Abstract
There is a global emergence of multidrug-resistant (MDR) strains of Klebsiella pneumoniae, a Gram-negative enteric bacterium that causes nosocomial and urinary tract infections. While the epidemiology of K. pneumoniae strains and occurrences of specific antibiotic resistance genes, such as plasmid-borne extended-spectrum β-lactamases (ESBLs), have been extensively studied, only four complete genomes of K. pneumoniae are available. To better understand the multidrug resistance factors in K. pneumoniae, we determined by pyrosequencing the nearly complete genome DNA sequences of two strains with disparate antibiotic resistance profiles, broadly drug-susceptible strain JH1 and strain 1162281, which is resistant to multiple clinically used antibiotics, including extended-spectrum β-lactams, fluoroquinolones, aminoglycosides, trimethoprim, and sulfamethoxazoles. Comparative genomic analysis of JH1, 1162281, and other published K. pneumoniae genomes revealed a core set of 3,631 conserved orthologous proteins, which were used for reconstruction of whole-genome phylogenetic trees. The close evolutionary relationship between JH1 and 1162281 relative to other K. pneumoniae strains suggests that a large component of the genetic and phenotypic diversity of clinical isolates is due to horizontal gene transfer. Using curated lists of over 400 antibiotic resistance genes, we identified all of the elements that differentiated the antibiotic profile of MDR strain 1162281 from that of susceptible strain JH1, such as the presence of additional efflux pumps, ESBLs, and multiple mechanisms of fluoroquinolone resistance. Our study adds new and significant DNA sequence data on K. pneumoniae strains and demonstrates the value of whole-genome sequencing in characterizing multidrug resistance in clinical isolates.
INTRODUCTION
The increasing clinical incidence of antibiotic-resistant bacteria is a major global health care issue. Characterization of antibiotic resistance determinants at the genomic level plays a critical role in understanding, and potentially controlling, the spread of multidrug-resistant (MDR) pathogens. Of particular concern is the spread of MDR strains of Klebsiella pneumoniae, a Gram-negative bacterium that causes nosocomial, urinary tract, and wound infections. K. pneumoniae can harbor both extended-spectrum β-lactamases (ESBL) and carbapenemases capable of hydrolyzing newer carbapenem drugs (25, 26, 42). Frequently associated with ESBL-producing K. pneumoniae is resistance to other antibiotics, including fluoroquinolones, aminoglycosides, trimethoprim, and sulfamethoxazoles. K. pneumoniae clinical isolates have been shown to manifest all three broad mechanisms of drug resistance in Gram-negative bacteria, which are the acquisition of novel antibiotic catalytic genes, mutations of antibiotic targets and membrane proteins, and differential expression of specific genes such as those for efflux pumps which mediate drug effects (43, 54).
K. pneumoniae, a gammaproteobacterium of the family Enterobacteriaceae, is a close relative of many familiar genera, such as Citrobacter, Escherichia, Enterobacter, and Salmonella (4). A distinguishing characteristic of K. pneumoniae is a thick polysaccharide coat which might facilitate its evasion of host defenses. The close genetic association of the members of the family Enterobacteriaceae facilitates the inter- and intraspecies transmission of plasmids and insertion elements, which are often vectors for the horizontal exchange of antibiotic resistance genes. K. pneumoniae multidrug resistance plasmids show a remarkable diversity and recombination history. For example, the recently reported plasmid pKP048 is a 151-kb sequence composed of multiple DNA segments with high nucleotide identities to other known plasmids and contains multiple drug resistance loci (21). Molecular surveillance studies have confirmed the spread of MDR isolates in the clinic due to commonly shared plasmids across K. pneumoniae, K. oxytoca, Enterobacter sp., Escherichia coli, and Salmonella sp (11, 37).
The intraspecific genetic variation among K. pneumoniae strains is considerable, with many strain-specific genes and genomic rearrangements being reported, often involving specific plasmids and mobile elements (29, 53, 55). K. pneumoniae is also very environmentally adaptable, with strains known to colonize plants as mutualistic nitrogen-fixing endophytes (13). At present, publicly available genome sequences exist for four strains of K. pneumoniae (see Table S1 in the supplemental material). K. pneumoniae MGH 78578 (Genome Sequencing Center of Washington University [http://genome.wustl.edu/genomes/view/klebsiella_pneumoniae/]) is a virulent human pathogen isolated from sputum and is resistant to several antibiotics. K. pneumoniae NTUH-K2044 (53) was isolated from a Taiwanese patient with a liver abscess and shows sensitivity to most clinically used antibiotics. K. pneumoniae subsp. rhinoscleromatis (ATCC 13884) is a human commensal species that had its DNA sequenced as part of the Human Microbiome Project (50). K. pneumoniae 342 is a nitrogen-fixing endophyte strain and a model for studying plant-bacterium associations (13). While it is not a clinical human pathogenic strain, in vivo infections of mice show that K. pneumoniae 342 has the potential to be virulent (13). The genome sizes of the different K. pneumoniae strains range from 5.1 to 5.6 Mb. In addition, five plasmids have been determined for K. pneumoniae MGH 78578, which range in size from 3.4 to 175.9 kb, while one and two different plasmids from K. pneumoniae strains NTUH-K2044 and 342, respectively, have been sequenced (13, 53).
Recent advances in DNA pyrosequencing or next-generation sequencing technology now facilitate the rapid determination of whole-genome sequences from microbial species (36). Here, we applied this technology to study the spectrum of genetic factors involved in K. pneumoniae antibiotic resistance. We selected for whole-genome sequencing, two K. pneumoniae clinical isolates with disparate antibiotic response phenotypes. One is highly susceptible to most clinically used antibiotics (strain JH1), while the other has a wide multidrug resistance phenotype (strain 112281). Comparative genomic analyses of these two strains, as well as four other published K. pneumoniae genomes, show the scope of the genomic variation which might underlie these diverse phenotypes, thereby shedding light on the determinants of antibiotic resistance in this bacterial species.
MATERIALS AND METHODS
Bacteriological experiments.
K. pneumoniae strains JH1 and 1162281 are clinical isolates from the culture collection of GlaxoSmithKline (Collegeville, PA). MICs were determined using broth microdilution methods according to Clinical and Laboratory Standards Institute guidelines (7). The MIC was the lowest concentration of an antibacterial compound that showed no visible growth after incubation at 37°C for 18 to 24 h, with a starting inoculum of ∼5.5 × 105 CFU/ml. Genomic DNA was purified from overnight cultures grown in Luria or cation-adjusted Mueller-Hinton broth by using a DNeasy kit from Qiagen (catalogue no. 69504) and following the manufacturer's instructions. Total RNA was isolated from mid-log-phase (optical density at 600 nm of 0.5) or overnight cultures using the RNeasy Miniprep kit from Qiagen (catalogue no. 74104). The RNA samples were then DNase I treated and used to synthesize double-stranded cDNA with the SuperScript Double-Stranded cDNA Synthesis Kit (catalogue no. 11917-020) and random primers (catalogue no. 48190-011) from Invitrogen.
Genome sequencing.
Genome sequencing was performed by contract with 454 Life Sciences/Roche (Branford, CT). Two runs were performed for separate DNA preparations of each K. pneumoniae strain, the numbers of reads were 271,198 and 287,404 for strain JH1 and 310,151 and 310,231 for strain 1162281, and the average read length was about 240 bp. The estimated average coverages of the JH1 and 1162281 genomes were 24-fold and 27-fold, respectively.
Genome assembly and annotation.
