Abstract
The long-standing conclusion that the Cdc7 kinase of Saccharomyces cerevisiae is required only to trigger S phase has been challenged by recent data that suggests it acts directly on individual replication origins. We tested the possibility that early- and late-activated origins have different requirements for Cdc7 activity. Cells carrying a cdc7ts allele were first arrested in G1 at the cdc7 block by incubation at 37°C, and then were allowed to enter S phase by brief incubation at 23°C. During the S phase, after return to 37°C, early-firing replication origins were activated, but late origins failed to fire. Similarly, a plasmid with a late-activated origin was defective in replication. As a consequence of the origin activation defect, duplication of chromosomal sequences that are normally replicated from late origins was greatly delayed. Early-replicating regions of the genome duplicated at approximately their normal time. The requirements of early and late origins for Cdc7 appear to be temporally rather than quantitatively different, as reducing overall levels of Cdc7 by growth at semi-permissive temperature reduced activation at early and late origins approximately equally. Our results show that Cdc7 activates early and late origins separately, with late origins requiring the activity later in S phase to permit replication initiation.
Keywords: Cdc7, kinase, origin, replication, S phase, yeast
At the core of the cell cycle is the chromosome cycle, in which the chromosomes are first faithfully replicated during the DNA synthesis or S phase, and then precisely distributed to the daughter cells at mitosis. Our knowledge of the mechanisms by which these processes are controlled has advanced greatly in recent years. The major cell-cycle events are coordinated by members of the family of cyclin-dependent protein kinases (CDKs), which are activated mainly by cell-cycle regulated association with cyclins (for review, see Nasmyth 1996). The process of chromosome replication requires the products of a large number of additional genes, many of which were originally identified by studies in the yeast Saccharomyces cerevisiae. One such protein is the product of the gene CDC7, first identified by Culotti and Hartwell (1971) and since shown to encode a nuclear serine/threonine protein kinase (Bahman et al. 1988; Hollingsworth and Sclafani 1990; Yoon and Campbell 1991). Although the levels of CDC7 transcript and Cdc7 protein do not appear to vary significantly during the cell cycle (Sclafani et al. 1988; Jackson et al. 1993), Cdc7 kinase activity is regulated by its physical association with the protein Dbf4 (Jackson et al. 1993). Unlike CDC7, DBF4 transcript levels are cell-cycle regulated, peaking at the G1/S boundary (Chapman and Johnston 1989). Consistent with the regulation of DBF4 transcription, the kinase activity of Cdc7 shows cyclical fluctuation and peaks close to the start of S phase (Jackson et al. 1993). The regulation of Cdc7 kinase activity by Dbf4 association has been compared with the activation of CDKs by their association with cyclins (Sclafani and Jackson 1994), although Cdc7 kinase does not belong to the CDK family, and Dbf4 shows no sequence similarity to the cyclins. Like members of the CDK family, Cdc7 kinase activity is probably also regulated by changes in its phosphorylation state (for review, see Sclafani and Jackson 1994).
Cdc7 kinase activity has long been known to be required for the initiation of S phase, but, on the basis of previous data (Hartwell 1973, 1976), has been thought to be dispensable for normal S-phase continuation and completion. When the requirement for Cdc7 was first investigated, however, nothing was known about the nature of S. cerevisiae replication origins or the control of their activation. During the intervening quarter century, it has been shown that DNA replication in budding yeast initiates at well-defined replication origins that were first identified as autonomous replication sequences (ARS elements)—named for their ability to promote plasmid DNA replication (Hsiao and Carbon 1979; Stinchcomb et al. 1979; Huberman et al. 1988; Brewer and Fangman 1991; Ferguson et al. 1991; Newlon and Theis 1993). Yeast replication origins are not all activated at the beginning of S phase, but instead different origins fire in a reproducible temporal sequence throughout S phase. Some origins (e.g., ARS305; Fig. 1A) initiate replication at the start of S phase (Reynolds et al. 1989), others fire slightly later during S (e.g., ARS1; Brewer et al. 1993), whereas still others (e.g., the telomeric ARS501 and a cluster of origins on chromosome XIV; Ferguson et al. 1991; Friedman et al. 1996) fire during the second half of S phase.
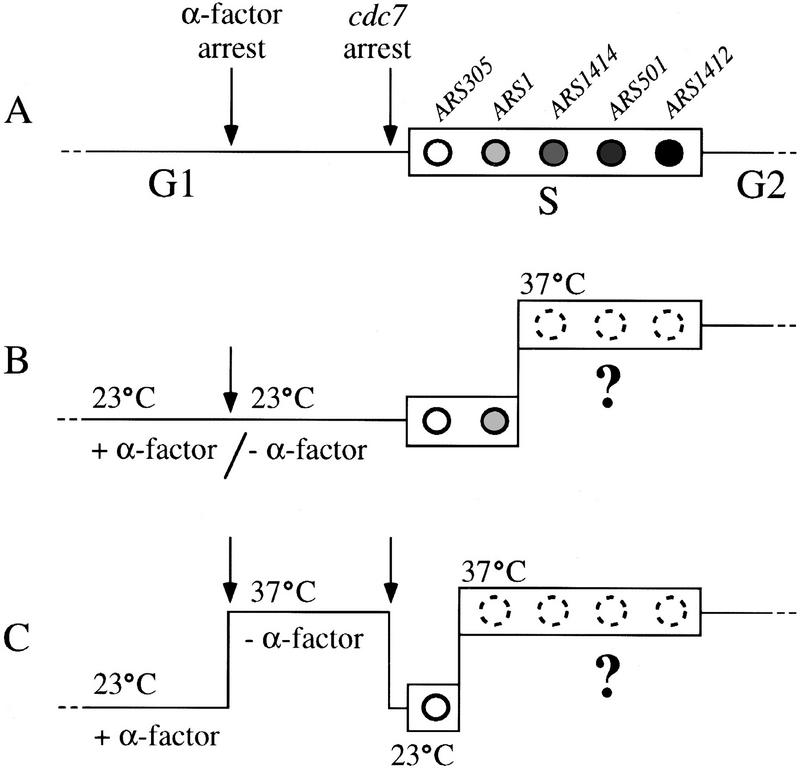
Figure 1.
