Abstract
We have investigated the mechanism, structural correlates, and cis-acting elements involved in chromatin opening and gene activation, using the human β-globin locus as a model. Full transcriptional activity of the human β-globin locus requires the locus control region (LCR), composed of a series of nuclease hypersensitive sites located upstream of this globin gene cluster. Our previous analysis of naturally occurring and targeted LCR deletions revealed that chromatin opening and transcriptional activity in the endogenous β-globin locus are dissociable and dependent on distinct cis-acting elements. We now report that general histone H3/H4 acetylation and relocation of the locus away from centromeric heterochromatin in the interphase nucleus are correlated and do not require the LCR. In contrast, LCR-dependent promoter activation is associated with localized histone H3 hyperacetylation at the LCR and the transcribed β-globin-promoter and gene. On the basis of these results, we suggest a multistep model for gene activation; localization away from centromeric heterochromatin is required to achieve general hyperacetylation and an open chromatin structure of the locus, whereas a mechanism involving LCR/promoter histone H3 hyperacetylation is required for high-level transcription of the β-globin genes.
Keywords: LCR, globin, acetylation, nuclear compartmentalization, gene activation
In higher eukaryotes, only a small subset of the genome is expressed in any given differentiated cell type; the great majority of genes are maintained in a stable inactive state. The actively transcribed and stably inactive states are generally characterized by distinct chromatin structures, as manifested in the DNase I sensitivity (open state) of active genes and DNase I resistance (closed state) of stably silent genes (Weintraub and Groudine 1976). However, the molecular basis for the dynamic alteration of chromatin structure and the influence of chromatin on transcriptional activity are not well understood (for review, see Gross and Garrard 1988; Bulger and Groudine 1999). Two facets of the phenomenon of chromatin opening and gene activation that have become apparent in recent years are acetylation of histones and localization of a gene in a nuclear compartment permissive for transcription.
Lysines at the amino-terminal tail of histones H3 and H4 can be acetylated in vivo, and several studies have reported a correlation between hyperacetylation of histones and generalized nuclease sensitivity (Hebbes et al. 1988, 1994; Madisen et al. 1998). Moreover, the recent observations that histone acetyltransferases (HATs) can interact with transactivators have provided the molecular basis for a link between chromatin modification and gene activation (Imhof and Wolffe 1998). Analysis of promoter activation in yeast has revealed that histone acetylation can be targeted locally and is associated with the activation of many promoters (for review, see Struhl 1998). Thus, histone acetylation could be involved in both chromatin opening and promoter-specific activation.
Silencing of gene expression correlates in several systems with the location of a gene in the interphase nucleus, close to heterochromatic compartments repressive for transcriptional activity (for review, see Cockell and Gasser 1999). In addition, we have shown recently that one component of the human β-globin locus control region (LCR), a transcriptional enhancer termed 5′HS2, can suppress silencing of a transgene, and maintain it at a distance from centromeric heterochromatin (Francastel et al. 1999). This result suggests that cis-acting transcriptional control elements may act to maintain gene expression and an open chromatin structure, by maintaining endogenous loci in a nuclear compartment in which these states are favored.
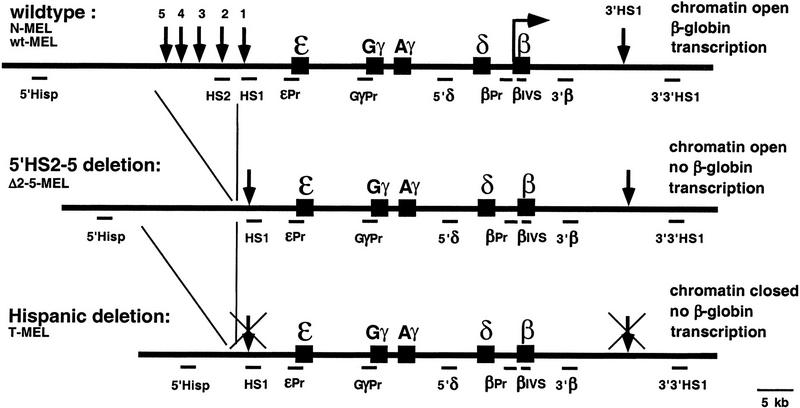
The human β-globin locus contains five genes that are arranged from 5′ to 3′ in the order of their expression during development. Upstream of the gene cluster are five DNase I hypersensitive sites (5′HS1–5; Fig. 1) within a 20-kb region referred to as LCR. Analysis of a naturally occurring deletion (Hispanic thalassemia), which removes 5′HS2 to HS5 and an additional 27 kb of upstream sequence, revealed that this region is required for the activity of the human locus. In the wild-type human locus in erythroid cells, the β-like globin genes are transcriptionally active, the hypersensitive sites at the LCR (5′HS1–5) and at the 3′ end of the locus (3′HS1) are present, the locus is nuclease sensitive, and it replicates early in S phase. In contrast, the Hispanic deletion locus is transcriptionally inactive, no hypersensitive site is formed, and the locus is nuclease insensitive and replicates late in S phase (Forrester et al. 1990). The phenotype of the Hispanic thalassemia chromosome and experiments with the human β-globin locus in transgenic mice (for review, see Fraser and Grosveld 1998; Bulger and Groudine 1999) led to the view that the LCR is required for chromatin opening and transcriptional activity of the endogenous human β-globin locus in an erythroid background.
Figure 1.
Different alleles of the human β-globin locus. Shown are the wild-type locus, present in the N-MEL and wt-MEL cell lines, the 5′HS2–5 deletion present in Δ2–5-MEL, and the Hispanic deletion allele present in T-MEL. Position of the five genes is represented by solid boxes, and strong hypersensitive sites by vertical arrows (for details, see Bulger et al. 1999). Transcription of the β gene in the wild-type allele is indicated by a horizontal arrow. Sequences analyzed in this study are indicated below each allele, and the corresponding primers for their amplification are listed in Materials and Methods.
To further define the cis-acting elements required for chromatin opening and gene activation, we generated (by gene targeting) a smaller deletion, which removes only 5′HS2–5 of the LCR (Δ2–5-MEL) (Reik et al. 1998). Surprisingly, whereas the globin promoters are not activated in Δ2–5-MEL, the locus is in an open (nuclease sensitive) conformation (Reik et al. 1998) and replicates early in S phase (D.M. Cimbora, D. Schübeler, A. Reik, J. Hamilton, and M. Groudine, submitted). Deletion of 5′HS1–6 from the mouse β-globin locus produces a similar result in that the locus is open, but in this case, a low level of developmental, stage-appropriate transcription is detectable (Epner et al. 1998; Bender et al. 2000). Thus, these systems separate an open chromatin structure from high level β-like globin gene transcription and provide a model to independently investigate the molecular mechanisms that mediate chromatin opening and transcriptional activity. To investigate the involvement of nuclear localization and histone acetylation in the processes of chromatin opening and gene activation of the globin locus, we have analyzed the wild-type, Hispanic thalassemia, and Δ2–5 human β-globin loci after their transfer from a nonerythroid into an erythroid background. Consistent with its silent and nuclease-resistant state, the Hispanic deletion allele is associated with centromeric heterochromatin and hypoacetylated histones H3 and H4. In contrast, both the open wild-type and Δ2–5 alleles localize away from centromeric heterochromatin and show hyperacetylation of both H3 and H4 throughout the locus. Thus, neither process requires the LCR or active transcription. Furthermore, as only the wild-type locus is transcriptionally active, these results suggest that localization of a gene away from centromeric heterochromatin may mediate an open chromatin structure and general hyperacetylation of the β-globin locus, but is not sufficient to activate globin gene transcription. However, H3 acetylation in the vicinity of the LCR and at the transcribed β-globin gene is significantly greater in the transcriptionally active wild-type allele, suggesting that this localized H3 hyperacetylation is linked to globin gene activation, and that both require the LCR.