The 454-assembled genomic contigs were ordered and oriented into scaffolds by using NUCMER of the MUMMER software package (27) against K. pneumoniae MGH 78578, the chosen reference strain. For the alignment of contigs with the reference genome, we set the NUCMER minimum exact match to 20 and the minimum cluster size to 100 to avoid short-repeat-induced matches. A tiling path was then constructed out of the query contigs as mapped to the reference sequences, with a threshold accuracy of at least 90%, resulting in a final alignment assembly consisting of a total of 75 scaffolds from strain JH1 and 62 scaffolds from strain 1162281.
Genes were initially identified by TBLASTN searches using two JH1 and 1162281 genome sequences as databases and all of the other K. pneumoniae genome sequences as queries with separate searches for chromosomal and plasmid genes. For three-way genome comparisons, homologous genes from JH1 and 1162281 were identified by choosing the top hit from the K. pneumoniae MGH 78578 genome if the TBLASTN (1) hit showed ≥90% identity. However, in those instances that showed multiple hits, the top hit was chosen if it started within the first 10 amino acids of the K. pneumoniae MGH 78578 query. Both genomes were also annotated by using the xBASE2 server (http://www.xbase.ac.uk/annotation/) (5), which is an automated pipeline of multiple genome annotation packages including GLIMMER (9), tRNAScan-SE (32), RNAmmer (28), and protein BLAST (1) for searches against a selected reference genome, K. pneumoniae MGH 78578. The annotations of the predicted open reading frames (ORFs) generated from the xBASE2 server were identified from BLASTP searches using an expectation (E) value of 10e−05 having ≥80% identity and a match length of ≥30%. For predicted ORFs not found in any of the published K. pneumoniae genomes, BLASTX and BLASTP (1) searches were performed against the entire GenBank nonredundant (nr) database. Gene functional categories were determined using the Comprehensive Microbial Resource (CMR) (44) and NCBI Protein Clusters (24) databases.
mRNA expression analysis.
To determine the completeness of genome sequence coverage, mRNAs from sequenced strain 1162281 and control strain K. pneumoniae MGH 78578 were purified from mid-log-phase and overnight cultures. Double-stranded cDNA generated from mRNA was hybridized to a K. pneumoniae MGH 78578 genomic microarray (NimbleGen A10103-00-01) by using the manufacturer's protocols. Four biological replicates were collected for each strain per growth condition. A Bayesian t test was used to compare K. pneumoniae MGH 78578 to 1162281 data to identify differentially expressed (DE) genes (14). Raw P values were adjusted for multiple-hypothesis testing using the FDR-BH procedure (3). DE genes with adjusted P values of <0.05 and >1.5-fold changes were selected for all subsequent analyses.
Phylogenetic analysis.
The evolutionary relationships among all of the sequenced K. pneumoniae genomes, as well as an outgroup species, K. variicola At-22 (45), were determined from a concatenated alignment of all single-copy orthologous proteins (see Table S1 in the supplemental material). Orthologous proteins were identified from reciprocal BLASTP searches after optimization by incremental testing of parameters for identity (0.8), match length (0.3), and E value (10e−5). Customized computer scripts and the sequence alignment program MUSCLE (10) were used to generate the initial concatenated alignments, which were subsequently edited manually. A final alignment of 3,631 concatenated proteins (1,190,596 amino acids) was used in phylogenetic analyses. Phylogenetic trees were reconstructed by two methods. The maximum-likelihood method was implemented in PROML (PHYLIP 3.69) (11a) with 100 bootstraps, the gamma distribution in six discrete rate categories, and K. variicola as the outgroup. Bayesian posterior probability trees were constructed by MrBayes 3.1.2 (18, 48) with parameter settings as gamma-shaped rate variation with a proportion of invariable sites (rates = invgamma), six discrete rate categories (Nst = 6), prior probability set to a mixed fixed-rate model (aamodelpr = mix), and 20,000-generation Markov Chain Monte Carlo with Nchains set to 16. Separate phylogenetic trees of dihydrofolate reductases (DHFRs) were also reconstructed using MrBayes (18, 48) and the Phylip programs PROTDIST and NEIGHBOR (Felsenstein, unpublished). All trees were visualized with the program TREEVIEW (40).
Identification of drug resistance genes.
Amino acid sequences of known K. pneumoniae antibiotic resistance genes (405 genes) were downloaded from the Antibiotic Resistance Genes Database (ARDB; http://ardb.cbcb.umd.edu/index.html) (31), which is a comprehensive, well-curated resource of antibiotic resistance genes of bacterial pathogens. Additional efflux proteins and beta-lactamases obtained from GenBank were added to our list of antibiotic genes. These sequences were used as TBLASTN queries against the JH1 and 1162281 genomes, as well as the published K. pneumoniae genomes and other same-species entries in GenBank (August 2010).
Nucleotide sequence accession numbers.
Whole-genome shotgun projects for K. pneumoniae JH1 and 1162281 have been deposited in DDBJ/EMBL/GenBank under accession numbers AFQK01000001-AFQK01000118 and AFQL01000001-AFQL01000116, respectively.
RESULTS
Antibiotic susceptibility profiles.
MICs across 22 antibiotics show differential susceptibilities of strains JH1 (high susceptibility) and 1162281 (low susceptibility) (Table 1). Strain 1162281 shows greater in vitro resistance to the aminoglycoside, chloramphenicol, quinolone, and cephalosporin drug classes. In contrast, JH1 was highly susceptible to most of these antibiotic classes. Both strains were sensitive to polymyxins and carbapenems, the latter presumably due to a lack of carbapenem-hydrolyzing β-lactamase or carbapenemase (8) genes in either genome (see below). For both strains, comparable high resistance profiles were observed for macrolides (azithromycin, erythromycin, and telithromycin), fusidanes, and glycopeptides.
Table 1.
Antibiotic MIC profiles of K. pneumoniae strains 1162281 and JH1
| Drug | Class | MIC (μg/ml) |
|
|---|---|---|---|
| 1162281 | JH1 | ||
| Gentamicin | Aminoglycoside | 8 | 1 |
| Chloramphenicol | Chloramphenicol | >32 | 4 |
| Trimethoprim | Diaminopyrimidine | >32 | 0.5 |
| Ciprofloxacin | Quinolone | 32 | 0.016 |
| Levofloxacin | Quinolone | 32 | ≤0.032 |
| Moxifloxacin | Quinolone | 32 | 0.032 |
| Norfloxacin | Quinolone | >32 | 0.063 |
| Ofloxacin | Quinolone | 32 | 0.063 |
| Azithromycin | Macrolide | 16 | 8 |
| Telithromycin | Macrolide | 32 | 32 |
| Erythromycin | Macrolide | >32 | >32 |
| Tetracycline | Tetracycline | 4 | 2 |
| Cefotaxime | Cephalosporin | 1 | 0.063 |
| Ceftriaxone | Cephalosporin | 4 | 0.063 |
| Ceftazidime | Cephalosporin | >32 | 0.125 |
| Cephalexin | Cephalosporin | 8 | 2 |
| Cefuroxime | Cephalosporin | 16 | 4 |
| Imipenem | Carbapenem | 0.125 | 0.25 |
| Meropenem | Carbapenem | ≤0.032 | ≤0.016 |
| Fusidic acid | Fusidane | >32 | >32 |
| Vancomycin | Glycopeptide | >32 | >32 |
| Polymyxin B | Polymyxin | 0.5 | 2 |
JH1 and 1162281 genome coverage.
