Abstract
The current standard of care for hepatitis C virus (HCV) patients is cotreatment with human alpha interferon (IFN-α) and ribavirin. The host factor USP18 functions to regulate the interferon signaling pathway by acting as an off-switch. In order to understand whether the inhibition of USP18 represents a valid target for the enhancement of interferon treatment for chronic viral diseases, we have used a wide range of RNA interference (RNAi) reagents to suppress USP18 gene expression in Huh7 cell lines. We demonstrate that a USP18 knockdown results in IFN-α2a signaling (measured by increased IFN-stimulated response element [ISRE] reporter gene activity, 2′,5′-oligoadenylate synthetase [2-5 OAS] expression, and ISG15 induction) that is increased by ∼100-fold, whereas the antiviral (AV) potency in both the Huh7 HCV subgenomic replicon assay and the Huh7.5 HCV infectious virus assay increased by ∼3-fold. While the degree of the USP18 knockdown of USP18 elicited by the different RNAi reagents correlated with the enhancement of IFN-α2a signaling, it did not correlate with the enhancement of AV activity. The failure of increased IFN-α2a signaling to fully translate into increased AV potency was also observed for encephalomyocarditis virus (EMCV) assays using Huh7.5 cells. These data suggest that the IFN-mediated AV response in Huh7.5 cells has only a limited dependence on USP18 activity.
INTRODUCTION
Hepatitis C virus (HCV) infection is a major cause of liver disease, with more than 170 million infected people worldwide at risk of liver failure and hepatocarcinoma (19). The current standard of care (SOC), using pegylated alpha 2a interferon (IFN-α2a) and ribavirin, is failing 40 to 50% of treated HCV patients, so new therapies are urgently required (18). Novel investigational directly acting antivirals (DAAs) targeting HCV protease (e.g., telaprevir and boceprevir) and polymerase (e.g., RG7128 and filibuvir) are progressing through clinical trials. However, these reagents will initially be added to the current SOC, and drug-resistant mutations will likely be selected in patients whose virus is not fully suppressed (8). One strategy for reducing the probability of the emergence of resistant virus would be to target host functions, because these genetic loci are not under the control of the promiscuous HCV replication machinery and therefore could increase the barrier to developing drug resistance.
IFN-α2a plays a crucial role in innate immunity against viral infections such as HCV (29). After binding to a specific receptor complex (composed of IFNAR1 and IFNAR2), signaling is initiated through the phosphorylation of STAT-1 and STAT-2 by JAK and Tyk2 kinases, and the subsequent heterodimerization of the phosphorylated STATs leads to an association with interferon regulatory factor 9 (IRF-9) to form IFN-stimulated gene (ISG) factor 3 (ISGF-3). This complex can bind to IFN-stimulated response elements (ISREs) found in the promoter regions of many ISGs. As a result, around 300 ISGs are transcriptionally upregulated, including canonical antiviral (AV) genes such as 2′,5′-oligoadenylate synthetase (2-5 OAS) and the ubiquitin-like protein ISG15 (5). ISG15 is one of the genes most strongly induced after IFN treatment and is known to play a role in the AV activity of alphaviruses and HCV (4, 20, 21, 36). The multiple upregulated ISG products then initiate multiple secondary effector pathways to further execute the AV activity.
Ubiquitin-specific peptidase 18 (USP18; known as UBP43 for the mouse ortholog) is a 43-kDa protein and exhibits homology to the catalytic domain of ubiquitin-specific proteases. The function of USP18/UBP43 is to modulate the conjugation of ISG15 to host proteins and is the major source for ISG-deconjugating protease activity (23). USP18/UBP43 knockout (KO) experiments in mice confirmed that the level of ISG15-ylation of host proteins is markedly high (32).
Recent studies have suggested that USP18/UBP43 is potentially an attractive host target for an anti-HCV therapeutics. For example, it has been shown that a reduced level of expression of USP18/UBP43 results in increased AV activity against Sindbis virus, hepatitis B (HepB) virus, and vesicular stomatitis virus (VSV) in knockout mice (17, 20, 24, 25, 31). USP18/UBP43 can also negatively regulate IFN signaling by binding to IFNAR2 and downregulating the JAK-STAT pathway (24); this property is distinct from its ISG15 deconjugation activity described above. Furthermore, it was reported previously that USP18/UBP43 is responsible for the IFN-induced, long-lasting, refractory mechanism of JAK-STAT signaling in mouse liver (34). Finally, several groups have reported that the upregulation of human USP18 is predictive of a nonsustainable viral response to interferon treatment (1, 3, 33). These findings suggest that an antagonism of USP18 could block its negative regulatory effect upon IFN signaling, resulting in a net increase in induced JAK-STAT activity and, therefore, AV activity.
Two groups have previously reported the effect of reduced human USP18 gene expression levels on IFN activity in HCV replication systems in vitro and found either modest (<10-fold) or substantial (>10-fold) increases in AV activity (4, 28), leaving this question unresolved. Our studies shed further light on the potential clinical utility of a USP18 antagonist as an HCV therapeutic. We describe the impact of a USP18 gene knockdown on IFN-α2a signaling. Our study uses improved RNA interference (RNAi) reagents containing a 2′ O-methyl ribosyl substitution to reduce the “off-target” knockdown of unrelated transcripts (11), and we have used multiple small interfering RNA (siRNA) and short hairpin RNA (shRNA) USP18-targeting RNAi tools, a recognized tactic to reduce the risk of potential nonspecific RNAi activity (10). We find that the suppression of USP18 expression leads to >100-fold increases in IFN-α2a signaling but only 3-fold increases in AV activity in the replicon system. Our results also suggest that canonical AV biomarkers for IFN-α2a signaling do not track with AV activity in replicon cells.
MATERIALS AND METHODS
Cell lines.
Huh7.5 cells are derived from human Huh7 hepatoma cells (2). HCV JFH is a full-length infectious genotype 2a HCV with the Renilla luciferase reporter cloned downstream of the internal ribosome entry site (IRES) (37). The 9B subgenomic HCV replicon line was described previously (7). All cells were maintained in Dulbecco's modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum, 22 IU/ml penicillin, 2 mM l-glutamine (Invitrogen), and 0.1 mM nonessential amino acids (Invitrogen) at 37°C in 5% CO2. The culture medium for some cells lines was further supplemented with 500 μg/ml Geneticin (Invitrogen) as appropriate.