Scheme of experimental procedures. A,B,and C show time lines representing the course of G1 and S phase under different experimental conditions. (A) Entry into and passage through a normal S phase. Before entry into S phase, cells transmit the α-factor arrest point and cdc7 arrest point, indicated by arrows. The rectangular box represents S phase. Replication origin activation events are drawn as shaded circles, with lighter shading indicating early-activated replication origins and darker shading for later origins. (B) Scheme of the experimental approach of Hartwell (1973) employing a cdc7-4 mutant. Vertical offset in lines represents temperature shift. Broken circles and question mark indicate replication origins that may not have been active under the particular experimental conditions. Although the temperature shift is drawn as having taken place two-fifths of the way through S phase, it should be noted that this is arbitrary; it was not possible to determine the time in S phase at which cells were shifted. (C) Illustration of experimental approach taken in this study, employing a cdc7-1 mutant. Broken circles as in B. See text for further explanation.
The finding that origins are activated throughout S phase has complicated the interpretation of Hartwell’s original data. Dbf4 is now known to interact physically with replication origins (Dowell et al. 1994). This observation suggested that the effects of the Cdc7–Dbf4 complex may be mediated at the level of individual origins, rather than acting as an upstream global regulator of S-phase initiation. If Cdc7 kinase acts on individual replication origins, then a question arises from the original suggestion that the CDC7 gene product appeared to be required only at the beginning of S phase. If, as originally concluded by Hartwell (1973), Cdc7 activity is required for entry into S phase and is not used during S phase, then either all origins are acted on by Cdc7 kinase at the beginning of S phase or late-firing origins do not require Cdc7 kinase for their activation. Here, we present data showing that Cdc7 kinase acts on individual origins of replication, with initiation of DNA replication at the different temporal categories of origins requiring separate activation by Cdc7 kinase.
Results
Failure to activate late origins after a brief pulse of Cdc7 activity
On the basis of the results of Hartwell (1973, 1976), the product of the gene CDC7 has been thought to be required for entry into S phase, but not to have a continuing role during S phase. The design of Hartwell’s original experiment (Hartwell 1973) is summarized in Figure 1B. A synchronized culture of temperature-sensitive cdc7-4 cells was shifted to the restrictive temperature after S phase had begun. Measurement of the incorporation of radioactive uracil into DNA indicated that the DNA content of the cells doubled, suggesting that S phase was completed. Inspection of these early data, however, reveal that the rate of DNA synthesis was slower at 37°C than in cells incubated at 23°C, consistent with the possibility that S phase was not completely normal at the restrictive temperature. These experiments did not address whether the usual set of replication origins was active under conditions where Cdc7 activity had been depleted (represented by broken circles in Fig. 1B). Additionally, Hartwell was not able to investigate the kinetics with which functional Cdc7 protein disappeared from the cells, leaving open the possibility that some Cdc7 activity persisted after the temperature shift. As was discussed (Hartwell 1973), slow loss of active gene product could lead to the misclassification of cdc7 mutants as defective only for entry into S phase, whereas in reality the kinase activity might be used throughout the DNA synthesis phase of the cell cycle. A second study (Hartwell 1976) employed a hydroxyurea block followed by release at the restrictive temperature for cdc7-4 cells. Cells subjected to this regime appeared able to complete nuclear division, confirming the conclusion from the earlier experiment that Cdc7 activity is not required beyond the beginning of S phase—however, in this second study, the kinetics of S phase were not examined.
To reinvestigate the requirement for Cdc7 kinase during S phase, we asked whether late origins are able to initiate replication after temperature inactivation of the Cdc7 kinase in early S phase. Conditions were sought under which cells are limited by the available Cdc7 activity (Fig. 1C). Cultures of a cdc7-1 strain were synchronized first using α-factor, and then released from α-factor at the restrictive temperature (37°C) and allowed to accumulate at the cdc7-1 block. The cultures were transferred to permissive temperature (23°C) for an interval of minutes, and then returned to 37°C. Flow cytometry analysis suggested that in cultures transferred to 23°C for between 3 and 6 min, the majority of cells were subsequently able to enter S phase but appeared to complete it slowly or not at all (data not shown).
To determine which replication origins are activated under these conditions of limiting Cdc7 activity, S-phase DNA was analyzed by two-dimensional gel electrophoresis (Brewer and Fangman 1987; Friedman and Brewer 1995). This technique allows active replication origins to be identified from the distinctive “bubble” arc formed by DNA fragments containing replication initiation structures (see cartoon, Fig. 2A). In contrast, fragments duplicated passively by replication forks originating outside the fragment form arcs of Y-shaped molecules. “37°C S-phase” DNA was prepared by briefly releasing a culture from the cdc7 block as described above (4.5 min at 23°C), and pooling samples taken every 2 min for 50 min following the initial transfer to 23°C. 23°C S-phase DNA was prepared in the same way from a culture left at 23°C after initial release from a cdc7 block; this culture completed S phase within 50 min. The early-activated chromosome III origin, ARS306, showed a strong arc of bubble structures in the 23°C S-phase preparation (Fig. 2B), indicating that this origin fired in a large majority of cells in the population. In the 37°C S-phase preparation, ARS306 gave an arc of bubbles that is somewhat reduced relative to the arc of Y-shaped molecules (Fig. 2C), reflecting some passive replication, presumably from one of the adjacent origins, ARS305 or ARS307.