Results
Experimental strategy
To determine the structural correlates of nuclease sensitivity and the transcriptional activity of the human β-globin locus, we examined the acetylation status of histones H3 and H4 and the subnuclear localization of the human locus in a red-cell environment, and assessed the effect of specific deletions of the human β-globin LCR and upstream sequences on those correlates.
Four different mouse erythroleukemia (MEL) cell hybrids were analyzed (Fig. 1). N-MEL contains a human chromosome 11 with the wild-type β-globin locus and was generated by chromosomal transfer from lymphocytes derived from a patient heterozygous for the Hispanic deletion, into an erythroid background. T-MEL contains the Hispanic deletion chromosome 11 from these lymphocytes (Forrester et al. 1990). Δ2–5-MEL contains a chromosome 11 from which 5′HS2–5 of the β-globin LCR were deleted by site-specific recombination. In contrast to our previously described Δ2–5 clones (Reik et al. 1998), this LCR deletion was performed in mouse ES cells after transfer of the human chromosome 11 from the DT40 chicken cell line into ES, but prior to transfer into MEL. Thus, all chromosome modifications (marking and deletion) have been performed in nonerythroid cells prior to transfer into the MEL cell background. In addition, the ES to MEL transfer strategy generates hybrids with a complete human chromosome 11, whereas a direct transfer from DT40 into MEL is associated with chromosome fragmentation (see Reik et al. 1998; Material and Methods). To control for a possible influence of the chromosomal history on the acetylation pattern and/or nuclear location of the β-globin locus, we also included wt-MEL in our analysis. Like N-MEL, the wt-MEL line contains an intact chromosome 11 with a wild-type human β-globin locus, but the chromosome underwent the same series of transfers and selections used to generate Δ2–5-MEL.
Analysis of histone acetylation in MEL hybrids
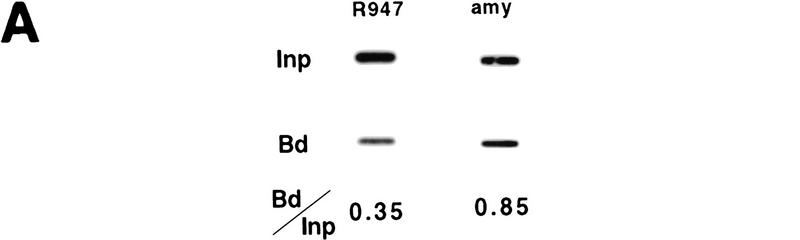
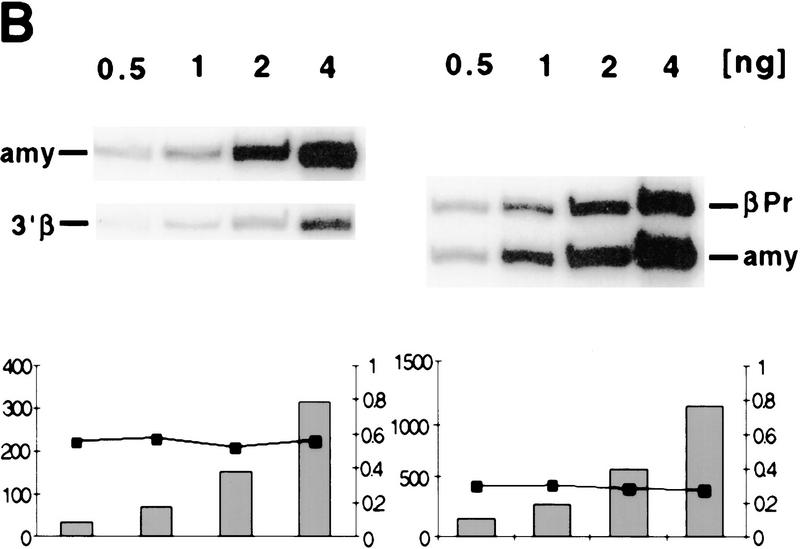
Formaldehyde cross-linked chromatin from all four lines was purified by isopycnic centrifugation (Orlando et al. 1997) and subsequently immunoprecipitated with antisera against acetylated isoforms of histone H3 and H4. Western blot analysis demonstrated that the antibodies against all acetylated isoforms of H4 (αH4–Ac) and against acetylated H3 (αH3–Ac) immunoprecipitate the expected acetylated histones from purified cross-linked chromatin (data not shown). To establish whether putative hypoacetylated constitutive heterochromatin is excluded from chromatin enriched for acetylated H4, we determined the ratio of murine centromeric DNA in the bound and input fractions. Slot blot analysis of input and αH4–Ac bound DNA was performed with a murine minor satellite-specific oligonucleotide probe. We find that this sequence is depleted in the bound fraction (Fig. 2A), in agreement with a previous study that utilized micrococcal nuclease-digested chromatin preparations (Keohane et al. 1996). As a result of the high sequence homology between the human and mouse β-globin loci present in the cell hybrids, globin sequences could not be analyzed by slot blot hybridization and therefore were analyzed in a quantitative PCR with reference to the mouse pancreatic amylase 2.1y gene. This gene is transcriptionally silent, late replicating, and nuclease insensitive in red cells (Dhar et al. 1988; Forrester et al. 1990). To determine the acetylation state of this control sequence, we rehybridized the slot blot with an amylase 2.1y probe; as shown in Figure 2A, this sequence is slightly less abundant in the chromatin enriched for acetylated histone H4 (Fig. 2A). This is consistent with a previous study showing that coding regions, even when they are inactive, are not as hypoacetylated for histone H4 as centromeric heterochromatin (O'Neill and Turner 1995). To analyze the acetylation state of sequences in the mouse globin locus, a duplex PCR was performed with one primer pair specific for a globin sequence, and a second pair specific for the amylase gene under conditions of linear amplification for both PCR products (see examples in Fig. 2B). The ratio of the two PCR products was determined for the antibody-bound fraction and normalized to the ratio in the input material to account for possible aneuploidy or loss of the human chromosome, which might occur during expansion of the hybrid cell lines.
Figure 2.
Immunoprecipitation and duplex PCR assays. (A) Depletion of centromeric (heterochromatic) sequences and an inactive mouse gene in chromatin enriched for acetylated histone H4. Chromatin from MEL cells was immunoprecipitated with an antibody that detects all acetylated isoforms of histone H4 (αH4–Ac). Input and antibody-bound DNA (500 ng) were slot blotted and hybridized with an oligomer corresponding to a murine centromeric minor satellite repeat (R947, Kipling et al. 1994). This sequence is 2.8-fold less abundant in the antibody bound fraction. The same blot was rehybridized with a probe from the mouse amylase gene (generated by PCR with the primer pairs amy4 and amy6, see Materials and Methods). This pancreatic-specific gene, which is inactive in a red cell background, is slightly less abundant in chromatin enriched for acetylated H4. (B) Abundance of human and mouse globin sequences in chromatin enriched for acetylated histones was determined relative to the mouse amylase gene using a duplex PCR assay (see text and Fig. 3). One primer pair amplifies a sequence from the mouse amylase gene, the other pair amplifies either a human or mouse β-globin locus sequence. To determine conditions of linear amplification, serial dilutions of chromatin containing 0.5–4 ng of DNA were used as template. Shown are products and quantification for two representative primer pairs (βPr with amy4 + 6 and 3′β with amy4 + 5), revealing linear amplification of the total signal (bars) and a constant ratio (line) for the two products under these PCR conditions (see Materials and Methods). For the quantitative analysis of immunoprecipitated material shown in Figs. 3 and 4, 1–2 ng of DNA were used per reaction to ensure amplification in the linear range.