RNAi techniques and HCV replicon luciferase assay.
Smart On-Target siRNAs for USP18 were obtained from Dharmacon (siUSP18-07 [CUGCAUAUCUUCUGGUUUAUU], siUSP18-08 [GGAAGAAGACAGCAACAUGUU], siUSP18-09 [GGACUACCCUCAUGGCCUGUU], and siUSP18-10 [GCAAAUCUGUCAGUCCAUCUU]), including a nontargeting control siRNA (siCONT [AUGUAUUGGCCUGUAUUAG]). Cells were transiently transfected with 25 to 150 nM siRNA in the presence of 0.4% siQUEST (Mirus) for 48 h prior to harvesting (by trypsinization), counting, and replating in 96-well plates. After 2 h, a dilution series of IFN-α2a (catalog number 11101-1; PBL) was added, and cells were incubated for a further 17 to 24 h. The luciferase activity was determined by using BriteLite reagent (Perkin-Elmer) and quantified by using an Analyst plate reader. The inhibition of replicon activity for each USP18 knockdown was calculated as a percentage relative to nontransfected cells treated with 10 IU/ml IFN-α2a by using the formula [RLUtest] − [RLUNT]0% inh/[RLUNT]100% inh − [RLUNT]0% inh ×100, where RLU is relative light units from the luciferase assays, [RLUNT]0% inh is zero percent inhibition for nontransfected cells, and [RLUNT]100% inh is 10 IU/ml IFN-α2a-treated nontransfected cells. Dose-response curves were generated by plotting the percent replicon inhibition for each USP18 knockdown against the IFN-α2a concentration. Nonlinear regression analysis was used to calculate 50% effective concentrations (EC50s) from each dose-response curve using GraphPad Prism software (version 5.01).
For stable transfected replicon and Huh7.5 cell lines, we obtained packaged lentiviruses containing shRNA for USP18 from Sigma Mission Library (Sigma-Aldrich) (shUSP18a [ACTGCATATCTTCTGGTTTACC], shUSP18b [CCTCATGGCCTGGTTGGTTTA], shUSP18c [AGCAACATGAAGAGAGAGCA], and shUSP18d [GAAGACAGCAACATGAAGAGA]), including a nontargeting control (shCONT [CAACAAGATGAAGAGCACCAA]). Adherent cells were transduced by the addition of ExpressMag-treated recombinant lentivirus (multiplicity of infection [MOI] of 3) to the supernatant, followed by a 15-min incubation over a magnet (ExpressMag 96-well magnetic kit, catalog number SHM02; Sigma). Two days later, 1.5 μg/ml puromycin was added to select for stable transduced cells over 1 week to be used for later assays.
RNAi and ISRE luciferase assay.
The pISRE-luc reporter plasmid (catalog number 631915; Clontech) was used to stably transfect Huh7.5 cells to generate “ISRE-luc cells.” The ISRE-luc cells were subsequently either transfected with USP18 siRNA or stably transduced with USP18 shRNA using the same siRNA and shRNA reagents described above. For USP18 siRNA experiments, cells were transiently transfected with 25 to 150 nM siRNA in the presence of 0.4% siQUEST (Mirus) for 48 h prior to harvesting (by trypsinization), counting, and replating in 96-well plates. After 1 h, a dilution series of IFN-α2a was added, and cells were incubated for 17 to 24 h. Luciferase activity was determined by using a BriteLite assay (Perkin-Elmer) and read with an Analyst plate reader. ISRE induction was calculated as a percentage of values for nontransfected IFN-α2a-treated controls, and dose-response curves were generated by plotting the percent ISRE induction against the IFN-α2a concentration. Nonlinear regression analysis was used to calculate EC50s from each dose-response curve using GraphPad Prism software (version 5.01).
HCV infectious assay.
The HCV infectious assay used was reported previously (37). Briefly, Huh7.5 cells (either transiently transfected with USP18 siRNA or stably transduced with USP18 shRNA) were plated onto tissue culture-treated 96-well plates (Perkin-Elmer) prior to the addition of serially diluted IFN-α2a for 17 to 24 h. The supernatants were aspirated and replaced with medium containing HCV strain JFH at an approximate MOI of 0.1, and cells were then incubated for 2 days. Plates were then assayed by using the Renilla luciferase assay (Promega), or cells were lysed by using RLT buffer (Qiagen) in preparation for RNA extraction for reverse transcription (RT)-PCR analysis. The percent inhibition of HCV JFH replication for each USP18 knockdown was calculated and plotted against the IFN-α2a dose. Nonlinear regression analysis was used to calculate each EC50 by using GraphPad Prism software (version 5.01).
RT-PCR assay.
For RT-PCR experiments, the IFN-treated, transfected cell monolayers were washed with ice-cold phosphate-buffered saline (PBS) and lysed in 300 μl RLT buffer (Qiagen). Total RNA was extracted (RNeasy kit; Qiagen), and first-strand cDNA was synthesized by using reverse transcriptase (cDNA Archive; Applied Biosystems). Quantitative PCR was performed by using TaqMan Universal PCR master mix (Applied Biosystems) using HCV replicon forward primer 5′-TCCCGGGAGAGCCATAGTG and reverse primer 5′-GGCATTGAGCGGGTTGATC and a 6-carboxyfluorescein (FAM)-labeled probe (5′-CCGGAATTGCCAGGACGACCG). Parallel reactions using the actin probe set (catalog number 4326315E; Applied Biosystems) were also set up. Reactions were performed by using an Applied Biosystems 7900 HT instrument.
Western blot assay.
Treated cells were washed with PBS and harvested by trypsinization. After further washes in cold PBS, the pelleted cells were lysed in 60 μl denaturing sample buffer (Invitrogen) and stored frozen until use. Samples were heat treated (70°C for 10 min), homogenized through 25-gauge needles, and loaded onto 4 to 12% Bis-Tris 1-mm PAGE gels, and the gels were run at 200 V in morpholineethanesulfonic acid (MES) running buffer. Wet transfer onto a polyvinylidene difluoride (PVDF) membrane was carried out at 30 V for 1 h, and the membrane was blocked with PBS–0.1% Tween 20–5% ECL blocking reagent (GE Healthcare) for 1 h at room temperature. The ISG15 antibody (3E5, catalog number ab48020; Abcam) was diluted 1/2,000 in PBS–0.1% Tween 20–5% ECL blocking reagent, and the secondary antibody [goat anti-mouse IgG(H+L), catalog number 626520; Zymed] was incubated for 1 h at room temperature. The membrane was washed with PBS–0.1% Tween 20–5% ECL blocking reagent and developed by using ECL Plus detection reagent (GE Healthcare).