Two DNA sequencing runs were performed for each genome. For JH1, total read lengths of 65,165,285 and 69,039,891 bp were assembled into 143 contigs, with 100 contigs larger than 500 bp. A total of 74 contigs were included in the assembly of the main chromosome, resulting in an estimated genome size of 5,187,438 bp (the remaining contigs either mapped to plasmids or too small for confident alignment with the genome). For 1162281, total read lengths runs of 73,730,000 and 73,951,923 bp were assembled into 136 contigs, of which 94 were larger than 500 bp. Of these large contigs, 61 were used to assemble the main chromosome, resulting in an estimated genome size of 5,159,649 bp. While these size estimates are smaller than those published for the complete genomes of K. pneumoniae strains MGH 78578 (5,315,120 bp), NTUH-K2044 (5,248,520 bp), and 342 (5,641,239 bp), the overall size differences are comparable.
In order to determine the extent of DNA sequencing coverage, we performed TBLASTN searches against the whole-genome sequences of JH1 and 1162281 using the published K. pneumoniae chromosomal genome and plasmid protein sequences (see Table S1 in the supplemental material). Overall, comparable numbers of chromosomal genes were detected in the two new genome sequences relative to previously published genomes (Table 2). Coverage of the plasmid genes was more variable. However, the plasmid content of particular K. pneumoniae strains is known to vary and high nucleotide sequence homology across different plasmid types can confound the proper assembly and identification of individual plasmids (55).
Table 2.
Summary of gene identities between 1162281 and JH1 sequences and published K. pneumoniae genomes
| Sequencea | No. of genes | 1162281 |
JH1 |
||||
|---|---|---|---|---|---|---|---|
| No. of genes identifiedb | Mean identity (%) | Mean MLc (%) | No. of genes identified | Mean identity (%) | Mean ML (%) | ||
| 342 chromosome | 5,425 | 4,827 | 97.3 | 99.2 | 4,801 | 97.3 | 99.1 |
| pKP187 | 113 | 44 | 91.4 | 81.1 | 28 | 91 | 90.8 |
| pKP91 | 230 | 12 | 88.3 | 76.8 | 23 | 89.1 | 71.7 |
| MGH chromosome | 4,776 | 4,613 | 99.3 | 99.3 | 4,731 | 99.3 | 99.1 |
| pKPN3 | 123 | 94 | 97.0 | 91.5 | 86 | 96.1 | 88.5 |
| pKPN4 | 98 | 38 | 95.3 | 95.1 | 16 | 93.7 | 90.4 |
| pKPN5 | 5 | 28 | 95.3 | 94.0 | 23 | 97.3 | 91.6 |
| pKPN6 | 178 | 0 | 4 | 99.4 | 36 | ||
| pKPN7 | 5 | 0 | 4 | 94 | 36 | ||
| NTUH chromosome | 4,992 | 5,029 | 99.2 | 99.3 | 4,920 | 99.2 | 99.3 |
| pK2004 | 270 | 62 | 92.0 | 83.0 | 62 | 92 | 83.1 |
| Rh chromosome | 5,671 | 5,199 | 98.9 | 98.1 | 5,246 | 98.8 | 98.1 |
| Overall mean | 98.6 | 98.8 | 98.5 | 98.7 | |||
The strains are K. pneumoniae 342 (342), K. pneumoniae MGH 75858 (MGH), K. pneumoniae NTUH-K2044 (NTUH), and K. pneumoniae subsp. rhinoscleromatis ATCC 13884 (Rh).
Genes identified by TBLASTN searches of all strain 1162281 or JH1 nucleotide contigs with protein sequences from respective public K. pneumoniae large chromosomes and plasmids.
ML, match length.
In another evaluation of genome coverage, we extracted all of the genes from K. pneumoniae MGH 78578 that were missing from either new genome according to BLASTP searches. We then searched the other three complete K. pneumoniae genomes in order to determine if these “missing” genes are, in fact, universally conserved across K. pneumoniae species and thereby represent genomic segments with poor sequencing coverage. In total, only 5 and 6 K. pneumoniae MGH 78578 genes missing from 1162281 and JH1, respectively, are also present in the other three K. pneumoniae genomes (see Table S2 in the supplemental material). For 1162281, two pairs of genes are adjacent in the K. pneumoniae MGH 78578 genome, while three missing genes in JH1 are tandemly colocated. Microarray experiments hybridizing 1162281 mRNA to K. pneumoniae MGH 78578 genome arrays showed that 3 out of 5 missing genes were expressed at higher levels in MGH 78578 (≥2-fold change), consistent with the notion that they may not be present in the 1162281 genome or, if they are present, they are expressed below the limit of detection. The remaining 2 missing genes do not show expression changes between MGH 78578 and 1162281. However, expression intensity values for these two genes are low in both strains, suggesting that, at least in MGH 78578, they may not be expressed under the conditions under which they were tested. PCR amplification analysis with primers from the flanking ORFs of the missing genes showed that no PCR products were obtained with genomic DNA from 1162281 or JH1, whereas the expected-size PCR products were amplified from the genomic DNA of control strain MGH 78578 for all of the missing genes except kpn_01329. In summary, it appears that genome coverage of the two new strains by DNA sequencing was high and largely complete for both strains.
Genome comparisons of K. pneumoniae JH1, 1162281, and reference strain MGH 78578.
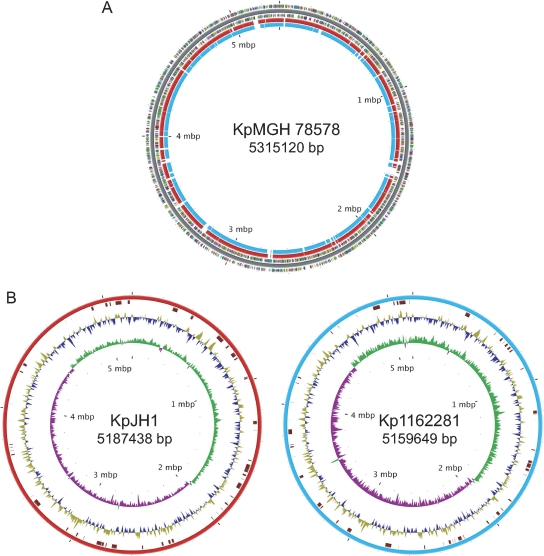
We used K. pneumoniae MGH 78578 as the basis for comparative genomic analyses with JH1 and 1162281 since it arguably has the most complete annotation of the main chromosome and accompanying plasmids for this species. Also, as the first published complete K. pneumoniae genome, strain MGH 78578 was widely used as the comparator sequence in other recent genomic studies (13, 53). Alignment of the JH1 and 1162281 genomic contigs with the genome of K. pneumoniae MGH 78578 revealed multiple deleted and inserted DNA segments in both strains (Fig. 1A). Although JH1 and 1162281 appear to have smaller genomes than K. pneumoniae MGH 78578, they also have between 60 and 62 inserted or novel regions, ranging from 107 to 88,316 nucleotides (nt) in size (Fig. 1B). In total, JH1 and 1162281 have estimated 690,434- and 618,802-nt inserted sequences relative to K. pneumoniae MGH 78578. Similar magnitudes of variation in genome content, as well as a smaller genome size, were also reported by Wu et al. when they compared the genome of pathogenic strain K. pneumoniae NTUH-K2044 with that of K. pneumoniae MGH 78578 (53).
Fig. 1.
Genomic maps of the K. pneumoniae JH1 (KpJH1) and 1162281 (Kp1162281) chromosomes. (A) JH1 (red) and 1162281 (blue) chromosomes mapped to the K. pneumoniae MGH 78758 (KpMGH 78578) reference genome showing gapped regions. Predicted protein-coding regions on the plus and minus strands were colored using the clusters of orthologous groups functional categories (http://www.ncbi.nlm.nih.gov/COG/grace/fiew.cgi). (B) Maps of the JH1 and 1162281 genomes. From the outer to the inner circles are the locations of inserted regions relative to K. pneumoniae MGH 78758 and AT skew and GS skew on the plus and minus strands.