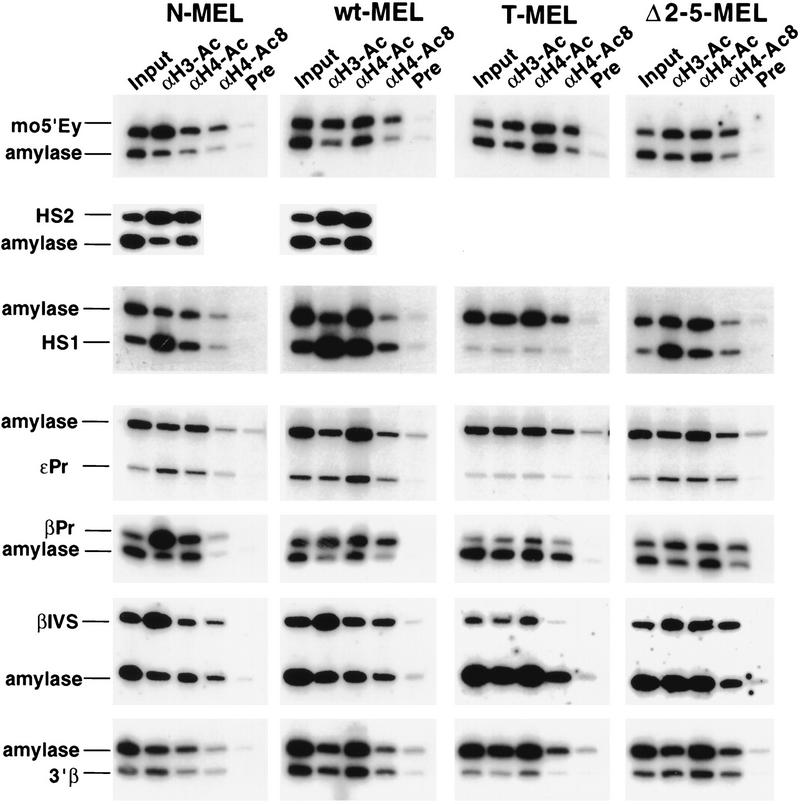
By use of this methodology, the relative enrichment or depletion of β-globin sequences in independent chromatin preparations and different cell lines can be compared. Ten different sequences throughout the human locus, spread over a distance of 129 kb (Fig. 1), were analyzed. Five of these are located in putative nonregulatory sequences, three at different promoters in the locus, and two in the LCR. To compare different chromatin preparations and to exclude clonal differences among the cell lines, a control PCR for an intergenic β-globin sequence (5′Ey) in the mouse locus was performed for each immunoprecipitation. PCR products of a representative immunoprecipitation experiment are shown in Figure 3, and the ratios to amylase for each sequence in Figure 4.
Figure 3.
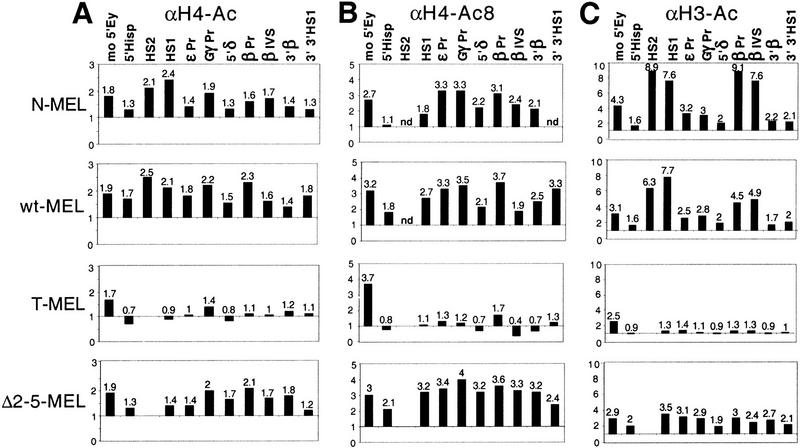
Comparative analysis of histone H3 and H4 acetylation at specific sequences in different alleles of the globin locus. Duplex PCR was performed on the input and bound fractions from the chromatin immunoprecipitation experiments. (see Fig. 2 and text). Three antibodies were used for immunoprecipitation. αH3–Ac recognizes histone H3 acetylated at lysines 9 and 14; αH4–Ac recognizes any acetylated isoforms of H4; and αH4–Ac8 is specific for H4 acetylated at lysine 8. To control for nonspecific binding, immunoprecipitation experiments were performed in parallel with rabbit preimmune serum (Pre), and a background signal of 10% or less compared with any antibody containing immunoprecipitation was observed. The figure shows PCR reactions from one representative experiment. Chromatin immunoprecipitation and PCR analysis were repeated at least twice with consistent results. Quantification reveals up to ninefold enrichment for a human globin sequence in N-MEL and wt-MEL (see Fig. 4) compared with the mouse amylase gene. HS2 is deleted in Δ2–5-MEL and T-MEL (see Fig. 1) and therefore could not be analyzed in these hybrids.
Figure 4.
Quantification of duplex PCR results. The ratio of products obtained with β-globin and amylase primers was determined for each input and antibody-bound sample for each cell line. The globin/amylase ratio from each bound fraction was standardized by dividing by the globin/amylase ratio from the input material to determine enrichment or depletion of a globin sequence during the immunoprecipitation. Enrichment of the globin sequence over amylase is therefore reflected by a number >1 and a depletion <1. The X-axis is drawn at 1, which reflects no enrichment. (A) Results obtained with the αH4–Ac antibody, (B) the αH4–Ac8 antibody, and (C) the αH3–Ac antibody. The sequence for the HS2 primer set is deleted in T-MEL and Δ2–5. (nd) Not done.
Histone H4 hyperacetylation of the globin locus correlates with an open chromatin structure and does not require 5′HS2–5
Using antisera that recognize all four acetylated lysine residues, and therefore all acetylated isoforms of histone H4 (αH4–Ac), we first compared the overall level of H4 acetylation in the human β-globin locus of N-MEL, T-MEL, wt-MEL, and Δ2–5-MEL. Quantitative PCR analysis reveals that all sequences in the human β-globin locus are enriched in the bound fraction in the two wild-type and the Δ2–5 alleles (Fig. 4A). The total enrichment relative to the amylase gene ranges from 1.4- to 2.4-fold and is comparable with that of a mouse globin control sequence (1.7–1.9), suggesting that the mouse and human β-globin loci share the same degree of total H4 acetylation in these hybrids. In contrast, β-globin sequences in the Hispanic allele in T-MEL display little or no enrichment relative to amylase in the bound fraction, suggesting that this nuclease-resistant and transcriptionally inactive locus is as hypoacetylated as the amylase gene (Fig. 4A). Because the mouse globin control sequence is similarly enriched in the bound fraction in all cell lines, clonal variation or the quality of the chromatin preparation does not account for the differences in the human loci.
Although reproducible and significant, the total enrichment of globin sequences (mouse and human) compared with the amylase gene is modest using this αH4–Ac antibody. Therefore, we repeated the immunoprecipitations using a polyclonal antibody that specifically recognizes histone H4 acetylated at lysine 8 (αH4–Ac8). As this modification is present only in di- to tetra-acetylated isoforms of H4 (Johnson et al. 1998), we reasoned that the αH4–Ac8 antibody may reveal a greater enrichment for hyperacetylated chromatin. In all cell lines, quantitative PCR analysis of the αH4–Ac8 bound fraction shows a higher enrichment for the mouse β-globin control (2.7- to 3.7-fold) than that obtained with the αH4–Ac antibody (1.7- to 1.9-fold). A more pronounced enrichment was also observed for sequences in the human β-globin locus in the two wild-type and Δ2–5 lines compared with T-MEL. As with the αH4–Ac antibody, this enrichment is nearly uniform throughout the locus, although the absolute level of enrichment is slightly more variable (Fig. 4, cf. A and B). Consistent with the results obtained with the serum against all acetylated isoforms of H4, the Hispanic allele shows little or no enrichment in the bound fractions compared with the two wild-type and Δ2–5 alleles.