Antiviral assay for EMCV.
Encephalomyocarditis virus (EMCV) (ATCC VR129B) was cultured in Vero cells (ATCC CCL-81), and the optimal dilution of the virus to infect the cells and cause death was determined empirically (an approximately 1/32,000 dilution with medium). The estimated titer of the virus was ∼107 PFU/ml. Huh7.5 cells were plated at 6 × 104 cells/well in 90 μl of medium (DMEM containing 2% fetal bovine serum, 22 IU/ml penicillin, 2 mM l-glutamine, and 0.1 mM nonessential amino acids) in clear flat-bottom plates. Serial dilutions of IFN-α2a were added, and the mixtures were incubated for 24 h. After aspiration, 100 μl fresh medium containing a 1/32,000 dilution of EMCV was added to the cells. The plates were incubated for a further 22 h at 37°C in 5% CO2. The cytotoxicity of successfully infected cells was assayed by the addition of 10 μl of WST-1 (catalog number 11644807001; Roche) to the mixture, and the mixture was incubated at 37°C in 5% CO2 for 2 to 4 h. Plates were read at 450 nm on a Victor plate reader. Nonlinear regression analysis was used to calculate each EC50 using GraphPad Prism software (version 5.01).
Statistical analysis.
The IFN EC50s obtained from both HCV assays and the EMCV assay were analyzed on the logarithmic scale by using a linear mixed model, comparing the nontargeting shCONT and test shUSP18 shRNA stable cell lines within each assay type and accounting for experimental variation. The control test EC50 comparisons were summarized by using fold change (FC) values (control/test) with 95% confidence limits. P values of less than 0.05 were considered statistically significant.
RESULTS
USP18 knockdown correlates with increased IFN-α2a signaling in ISRE reporter cells but not with increased AV activity in HCV subgenomic replication.
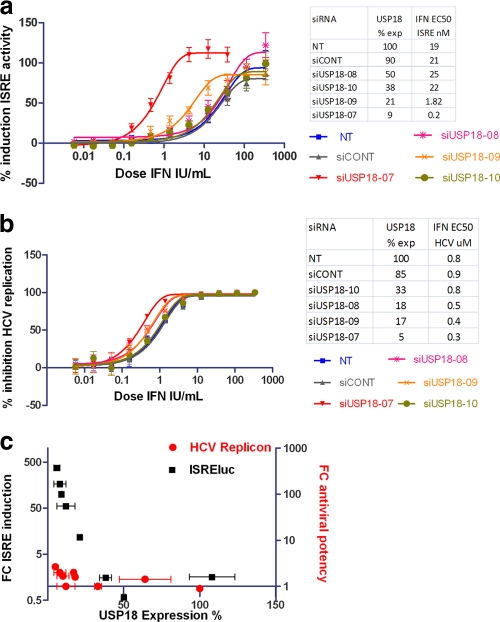
The effect of USP18 activity on the regulation of IFN receptor signaling was previously reported (24, 28). We confirmed and extended these observations with an assay utilizing a stable transfected Huh7.5 cell line harboring an ISRE-luciferase reporter gene (ISRE-luc cells). We used 4 different siRNA reagents to knock down USP18 expression in transient-transfection assays with ISRE-luc cells, which were subsequently treated overnight with serial IFN-α2a dilutions, and the resulting ISRE activity was measured by luciferase assays (Fig. 1a). The percent knockdown of USP18 expression for each siRNA (measured by RT-PCR) is also shown. We observed a 100-fold increase in potency for the IFN-α2a induction of ISRE activity at 9% USP18 expression (EC50 of 0.2 IU IFN/ml, compared to 21 IU IFN/ml for the control) and an increase in the maximum response relative to that of control cells. These siRNA reagents were unable to increase IFN-α2a AV potency beyond 3-fold when tested with the Huh7 HCV subgenomic replicon cell line despite showing a similar USP18 knockdown (Fig. 1b).
Fig. 1.
(a) Huh7.5 cells stably expressing an ISRE-luciferase reporter gene were transiently transfected with 25 nM siRNAs targeting USP18 as described in Materials and Methods. After 48 h, a dilution series of IFN-α2a was added to the cells, the cells were incubated for a further 17 to 24 h, and the percent induction of ISRE-luciferase reporter activity by IFN-α2a was measured relative to non-IFN-treated baseline ISRE-luciferase controls and plotted against IFN-α2a concentrations. Each condition was tested at n ≥ 2 replicates, and each experiment was independently repeated at least 3 times. Nonlinear regression analysis of the IFN-α2a dose-response curve was used to calculate each EC50 by using GraphPad Prism software (version 5.01). A representative experiment is shown. Error bars represent the ranges of replicate data points. The inset table shows the degree of USP18 expression calculated by RT-PCR (corrected for actin) and the IFN-α2a EC50 for ISRE induction. (b) Huh7 cells harboring the 9B subgenomic HCV replicon were transiently transfected with the same USP18 siRNA reagents described above and as described in Materials and Methods. After 48 h, a dilution series of IFN-α2a was added to the transfected cells and incubated for a further 17 to 24 h. Luciferase activity was measured by using an Analyst plate reader, and the percent inhibition of replicon activity relative to nontransfected cells treated with 10 IU/ml IFN-α2a was measured for each USP18 knockdown. Dose-response curves were generated by plotting the percent replicon inhibition for each USP18 knockdown against the IFN-α2a concentration. Nonlinear regression analysis was used to calculate EC50s from each dose-response curve by using GraphPad Prism (version 5.01). Each condition was tested at n ≥ 2 replicates, and each experiment was independently repeated at least 3 times. A representative experiment is shown. Error bars represent the ranges of replicate data points. The inset table shows the degree of USP18 expression calculated by RT-PCR (corrected for actin) and the IFN antiviral (AV) EC50. (c) The fold changes (FCs) in ISRE induction were calculated by dividing the EC50 values for siCONT by the EC50 values for siUSP18. Similarly, FCs in IFN-α2a AV potency were calculated by dividing the EC50 values for siCONT values by the EC50 values for siUSP18 from the replicon assay. The USP18 expression levels were measured by RT-PCR and calculated for each siRNA after correction for actin. Error bars represent standard deviations (SD) of pooled EC50 differences generated by GraphPad Prism (version 5.01). NT, not transfected.
