field gel electrophoresis (PFGE) (20) was then performed on both the DVR22 strain and selected transconjugants, revealing the presence of a single plasmid of ca. 300 kb. Digoxigenin-labeled probes for the blaNDM-1, blaCTX-M-15, blaTEM-1, blaDHA-1, and armA genes were hybridized against blotted nylon membranes from the S1-PFGE gels. All probes matched the band corresponding to the 300-kb plasmid. Altogether, these results indicate that all β-lactamases plus the armA gene are located in a single conjugative plasmid. Replicon typing classified this plasmid within the incompatibility group IncHI1 (3).
Previous reports have described the concurrence of blaNDM-1 together with additional ESBLs, mainly CTX-M-15 (1, 16–19), but this is the first time that they seem to be located on the same plasmid. Then again, this is also the first time that a blaNDM-1 gene has been found on such a large plasmid (NDM enzyme genes are typically found in plasmids ranging from 50 to 200 kb) (1, 5, 16–18) and could reflect the formation of a cointegrate from two or more smaller plasmids carrying individual β-lactamase genes, such as blaCTX-M-15 or blaNDM-1. On the other hand, the armA gene appears to be commonly linked to blaNDM-1 genes (1, 10, 13, 16, 19).
Multilocus sequence typing (MLST) (http://mlst.ucc.ie/mlst/dbs/Ecoli) and PCR-based phylogroup analysis (6) revealed that E. coli DVR22 belonged to sequence type 156 (ST156) and phylogroup B1, respectively, differing from previously described NDM-carrying E. coli sequence types (16, 19, 22). PCR analysis to detect the presence of heat-stable (ST) and heat-labile (LT) toxins, verotoxins (VT), and enteroaggregative E. coli virulence factors were all negative.
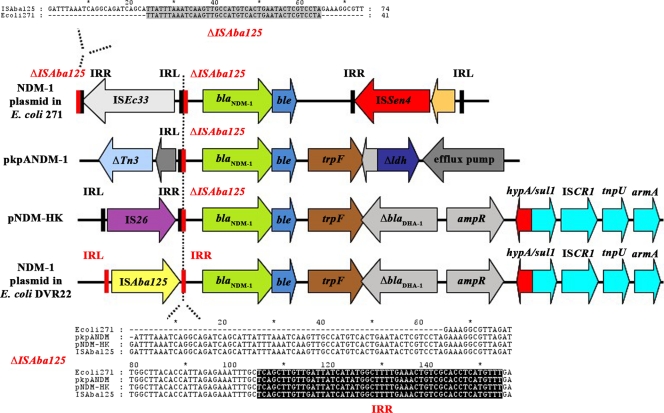
In order to characterize the genetic environment of the blaNDM-1 gene, outward NDM primers (NDMinv-R2, 5′-GGTCGCCAGTTTCCATTTGC-3′, and NDMinv-F2, 5′-TGCCGACACTGAGCACTAC-3′) were used to perform an inverse PCR over genomic DNA from strain DVR22 partially digested with Sau3AI and ligated with T4 DNA ligase (New England BioLabs, Ipswich, MA). Sequencing of the PCR products revealed that the region flanking the 3′ end of blaNDM-1 was very similar to that described for plasmid pNDM-HK, with a downstream trpF gene, encoding the N-(5′-phosphoribosyl)anthranilate isomerase, followed by a truncated blaDHA-1 gene (10). The published sequence of pNDM-HK was then used as a template to design specific primers to further span the downstream region. The PCR amplicons obtained with these primers always concurred with the expected size from the pNDM-HK sequence and allowed the blaNDM-1 gene to link up to the armA gene (Fig. 1). Sequencing of the blaNDM-1 upstream region identified the conserved putative promoter sequence described by Poirel et al. (16), as well as the presence of an ISAba125 insertion sequence (http://www-is.biotoul.fr). Although the presence of insertion sequences upstream from the blaNDM-1 gene has already been reported in Enterobacteriaceae (10, 16, 22), DVR22 is unique in the sense that ISAba125 (IS30 family) has only been described in Acinetobacter spp. (9, 11, 13, 15, 24) and has not been found to be linked to blaNDM-1 before, suggesting horizontal transfer between Acinetobacter and Enterobacteriaceae. To further support this hypothesis, sequence comparison of the blaNDM-1 upstream sequences from E. coli strain 271 (16) and plasmids pNDM-HK and pKpANDM-1 (10, 22) identified a fragment of variable length containing the right-end repeat from ISAba125 in between the blaNDM-1 gene and the corresponding IS element as a remnant of ISAba125 insertion in all of these sequences. Moreover, the insertion sequence element ISEc33 from E. coli strain 271 is bracketed by the sequence upstream from the ISAba125 right end (Fig. 1). Reports describing NDM enzymes in Acinetobacter have already been published, but there is no information available regarding their genetic surroundings (5, 12, 13). In view of these results and also taking into account current investigations by our group concerning the identification of a chromosomally encoded ISAba125-blaNDM-2 in A. baumannii (Paula Espinal, personal communication), it may be speculated that Acinetobacter is the source of the NDM enzymes found in Enterobacteriaceae that were originally spread by ISAba125-mediated mobilization.