Figure 2.
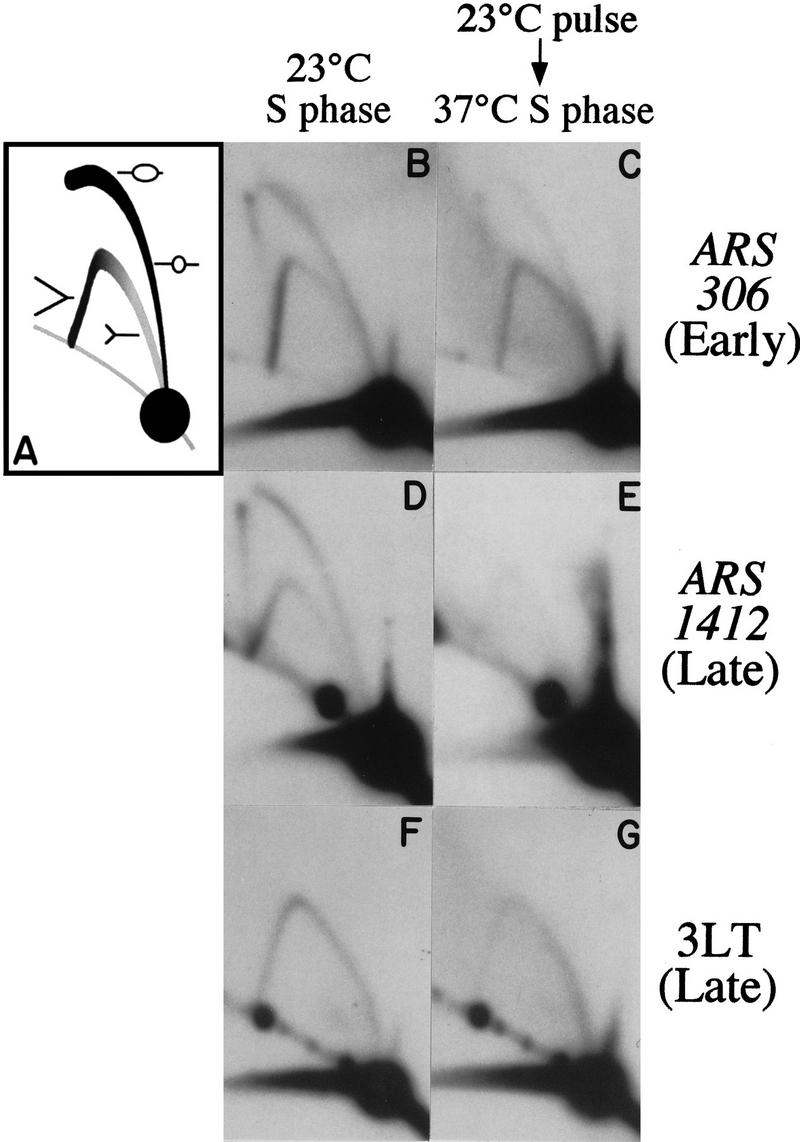
Two-dimensional gel analysis of early and late replication origin activation during S phase after a pulse of Cdc7 activity. (A) The cartoon depicts the structure of replication intermediates in the arcs of Y and bubble-shaped molecules. (B,D,F) Cells released from the cdc7 block and kept at 23°C (23°C S phase). (C,E,G) Cells released from a cdc7 block, and returned to 37°C after 4.5 min at 23°C (37°C S phase). B and C were probed to detect a fragment containing the early replication origin ARS306. D and E show a fragment containing the late origin ARS1412. F and G were probed for a restriction fragment (3LT) close to the left end of chromosome III.
The late replication origin we chose to examine was ARS1412. It lies on chromosome XIV, centrally located within a cluster of five late-activated replication origins (Friedman et al. 1996; C. Lau, B. Brewer, and W. Fangman, unpubl.). The closest early replication origins to ARS1412 have not yet been mapped, but must lie more than 80 kb away. Like ARS306, ARS1412 is efficiently activated during a normal, 23°C S phase (Fig. 2D). In contrast, very few replication intermediates (either Y or bubble structures) were present at ARS1412 in the 37°C S-phase preparation (Fig. 2E). Because identical quantities of DNA were loaded in each of the gels shown in Figure 2, the low level of replication intermediates at ARS1412 in the 37°C S-phase preparation suggested that few cells had replicated the ARS1412 region. Long exposures of the ARS1412 blot showed a faint Y arc (indicative of passive replication) but no bubble arc, in either this DNA preparation, or in a pooled DNA sample made from cells harvested between 70 and 120 min after the brief release (data not shown). One possible explanation for the dearth of replication intermediates at ARS1412 is that late replication origins did not fire during the 37°C S phase. An alternative possibility is that the late origins were activated, but that replication forks could not progress at their normal rate late in the 37°C S phase. To distinguish between these possibilities, we examined replicating structures in a region that duplicates as late in a normal S phase as ARS1412, but is replicated by a fork initiated from a distant early-firing replication origin. The 3LT sequence is within 8 kb of the left telomere of chromosome III and is replicated from the early-activated replication origin ARS305, which lies more than 30 kb away. Under normal conditions, the replication fork from ARS305 slows as it approaches the left telomere, arriving at 3LT late in S phase (Reynolds et al. 1989; Newlon et al. 1993). We reasoned that if the lack of replication intermediates at ARS1412 in the 37°C S phase were attributable to a defect in replication fork progression late in the 37°C S phase, then replicating structures should similarly be absent from the 3LT region. Two-dimensional gel analysis of a 3LT fragment showed a complete Y arc of replicating structures in both the 23°C S-phase and the 37°C S-phase preparations (Fig. 2F,G). This result suggests that the absence of replicating structures from the ARS1412 region is not primarily attributable to a defect in replication fork progression late in S phase, but is more likely the consequence of failure to fire late replication origins, such as ARS1412.