We wanted to directly compare the effects of the USP18 knockdown on both ISRE activity and AV activity. We measured the increase in IFN signaling by calculating the fold change in the IFN-α2a EC50 for each siRNA in ISRE-luc cells and also measured the degree of USP18 knockdown using RT-PCR. The correlation between IFN-α2a signaling and USP18 knockdown is shown in Fig. 1c and demonstrates that an increased USP18 knockdown tracks with increased IFN-α2a signaling. We similarly measured the fold change in the IFN AV EC50 in the siRNA-transfected subgenomic HCV assay and plotted these values against the degree of USP18 knockdown (Fig. 1c). Clearly, the observed increase in IFN-α2a signaling activity induced by the USP18 knockdown does not fully translate to AV efficacy in the HCV subgenomic assay system.
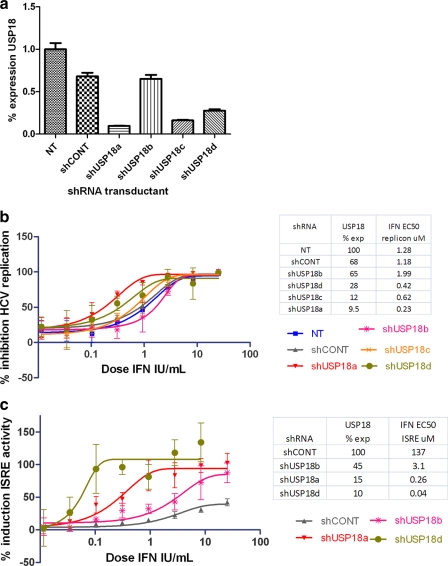
We were concerned that the relatively limited increase in AV potency may reflect the nonhomogenous nature of siRNA transfections, reducing the impact of the USP18 knockdown on IFN-α2a AV activity. Thus, we generated HCV subgenomic replicon lines and ISRE-luc cells stably transfected with shRNA directed toward USP18 to obtain a more homogenous population of USP18-suppressed cells. We observed that these shRNAi reagents had a consistent efficacy regarding the knockdown of USP18 expression levels, with three reagents reducing expression by 80 to 90% but a fourth reagent (shUSP18b) reducing USP18 expression in Huh7.5 cells by only approximately 50% (Fig. 2a and Table 1). We then measured the effect of IFN-α2a on ISRE induction and the IFN AV EC50 and compared it to that of the nontargeting shRNA control (Fig. 2b and c). We demonstrated that a 90% knockdown of USP18 in our stable cells resulted in an increase of IFN-α2a-induced ISRE activity of up to 300-fold, whereas the IFN-α2a-induced AV potency showed an increase up to 5-fold relative to control values.
Fig. 2.
(a) RT-PCR data measuring the level of USP18 expression in shRNA stable transfected subgenomic replicon cells, corrected for actin. Error bars are SD calculated by GraphPad Prism software. (b) The 9B HCV subgenomic replicon cells were stably transfected with the shRNA reagents targeting USP18. After 24 h of treatment with a serial dilution of IFN-α2a, the percent inhibition of replicon activity was measured as described in the legend of Fig. 1. Data from a representative experiment are shown. Error bars represent the ranges of replicate data points. The inset table shows the degree of USP18 expression calculated by RT-PCR (corrected for actin) and EC50 values. (c) Similarly, the ISRE-luc stable shRNA-transfected clones were treated with IFN-α2a for 24 h, and the percent ISRE induction was measured relative to IFN-α2a shCONT stable cells. Data from a representative experiment are shown. Error bars represent the ranges of replicate data points. The inset table shows the degree of USP18 expression calculated by RT-PCR (corrected for actin) and the corresponding EC50s.
Table 1.
Fold change in IFN antiviral EC50 values for shUSP18 stable cell lines
| Cell line | % USP18 expression (95% confidence limit)a | HCVcc assay |
HCV replicon |
EMCV assay |
|||
|---|---|---|---|---|---|---|---|
| FC in IFN antiviral potency (95% confidence limit)b | P value | FC in IFN antiviral potency (95% confidence limit)b | P value | FC in IFN antiviral potency (95% confidence limit)b | P value | ||
| Control | 70.4 (63–77) | 1.1 (0.5–2.1) | 0.90 | 0.7 (0.4–1.3) | 0.29 | 0.7 (0.3–1.4) | 0.26 |
| shUSP18a | 13.9 (11–17) | 2.9 (1.4–5.7) | 0.003 | 2.9 (1.5–5.7) | 0.002 | 2.6 (1.3–5.1) | 0.0076 |
| shUSP18b | 49.4 (37–62) | 1.4 (0.7–3.0) | 0.37 | 0.7 (0.3–1.5) | 0.33 | 0.7 (0.4–1.4) | 0.32 |
| shUSP18c | 10.9 (8–14) | 1.1 (0.6–2.2) | 0.73 | 1.4 (0.8–2.6) | 0.25 | 0.9 (0.5–1.8) | 0.77 |
| shUSP18d | 16.5 (10–23) | 1 (0.5–2.1) | 1.00 | 2.9 (1.5–5.6) | 0.002 | 0.4 (0.2–0.8) | 0.013 |
The USP18 mRNA levels were assayed by using RT-PCR, and values were corrected for actin expression by adjusting the USP18 arbitrary copy number in each well by the relative amount of actin present. The corrected values were then normalized to the nontransfected cell control to give the percent expression (percent USP18). Values from Huh7.5 cells (n = 2), Huh7 cells (n = 1), and HeLa cells (n = 4) were averaged, and the 95% confidence limits are given in parentheses.
The fold change (FC) is the difference in EC50s between the nontargeting shRNA control stable cells and USP18 shRNA stable cell line (note that the control FC values were calculated relative to nontransfected cells). The FC values and probabilities were calculated by generating control test pairwise comparisons The shUSP18 IFN EC50s were analyzed on the logarithmic scale by using a linear mixed model, comparing the nontargeting shRNA control stable cells and USP18 shRNA stable test cell lines within each assay type and accounting for experimental variation. The comparisons were summarized with FC values, 95% confidence limits (in parentheses), and P values.