Taken together, the results obtained with two different antisera against acetylated H4 suggest that the nuclease-insensitive and transcriptionally inactive Hispanic allele is hypoacetylated, and that the human globin locus is equally hyperacetylated throughout the two wild-type alleles and in the nuclease-sensitive Δ2–5 allele. Thus, H4 hyperacetylation correlates with an open chromatin structure at the human β-globin locus and requires neither globin transcription nor the LCR.
Histone H3 hyperacetylation marks open chromatin but is more prominent at the LCR and the transcribed gene
To further characterize the histone acetylation status of the human β-globin locus, we repeated the immunoprecipitation experiments using an antibody that recognizes histone H3, specifically acetylated at lysine 9 and 14 of the four potentially acetylated lysines (Braunstein et al. 1996). Chromatin from T-MEL immunoprecipitated with serum against acetylated histone H3 displays little or no enrichment of any sequences in the human globin locus as compared with the mouse β-globin locus (Fig. 4C). This suggests that the nuclease-insensitive chromatin in T-MEL is hypoacetylated for histone H3, consistent with the observed hypoacetylation for H4.
In contrast, both wild-type lines and the Δ2–5 allele show at least two to threefold enrichment for all sequences in the human locus after immunoprecipitation with the serum against acetylated H3. The level of enrichment is similar among these three alleles at six positions in the locus, 5′ of the Hispanic breakpoint (5′Hisp), the inactive ε and Gγ promoters (εPr and GγPr), within the intergenic region between Aγ and δ (5′δ), downstream of the adult gene (3′β), and downstream of 3′HS1 (3′3′HS1). These results indicate that open chromatin in the β-globin locus is marked by an increased H3 acetylation, as was observed for H4.
However, although the LCR (HS1) and the β-globin promoter and gene (βPr, βIVS) in the Δ2–5 allele display a level of enrichment similar to the other positions in the locus, the two wild-type lines show a significantly higher degree of H3 acetylation at these positions. At HS1, both wild-type lines show a twofold higher enrichment than Δ2–5. At the β promoter and the first intron, both wild-type lines show a higher enrichment than the Δ2–5 allele, but the degree of enrichment differs, fivefold for wt-MEL and eightfold for N-MEL. The absolute level of enrichment at the LCR and the β-globin gene in these lines is the highest that we have observed, suggesting that histone H3 is significantly more acetylated at the complete LCR and the active gene than at other sequences in the locus or the same sequences in the Δ2–5 allele.
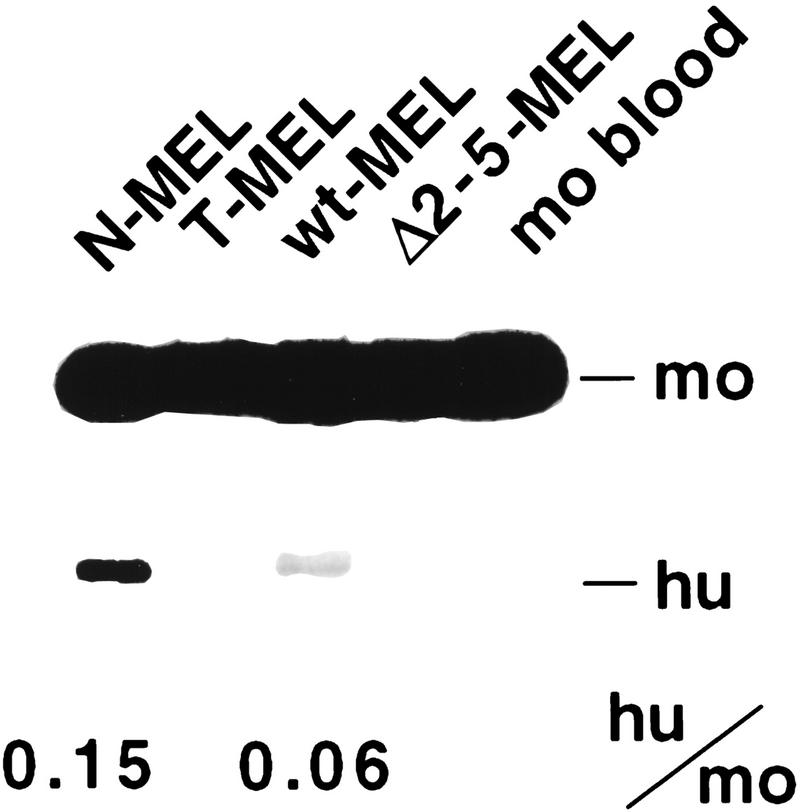
As the difference in H3 acetylation observed between wt-MEL and N-MEL is localized to a region including the β promoter and the first intron of this gene, we asked whether this difference correlates with the level of transcriptional activity as measured by RT–PCR. As expected, T-MEL and Δ2–5-MEL express no human β-globin mRNA, whereas both wild-type lines do. However, N-MEL expresses a twofold higher level of β-globin than wt-MEL (Fig. 5), a result that correlates well with the degree of enrichment for acetylated H3 at the promoter and at the transcribed region.
Figure 5.
Expression of β-globin mRNA. RT–PCR was performed with a primer pair which coamplifies the human and murine adult β-globin mRNA, followed by restriction enzyme digestion specific for the human product, allowing separation of the human and murine products by gel electrophoresis. Bands are quantified and signal intensities are corrected for the size difference between the digested human (hu) product and the undigested mouse (mo) product, but not for the number of alleles (mouse 2, human 1).
In summary, the general pattern of histone H3 acetylation is consistent with that of histone H4. The nuclease-insensitive chromatin in the Hispanic deletion is characterized by H3 and H4 hypoacetylation, whereas the nuclease-sensitive chromatin in the Δ2–5 deletion shows considerable H3 and H4 hyperacetylation throughout the locus. Taken together, these results reveal that an open chromatin structure is marked with acetylated H3 and H4. However, in contrast to the homogeneous pattern of histone H4 acetylation observed throughout the locus, the two wild-type alleles show increased hyperacetylation of histone H3 at the LCR and the transcribed β-globin gene. This localized H3 hyperacetylation is dependent on the presence of the LCR and may either be a requirement for globin gene activation or a consequence thereof.
Localization of the human β-globin locus relative to murine centromeres correlates with its open and hyperacetylated configurations and does not require the LCR
Several studies have suggested a correlation between silencing of a gene and its proximity to centromeric heterochromatin in the interphase nucleus (Csink and Henikoff 1996; Brown et al. 1997, 1999). Moreover, we showed recently that suppression of transgene silencing and maintenance of its open chromatin configuration require a functional enhancer and distance away from centromeric heterochromatin (Francastel et al. 1999). Thus, we asked whether the β-globin locus in its endogenous location is also subjected to changes in its nuclear location, if its open chromatin configuration and/or transcriptional activity are associated with distance away from centromeres, and whether the LCR plays a role in these processes.
To address these questions, we performed FISH experiments using a probe specific for the human β-globin genes and a probe specific for murine centromeres. We chose to define the location of the human β-globin locus relative to the centromeres of the host murine cell because these centromeres cluster in the interphase nucleus and are part of the dominant heterochromatin fraction in murine cells. The cell hybrids used here contain between 1 and 8 human chromosomes, including human chromosome 11. We found that, in contrast to the murine centromeres, human centromeres do not cluster in the interphase nucleus of the murine hybrids; rather, they are scattered throughout the nucleus, and no association of human and murine centromeres is observed. Moreover, in the MEL hybrids, we do not observe a colocalization of the human globin locus with its own centromere or any other human centromere (data not shown).
Next, we determined the proximity of the human locus to murine centromeres and observed that the silent/closed chromatin locus in T-MEL cells is closer to murine centromeric heterochromatin than is the active/open chromatin locus in the N-MEL hybrids (Fig. 6A). To confirm this observation, distances between the human β-globin locus and murine centromeres from at least 35 interphase nuclei were measured and compared between the different cell hybrids. Statistical analysis of these results (Fig. 6B) shows that the β-globin locus is significantly closer to murine centromeric heterochromatin in T-MEL than it is in N-MEL.