Results similar to those for ARS306 and ARS1412 were obtained by two-dimensional gel analysis of two chromosome VI origins (Friedman et al. 1997; Yamashita et al. 1997). Bubble and Y structures were present at the early origin ARS607 in both the 23°C and the 37°C S-phase DNA preparations. The late origin ARS603 was active in the 23°C S phase, but showed low levels of replication intermediates, with no bubble structures, in the 37°C S phase (data not shown). These results suggest that, after a pulse of Cdc7 activity (caused by brief incubation at 23°C) sufficient for most cells to begin S phase, early-replicating origins are activated in many cells, but replication fails to be initiated at normally late-firing origins.
Delayed replication of late chromosomal regions after a brief pulse of Cdc7 activity
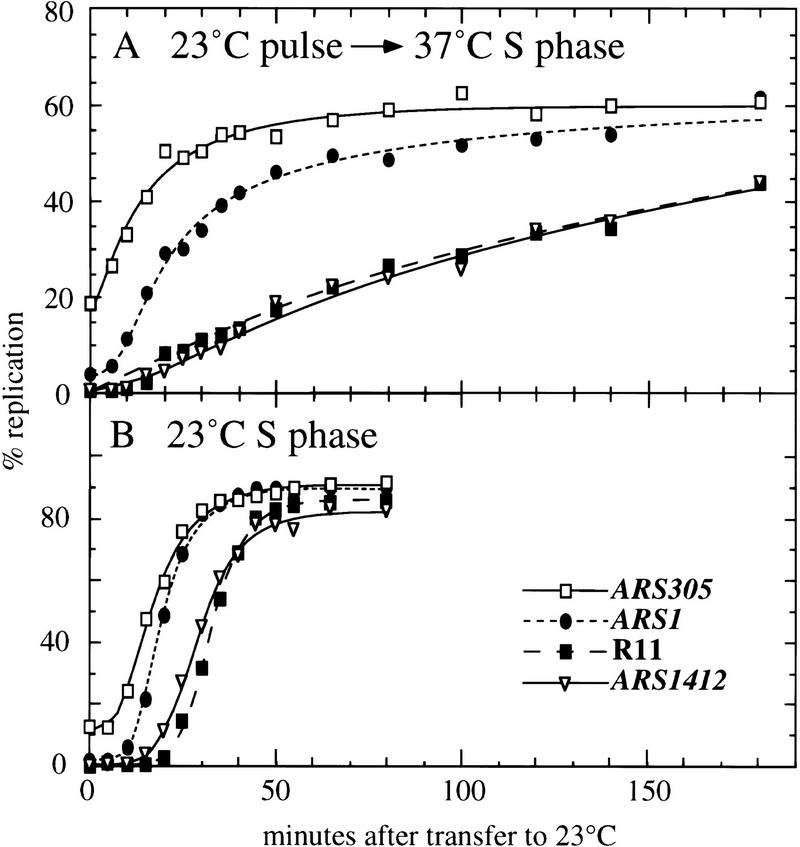
To determine whether the kinetics of DNA replication were consistent with a failure to fire late origins, we carried out dense isotope transfer experiments (McCarroll and Fangman 1988). Such density transfer experiments allow the replication kinetics of particular DNA regions to be analyzed, by following the time during S phase when the restriction fragment of interest moves from the heavy–heavy to the heavy–light DNA density peak in a cesium chloride gradient. Replication kinetics of four EcoRI fragments were examined. The first was an early-replicating marker fragment containing the ARS305 origin. ARS305 is the next origin located telomere-proximal to ARS306 on chromosome III; both origins are activated at the same early time during S phase (Reynolds et al. 1989). The second fragment analyzed is replicated soon after the ARS305 fragment, by the early/mid-S phase-activated origin ARS1. The two late-replicating sequences analyzed were the fragment containing the chromosome XIV late origin ARS1412, and the late-replication standard fragment R11. R11 lies on the right arm of chromosome V, ∼15 kb centromere-proximal to the late origin ARS501 (Ferguson et al. 1991).
Replication kinetics were determined for these four fragments during a 37°C S phase, where the culture had been released from a cdc7 block for 3.5 min at 23°C (Fig. 3A). The kinetics of a normal 23°C S phase are shown in Figure 3B for comparison (data from Friedman et al. 1996). The data for the two experiments differ in several respects. First, in the 37°C S phase, the fragments containing the early-activated origins ARS305 and ARS1 attained a final value of ∼60% as compared with the 80%–90% levels for these fragments in the 23°C S-phase culture. The lower final values indicate that ∼40% of the cells failed to begin S phase at all (compared with ∼10% of cells in the 23°C S phase). This elevation in the proportion of noncycling cells presumably reflects the stringency of the release conditions used; the 3.5-min pulse provided sufficient Cdc7 activity for many, but not all, cells to begin S phase. Second, the rates at which the early and late fragments replicated differed between the two experiments. All the fragments in the 23°C S-phase experiment completed replication by 50 min after release from the cdc7 block. In the 37°C S phase, the early-replicating fragments had also largely completed their replication within a 50-min interval. In contrast, the rate of replication of the R11 and ARS1412 late fragments was very slow in the 37°C S-phase experiment, and the level of their replication at 180 min was significantly below the 60% level seen for early fragments. During the 50-min interval following the brief release, only ∼20% of cells replicated the ARS1412 fragment. This observation explains the low level of replication intermediates found at ARS1412 by two-dimensional gel analysis—DNA samples for the two-dimensional gels shown in Figure 2 were taken during the first 50 min of the experiment, over which time period only a small fraction of the ARS1412 fragments would have been replicated.