USP18 knockdown causes upregulation of canonical IFN-α2a-induced AV ISG activity.
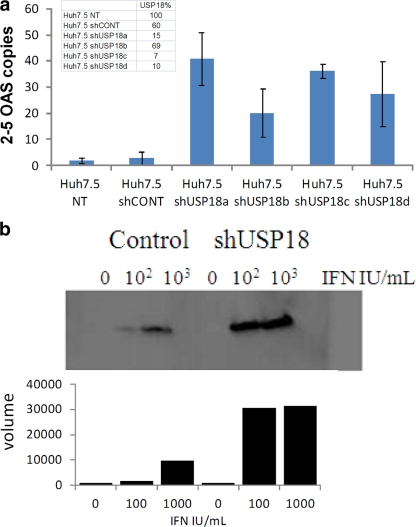
To further investigate the apparent disconnect between IFN-α2a signaling and AV potency, we studied the effects of the USP18 knockdown on two canonical antiviral ISGs, namely, 2-5 OAS and ISG15. We used the above-described shRNA reagents to stably transfect Huh7.5 cells to ensure uniform USP18 suppression across the cell population. All four stable shRNA cell lines were treated with IFN-α2a for 2 days, and the degrees of 2-5 OAS and actin expression were measured by using TaqMan RT-PCR and compared to those of control cells. We observed an increase in the IFN-α2a induction of 2-5 OAS (corrected for actin) by 10- to 100-fold in all cell lines, with the least effective knockdown (reagent shUSP18b) showing the lowest level of induction, as expected (Fig. 3a). Thus, we saw similar correlations between the degree of USP18 suppression and IFN-α2a activity, as described above for the siRNA system. We note that the USP18 knockdown instigated by these RNAi tools does not result in overt increased endogenous IFN-α2a secretion, as evidenced by the lack of 2-5 OAS induction in the absence of added IFN (data not shown).
Fig. 3.
(a) Huh7.5 cells stably transfected with USP18 shRNA were treated with 210 IU/ml IFN-α2a for 17 to 24 h prior to RNA extraction and TaqMan determinations of both 2-5 OAS and actin. The degree of expression of USP18 was corrected for actin expression and is shown in the inset table. Error bars show ranges of duplicate data points as plotted with GraphPad Prism software (version 5.01). This experiment was performed 3 times (with different IFN concentrations) and showed the same degree of induction. Data from a representative experiment are shown. (b) Huh7.5 cells stably transfected with USP18 shRNA were treated with IFN-α2a as indicated for 17 to 24 h and harvested for Western blot analysis. Control cells were nontransfected and IFN-α2a treated as shown in the figure. The filters were incubated with an anti-ISG15 antibody as a primary antibody and a horseradish peroxidase (HRP)-conjugated secondary antibody prior to development. The bar graph represents an estimate of the pixel raw volume of each band in the Western blot, quantified by using a SynGene Genius BioImaging system using GeneTools software, version 3.08.
IFN-α2a stimulation leads to the upregulation of ISG15, a ubiquitin-like protein which conjugates a variety of cellular proteins (22, 25, 27, 31). This conjugation is reversible, and deconjugation is mediated by USP18 (23). Furthermore, ISG15 was shown previously to play a role in the AV mechanism of action of IFN-α2a and is critical for the host response to a number of viral infections (21). Therefore, we investigated whether this principal substrate for USP18 was superinduced by IFN-α2a in our stable shUSP18a Huh7.5 cell lines. As expected, we observed much higher levels of free unbound ISG15 protein in USP18 knockdown cell lines than in nontransfected Huh7.5 parental cells (Fig. 3b). Thus, we conclude that the knockdown of USP18 expression causes the increased IFN-α2a induction of two genes with known AV activities.
USP18 suppression mediates superinduction of 2-5 OAS within the time frame for AV activity.
We reasoned that the lack of a translation of increased IFN-α2a signaling to AV efficacy in our USP18 knockdown cells may be due to delayed kinetics of ISG induction during the time frame of our assay. We decided to perform a time course for IFN-α2a-induced 2-5 OAS expression in the HCV subgenomic cell line. The HCV subgenomic replicon cell line stably transfected with USP18 shRNA was treated with increasing doses of IFN-α2a (50-IU/ml top dose), and total RNA was harvested at 4 h, 8 h, and 24 h and assayed for 2-5 OAS, HCV, and actin gene expression by using RT-PCR. The IFN-α2a AV EC50 was determined from the replicon-IFN dose-response curve by using the RT-PCR readout for HCV RNA at 24 h and was calculated to be approximately 1 IU/ml (Fig. 4a). This value is consistent with the HCV replicon IFN-α2a AV EC50s obtained from the luciferase format for this assay (Fig. 1). Figure 4b shows the time course for 2-5 OAS mRNA induction in these cells at the AV IFN-α2a EC50 dose and shows the rapid, early, >100-fold induction of 2-5 OAS for all three effective RNAi knockdown reagents at between 4 and 8 h. As expected, the poorly efficacious RNAi (shUSP18b) showed kinetics of 2-5 OAS induction similar to those for the two negative controls (nontransfected controls and the nontargeting shRNA control). Thus, we conclude that the USP18 knockdown in our assay system leads to a timely >100-fold induction of AV ISGs within the time frame of our assay.
Fig. 4.
(a) Huh7 cells harboring the 9B HCV subgenomic replicon were stably transfected with USP18 shRNA as shown. After 24 h of treatment with a serial dilution of IFN-α2a, the number of replicon RNA genomes was measured by using RT-PCR and corrected to actin. The numbers in square brackets represent the EC50s calculated from the IFN dose-response curves generated with GraphPad Prism software (version 5.01). Error bars represent ranges of replicate data points. (b) The same cells as those described above were treated with 1.8 IU/ml IFN-α2a for 4, 8, and 24 h, and RNA was extracted for RT-PCR analysis for actin and 2-5 OAS. The extent of induced 2-5 OAS expression corrected for actin for each shUSP18 stable cell line is plotted against time. The numbers in square brackets represent USP18 expression levels.