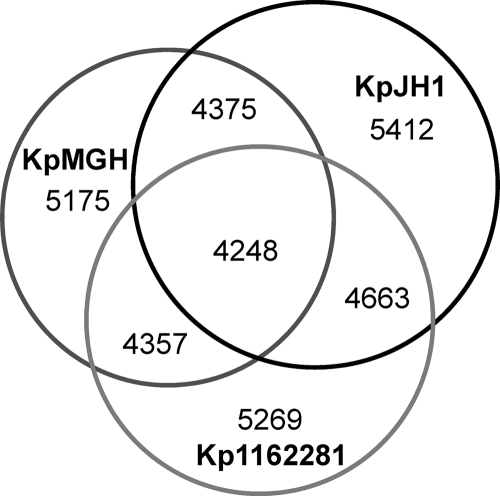
We compared the complement of all of the predicted protein-coding ORFs greater than 30 amino acids in length among the three strains (Fig. 2). 1162281 and JH1 shared more proteins between them than with K. pneumoniae MGH 78578. BLASTP searches of the GenBank nr database revealed that most of the genes common to both 1162281 and JH1 also occurred in other K. pneumoniae strains, suggesting lineage-specific gene losses in K. pneumoniae MGH 78578 (see Tables S3 and S4 in the supplemental material). Examples include a 12-gene cluster encoding the siderophore-independent FbpABC iron(III) transport system (2) and an anaerobic sugar metabolism phosphotransferase system (46) found in both new genomes yet absent from K. pneumoniae MGH 78578.
Fig. 2.
Venn diagram of the tally of homologous proteins shared by or unique among K. pneumoniae strains MGH 78758, JH1, and 1162281. Pairwise comparisons of proteins ≥30 amino acids in length were made between each pair of genomes using BLASTP (1) and deemed homologous with set parameter cutoffs (E value, ≤10e−05; match length, ≥30%; identity, ≥80%).
Interestingly, the 1162281 and JH1 chromosomes have a gene cluster that is unique among Klebsiella spp. and appears to have Gram-positive bacterial origins. Five genes encoding a sugar isomerase, a ribokinase, an amidohydrolase, a C4-dicarboxylate anaerobic carrier, and a GntR transcriptional regulator have highly similar homologs in Enterococcus faecium and the soil bacterium Clostridium beijerinckii. BLASTP searches with this gene cluster, which is probably involved in anaerobic sugar metabolism, did not identify any closely related homologs in members of the family Enterobacteriaceae, including other K. pneumoniae strains.
Compared to K. pneumoniae MGH 78578 and 1162281, JH1 had about 444 unique genes with homologs in the GenBank nr database (see Table S5 in the supplemental material). While most matches were to one of the other sequenced K. pneumoniae strains, NTUH-K2044 or 342, JH1 also had several genes that are possible first reports for this species. For example, JH1 has an AtoS-AtoC two-component signal transduction system which regulates the expression of the atoDAEB operon required for short-chain fatty acid catabolism (12, 19). Although found in other enteric bacteria such as E. coli and Citrobacter sp., ato gene sequences from other K. pneumoniae strains were not detected in BLASTP searches of the GenBank database. In addition to the fbpABC transporter shared with 112281, JH1 has the plasmid-borne, siderophore-dependent ferric-citrate operon fecABCDE (33). However, the siderophore transport iroNBCD cluster found in K. pneumoniae NTUH-K2044 and reportedly missing from K. pneumoniae MGH 78578 is also absent from both JH1 and 1162281 (53). Another unique feature of JH1 is a large prophage element of 39 genes integrated into the chromosome.
Relative to K. pneumoniae MGH 78578 and JH1, 1162281 had 409 specific genes which also had significant GenBank BLAST hits (see Table S6 in the supplemental material). The majority of the unique gene clusters had homologs in other K. pneumoniae strains, including the acetoin catalysis aco operon and the capsular polysaccharide and lipopolysaccharide gene clusters, which have the highest similarity to those of K. pneumoniae NTUH-K2044 (53). Strain 1162281 has a five-gene fimbrial lpf operon which is nearly identical to that of soil/plant epiphyte strains K. pneumoniae 342 and K. variicola At-22 but lacks significant homologs in any other pathogenic K. pneumoniae strain. Three major prophage clusters were also detected in the 1162281 genome. About half of the unique proteins attributed to 1162281 were not in the main chromosomal sequence assembly and thus are probably plasmid encoded. Several individual contigs have high-homology hits to enteric IncN plasmid R46 and multidrug resistance plasmids from Yersinia pestis (pIP202) (52) and the fish pathogen Photobacterium damselae (pP91278) (23). We also detected a putative mercury resistance operon, a hallmark of antibiotic resistance plasmids, embedded in a large contig (38 ORFs) that is highly similar to plasmid pHCM1 of MDR Salmonella enterica serovar Typhi CT18 (41). The genomic elements contributing to the multidrug resistance phenotype of 1122681 are further discussed below.
K. pneumoniae core genome and phylogeny.
Using conservative reciprocal BLASTP criteria, we identified across all six K. pneumoniae genomes protein-coding genes with a 1:1 orthologous relationship to each other that were not species-specific gene duplications. A total of 3,631 common proteins were identified which could be considered the K. pneumoniae core set of orthologous genes, at least for those strains with complete genome sequences. The proteins in K. pneumoniae 342 with assigned biological roles were downloaded from the CMR database and mapped to the 3,631 common protein orthologs (Table 3). The largest core gene category is transport and binding proteins, with 674 genes, followed by energy metabolism, with 530 genes. The third and fourth biggest categories are regulatory functions and cell envelope, respectively. Collectively, these four categories have 1,892 proteins composing 52.11% of the common orthologous protein-coding genes among the complete K. pneumoniae genomes. We also downloaded the curated protein clusters for K. pneumoniae 342 from the NCBI Protein Clusters Database and similarly mapped them to the common orthologs (see Table S7 in the supplemental material). In general, agreement with the CMR database assignments, the three largest protein cluster categories are metabolism, cellular process and signaling, and information storage and processing.
Table 3.