Figure 6.
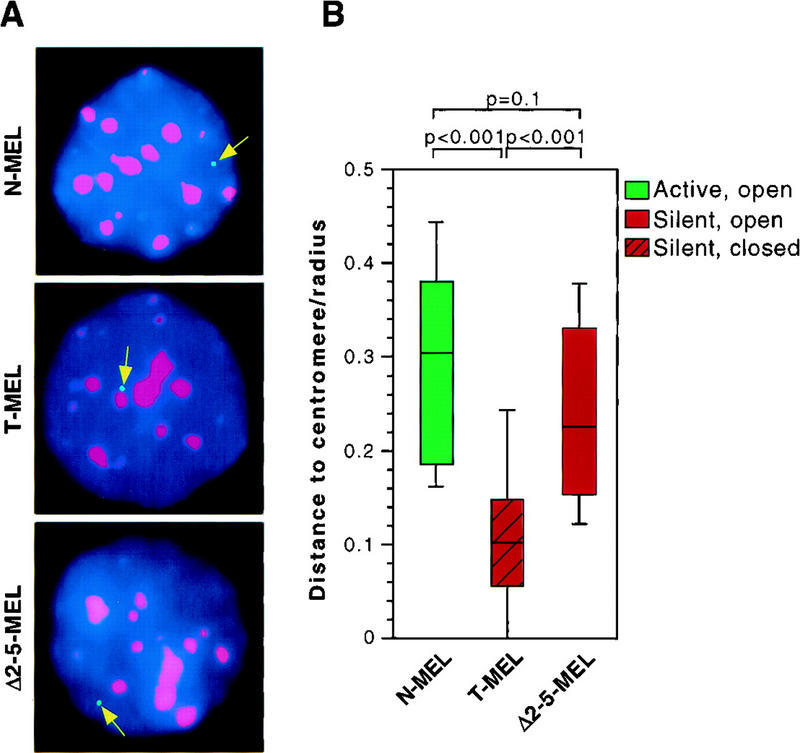
FISH analysis of the localization of the human β-globin locus in interphase MEL nuclei. (A) Examples of interphase nuclei from MEL cell hybrids carrying a human chromosome 11 containing a wild-type β-globin locus (N-MEL), Hispanic deletion (T-MEL), or deletion of 5′HS2–5 of the LCR (Δ2–5-MEL). DAPI is shown in blue, centromeres in red, and the β-globin locus in green. In MEL hybrids containing an open β-globin locus (N-MEL and Δ2–5-MEL), the green signal for the globin locus is found away from a red signal for centromeres, regardless of β-globin gene transcription. In contrast, the locus is in close proximity to centromeres in hybrids harboring a silent, closed locus (T-MEL). (B) Box plot representing the distance between the globin locus and the closest murine centromere cluster. Data were collected for each cell line from at least 35 interphase nuclei. Horizontal bars represent the 10th, 25th, 50th (median), 75th, and 90th percentiles, and p values for pairs of samples are indicated (see Materials and Methods). The distance between the globin locus and the closest murine centromere cluster is significantly higher when the locus is in an open configuration (N-MEL and Δ2–5-MEL), than when it is in a closed configuration (T-MEL).
These results reveal an association between the closed and transcriptionally silent state of the β-globin locus and proximity to centromeric heterochromatin. They also demonstrate that the endogenous β-globin locus can be located in two different compartments of the interphase nucleus, as measured by its positioning relative to centromeric heterochromatin. The differences in localization could be due to transcriptional activity, or chromatin configuration, or the presence of the LCR. To distinguish these possibilities, we analyzed the positioning of the β-globin locus in the Δ2–5-MEL hybrid, in which transcriptional activity and open chromatin structure of the locus are dissociated. As shown in Figure 6B, the Δ2–5 locus localizes far from murine centromeres, and shows no significant difference in the distance to centromeric heterochromatin compared with that of the wild-type allele in N-MEL. This result suggests that open chromatin configuration, rather than transcriptional activity per se, is associated with distance from centromeric heterochromatin. This result also demonstrates that LCR 5′HS2–5, which are not required for opening of the locus, are also not required for positioning of the β-globin locus away from centromeric heterochromatin.
Discussion
Activation of gene expression is believed to be a multistep process involving changes in chromatin structure and promoter activation (Felsenfeld 1996). However, the sequence of events leading to the active transcriptional state and the cis-acting elements involved in these processes remain unclear. Previously, we demonstrated that chromatin opening and transcriptional activity in the β-globin locus are dissociable processes, and that the LCR is not necessary for an open chromatin structure. We now report that general histone acetylation in the locus, and its placement at a distance from centromeric heterochromatin, are both LCR independent. In contrast, promoter activity, which is LCR dependent, correlates with localized histone H3 hyperacetylation. These findings may reflect a multistep process by which transcriptional activation at the globin locus is achieved.
Chromatin structure and global histone acetylation at the human β-globin locus
Acetylation of lysine residues at the amino-terminal tails of histones is a reversible modification that has been shown to mark open (nuclease sensitive) chromatin and to be involved in promoter activation (for review, see Struhl 1998). Our analysis of histone acetylation across the human β-globin locus reveals a strict correlation between the level of H4 acetylation and the nuclease sensitivity of the locus: The closed, transcriptionally inactive locus carrying the Hispanic thalassemia deletion is hypoacetylated compared with the open wild-type or 5′HS2–5 deleted loci. The transcriptionally active wild-type locus and the silent 5′HS2–5 deletion locus are hyperacetylated to a similar extent, consistent with a previous observation that genes are hyperacetylated in the cell type in which they are potentially active, independent of their transcriptional status (O'Neill and Turner 1995). The histone H3 acetylation state is comparable with that of H4 in each of the alleles, with the exception of the localized hyperacetylation discussed below.
A previous study that used an anti-acetyl-lysine antibody with no specificity for histones revealed domain-wide hyperacetylation in the chicken β-globin locus in mature erythrocytes (Hebbes et al. 1994). This hyperacetylated domain comapped with the region of nuclease sensitivity and was flanked by DNase I-insensitive and hypoacetylated chromatin (Stalder et al. 1980; Hebbes et al. 1994). Such borders of nuclease sensitivity have not yet been identified in the mouse and human β-globin loci, and extended sequence analysis suggests that the domain in which these loci reside may be larger and more complex than thought previously (Bulger et al. 1999). Our observation of hyperacetylation at the most 5′ and 3′ regions analyzed (34 kb upstream of 5′HS5 and 9 kb downstream of 3′HS1, respectively) support this hypothesis. Thus, general histone acetylation correlates with an open chromatin structure in the human β-globin locus in erythroid cells, and this locus-wide acetylation is independent of both globin gene transcription and the presence of the LCR.
H3 acetylation in LCR-mediated activation of the human β-globin gene
We observed a striking localized increase in H3 hyperacetylation in the LCR and the β-globin promoter and gene in the two β-globin expressing wild-type alleles (N-MEL and wt-MEL) compared with the silent but open Δ2–5 allele. The localized H3 hyperacetylation at the LCR was similar in both wild-type alleles, whereas the β-globin promoter/gene hyperacetylation was two-fold higher in N-MEL compared with wt-MEL (Fig. 4C). Interestingly, N-MEL also has a twofold higher level of β-globin mRNA than wt-MEL (Fig. 5). This correlation of the level of transcription with the degree of localized acetylation suggests that if H3 hyperacetylation is involved in promoter activation it may be a rate-limiting step.