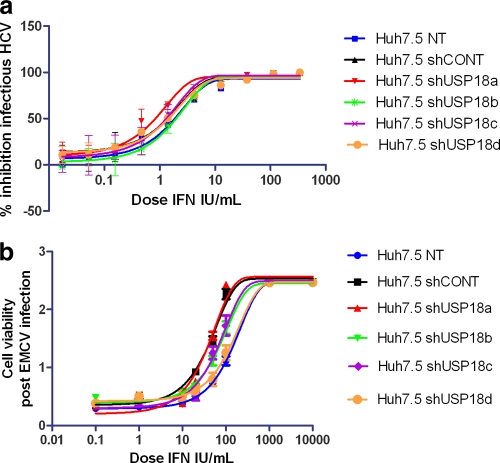
Increased IFN signaling caused by USP18 knockdown does not translate to AV activity in fully infectious virus assay systems.
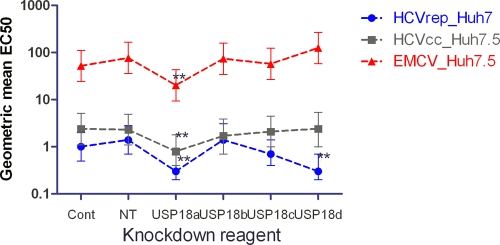
We reasoned that the relatively limited effect on AV activity in the IFN-α2a-treated USP18 knockdown HCV subgenomic replicon cell lines may be due to the lack of a USP18-sensitive component normally active during the complete life cycle of an infectious virus. To investigate this further, we used the stable USP18 shRNA-transfected Huh7.5 cell lines described above as host cells in the HCV infectious virus assay. We pretreated the cells with various doses of IFN-α2a overnight and subsequently infected them with HCV strain JFH (MOI of ∼0.1) for 2 days. HCV strain JFH encodes a luciferase reporter gene, enabling a rapid, easy, and quantitative readout for HCV replication, as shown in Fig. 5a. In addition, we wished to check whether the increased IFN signaling in the USP18 knockout hosts could result in increased AV activity in different host-virus experimental systems using non-HCV viruses. The picornavirus encephalomyocarditis virus (EMCV) was chosen because it is a well-characterized, commercially available viral stock with a broad host range and a convenient cytopathic endpoint, which can be used to measure IFN susceptibility. Infectious EMCV was assayed by use of shUSP18 stable transfected HeLa cell lines and shUSP18 stable transfected Huh7.5 cells, and we observed the same lack of an effect on AV activity; however, only the latter data are shown in Fig. 5b, to maintain consistency with infectious HCV. Figure 6 shows all the IFN AV EC50 data generated by the HCV replicon, HCV infectious virus, and EMCV assays plotted against the various USP18 knockdown reagents and shows that shUSP18a causes a 3-fold increase in AV potency with a high statistical significance (P = 0.002) across all the viral assays. Table 1 shows the fold changes (FCs) in IFN-α2a EC50s relative to the percentage of USP18 expression in each knockdown cell line. It is clear that shUSP18c has an equally efficacious knockdown effect compared to that of shUSP18a but has little or no effect on IFN-α2a AV potency, an observation suggesting that the 3-fold AV increases seen with shUSP18a may not necessarily be caused specifically by USP18 suppression but could be manifested via nonspecific properties of shUSP18a stable transfection.
Fig. 5.
(a) Huh7.5 cells stably transduced with USP18 shRNA were treated with serially diluted IFN-α2a for 17 h. Infectious HCV (JFH) was added to an MOI of ∼0.1 and incubated for 2 days. Luciferase levels were measured and plotted against IFN-α2a concentrations as the percent inhibition relative to non-IFN-α2a-treated cells. Each condition was tested at n ≥ 2 replicates, and each experiment was independently repeated at least 3 times. Nonlinear regression analysis of the IFN-α2a dose-response curve was used to calculate each EC50 by using GraphPad Prism (version 5.01). Data from a representative experiment are shown. Error bars represent the ranges of replicate data points. (b) Huh7.5 cells stably transduced with USP18 shRNA were treated with serially diluted IFN-α2a for 17 h. EMCV was added, and the mixture was incubated for 24 h. Cell viability was measured by using the WST-1 assay, and cell viability was plotted against the IFN dose. Each condition was tested at n ≥ 2 replicates, and each experiment was independently repeated at least 3 times. Nonlinear regression analysis of the IFN-α2a dose-response curve was used to calculate each EC50 by using GraphPad Prism (version 5.01). Data from a representative experiment are shown. Error bars represent the ranges of replicate data points.
Fig. 6.
The geometric mean for IFN AV EC50s was calculated for each USP18 knockdown reagent for EMCV, HCVcc, and HCV replicon assays. The error bars are upper and lower 95% confidence limits. ** indicates a P value of <0.01. Cont, nontargeting shRNA; NT, not transfected.
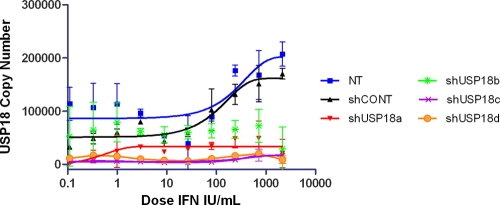
We also measured the induction of basal USP18 expression during the IFN-α2a treatment period for nontransfected controls, scrambled nontargeting controls, and shUSP18-transfected cells (Fig. 7). The data confirm that USP18 gene expression was reduced to ∼10% for the Huh7.5shUSP18a, Huh7.5shUSP18b, and Huh7.5shUSP18d stable clones, whereas USP18 expression in the Huh7.5shUSP18b stable clone was ∼50%, again as expected. We observed that the basal level for USP18 in control nontransfected Huh7.5 cells is very low and that the IFN-α2a induction of USP18 gene expression occurred only at >100 IFN IU/ml, which is much higher than the AV EC50s of ∼1 IFN IU/ml.
Fig. 7.
Huh7.5 cells stably transduced with USP18 shRNA were treated with serially diluted IFN-α2a for 17 h. Infectious HCV (JFH) was added to an MOI of ∼0.1 and incubated for 2 days. RNA was extracted, and USP18 levels were measured and corrected for actin. Nonlinear regression analysis of the IFN-α2a dose-response curve using GraphPad Prism (version 5.01) was performed. Error bars represent the ranges of replicate data.