Biological role categories of orthologous genes conserved across K. pneumoniae genomesa
| Biological role categoryb | No. of genes | Proportion (%) of total no. |
|---|---|---|
| Transport and binding proteins | 674 | 18.56 |
| Energy metabolism | 530 | 14.60 |
| Regulatory functions | 363 | 10.00 |
| Cell envelope | 325 | 8.95 |
| Hypothetical | 243 | 6.69 |
| Cellular process | 186 | 5.12 |
| Protein fate (secretion, trafficking, folding, etc.) | 184 | 5.07 |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 176 | 4.85 |
| Central intermediary metabolism | 175 | 4.82 |
| Protein synthesis | 167 | 4.60 |
| DNA metabolism | 147 | 4.05 |
| Amino acid biosynthesis | 114 | 3.14 |
| Purines, pyrimidines, nucleosides, and nucleotides | 80 | 2.20 |
| Fatty acid and phospholipids metabolism | 69 | 1.90 |
| Transcription | 47 | 1.29 |
| Signal transduction | 45 | 1.24 |
| Mobile and extrachromosomal element functions | 11 | 0.30 |
| Unclassified | 95 | 2.62 |
| Total | 3,631 |
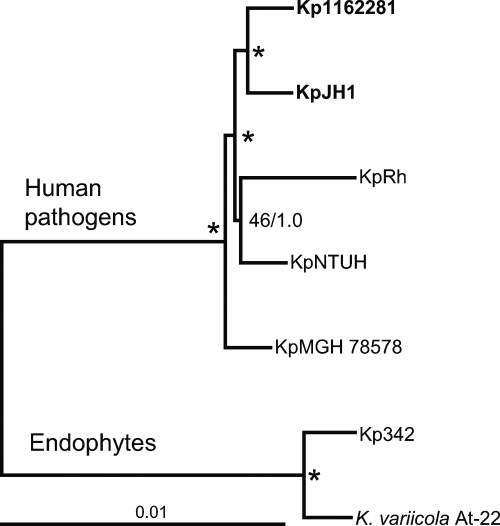
For phylogenetic analysis, orthologous proteins were identified in the genome of the outgroup species K. variicola and added to the core set of K. pneumoniae proteins. Subsequently, individual data sets of each of the 3,631 proteins for all seven species were aligned and edited for gaps. These individual alignments were concatenated into a mega-alignment totaling 1,190,596 amino acids and used for phylogenetic tree reconstruction. The Bayesian and maximum-likelihood methods produced similar tree topologies (Fig. 3). The two endophyte species K. pneumoniae 342 and K. variicola form a clade separate from that composed of K. pneumoniae strains isolated from human hosts. Among the pathogenic K. pneumoniae strains, the newly sequenced genomes of JH1 and 1162281 were significantly supported as a single clade. The cluster of K. pneumoniae NTUH-K2044 and K. pneumoniae subsp. rhinoscleromatis ATCC 13884 had weaker support. However, both phylogenetic tree reconstruction methods strongly supported K. pneumoniae MGH 78578 at the basal position of the pathogenic strain clade.
Fig. 3.
Protein maximum-likelihood phylogeny of K. pneumoniae strains. The strain name abbreviations used are defined in Table 2, footnote a. Phylogeny is based on the alignment of 3,631 conserved orthologous proteins concatenated into an alignment totaling 1,190,596 amino acids. The Bayesian (MrBayes) (18, 48) and maximum-likelihood (PROML of PHYLIP package) (11a) methods produced similar tree topologies. Nodes supported by >70% of 100 bootstrap replicates and posterior Bayesian probabilities of ≥0.95 are marked by asterisks. The scale bar represents an estimated 0.01 amino acid substitution per site.
Overview of antibiotic resistance genes.
The DNA sequences of JH1 and 1162281, as well as other publicly available K. pneumoniae genomes, were searched for homologs to known K. pneumoniae antibiotic resistance proteins obtained from the ARDB (http://ardb.cbcb.umd.edu/index.html) (31; see Table S8 in the supplemental material), supplemented with additional Gram-negative efflux pump proteins and β-lactamases from the GenBank database (August 2010).
According to the ARDB classification system, there are 12 different classes of genomic and plasmid-borne antibiotic resistance genes reported at least once for K. pneumoniae (Table 4). We found that all 12 classes occurred in at least one of the complete K. pneumoniae genomes, with the exception of a ribosomal protection protein, which is rare for this species, with only a single reported occurrence in the GenBank database. Among the strains with genome level sequences, K. pneumoniae MGH 78578 had the largest complement of putative antibiotic resistance genes (28 genes), followed by 112281 (24 genes). Antibiotic-sensitive JH1 had a much lower complement of resistance genes (16 genes), which was similar to the three remaining K. pneumoniae strains. Comparison of K. pneumoniae MGH 78578 and 1162281 showed that the former had several aminoglycoside-related resistance genes, as well as one additional efflux pump gene and a beta-lactamase gene. In contrast, 1162281 had several additional genes encoding trimethoprim-insensitive DHFR, a pentapeptide repeat protein, and sulfonamide-resistant dihydropteroate synthase. Specific drug resistance genes are further discussed below.
Table 4.
Inventory of antibiotic resistance genes identified in whole K. pneumoniae genomes
| Groupa | No. of genesb |
||||||
|---|---|---|---|---|---|---|---|
| 1162281 | JH1 | MGH | NTUH | Rh | 342 | Other Klebsiella spp.c | |
| Aminoglycoside acetyltransferase | 0 | 0 | 4 | 0 | 0 | 0 | 130 |
| Aminoglycoside phosphotransferase | 1 | 0 | 1 | 0 | 0 | 0 | 15 |
| Beta-lactamase | 3 | 1 | 4 | 1 | 1 | 2 | 915 |
| Chloramphenicol acetyltransferase | 1 | 0 | 1 | 0 | 0 | 0 | 33 |
| Drug-insensitive DHFR | 1 | 0 | 0 | 0 | 0 | 0 | 70 |
| Specific demethylase | 1 | 1 | 1 | 1 | 1 | 1 | 3 |
| Efflux pump | 12 | 11 | 13 | 11 | 11 | 11 | 44 |
| Pentapeptide repeat | 1 | 0 | 0 | 0 | 0 | 0 | 49 |
| Sulfonamide-resistant dihydropteroate synthase | 1 | 0 | 1 | 0 | 0 | 0 | 60 |
| Ribosomal protection protein | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Undecaprenyl pyrophosphate phosphatase | 1 | 1 | 1 | 1 | 1 | 1 | 2 |
| Unclassified | 2 | 2 | 2 | 2 | 2 | 2 | 4 |
| Total | 24 | 16 | 28 | 16 | 16 | 17 | 1,326 |
Antibiotic resistance gene categories were adapted from the ARDB and identified by TBLASTN searches.
Species name abbreviations are the same as those in Table 2.
Other Klebsiella sp. hits from the GenBank database.
β-Lactamases.
Three β-lactamase genes were identified in 1162281, while only one was found in JH1 (Table 5). Both strains had an SHV family non-ESBL located on a contig mapped to the main chromosome of the respective strain. Strain 1162281 had two additional β-lactamase genes on separate putative plasmid contigs. BlaP1 of the PSE family has been observed in other K. pneumoniae isolates (35). The third enzyme is also an ESBL with 100% amino acid homology to TEM-12 of K. oxytoca (17). However, it differed by one amino acid (L19F) to the closest K. pneumoniae enzyme, TEM-53, and might be the first report of this particular ESBL in a K. pneumoniae clinical isolate.
Table 5.
β-Lactamases identified in K. pneumoniae strains and closest known enzymes
| Strain and locationa | Family | Name | Top hit accession no.b | Typec | Species | Identity (%) |
|---|---|---|---|---|---|---|
| 1162281 | ||||||
| Chromosome | SHV | SHV-75 | CAJ47130.2 | BLA | K. pneumoniae | 100 |
| Plasmid | TEM | TEM-12 | Q48406.1 | ESBL | K. oxytoca | 100 |
| Plasmid | PSE | BlaP1 | ABI50466.1 | BLA | K. pneumoniae | 100 |
| JH1, chromosome | SHV | SHV-60 | CAI30649.2 | BLA | K. pneumoniae | 100 |
Contig in main chromosome assembly or putative plasmid.
Highest-homology hit from BLASTP searches of the GenBank database.
BLA (β-lactamase) and ESBL are as annotated in reference 35.
Efflux pumps.
K. pneumoniae 1162281 and MGH 78578 had one and two additional efflux pumps, respectively, relative to JH1 and the other three K. pneumoniae genomes (Table 6). Strain 1162281 lacked tetD but otherwise has the same efflux pump complement as K. pneumoniae MGH 78578. The recently discovered RND multidrug efflux pump OqxAB is absent from the ARDB list but was added to our search and found to be conserved in all K. pneumoniae genomes (22). A florfenicol/chloramphenicol resistance protein or FloR efflux pump was absent from all six sequenced genomes.