Fig. 1.
Schematic drawing showing the genetic elements surrounding the blaNDM-1 genes in E. coli 271, pkpANDM-1, pNDM-HK, and E. coli DVR22. Adapted from reference 10. The lengths of the arrows are proportional to the lengths of the genes or open reading frames (ORFs) except for the region spanning hypA to armA in both pNDM-HK and DVR22, which has been compressed to fit in the figure. The partial downstream region of ISAba125 containing the right inverted repeat (highlighted in black) found in all four sequences is shown in the lower alignment. The upper alignment shows the 42 missing base pairs from the downstream region of ISAba125 that are located upstream from the ISEc33 insertion element in the sequence from E. coli 271. Δ, truncated gene; ampR, LysR family blaDHA-1 regulator; armA, 16S rRNA methylase gene; blaDHA-1, class C β-lactamase gene; blaNDM-1, New Delhi metallo-β-lactamase gene; ble, bleomycin resistance gene; hypA, putative hydrogenase nickel-incorporating gene; IRL, inverted repeat left; IRR, inverted repeat right; IS, insertion sequence; ldh, lactate dehydrogenase gene; sul1, sulfonamide resistance gene; Tn, transposon; trpF, phosphoribosylanthranilate isomerase gene. The GenBank accession numbers of the sequences are as follows: plasmid encoding NDM-1 in E. coli 271 (HQ162469), pkpANDM-1 (FN396877), pNDM-HK (HQ451074), and plasmid encoding NDM-1 in E. coli DVR22 (JF922606.1).
It is also worth mentioning that after a 4-month period, another stool specimen from the same patient was screened for blaNDM-1 carriage, but neither the DVR22 strain nor additional carbapenem-resistant strains could be isolated. These findings suggest that while carriage of multiple resistance determinants in a single plasmid might be beneficial under certain circumstances, in the absence of selective pressure, the burden associated with the replication and expression of extrachromosomal DNA involves a fitness cost that is not affordable, leading to the eradication of NDM-carrying bacteria (4). However, the simple clearance of DVR22 unrelated to its plasmid burden cannot be excluded.
This study reports the first identification of an E. coli strain producing NDM-1 in Spain and highlights the tremendous plasticity and disseminating potential of the blaNDM-1 gene.
Nucleotide sequence accession number.
The sequence spanning from the ISAba125 to the ampR gene from the E. coli strain DVR22 has been submitted to GenBank and assigned sequence accession number JF922606.1.
Acknowledgments
This study was supported by grant 2009-SGR1256 from the Departament de Universitats, Recerca i Societat de la Informació de la Generalitat d'Catalunya, and by the Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III, Spanish Network for Research in Infectious Diseases (REIPI 06/0008). This work was also supported by funding from the European Community (TROCAR contract HEALTH-F3-2008-223031).
We thank Alessandra Carattoli for kindly providing the control plasmids for replicon typing of incompatibility groups.
Footnotes
Published ahead of print on 5 July 2011.