Localized hyperacetylation of H3 and/or H4 has been reported for several inducible mammalian genes (Chen et al. 1999; Parekh and Maniatis 1999). However, our study is the first description of localized hyperacetylation at a cis-acting element distal from the activated promoter. The preferential localized hyperacetylation of H3 during gene activation in the globin locus argues that HATs with H3 preference are recruited to the LCR and the globin gene. In vitro studies suggest that HATs vary widely in their histone specificity (for review, see Davie 1998; Kuo and Allis 1998); for example, the yeast transactivator GCN5 (the HAT activity of the SAGA complex) preferentially acetylates H3 in vivo (Burgess et al. 1999). Recently, it was shown that activation of the yeast HO promoter is a sequential process (Cosma et al. 1999), in which binding of the transcription factor Swi5P leads to recruitment of the SWI/SNF chromatin remodeling complex, followed by the HAT-containing SAGA complex, and finally by the binding of another transactivator (SBF). It is now clear that a variety of transcriptional coactivators with intrinsic HAT activity can be recruited by DNA-binding transcriptional activators (for review, see Kuo and Allis 1998; Struhl 1998). For example, in the erythroid lineage, GATA-1 (Blobel et al. 1998), p45 (the transactivating subunit of NF-E2; Cheng et al. 1997), and the erythroid Krüppel-like factor (EKLF; Zhang and Bieker 1998) can interact with the HAT CBP/P300. In addition, EKLF and a SWI/SNF-related chromatin remodeling complex (E-RC1) are required for chromatin remodeling of the β-globin promoter in vitro (Armstrong et al. 1998). Thus, as in the case of the HO promoter, a sequence of initial transcription factor binding, chromatin remodeling, and acetylation may occur at the LCR and the β-globin gene.
Localized H3 hyperacetylation at the LCR and β-globin promoter/gene is consistent with multiple current models of long-range gene activation by the LCR. For example, in the case of a looping mechanism (Choi and Engel 1988; Fraser and Grosveld 1998), promoter/gene H3 acetylation could be the consequence of a HAT-containing complex recruited by the LCR and a direct physical association between these regions. On the other hand, if LCR-mediated activation occurs by a spreading or linking mechanism (Bulger and Groudine 1999), localized acetylation at the promoter, along with active transcription, would not require a direct LCR/promoter interaction. Regardless, our studies reveal separable processes, chromatin opening marked by a domain-wide H3/H4 acetylation, and gene activation marked by an additional localized H3 hyperacetylation at the LCR and active gene. The localized H3 change is most simply explained by the specific recruitment of a HAT activity to the LCR and to the active gene by an erythroid-specific transcriptional activator. In contrast, the domain-wide change in acetylation may result from the recruitment of HAT activity via protein(s) with binding sites throughout the locus. This locus-wide change in acetylation may also be the consequence of the removal of the β-globin locus to a nuclear compartment enriched in such factors.
Nuclear compartmentalization as a mediator of chromatin opening and histone acetylation?
Our FISH analyses reveal that the β-globin locus can be found in distinct locations in the interphase nucleus, relative to the heterochromatin compartment defined by the centromeres of the murine host cell. The open and transcriptionally active wild-type globin locus is located away from centromeric heterochromatin, whereas the silent and closed Hispanic deletion allele localizes close to heterochromatin. However, the Δ2–5 allele, which is transcriptionally silent but has an open chromatin structure, demonstrates the same nuclear localization as the wild-type locus, that is, away from centromeres. Together, these results show that the different locations of the endogenous β-globin locus in the erythroid nucleus correlate with the chromatin and general acetylation configurations of the locus, but not with transcription of the β-globin genes. These results are in agreement with our previous work on transgene silencing, which demonstrated that stability of the open chromatin configuration requires positioning of the transgene away from the centromeric heterochromatin compartment (Francastel et al. 1999).
Several mechanisms could account for how structural modifications of the β-globin locus, such as histone acetylation and chromatin opening, are achieved over broad regions. The localization of a gene in a specific nuclear compartment may be a prerequisite for propagation of an open or closed chromatin structure, or a consequence thereof. Silencing at the mating-type loci in yeast, for example, is associated both with a silenced chromatin structure and with localization of silenced regions near the nuclear periphery, suggesting that both nuclear compartments and spreading of specific chromatin structures are involved in the mechanism of regulation of the silenced domains (Chien et al. 1993; Gotta and Gasser 1996; Maillet et al. 1996). Furthermore, the targeting of a locus to the nuclear periphery can facilitate SIR-mediated silencing (Andrulis et al. 1998). Other studies suggest that the nucleus is divided into compartments in which specific proteins and specific sequences concentrate. For example, Ikaros, a DNA-binding protein, associates with centromeres and silent genes in murine lymphocytes (Brown et al. 1997, 1999). Other proteins involved in the silencing of transcription (e.g., MeCP2, MBDs, or HDACs) also associate with constitutive heterochromatin in the repressive centromeric compartment (Lewis et al. 1992; Hendrich and Bird 1998; Kim et al. 1999). Thus, sequestration and/or recruitment of a locus to such a compartment could lead to broad deacetylation and chromatin compaction.
The mechanisms by which a locus is recruited and/or sequestered into specific compartments remain to be determined. However, our recent demonstration that 5′HS2, an enhancer that is a component of the β-globin LCR, is sufficient for localization of a transgene away from centromeric heterochromatin and suppression of transgene silencing (Francastel et al. 1999) suggests that enhancers or LCRs may maintain gene expression by preventing its localization close to the repressive heterochromatic compartment. The present study finds that the LCR is unnecessary to relocate the β-globin locus away from heterochromatin; the contrast between these and our earlier results emphasizes the likelihood that the large number of factor-binding sites, similar to those found in the LCR, scattered throughout the native locus function to alter subnuclear location and chromatin structure. Thus, specific cis-acting elements other than the LCR may maintain the β-globin locus in an open chromatin/acetylated configuration by disrupting or preventing its association with a nuclear compartment enriched in heterochromatin proteins and HDACs. This disruption could result from the recruitment of HATs or other trans-acting factors, leading to chromatin opening and acetylation of the locus. It is also possible that the closed chromatin structure and pericentromeric localization of the Hispanic allele may be due to the deletion of specific sequences upstream of the LCR that confer chromatin opening or protect against a silencing activity located upstream of the domain.
Previously, we demonstrated that the β-globin LCR, although required for high-level expression from the locus, is not required for establishment or maintenance of its open chromatin structure (Epner et al. 1998; Reik et al. 1998; Bender et al. 2000). We now show that deletion of the LCR affects only globin transcription and localized H3 hyperacetylation, and has no affect on either general acetylation or nuclear localization of the locus. Thus, we postulate that the LCR-independent localization of the β-globin cluster away from centromeric heterochromatin is not sufficient, but is most likely necessary, for the LCR-dependent transcriptional activation of the β-globin genes.
Gene activation at the β-globin locus: A multi-step process?
Our previous demonstration that chromatin opening and transcriptional activation of the β-globin locus are dissociable, in combination with our present demonstration that these two processes are associated with distinct structural correlates, strongly suggests that chromatin opening and transcriptional activation of the locus are achieved through distinct mechanisms. We propose a sequential model of gene activation, the first step involving relocation away from centromeric heterochromatin and the establishment of an open chromatin configuration marked by locus wide acetylation, and the second step involving local histone H3 hyperacetylation and promoter activation. The configuration of the chromatin structure in a multipotent stem cell is not clear, but commitment to the red cell lineage may be characterized by an LCR-independent preactivation step in which the globin locus is open, acetylated, and localized away from centromeric heterochromatin in the nucleus of a red cell precursor, with terminal differentiation leading to an LCRdependent local hyperacetylation and gene activation.