Figure 3.
Replication kinetics after a pulse of Cdc7 activity. (A) Cells released at 23°C from a cdc block and returned to 37°C after 3.5 min at 23°C. (B) Control cells released from the cdc7 block and kept at 23°C. Replication kinetics of two early-replicating standard fragments, containing ARS305 (solid line with open rectangles) and ARS1 (broken line with filled ovals), and two late-replicating standard fragments, R11 (broken line with filled rectangles) and a fragment containing ARS1412 (solid line with open triangles) are shown.
A more quantitative comparison of the replication kinetics of each fragment can be made from the time of replication, or Trep value, defined as the time at which half of the replication has taken place (McCarroll and Fangman 1988). Trep values of sequences in the 23°C and 37°C S phases shown in Figure 3 are listed in Table 1, together with replication levels for each of the DNA sequences at the ends of the experiments. For the early fragments ARS305 and ARS1, the replication times were not significantly different between the two experiments. The extent to which replication of normally late sequences ARS1412 and R11 was delayed in the 37°C S phase is highlighted by >30-min increases in their Trep values. The real replication times of ARS1412 and R11 in this experiment may have been even later, as their levels of replication were still increasing when the last sample was taken at 180 min. As a comparison, replication kinetics of the chromosome III 3LT fragment were determined in the same experiments. Replication of 3LT, which is normally replicated late from an early origin, was not significantly delayed in the 37°C S phase when compared with the 23°C S phase (Table 1). In this respect, the 3LT fragment behaved similarly to the early fragments, and very differently from the other late fragments ARS1412 and R11, which are normally replicated from late origins.
Table 1.
Times of replication (Trep values) and final replication levels of chromosomal sequences
In general, the replication kinetics observed in this 37°C S phase are consistent with the origin activation profile suggested by the two-dimensional gels. The relatively normal Trep values obtained for the early fragments show that a brief exposure to Cdc7 activity is adequate to begin the program of early origin activation. The normal replication time of the 3LT fragment, in comparison with ARS1412 and R11, indicates that the primary defect in late S phase is in activation of origins rather than elongation at replication forks. Therefore, both the kinetics of replication and the two-dimensional gel analysis suggest that many cells fire early origins, but not late origins, after a brief pulse of Cdc7 kinase activity. The abnormally slow replication of the ARS1412 and R11 fragments during the 37°C S phase suggests that these sequences are duplicated passively by forks proceeding from early origins.
Replication of early and late plasmids after a pulse of Cdc7 activity
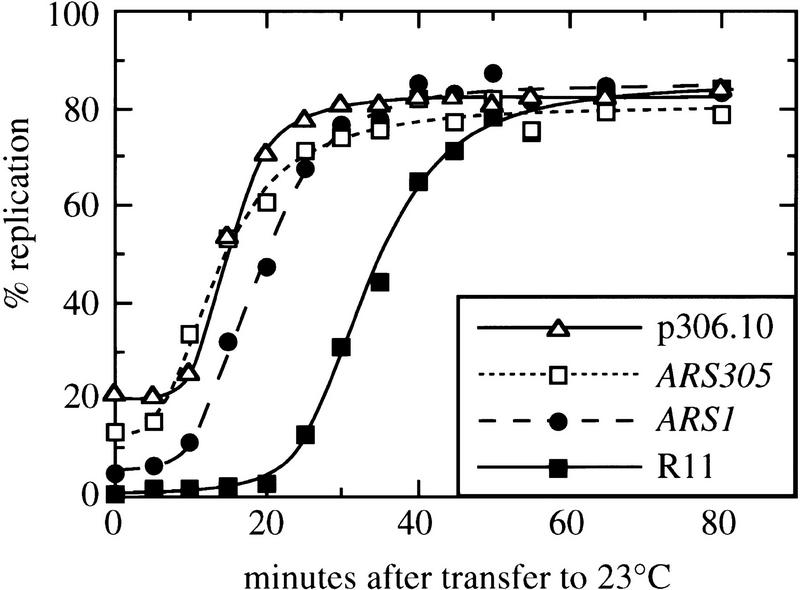
As an independent confirmation of the differences in origin activation under minimal release conditions, we analyzed replication levels of plasmids containing either an early- or a late-activated replication origin. Because each plasmid contains only a single origin, replication of such plasmids solely reflects initiation events at that origin. For consistency, we wished to use plasmids containing the same replication origins as were examined by two-dimensional gel electrophoresis. To obtain a suitable early-replicating plasmid, a library of yeast DNA fragments contained in a centromeric vector (Ferguson et al. 1991) was screened for inserts containing the early-activated chromosome III origin ARS306. One of the plasmids obtained, called p306.10, contains ARS306 on a 13.3-kb insert. This plasmid was transformed into yeast and its replication kinetics were analyzed (Fig. 4). Like the chromosomal copy of ARS306, p306.10 replicates very early during S phase, having a Trep of 14.8 min. In the same experiment, the early chromosomal origin ARS305 replicated at 13.4 min. A late-replicating centromeric plasmid named p12, which contains a 17.4-kb insert with ARS1412 as its single replication origin, has been described previously (Friedman et al. 1996).
Figure 4.