Table 6.
Inventory of efflux pump genes in different K. pneumoniae genomes
| Genea | Product | No. of genesb |
|||||
|---|---|---|---|---|---|---|---|
| 1162281 | JH1 | MGH | NTUH | Rh | 342 | ||
| cmlA1 | Chloramphenicol resistance protein | 1 | 0 | 1 | 0 | 0 | 0 |
| floR | Florfenicol/chloramphenicol resistance protein | 0 | 0 | 0 | 0 | 0 | 0 |
| tetD | Tetracycline resistance protein, efflux | 0 | 0 | 1 | 0 | 0 | 0 |
| yceE (mdtG) | Drug efflux system protein MdtG | 1 | 1 | 1 | 1 | 1 | 1 |
| norA (norM/mdtK) | Multidrug efflux protein | 1 | 1 | 1 | 1 | 1 | 1 |
| yceL (mtdH) | Multidrug resistance protein MdtH | 1 | 1 | 1 | 1 | 1 | 1 |
| emrD | Multidrug resistance protein D | 1 | 1 | 1 | 1 | 1 | 1 |
| tolC | Outer membrane channel protein, TolC subunit | 1 | 1 | 1 | 1 | 1 | 1 |
| acrA | Acridine/acriflavine resistance protein | 1 | 1 | 1 | 1 | 1 | 1 |
| acrB | Acridine/acriflavine resistance protein | 2 | 2 | 2 | 2 | 2 | 2 |
| macA | Macrolide-specific efflux protein | 1 | 1 | 1 | 1 | 1 | 1 |
| macB (ybjZ) | Macrolide transporter ATP-binding protein | 1 | 1 | 1 | 1 | 1 | 1 |
| oqxAB | RND multidrug efflux membrane fusion/permease proteins OqxAB | 1 | 1 | 1 | 1 | 1 | 1 |
| Total | 12 | 11 | 13 | 11 | 11 | 11 | |
K. pneumoniae efflux proteins obtained from the ARDB and literature sources.
Species name abbreviations are the same as those in Table 2.
Fluoroquinolone resistance.
Strain 1162281 is notably distinguished from JH1 by its high resistance to quinolones and fluoroquinolones, including ciprofloxacin. In addition to an enhanced complement of efflux pumps and >60-fold-elevated transcription in the OqxAB quinolone efflux pump (22) revealed by transcriptome analysis (J.R.B., unpublished data), 1162281 has two additional mechanisms of quinolone resistance. Comparative sequence analysis of the gyrA and parC genes revealed three nonsynonymous variants in the protein topoisomerase, the target of quinolone class drugs. One variant was Ser83Phe in the quinolone resistance-determining region (QRDR) of gyrase A, which is known to convey resistance to fluoroquinolone class drugs (51). This variant also occurred in the gyrA gene of K. pneumoniae MGH 78578 but not in JH1 or the other K. pneumoniae genomes. Strain 1162281 also had two novel nonsynonymous changes found outside the QRDR of the parC gene, Ala339Gly and Asp641Tyr. These variants have not been previously reported, and their effects on fluoroquinolone susceptibility are unknown.
A third fluoroquinolone resistance mechanism of 1162281 is the presence of a pentapeptide repeat family protein encoded by a homolog of plasmid-encoded quinolone resistance gene qnrA1 (34). The 1162281 putative qnrA1 gene had 100% amino acid homology with that reported for plasmid pKP96, which was isolated from a sputum specimen in China in 2002 (49). The 1162281 putative qnrA1 gene was located on a short 1,525-bp contig, so flanking DNA regions could not be fully characterized. However, the entire contig had nearly complete nucleotide sequence identity with other reported qnr integrons associated with conjugal plasmids sequenced from Asian hospital infections with members of the family Enterobacteriaceae (20).
Trimethoprim/diaminopyrimidine resistance.
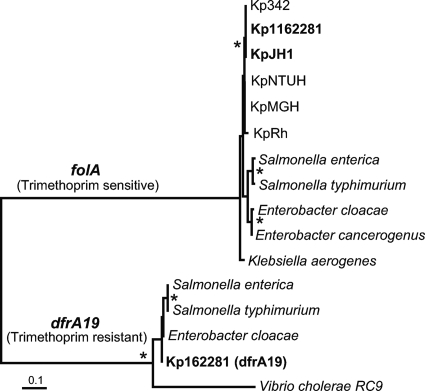
The antibiotic trimethoprim and other diaminopyrimidines inhibit the enzyme DHFR. Sequence analysis revealed that 1162281 has dual DHFR-coding genes, which might account for its high trimethoprim resistance. The main chromosome has the gene folA, which encodes a trimethoprim-sensitive DHFR. The folA gene is highly conserved and universally found in all bacteria, including the other Klebsiella species studied here. However, 1162281 has a second, divergent DHFR (dfrA19) previously found on plasmid-borne integrons and known to convey trimethoprim resistance (Fig. 4) (30). Similar to the qnrA1 gene, 1162281 dfrA19 was found on a short 1,503-bp contig with nearly perfect nucleotide identity to several reported Gram-negative plasmids (6). Phylogenetic analyses show the divergence between the two types of DHFR proteins, as well as the close relationships among dfrA19 genes from different members of the family Enterobacteriaceae, suggesting frequent and recent horizontal gene transfer of this gene, likely mediated by plasmids and transposable elements.
Fig. 4.
Phylogeny of DHFRs from K. pneumoniae strains and other members of the family Enterobacteriaceae. The strain name abbreviations used are defined in Table 2, footnote a. Clades of genes encoding a DHFR that is either trimethoprim sensitive (folA) or resistant (dfrA19) are labeled. Phylogenetic reconstruction was done using MrBayes (18, 48), as well as PROTDIST and NEIGHBOR from the PHYLIP package (11a) (tree shown). Nodes supported by >70% of 1,000 bootstrap replicates and posterior Bayesian probabilities of ≥0.95 are marked by asterisks. The scale bar represents an estimated 0.01 amino acid substitution per site.
Chloramphenicol and sulfonamide resistance.
The gene set from ARDB contained 15 chloramphenicol acetyltransferase genes. Among the complete K. pneumoniae genome sequences, chloramphenicol acetyltransferase genes were found only in K. pneumoniae 116281 and MGH 78578. Both strains had single genes but of different types. K. pneumoniae MGH 78578 has a plasmid-borne cat gene, while 116281 has a catB3-type gene, also likely plasmid borne.
Similar to K. pneumoniae MGH 78578 plasmid pKN5, the gene sul1, encoding the sulfonamide resistance protein, was found on a 1162281 contig with high overall nucleotide identity to known plasmids (47). Although we could not assemble individual plasmids, finding plasmid-borne resistance genes such as qnrA1, dfrA19, catB3, and sul1, as well as the mec operon for mercury resistance, suggests that most of the chromosomal and plasmid-borne drug resistance genes were identified in strain 116281.
DISCUSSION
Genome plasticity is an important facilitator in the spread of antibiotic-resistant pathogenic bacteria. K. pneumoniae and other enteric bacteria have extensive genes in common that are responsible for resistance to clinically used antibiotics, leading to the increasing prevalence of drug resistance phenotypes in the clinic. The availability of two new genome sequences from K. pneumoniae strains with differential antibiotic resistance profiles furthers our understanding and appreciation of drug resistance mechanisms and their evolution.