Materials and methods
Cell lines and culture conditions
Human chromosome 11 hybrids wt-MEL and Δ2–5-MEL were generated by microcell transfer (Dieken et al. 1996) of the human chromosome 11 as described (Reik et al. 1998), except that the chromosome was transferred into murine ES cells and then into MEL cells. This transfer scheme avoids the chromosomal fragmentation we have observed in direct transfer of chromosome 11 from DT40 into the MEL background (Dieken et al. 1996; Reik et al. 1998). Deletion of 5′HS2-5 was accomplished in ES cells prior to transfer into MEL cells. The β-globin locus on the complete chromosomes displays the same chromatin and transcription phenotype as the previously described large fragments spanning several megabase bairs (A. Reik, unpubl.). Specifically, both loci are DNase I sensitive, and the β-like globin genes are inactive in the Δ2–5 deleted locus (Fig. 5, A. Reik, unpubl.). MEL cell lines were maintained in DME supplemented with 10% bovine calf serum. The wild-type and Δ2–5 chromosomes contain the Hygro gene inserted into the ras locus; to ensure maintenance of these chromosomes, wt-MEL and Δ2–5-MEL were selected periodically with hygromycin. N-MEL and T-MEL were derived by transfer of the wild-type and thalassemic chromosome 11, respectively, from lymphocytes of the patient with Hispanic thalassemia into MEL cells (Forrester et al. 1990). Cells containing the human chromosome were enriched by antibody-mediated culture dish binding (panning) with an antibody for an expressed surface antigen encoded by human chromosome 11, as described previously (Forrester et al. 1990).
DNA FISH
FISH was performed as described previously (Francastel et al. 1999). Briefly, MEL cell hybrids were resuspended in 0.075 m KCl for 20 min at room temperature, fixed in 3:1 methanol/acetic acid, and deposited on a slide. Slides were treated with 100 μg/ml RNase in 2× SSC for 1 hr at 37°C, postfixed in 4% paraformaldehyde/5% acetic acid/PBS for 20 min at room temperature, equilibrated in 70%formamide/2× SSC at room temperature, and denatured for 3 min in 70% formamide/2× SSC at 72°C. The slides were then hybridized overnight in 50% formamide/10% dextran sulfate/2× SSC containing murine (Cambio, UK) or human (Oncor) biotin-labeled pan-centromeric DNA probes and 20–40 ng of a digoxygenin (DIG)-labeled probe covering 15 kb of the human β-globin locus (ClaI fragment that spans the adult δ- and β-genes), in the presence of 10 μg of human Cot-1 repetitive DNA (BRL). After hybridization, slides were washed twice for 5 min in 50% formamide/2× SSC at 42°C and twice for 5 min in 2× SSC at 42°C. Slides were then treated with streptavidin–Texas red (Vector) and anti-DIG-fluorescein Fab fragments (Boehringer Mannheim) in 4× SSC/5% milk for 30 min at room temperature, washed three times for 5 min in 2× SSC/0.005% chaps, and mounted in Vectashield (Vector) containing 0.1 μg of DAPI. Plasmid probe for the globin genes was DIG-labeled by nick translation (Boehringer Mannheim). Nuclei were visualized with a Deltavision SA3 microscope (Applied Precision) with a cooled CCD camera. Each of the three wavelengths was corrected using the Deltavision 2D deconvolution program (Applied Precision) and merged using Adobe Photoshop. Distances between the human β-globin genes signal and the closest centromeric signal were measured in Adobe Photoshop, between the middle of a green dot and the middle of the closest red spot, and divided by the radius of the cell. For each population studied, at least three independent series of slides were assessed. At least 35 nuclei were counted, and the 10th, 25th, 50th (median), 75th, and 90th percentiles were calculated. The P value for a pair of samples was determined by a two-tailed U test for comparison of two unpaired groups.
Chromatin immunoprecipitation
Chromatin fixation and purification were performed as described in Orlando et al. (1997) with minor modifications. Exponentially growing cells (2 × 108) were fixed in 150 ml of DME with 1% formaldehyde for 3 min at room temperature. DNA content of cross-linked chromatin was quantified using a Hoefer Instruments fluorometer. Polyclonal antibodies against all acetylated isoforms of H4 (αH4–Ac) and against H3 acetylated at Lysine 9 and 14 (αH3–Ac) were purchased from Upstate Biotechnology. Antisera recognizing histone H4 acetylated at Lysine 8 (αH4–Ac8) was purchased from Serotec and rabbit preimmune serum from Jackson Immunoresearch Laboratories. Immunoprecipitation conditions for all antisera followed the protocol suggested by one of the manufacturers (Upstate Biotechnology) with minor modifications. Dialyzed cross-linked chromatin (∼20 μg in each immunoprecipitation) was adjusted to 1× RIPA buffer before immunoprecipitation by adding 2× RIPA buffer.
DNA analysis
For slot blot analysis, 500 ng of input and antibody-bound DNA were applied to a membrane using the manufacturer's protocol (GeneScreen). The filter was hybridized to an end-labeled oligomer (R947) corresponding to a mouse centromeric minor satellite repeat (Kipling et al. 1994). A PhosphorImager and Image Quant software were used for quantification. Quantitative PCR of input and bound chromatin was performed with 1–2 ng of DNA as a template in a total volume of 25 μl with the appropriate primer pairs. Primers for β-globin were designed and tested to be either human or mouse specific and to give a product size between 340 and 400 bp. Three different primers for the mouse amylase gene were designed to amplify from the same sequence but give two products of different sizes (primers amy4 + amy5 = 350 bp, primers amy4 + amy6 = 400 bp) to allow duplex PCR with any of the globin primer sets. A total of 0.1 μl of [32P]dCTP (NEN) was added to each reaction. For each sequence, PCR reactions were performed in parallel under conditions of linear amplification in a Perkin Elmer 9600 thermocycler, for 27 cycles, using identical temperature profiles for all primer pairs. One-third of the reaction was subjected to electrophoresis on a 5% polyacrylamide gel and quantified with a PhosphorImager and the Image Quant software.
Primers
Mouse amylase 2.1y gene. Primer pairs amy4 + 5 yield a 348-bp, and primers 4 + 6 a 401-bp product. Amy4, TCAGTTGTAATTCTCCTTGTACGG; Amy5, CCTCCCATCTGAAGTATGTGGGTC; Amy6, CATTCCTTGGCAATATCAACC. For primers amplifying mouse or human globin sequences, the name of the product based on location, predicted size in base pairs, and sequence of the primers are listed. The location of the amplified sequences from the human locus are shown in Figure 1.
Mouse: 5′Ey, (located 1.1 kb 5′ of the Ey start codon), 376 bp; 5Ey-3, GCACATGGATGCAGTTAAACAC; 5Ey-4, GAGTGACAGTGTAGAGAAGATG. Human: 5′Hisp, 376 bp; hu5hisp1, TATCTAGCTCTCCTAGAATCC; hu5hisp2, AGATTTCCAGAGCACAAGTAC. HS2, 395 bp; huHS2-1, TTCCAGCATCCTCATCTCTGA; huHS2-2, TCACATTCTGTCTCAGGCATC. HS1, 355 bp; huHS1-3, CCTGCAAGTTATCTGGTCAC; huHS1-4, CTGGGCAGCGTCAGAAACTG. εPr, 342 bp; huEp-5, TTTTAAGTACCATGGAGAACAGG; huEp-6, ATGAAATGACACCATATCAGATAC. GγPr, 336 bp; huGam1, GAGATTGACAAGAACAGTTTGAC; huGam2, ATCCAGTGAGGCCAGGGGC. 5′δ, 350 bp; 5delta-1, GTAACCAGATCTCCCAATGTG; 5delta-3, ATATGTGGATCTGGAGCTCAG. βPr, 395 bp; hubPr1, TGCTTACCAAGCTGTGATTCC; hubPr2, AACGGCAGACTTCTCCTCAGG. βIVS, 419 bp; BIVS-1, GGAAGGGGAGAAGTAACAGGG; BIVS-3, TACCCTGATTTGGTCAAT GTG. 3′β, 367 bp; h3beta-1, AGTTCATGTCCTTTGTAGGGAC. h3beta-2, GCTCTACGGATGTGTGAGATC. 3′3′HS1, 377 bp; hu3HS1-2, ATTGATTCCTCAGTTCTGGCTG; hu3HS1-1, TCTACTTGAGGTTGTGTCTCC.