Replication kinetics of an early-duplicating plasmid containing ARS306. Kinetics of replication of the plasmid p306.10 (solid line with open triangles), of early chromosomal fragments containing ARS305 (dotted line with open squares) and ARS1 (dashed line with solid circles), and the late chromosomal fragment R11 (solid line with solid squares) are shown.
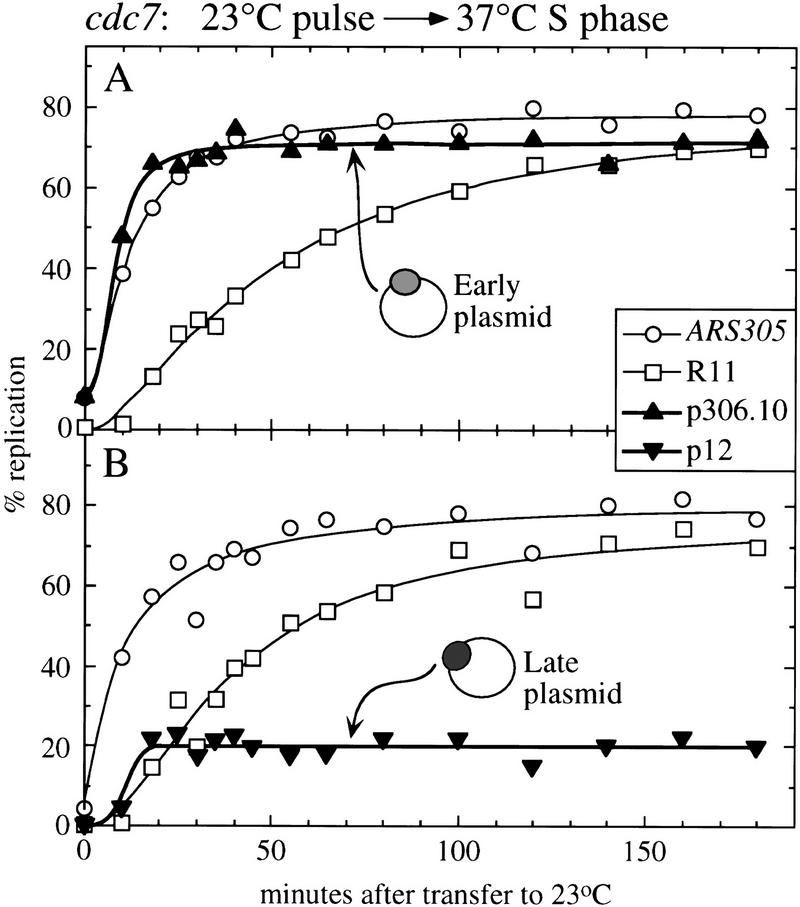
Because the early-replicating plasmid p306.10 and the late-replicating plasmid p12 each contain only a single ARS or replication origin, the plasmid sequences cannot be subject to passive replication from other origins. For this reason, duplication levels seen for either plasmid can be used as a measure of the extent of initiation of the ARS it contains. If early, but not late, origins are activated after a brief pulse of Cdc7 activity (as suggested by the experiments in Figs. 2 and 3), then we would expect that the early, but not the late, plasmid would be duplicated in an S phase under the same conditions.
Cultures of cdc7-1 cells containing either p306.10 or p12 were synchronized as usual with α-factor at 23°C, then incubated at 37°C without α-factor, and finally given a 7.5-min pulse at 23°C before being returned to 37°C. The slightly longer release length used in this experiment allowed a larger proportion of cells (>75% in each case) to begin S phase. Kinetic data obtained from the two cultures are shown in Figure 5. The replication kinetics of chromosomal fragments ARS305 and R11 were almost identical for the two cultures, and similar to those shown in Figure 3A, except that R11 showed more complete replication with less delay in these two experiments (Trep values 43.0 and 37.0 min compared with 63.3 min in Fig. 3A). This decreased delay in the replication of R11 was presumably the result of more efficient firing of early or mid-S-phase chromosome V origins, attributable to the longer, 7.5-min release time used.
Figure 5.
Replication kinetics of early and late plasmids after a pulse of Cdc7 activity. Cultures held at the cdc7 block were given a 7.5-min pulse at 23°C, then returned to 37°C. (A) Cells containing the early-replicating plasmid p306.10. (B) Cells containing the late-replicating plasmid p12. Plasmid replication levels in each culture are indicated by solid triangles. The replication curves of chromosomal early marker ARS305 (open circles) and late marker R11 (open squares) in the two cultures are also shown.
In these experiments, the early p306.10 plasmid (Fig. 5A) replicated to 71% early in the S phase. The late plasmid p12 (Fig. 5B), which is dependent on a single late-activated origin for duplication, in contrast, replicated to only 20%. In a normal S phase at 23°C, both p306.10 and p12 replicate to high levels (Fig. 4, and data for p12 contained in Friedman et al. 1996). The data in Figure 5 confirm that an early origin is activated efficiently but a late one is very defective in activation during an S phase initiated by a pulse of 7.5 min at 23°C. The plasmid replication kinetics also highlight other features of the S-phase defect. The 20% replication of the normally late plasmid p12 that did occur took place early (Trep 11.0 min). The activation time for any replication origin (during a normal S phase) is not precisely the same in each cell of a synchronized culture, but is spread over an interval—the time of origin firing as judged from the Trep value represents the average firing time for all cells in the population. The 20% replication of the normally late plasmid in Figure 5B presumably occurred in the fraction of cells that would have activated the p12 origin at a relatively early time in a normal S phase. It is also of note that the replication of p12 achieved its maximum value soon after the brief release from the cdc7 block, and did not continue to increase at later times. The origin on plasmid p12 cannot be passively replicated, unlike late chromosomal sequences such as R11 and ARS1412. In such experiments, these late chromosomal sequences increased slowly in percent replication at times long after the initial brief release (Figs. 3 and 5). This contrast between the replication kinetics of plasmid p12 and chromosomal fragment R11 (Fig. 5B) suggests that most of the replication of normally late chromosomal markers seen in these experiments was the result of passive duplication by forks proceeding from origins outside the normal late domain.