DISCUSSION
We have shown that RNAi-mediated USP18 knockdown results in increased IFN-α2a signaling, as measured by an ISRE reporter gene assay, 2-5 OAS gene expression, and ISG15 protein production. The observed effects on IFN signaling are in the ∼100-fold range for both ISRE-luciferase and 2-5 OAS expressions and demonstrate a direct correlation between the degree of USP18 knockdown and the potentiation of IFN-α2a signaling. However, we observed a much reduced effect on IFN-α2a AV activity (∼3-fold for shUSP18a) for both HCV and EMCV in Huh7.5 host cells. Importantly, the 3-fold increase in AV potency did not correlate with the extent of the USP18 knockdown because RNAi reagents inducing the same degree of USP18 knockdown did not show the same effect on AV IFN-α2a potency. Thus, taking into account all the RNAi tools used, the impact of shUSP18a on AV potency is seemingly unrelated to the USP18 knockdown observed and could be due to a nonspecific effect, including detrimental effects on general microRNA (miRNA) transport and/or nonspecific effects on host mRNA expression (10, 12). The lack of an effect of the USP18 knockdown on AV potency is wholly unexpected, because we observed a concomitant ∼100-fold increase in IFN signaling, and it was previously demonstrated with USP18/UBP43−/− mice that increased IFN sensitivity can translate to markedly increased antiviral potency against lymphocytic choriomeningitis virus (LMCV), VSV, and HepB virus (17, 31). Previous mechanistic studies using USP18/UBP43−/− mouse embryonic fibroblast (MEF) lines demonstrated that USP18 negatively regulates IFN signaling independent of the isopeptidase activity and functions specifically to regulate type I IFN at the receptor level (24).
The mechanism of IFN-mediated HCV inhibition in Huh7 cells is poorly defined but appears to be linked to a reduction in viral protein synthesis and the eventual inhibition of viral RNA amplification (9). Our data suggest that 2-5 OAS and ISG15 are excellent inducible biomarkers for monitoring changes in the primary IFN signaling cascade but are unreliable biomarkers for tracking subsequent AV activity during the later antiviral effector cascade. This could be due to either a very low threshold of 2-5 OAS or ISG15 activity being required to saturate the AV effector(s) or the AV effector mechanism in this experimental system being independent of 2-5 OAS and ISG15. Under these circumstances, it would not be possible to predict the consequences of enhanced IFN-α2a signaling for AV activity using these biomarkers. Indeed, it was reported that the overexpression of 2-5 OAS and ISG15 in HEK cells does not have an impact on HCV replicon activity, although this observation is confounded by the absence of interferon treatment (13). In addition, other reports noted that the 2-5 OAS/RNase L pathway does not play a major role in limiting HCV activity in IFN-α2a-treated Huh7 cells (35). Finally, our data clearly show that the IFN induction of 2-5 OAS occurs at a dose of >10 IFN IU/ml but that the AV effect occurs at a dose of <10 IFN IU/ml, further suggesting that these classical IFN induction biomarkers are not reporting on the appropriate AV mechanism of action in these popular experimental HCV models.
The inability to translate increased IFN signaling to anti-HCV efficacy is unlikely to be due to the loss of a host factor(s) in Huh7.5/Huh7 cells linking ISG upregulation to AV activity, because we find that USP18-suppressed HeLa cells also showed no enhanced IFN AV activity in EMCV assays relative to controls (data not shown), suggesting that the disconnect between IFN-α2a signaling and AV potency is not limited to Huh hepatoma cell lines. It is known that the 9B subgenomic replicon contains cell culture-adapted mutations in NS5A, which could be the basis for the lack of the response to IFN potentiation. However, the HCV JFH strain used for the infectious HCV complete cycle assay does not have these adaptations (26) and yet is also refractory to increased IFN-α2a signaling. Furthermore, EMCV in Huh7.5 cells is similarly resistant to USP18-mediated IFN-α2a potentiation, so we conclude that cell-adapted mutations in the replicon line are not responsible for the lack of AV sensitivity in USP18 knockdown replicon lines. Finally, we note that the IFN AV EC50 obtained for EMCV in Huh7.5 cells is relatively high (∼50 IU/ml) compared to that obtained by the HCVcc assay (∼1 IU/ml), but we believe that this does not diminish the utility of EMCV as a tractable control non-HCV experimental model system for investigating whether increased IFN signaling can translate into increased antiviral activity. We note that the IFN AV EC50 in HeLa cells was much lower (∼4 to 7 IU/IFN) than that in Huh7.5 host cells, but we still failed to see the translation of increased IFN signaling to AV activity in this HeLa system (data not shown).
Our data broadly agree with those reported previously by Chua et al. (4), who also showed a modest effect of the USP18 knockdown on AV potency, whereas the results reported by Randall et al. (28) suggest a more potent effect. In an attempt to understand why one study observed a >1-log effect on AV potency but we and Chua et al. saw only a <1-log effect, we can point to a few differences in experimental conditions which may be significant. The study by Randall et al. used a very high concentration of siUSP18 (i.e., 2,500 nM siRNA compared to 25 nM in this study and that of Chua et al.) using electroporation and a different infectious HCV clone at a much higher multiplicity of infection (MOI) (3 compared to our 0.1). It was previously documented that high concentrations of siRNA result in increased off-target effects mediated by seed region sequence complementarity (10, 12, 30). Crucially, the off-target gene suppression is specific for each siRNA reagent. This is evidenced by the different spectrum of suppressed genes for each siRNA, as shown by previous gene array studies (10). Thus, it is possible that other host targets are being modulated at 2,500 nM siRNA and could be contributing to the antiviral state in an IFN-α2a-independent or USP18-independent manner. Second, high HCV MOIs have been reported to induce ISG expression in the absence of added interferon (6). This could lead to altered consequences/outcomes of IFN signaling, in contrast to our current study, which used low MOIs and low RNAi levels.
The nonspecific RNAi effect driving the 3-fold increase in AV potency could be due to generic changes in the normal metabolism of endogenous miRNAs due to overexpression causing a bottleneck (30). It was established that endogenous miR122 is required for efficient HCV replication (14–16), so any bottlenecks in normal miR122 transport may have an indirect impact on HCV replicative activity. Clearly, other, non-miR122, mechanisms would have to be envisaged for the impact of shUSP18a upon EMCV. Alternatively, the shRNAs may be having an impact on the translation of other host mRNAs via mismatched seed regions in the 3′ termini (10–12).