REFERENCES
- 1. Bogaerts P., et al. 2011. Emergence of NDM-1-producing Enterobacteriaceae in Belgium. Antimicrob. Agents Chemother. 55:3036–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Calbo E., et al. 2011. Foodborne nosocomial outbreak of SHV1 and CTX-M-15-producing Klebsiella pneumoniae: epidemiology and control. Clin. Infect. Dis. 52:743–749 [DOI] [PubMed] [Google Scholar]
- 3. Carattoli A., et al. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 4. Chen T. L., Fung C. P., Lee S. D. 2011. Spontaneous eradication of a NDM-1 positive Klebsiella pneumoniae that colonized the intestine of an asymptomatic carrier. J. Chin. Med. Assoc. 74:104. [DOI] [PubMed] [Google Scholar]
- 5. Chen Y., Zhou Z., Jiang Y., Yu Y. 2011. Emergence of NDM-1-producing Acinetobacter baumannii in China. J. Antimicrob. Chemother. 66:1255–1259 [DOI] [PubMed] [Google Scholar]
- 6. Clermont O., Bonacorsi S., Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. CLSI document M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. Cornaglia G., et al. 2007. Metallo-β-lactamases as emerging resistance determinants in Gram-negative pathogens: open issues. Int. J. Antimicrob. Agents 29:380–388 [DOI] [PubMed] [Google Scholar]
- 9. Evans B. A., Hamouda A., Towner K. J., Amyes S. G. 2010. Novel genetic context of multiple blaOXA-58 genes in Acinetobacter genospecies 3. J. Antimicrob. Chemother. 65:1586–1588 [DOI] [PubMed] [Google Scholar]
- 10. Ho P. L., et al. 2011. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One 6:e17989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Iacono M., et al. 2008. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob. Agents Chemother. 52:2616–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaase M., et al. 2011. NDM-2 carbapenemase in Acinetobacter baumannii from Egypt. J. Antimicrob. Chemother. 66:1260–1262 [DOI] [PubMed] [Google Scholar]
- 13. Karthikeyan K., Thirunarayan M. A., Krishnan P. 2010. Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J. Antimicrob. Chemother. 65:2253–2254 [DOI] [PubMed] [Google Scholar]
- 14. Mendes R. E., et al. 2007. Rapid detection and identification of metallo-β-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. J. Clin. Microbiol. 45:544–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mussi M. A., Limansky A. S., Viale A. M. 2005. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of β-barrel outer membrane proteins. Antimicrob. Agents Chemother. 49:1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Poirel L., Lagrutta E., Taylor P., Pham J., Nordmann P. 2010. Emergence of metallo-β-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob. Agents Chemother. 54:4914–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Poirel L., Revathi G., Bernabeu S., Nordmann P. 2011. Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrob. Agents Chemother. 55:934–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poirel L., et al. 2011. Extremely drug-resistant Citrobacter freundii isolate producing NDM-1 and other carbapenemases identified in a patient returning from India. Antimicrob. Agents Chemother. 55:447–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Samuelsen O., et al. 2011. Identification of NDM-1-producing Enterobacteriaceae in Norway. J. Antimicrob. Chemother. 66:670–672 [DOI] [PubMed] [Google Scholar]
- 20. Sanchez-Cespedes J., et al. 2009. Two chromosomally located qnrB variants, qnrB6 and the new qnrB16, in Citrobacter spp. isolates causing bacteraemia. Clin. Microbiol. Infect. 15:1132–1138 [DOI] [PubMed] [Google Scholar]
- 21. Yamane K., Wachino J., Doi Y., Kurokawa H., Arakawa Y. 2005. Global spread of multiple aminoglycoside resistance genes. Emerg. Infect. Dis. 11:951–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yong D., et al. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zarfel G., et al. 2011. Emergence of New Delhi metallo-β-lactamase, Austria. Emerg. Infect. Dis. 17:129–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zarrilli R., et al. 2008. A plasmid-borne blaOXA-58 gene confers imipenem resistance to Acinetobacter baumannii isolates from a Lebanese hospital. Antimicrob. Agents Chemother. 52:4115–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]