Similar results were obtained for the two plasmids when slightly different 23°C pulse lengths were used. In these experiments, we chose to take samples 65 and 80 min after the beginning of the release, as by then replication in a normal S phase would have been completed (Fig. 3B). Table 2 shows data obtained 65 and 80 min after cultures were placed at 23°C for 5 or 6 min respectively, as well as the values at the same times in the 7.5-min release experiments shown in Figure 5. Data for ARS305 and R11 at 65 and 80 min in the 3.5-min release experiment (Fig. 3A) are also listed for comparison. The values in Table 2 confirm the defective replication of p12 as compared with p306.10 in these experiments. The data also highlight the general increase in replication levels with lengthening time of release at 23°C, and demonstrate the reproducibility of the data under slightly differing release conditions.
Table 2.
Percent replication of early and late plasmids and chromosomal sequences after a pulse of Cdc7 activity
| Length of release at 23°C
|
ARS305
|
R11
|
p306.10 (early)
|
p12 (late)
|
||||
|---|---|---|---|---|---|---|---|---|
| 65 min
|
80 min
|
65 min
|
80 min
|
65 min
|
80 min
|
65 min
|
80 min
|
|
| 3.5 min | 57 | 59 | 22 | 26 | — | — | — | — |
| 5 min | 56 | 60 | 26 | 32 | 63 | 60 | — | — |
| 5 min | 63 | 66 | 28 | 33 | — | — | 13 | 13 |
| 6 min | 64 | 66 | 33 | 41 | 65 | 63 | — | — |
| 6 min | 63 | 67 | 29 | 38 | — | — | 13 | 17 |
| 7.5 min | 72 | 76 | 48 | 53 | 71 | 71 | — | — |
| 7.5 min | 77 | 75 | 54 | 58 | — | — | 18 | 21 |
Samples were taken 65 and 80 min after brief release from a cdc7 block. The 3.5-min release length data are from the experiment shown in Fig. 3, in which the culture contained no plasmid. For 5-, 6-, and 7.5-min release lengths, the top values correspond to a culture containing p306.10, and the bottom values to a culture containing p12. The 7.5-min release data are taken from Fig. 5.
Defects in both early and late origin activation with constantly limiting Cdc7 activity
If early and late replication origins differ in their quantitative requirement for Cdc7, then activation of the temporal classes of origins will be affected to differing extents by maintaining Cdc7 at a constant low level throughout S phase. To obtain such conditions, we grew asynchronous cultures at a semi-permissive temperature for the cdc7-1 allele, and examined the effects on replication origin activation. We found that the cdc7-1 strain RM14-3a was able to grow at temperatures up to 27°C, although slightly more slowly than at 23°C. DNA preparations were made from asynchronous cultures grown at 23, 26.5, or 27°C, and chromosomal origin use was analyzed by two-dimensional gel electrophoresis (Fig. 6). Bubble arcs were diminished at both the early origin ARS306 and the late origin ARS1412 in cells grown at 26.5°C, and almost absent in cells grown at 27°C. These decreases in origin activation confirmed that RM14-3a cells are stressed by limited Cdc7 at 26.5 and 27°C. The bubble arc at ARS1412, however, was not more reduced as a result of the increase in temperature than that at ARS306. This finding suggests that the deleterious effects of constantly limiting Cdc7 are no worse for late origins than for early ones.
Figure 6.
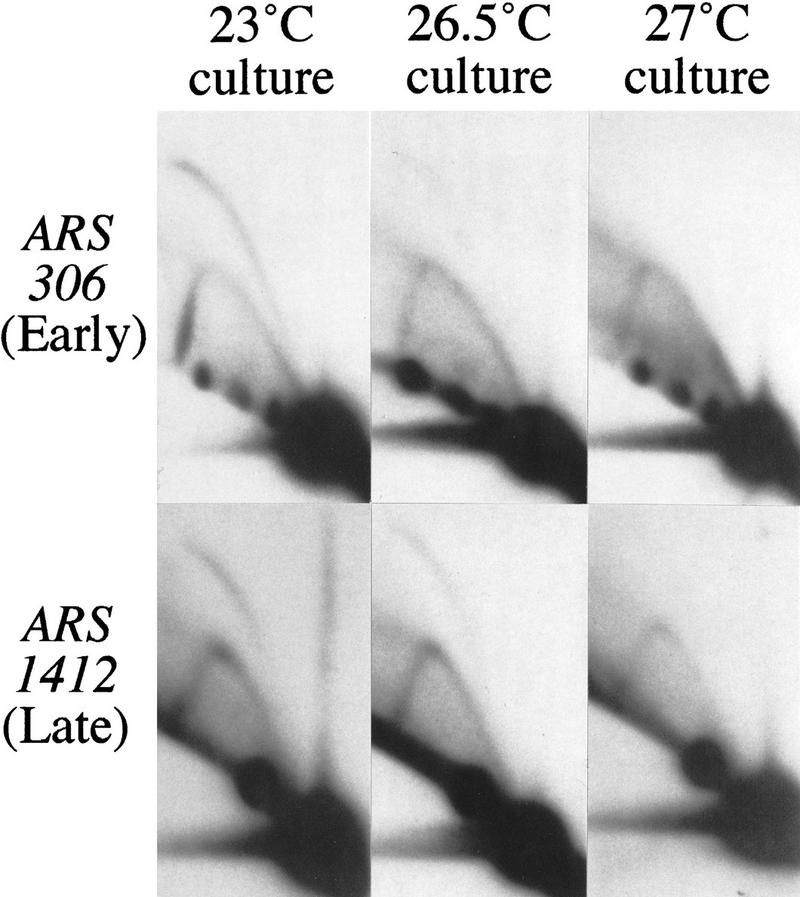
Two-dimensional gel analysis of early and late replication origin activation at semipermissive temperatures for the cdc7-1 allele. (Top) Blots probed for replication intermediates of a fragment containing early origin ARS306. (Bottom) Blots probed for the late origin ARS1412. Left panels show DNA from an asynchromous culture grown at permissive temperature (23°C). Center and right panels show blots made with DNA from cultures grown at semipermissive temperatures (26.5°C and 27°C).