To date, our conclusions are that the HCV replicon system and infectious HCVcc viral assays are very sensitive to exogenously added IFN-α2a but that increased receptor signaling has little or no impact on AV activity. Given the many published reports of USP18/UBP43 knockout models resulting in increased AV activity (see the introduction), it is unclear whether the widespread and commonly used HCV replicon surrogate assay can be reliably used as a predictor of clinical efficacy for USP18 inhibition as an HCV therapeutic mechanism.
Footnotes
Published ahead of print on 27 June 2011.
REFERENCES
- 1. Asahina Y., et al. 2008. Potential relevance of cytoplasmic viral sensors and related regulators involving innate immunity in antiviral response. Gastroenterology 134:1396–1405 [DOI] [PubMed] [Google Scholar]
- 2. Blight K. J., McKeating J. A., Rice C. M. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen L., et al. 2005. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology 128:1437–1444 [DOI] [PubMed] [Google Scholar]
- 4. Chua P. K., et al. 2009. Modulation of alpha interferon anti-hepatitis C virus activity by ISG15. J. Gen. Virol. 90:2929–2939 [DOI] [PubMed] [Google Scholar]
- 5. Der S. D., Zhou A., Williams B. R., Silverman R. H. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. U. S. A. 95:15623–15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fredericksen B., et al. 2002. Activation of the interferon-beta promoter during hepatitis C virus RNA replication. Viral Immunol. 15:29–40 [DOI] [PubMed] [Google Scholar]
- 7. Frese M., et al. 2003. Hepatitis C virus RNA replication is resistant to tumour necrosis factor-alpha. J. Gen. Virol. 84:1253–1259 [DOI] [PubMed] [Google Scholar]
- 8. Gelman M. A., Glenn J. S. 2011. Mixing the right hepatitis C inhibitor cocktail. Trends Mol. Med. 17:34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo J. T., Sohn J. A., Zhu Q., Seeger C. 2004. Mechanism of the interferon alpha response against hepatitis C virus replicons. Virology 325:71–81 [DOI] [PubMed] [Google Scholar]
- 10. Jackson A. L., et al. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21:635–637 [DOI] [PubMed] [Google Scholar]
- 11. Jackson A. L., et al. 2006. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA 12:1197–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jackson A. L., et al. 2006. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12:1179–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang D., et al. 2008. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J. Virol. 82:1665–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jopling C. L. 2008. Regulation of hepatitis C virus by microRNA-122. Biochem. Soc. Trans. 36:1220–1223 [DOI] [PubMed] [Google Scholar]
- 15. Jopling C. L., Schutz S., Sarnow P. 2008. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe 4:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jopling C. L., Yi M., Lancaster A. M., Lemon S. M., Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577–1581 [DOI] [PubMed] [Google Scholar]
- 17. Kim J. H., Luo J. K., Zhang D. E. 2008. The level of hepatitis B virus replication is not affected by protein ISG15 modification but is reduced by inhibition of UBP43 (USP18) expression. J. Immunol. 181:6467–6472 [DOI] [PubMed] [Google Scholar]
- 18. Kronenberger B., Zeuzem S. 2009. Current and future treatment options for HCV. Ann. Hepatol. 8:103–112 [PubMed] [Google Scholar]
- 19. Lavanchy D. 2009. The global burden of hepatitis C. Liver Int. 29(Suppl. 1):74–81 [DOI] [PubMed] [Google Scholar]
- 20. Lenschow D. J., et al. 2005. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J. Virol. 79:13974–13983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lenschow D. J., et al. 2007. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. U. S. A. 104:1371–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu G., et al. 2006. ISG15 enhances the innate antiviral response by inhibition of IRF-3 degradation. Cell. Mol. Biol. (Noisy-le-Grand) 52:29–41 [PubMed] [Google Scholar]
- 23. Malakhov M. P., Malakhova O. A., Kim K. I., Ritchie K. J., Zhang D. E. 2002. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 277:9976–9981 [DOI] [PubMed] [Google Scholar]
- 24. Malakhova O. A., et al. 2006. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 25:2358–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malakhova O. A., Zhang D. E. 2008. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J. Biol. Chem. 283:8783–8787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mateu G., Donis R. O., Wakita T., Bukh J., Grakoui A. 2008. Intragenotypic JFH1 based recombinant hepatitis C virus produces high levels of infectious particles but causes increased cell death. Virology 376:397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Osiak A., Utermohlen O., Niendorf S., Horak I., Knobeloch K. P. 2005. ISG15, an interferon-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis virus. Mol. Cell. Biol. 25:6338–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Randall G., et al. 2006. Silencing of USP18 potentiates the antiviral activity of interferon against hepatitis C virus infection. Gastroenterology 131:1584–1591 [DOI] [PubMed] [Google Scholar]
- 29. Randall R. E., Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–47 [DOI] [PubMed] [Google Scholar]
- 30. Rao D. D., Vorhies J. S., Senzer N., Nemunaitis J. 2009. siRNA vs. shRNA: similarities and differences. Adv. Drug Deliv. Rev. 61:746–759 [DOI] [PubMed] [Google Scholar]
- 31. Ritchie K. J., et al. 2004. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat. Med. 10:1374–1378 [DOI] [PubMed] [Google Scholar]
- 32. Ritchie K. J., et al. 2002. Dysregulation of protein modification by ISG15 results in brain cell injury. Genes Dev. 16:2207–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sarasin-Filipowicz M., et al. 2008. Interferon signaling and treatment outcome in chronic hepatitis C. Proc. Natl. Acad. Sci. U. S. A. 105:7034–7039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sarasin-Filipowicz M., et al. 2009. Alpha interferon induces long-lasting refractoriness of JAK-STAT signaling in the mouse liver through induction of USP18/UBP43. Mol. Cell. Biol. 29:4841–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang C., et al. 2003. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J. Virol. 77:3898–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Y., Burke C. W., Ryman K. D., Klimstra W. B. 2007. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J. Virol. 81:11246–11255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Y., Weady P., Duggal R., Hao W. 2008. Novel chimeric genotype 1b/2a hepatitis C virus suitable for high-throughput screening. Antimicrob. Agents Chemother. 52:666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]