RNA analysis
Expression analysis by RT–PCR was performed as described previously (Reik et al. 1998) using a single primer pair for the adult genes (HBG1 + 2) but without HMBA induction.
Acknowledgments
This work was supported by a fellowship from the Deutsche Forschungsgemeinschaft to D.S., a fellowship from the American Cancer Society to D.M.C., a fellowship from the Leukemia Research Foundation to A.R., and NIH grants HL57620 to D.M. and M.G. and DK44746 and CA54337 to M.G. We thank Linda Madisen and Tony Krumm for advice, Agnes Telling and Urszula Maliszewski for technical assistance, Mike Bulger for sequence data, Toshi Tsukiyama, Matthew Lorincz, and Mike Bulger for helpful comments on the manuscript, and the FHCRC Image Analysis Laboratory for assistance with FISH analysis.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL markg@fhcrc.org; FAX (206) 667-5894.
References
- Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- Armstrong JA, Bieker JJ, Emerson BM. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- Bender M, Bulger M, Close J, Groudine M. Globin gene switching and DNaseI sensitivity of the endogenous β-globin locus in mice do not require the Locus Control Region. Mol Cell. 2000;5:387–393. doi: 10.1016/s1097-2765(00)80433-5. [DOI] [PubMed] [Google Scholar]
- Blobel GA, Nakajima T, Eckner R, Montminy M, Orkin SH. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M, Sobel RE, Allis CD, Turner BM, Broach JR. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Brown KE, Baxter J, Graf D, Merkenschlager M, Fisher AG. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Looping versus linking: Toward a model for long-distance gene activation. Genes & Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- Bulger M, van Doorninck JH, Saitoh N, Telling A, Farrell C, Bender MA, Felsenfeld G, Axel R, Groudine M. Conservation of sequence and structure flanking the mouse and human beta-globin loci: The beta-globin genes are embedded within an array of odorant receptor genes. Proc Natl Acad Sci. 1999;96:5129–5134. doi: 10.1073/pnas.96.9.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SM, Ajimura M, Kleckner N. GCN5-dependent histone H3 acetylation and RPD3-dependent histone H4 deacetylation have distinct, opposing effects on IME2 transcription, during meiosis and during vegetative growth, in budding yeast. Proc Nat Acad Sci. 1999;96:6835–6840. doi: 10.1073/pnas.96.12.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- Cheng X, Reginato MJ, Andrews NC, Lazar MA. The transcriptional integrator CREB-binding protein mediates positive cross talk between nuclear hormone receptors and the hematopoietic bZip protein p45/NF-E2. Mol Cell Biol. 1997;17:1407–1416. doi: 10.1128/mcb.17.3.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CT, Buck S, Sternglanz R, Shore D. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell. 1993;75:531–541. doi: 10.1016/0092-8674(93)90387-6. [DOI] [PubMed] [Google Scholar]
- Choi OR, Engel JD. Developmental regulation of beta-globin gene switching. Cell. 1988;55:17–26. doi: 10.1016/0092-8674(88)90005-0. [DOI] [PubMed] [Google Scholar]
- Cockell M, Gasser SM. Nuclear compartments and gene regulation. Curr Opin Genet Dev. 1999;9:199–205. doi: 10.1016/S0959-437X(99)80030-6. [DOI] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- Csink AK, Henikoff S. Genetic modification of heterochromatic association and nuclear organization in Drosophila. Nature. 1996;381:529–531. doi: 10.1038/381529a0. [DOI] [PubMed] [Google Scholar]
- Davie JR. Covalent modifications of histones: Expression from chromatin templates. Curr Opin Genet Dev. 1998;8:173–178. doi: 10.1016/s0959-437x(98)80138-x. [DOI] [PubMed] [Google Scholar]
- Dhar V, Mager D, Iqbal A, Schildkraut CL. The coordinate replication of the human beta-globin gene domain reflects its transcriptional activity and nuclease hypersensitivity. Mol Cell Biol. 1988;8:4958–4965. doi: 10.1128/mcb.8.11.4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieken ES, Epner EM, Fiering S, Fournier RE, Groudine M. Efficient modification of human chromosomal alleles using recombination-proficient chicken/human microcell hybrids. Nat Genet. 1996;12:174–182. doi: 10.1038/ng0296-174. [DOI] [PubMed] [Google Scholar]
- Epner E, Reik A, Cimbora D, Telling A, Bender MA, Fiering S, Enver T, Martin DI, Kennedy M, Keller G, Groudine M. The beta-globin LCR is not necessary for an open chromatin structure or developmentally regulated transcription of the native mouse beta-globin locus. Mol Cell. 1998;2:447–455. doi: 10.1016/s1097-2765(00)80144-6. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin unfolds. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- Forrester WC, Epner E, Driscoll MC, Enver T, Brice M, Papayannopoulou T, Groudine M. A deletion of the human beta-globin locus activation region causes a major alteration in chromatin structure and replication across the entire beta-globin locus. Genes & Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- Francastel C, Walters MC, Groudine M, Martin DIK. A functional enhancer suppresses silencing of a transgene and prevents its localization close to centromeric heterochromatin. Cell. 1999;99:259–269. doi: 10.1016/s0092-8674(00)81657-8. [DOI] [PubMed] [Google Scholar]
- Fraser P, Grosveld F. Locus control regions, chromatin activation and transcription. Curr Opin Cell Biol. 1998;10:361–365. doi: 10.1016/s0955-0674(98)80012-4. [DOI] [PubMed] [Google Scholar]
- Gotta M, Gasser SM. Nuclear organization and transcriptional silencing in yeast. Experientia. 1996;52:1136–1147. doi: 10.1007/BF01952113. [DOI] [PubMed] [Google Scholar]
- Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Hebbes TR, Clayton AL, Thorne AW, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhof A, Wolffe AP. Transcription: Gene control by targeted histone acetylation. Curr Biol. 1998;8:R422–R424. doi: 10.1016/s0960-9822(98)70268-4. [DOI] [PubMed] [Google Scholar]
- Johnson CA, O'Neill LP, Mitchell A, Turner BM. Distinctive patterns of histone H4 acetylation are associated with defined sequence elements within both heterochromatic and euchromatic regions of the human genome. Nucleic Acids Res. 1998;26:994–1001. doi: 10.1093/nar/26.4.994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keohane AM, O'Neill LP, Belyaev ND, Lavender JS, Turner BM. X-Inactivation and histone H4 acetylation in embryonic stem cells. Dev Biol. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- Kipling D, Wilson HE, Mitchell AR, Taylor BA, Cooke HJ. Mouse centromere mapping using oligonucleotide probes that detect variants of the minor satellite. Chromosoma. 1994;103:46–55. doi: 10.1007/BF00364725. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Madisen L, Krumm A, Hebbes TR, Groudine M. The immunoglobulin heavy chain locus control region increases histone acetylation along linked c-myc genes. Mol Cell Biol. 1998;18:6281–6292. doi: 10.1128/mcb.18.11.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM. Evidence for silencing compartments within the yeast nucleus: A role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes & Dev. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- O'Neill LP, Turner BM. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- Parekh BS, Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- Reik A, Telling A, Zitnik G, Cimbora D, Epner E, Groudine M. The locus control region is necessary for gene expression in the human beta-globin locus but not the maintenance of an open chromatin structure in erythroid cells. Mol Cell Biol. 1998;18:5992–6000. doi: 10.1128/mcb.18.10.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder J, Larsen A, Engel JD, Dolan M, Groudine M, Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980;20:451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes & Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976;193:848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Zhang W, Bieker JJ. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc Natl Acad Sci. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]