Comparative analyses of the new genomes of strains JH1 and 1162281 with previously published genomes of four strains of K. pneumoniae revealed a core set of 3,631 conserved proteins or approximately 65 to 75% of the total number of predicted protein-coding genes in any given genome. Thus, there is considerable latitude for variation in the genomic content of this species. Although JH1 and 1162281 (both are clinical isolates from the United States) have highly different antibiotic profiles of high and low susceptibility, respectively, highly robust phylogenetic trees based on the alignment of all of the conserved core proteins positioned these two strains as very closely related. The only other sequenced strain with known multidrug resistance, K. pneumoniae MGH 78578 (a clinical isolate from Japan), was the most divergent among the human isolated strains and effectively rooted that clade (relative to the two environmental strains, K. pneumoniae 342 and K. variicola At-22). Thus, at the whole-genome level, antibiotic resistance is not clonal and can be acquired or lost by K. pneumoniae strains with highly different genomic backgrounds. This phylogenetic tree topology suggests that exogenous or horizontal gene transfer is a key mechanism for acquisition of drug resistance. This is not surprising, since many antibiotic resistance determinants are borne on transferrable plasmids or mobile elements, particularly for enteric bacteria (16). For strain 1162281, several genes involved in antibiotic resistance had high or nearly identical DNA sequence identity to plasmid-borne homologs in public databases, including the genes qnrA1, dfrA19, catB3, and sul1 and those for two β-lactamases, TEM-12 and BlaP1. The absence of these genes from antibiotic-susceptible strain JH1 suggests that it has a different plasmid complement and that any drug resistance determinants were either lost or never acquired.
Although our phylogenetic tree shows a clear separation of Klebsiella species originating from human and soil environments, we found evidence of at least one specific transfer event of genes found exclusively in environment isolates, the fimbrial lpf operon, into strain 1162281. The environmental demarcation of K. pneumoniae strains might be arbitrary. For example, endophyte strain K. pneumoniae 342 has been shown to be partially virulent in mouse models (13) and β-lactamase-resistant K. pneumoniae occurs extensively in nonhuman organisms such as swine (56). We found multiple plasmid sequences in 1162281 which were not of immediate K. pneumoniae origin, including multidrug resistance plasmids from the plague agent Y. pestis (pIP202) (52), the fish pathogen P. damselae (pP91278) (23), and MDR S. enterica serovar Typhi CT18 (41). We also detected in both strains 1162281 and JH1 a five-gene cluster implicated in anaerobic sugar metabolism with the highest similarity to homologs from the Gram-positive genera Clostridium and Enterococcus. Thus, the potential reservoir of antibiotic resistance genes accessible to K. pneumoniae strains is likely extensive and goes beyond solely human pathogens or closely related bacteria.
Extrachromosomal elements only partially account for phenotypic differences in drug resistance. We note that among the five K. pneumoniae genomes, that of MDR strain 1126681 has the smallest estimated size, which is about 27,289 bp smaller than that of drug-susceptible strain JH1. While these genomes were not sequenced to full closure, the presence of all but a few core genes conserved in other K. pneumoniae genomes and the finding of all of the genetic determinants of drug resistance suggest that the genome coverage obtained by pyrosequencing was nearly complete. Wu et al., using genome shotgun arrays, found extensive differences in the genome profiles of clinical K. pneumoniae isolates which were indicative of antibiotic susceptibilities (53). Therefore, several questions arise, such as whether antibiotic resistance is also promoted by loss of specific genes and whether the acquisition of drug resistance genes requires some genome streamlining to improve fitness. Comparative genomic analysis revealed that, in addition to several genes of unknown function, JH1 had several unique functional gene clusters, such as an AtoS-AtoC two-component signal transduction system and the atoDAEB operon required for short-chain fatty acid catabolism (12, 19). Whether drug susceptibility could be affected by the deletion of specific genes and disabling of certain biochemical pathways is an intriguing question.
Strain 1162281 is highly resistant to a number of different antibiotics but particularly refractory to the fluoroquinolone class. This phenotype can be attributed to the presence of three distinct resistance mechanisms, which are a specific Ser83Phe mutation in the QRDR of the targeted gyrase A, the presence of the plasmid-borne quinolone resistance gene qnrA1 and a multitude of efflux pumps, and elevated expression of the OqxAB pump. We also found two novel amino acid changes outside the QRDR of the parC gene, Ala339Gly and Asp641Tyr, with unknown effects on fluoroquinolone susceptibility that might warrant further investigation. The enhanced complement of efflux pumps relative to drug-susceptible strains (Table 6) might further account for the resistance profile of strain 1162281. Aminoglycoside resistance in this strain is likely efflux pump driven, since the 16S rRNA methylase genes armA and rmtC are absent. Variations in efflux pump regulation and gene expression can also contribute to changes in membrane permeability and drug flux (38, 39).
The application of pyrosequencing to study clinical isolates of bacterial pathogens is rapidly growing. Recent studies include the pyrosequencing of methicillin-resistant Staphylococcus aureus isolates from both outbreaks in a regional hospital and intercontinental collections (15). Here, we show the further contribution of whole-genome sequencing to a better understanding of the specific genetic mechanisms underlying phenotypic differences in drug susceptibility in K. pneumoniae. By being available in the public domain, these new genome sequences will be a resource for future genetic studies of enteric bacteria, especially K. pneumoniae. Our analyses engender a deeper appreciation of the genetic diversity and evolutionary dynamics of this important bacterial pathogen and provide further insights into the factors driving the clinical rise of MDR isolates.
Supplementary Material
ACKNOWLEDGMENTS
K.I., L.P., and J.H. were in part supported by GlaxoSmithKline, the Wellcome Trust Seeding Drug Discovery Initiative, and Transformational Medical Technologies program contract HDTRA1-07-9-0002 from the Department of Defense Chemical and Biological Defense program through the Defense Threat Reduction Agency (DTRA).
We thank Stephen Rittenhouse for his comments on the manuscript.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 11 July 2011.
REFERENCES
- 1. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson D. S., Adhikari P., Nowalk A. J., Chen C. Y., Mietzner T. A. 2004. The hFbpABC transporter from Haemophilus influenzae functions as a binding-protein-dependent ABC transporter with high specificity and affinity for ferric iron. J. Bacteriol. 186:6220–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 57:289–300 [Google Scholar]
- 4. Brisse S., Verhoef J. 2001. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int. J. Syst. Evol. Microbiol. 51:915–924 [DOI] [PubMed] [Google Scholar]
- 5. Chaudhuri R. R., et al. 2008. xBASE2: a comprehensive resource for comparative bacterial genomics. Nucleic Acids Res. 36:D543–D546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen Y. T., et al. 2009. Mobilization of qnrB2 and ISCR1 in plasmids. Antimicrob. Agents Chemother. 53:1235–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clinical and Laboratory Standards Institute 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. Cuzon G., et al. 2010. Worldwide diversity of Klebsiella pneumoniae that produce beta-lactamase blaKPC-2 gene. Emerg. Infect. Dis. 16:1349–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Delcher A. L., Bratke K. A., Powers E. C., Salzberg S. L. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Falagas M. E., Karageorgopoulos D. E. 2009. Extended-spectrum beta-lactamase-producing organisms. J. Hosp. Infect. 73:345–354 [DOI] [PubMed] [Google Scholar]
- 11a. Felsenstein J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164–166 [Google Scholar]
- 12. Filippou P. S., Kasemian L. D., Panagiotidis C. A., Kyriakidis D. A. 2008. Functional characterization of the histidine kinase of the E. coli two-component signal transduction system AtoS-AtoC. Biochim. Biophys. Acta 1780:1023–1031 [DOI] [PubMed] [Google Scholar]
- 13. Fouts D. E., et al. 2008. Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genet. 4:e1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fox R. J., Dimmic M. W. 2006. A two-sample Bayesian t-test for microarray data. BMC Bioinformatics 7:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harris S. R., et al. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hawkey P. M., Jones A. M. 2009. The changing epidemiology of resistance. J. Antimicrob. Chemother. 64(Suppl. 1):i3–i10 [DOI] [PubMed] [Google Scholar]
- 17. Heritage J., Hawkey P. M., Todd N., Lewis I. J. 1992. Transposition of the gene encoding a TEM-12 extended-spectrum beta-lactamase. Antimicrob. Agents Chemother. 36:1981–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huelsenbeck J. P., Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755 [DOI] [PubMed] [Google Scholar]
- 19. Jenkins L. S., Nunn W. D. 1987. Genetic and molecular characterization of the genes involved in short-chain fatty acid degradation in Escherichia coli: the ato system. J. Bacteriol. 169:42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jeong J. Y., et al. 2005. Detection of qnr in clinical isolates of Escherichia coli from Korea. Antimicrob. Agents Chemother. 49:2522–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiang Y., et al. 2010. Complete nucleotide sequence of Klebsiella pneumoniae multidrug resistance plasmid pKP048, carrying blaKPC-2, blaDHA-1, qnrB4, and armA. Antimicrob. Agents Chemother. 54:3967–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim H. B., et al. 2009. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob. Agents Chemother. 53:3582–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim M. J., et al. 2008. Complete DNA sequence and analysis of the transferable multiple-drug resistance plasmids (R plasmids) from Photobacterium damselae subsp. piscicida isolates collected in Japan and the United States. Antimicrob. Agents Chemother. 52:606–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klimke W., et al. 2009. The National Center for Biotechnology Information's Protein Clusters Database. Nucleic Acids Res. 37:D216–D223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ko W. C., Hsueh P. R. 2009. Increasing extended-spectrum beta-lactamase production and quinolone resistance among Gram-negative bacilli causing intra-abdominal infections in the Asia/Pacific region: data from the Smart Study 2002-2006. J. Infect. 59:95–103 [DOI] [PubMed] [Google Scholar]
- 26. Ko W. C., et al. 2002. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg. Infect. Dis. 8:160–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kurtz S., et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lagesen K., et al. 2007. RNAmmer: consistent and rapid annotation of rRNA genes. Nucleic Acids Res. 35:3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lai Y. C., Yang S. L., Peng H. L., Chang H. Y. 2000. Identification of genes present specifically in a virulent strain of Klebsiella pneumoniae. Infect. Immun. 68:7149–7151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Levings R. S., Lightfoot D., Elbourne L. D., Djordjevic S. P., Hall R. M. 2006. New integron-associated gene cassette encoding a trimethoprim-resistant DfrB-type dihydrofolate reductase. Antimicrob. Agents Chemother. 50:2863–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu B., Pop M. 2009. ARDB—Antibiotic Resistance Genes Database. Nucleic Acids Res. 37:D443–D447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lowe T. M., Eddy S. R. 1997. tRNAscan-SE: a program for improved detection of tRNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mahren S., Schnell H., Braun V. 2005. Occurrence and regulation of the ferric citrate transport system in Escherichia coli B, Klebsiella pneumoniae, Enterobacter aerogenes, and Photorhabdus luminescens. Arch. Microbiol. 184:175–186 [DOI] [PubMed] [Google Scholar]
- 34. Martínez-Martínez L., Pascual A., Jacoby G. A. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797–799 [DOI] [PubMed] [Google Scholar]
- 35. Mendonça N., Ferreira E., Louro D., Canica M. 2009. Molecular epidemiology and antimicrobial susceptibility of extended- and broad-spectrum beta-lactamase-producing Klebsiella pneumoniae isolated in Portugal. Int. J. Antimicrob. Agents 34:29–37 [DOI] [PubMed] [Google Scholar]
- 36. Metzker M. L. 2010. Sequencing technologies—the next generation. Nat. Rev. Genet. 11:31–46 [DOI] [PubMed] [Google Scholar]
- 37. Miró E., et al. 2010. Spread of plasmids containing the bla(VIM-1) and bla(CTX-M) genes and the qnr determinant in Enterobacter cloacae, Klebsiella pneumoniae and Klebsiella oxytoca isolates. J. Antimicrob. Chemother. 65:661–665 [DOI] [PubMed] [Google Scholar]
- 38. O'Regan E., et al. 2009. Multiple regulatory pathways associated with high-level ciprofloxacin and multidrug resistance in Salmonella enterica serovar Enteritidis: involvement of RamA and other global regulators. Antimicrob. Agents Chemother. 53:1080–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Padilla E., et al. 2010. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 54:177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Page R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357–358 [DOI] [PubMed] [Google Scholar]
- 41. Parkhill J., et al. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848–852 [DOI] [PubMed] [Google Scholar]
- 42. Paterson D. L., et al. 2004. International prospective study of Klebsiella pneumoniae bacteremia: implications of extended-spectrum beta-lactamase production in nosocomial Infections. Ann. Intern. Med. 140:26–32 [DOI] [PubMed] [Google Scholar]
- 43. Peleg A. Y., Hooper D. C. 2010. Hospital-acquired infections due to gram-negative bacteria. N. Engl. J. Med. 362:1804–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peterson J. D., Umayam L. A., Dickinson T., Hickey E. K., White O. 2001. The Comprehensive Microbial Resource. Nucleic Acids Res. 29:123–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pinto-Tomás A. A., et al. 2009. Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science 326:1120–1123 [DOI] [PubMed] [Google Scholar]
- 46. Reizer J., Reizer A., Saier M. H., Jr 1995. Novel phosphotransferase system genes revealed by bacterial genome analysis—a gene cluster encoding a unique enzyme I and the proteins of a fructose-like permease system. Microbiology 141(Pt. 4):961–971 [DOI] [PubMed] [Google Scholar]
- 47. Rodríguez-Martínez J. M., et al. 2007. Characterisation of integrons containing the plasmid-mediated quinolone resistance gene qnrA1 in Klebsiella pneumoniae. Int. J. Antimicrob. Agents 29:705–709 [DOI] [PubMed] [Google Scholar]
- 48. Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 49. Shen P., et al. 2008. Complete nucleotide sequence of pKP96, a 67 850 bp multiresistance plasmid encoding qnrA1, aac(6′)-Ib-cr and blaCTX-M-24 from Klebsiella pneumoniae. J. Antimicrob. Chemother. 62:1252–1256 [DOI] [PubMed] [Google Scholar]
- 50. Turnbaugh P. J., et al. 2007. The human microbiome project. Nature 449:804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weigel L. M., Steward C. D., Tenover F. C. 1998. gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrob. Agents Chemother. 42:2661–2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Welch T. J., et al. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wu K. M., et al. 2009. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J. Bacteriol. 191:4492–4501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zavascki A. P., Carvalhaes C. G., Picão R. C., Gales A. C. 2010. Multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii: resistance mechanisms and implications for therapy. Expert. Rev. Anti Infect. Ther. 8:71–93 [DOI] [PubMed] [Google Scholar]
- 55. Zhao F., et al. 2010. Sequencing and genetic variation of multidrug resistance plasmids in Klebsiella pneumoniae. PLoS One 5:e10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zou L. K., et al. 2011. Phenotypic and genotypic characterization of beta-lactam resistance in Klebsiella pneumoniae isolated from swine. Vet. Microbiol. 149:139–146 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.