Differences in origin activation frequency can be assessed quantitatively by measuring the rates at which plasmids are lost from dividing cells. Loss rates of the early-replicating plasmid p306.10 and the late-replicating plasmid p12 were measured at 23 and 26.5°C, in cdc7-1 and CDC7 strains (Table 3). Both plasmids were lost at elevated rates from cdc7-1 cells when grown at 26.5°C, confirming that the cells are limited in their capacity for origin activation at this temperature. Importantly, the extent of this elevation in loss rate was not significantly different for the early- and late-replicating plasmids. This result is consistent with the data obtained by two-dimensional gel analysis, in that it also suggests that initiation events at late replication origins are not more reduced than those at early origins as a consequence of growth at semi-permissive temperature for the cdc7-1 allele.
Table 3.
Rates of loss of early- and late-replicating plasmids at semipermissive temperature for cdc7-1
|
|
|
CDC7
|
cdc7-1
|
||
|---|---|---|---|---|---|
| 23°C
|
26.5°C
|
23°C
|
26.5°C
|
||
| p306.10 | (1) | <0.5 | <0.5 | <0.5 | 5.2 |
| (early plasmid) | (2) | 0.8 | 2.0 | <0.5 | 3.6 |
| p12 | (1) | 0.6 | 1.9 | <0.5 | 6.5 |
| (late plasmid) | (2) | <0.5 | 0.6 | <0.5 | 4.7 |
Values are percentage of cells that lose plasmid each generation, determined by a comparative Southern blot analysis (see Materials and Methods). The top values (1) for each plasmid were obtained by comparison with an rDNA restriction fragment; the bottom values (2) were obtained by comparison with a chromosome XIV fragment.
Discussion
Separate activation by Cdc7 of early and late replication origins
Cdc7 kinase activity has been known to be required for the initiation of S phase, but was believed not to be necessary for its normal continuation. Identification of different temporal classes of replication origins, combined with the discovery that Dbf4 interacts physically with ARS1, led us to investigate a possible role of Cdc7 in origin activation throughout S phase. We wished to ascertain whether the effects of the Cdc7–Dbf4 complex are in fact mediated at the level of individual origins, and, if so, whether Cdc7 is required for activation of both early and late origins.
To examine the requirement for Cdc7 kinase activity during S phase, we have monitored replication origin use when cells attempt S phase under conditions where Cdc7 activity is limiting. The first series of experiments described here employed conditions in which cells were synchronized by the use of α-factor followed by arrest at the temperature-sensitive cdc7-1 block. The cultures were then placed at permissive temperature for between 3.5 and 7.5 min before return to restrictive temperature. It is not known whether Cdc7 kinase activity produced when cells are released from a cdc7-1 block represents new protein synthesis or reactivation (perhaps by refolding) of existing thermolabile protein molecules. Regardless of how Cdc7 activity is recovered after return to the permissive temperature, we can infer that the short release times used in these experiments produce a pulse of kinase activity that is limiting, as briefly released cells do not complete S phase normally, whereas cultures left at permissive temperature do. From both two-dimensional gel analysis and measurement of plasmid replication using dense isotope transfer experiments, the results described here show that under these conditions, early, but not late, replication origins are activated in most cells. A significant proportion of the duplication seen for normally late chromosomal sequences appears to be carried out by replication forks that originated at distant early origins.
Three models for the action of Cdc7 in promoting S phase can be proposed. Each makes predictions for the consequences of subjecting cells to a limiting pulse of Cdc7 activity. The first model suggests that Cdc7 kinase acts only as a global switch that triggers S phase, controlling its initiation at a stage upstream of the firing of individual replication origins. This suggestion is consistent with Hartwell’s observation, where he found that cells that have begun S phase can complete it even if returned to the restrictive temperature. If this model were correct, however, then under limiting conditions of Cdc7 activity, all cells that initially produced levels of Cdc7 sufficient to trigger S phase would also be able to complete it normally, and the use of both early- and late-activated origins would be expected to be equal. Contrary to this expectation, the data in Figures 2 and 5 show that most cells activate early, but not late, origins in response to the Cdc7 pulse. We cannot rule out a role for Cdc7 in the G1–S transition (independent of its action on individual origins), but our results do exclude the possibility that this first model illustrates the only mechanism of action of Cdc7 in promoting S phase.
The second model proposes that Cdc7 acts separately on individual origins, with each of the different temporal categories of origins having identical requirements for Cdc7 activity early in S phase. This idea is also consistent with Hartwell’s data, because by this model, both early and late replication origins could fulfill their requirement for Cdc7 for activation at the same early time (presumably the so-called CDC7 execution point; Culotti and Hartwell 1971). The predictions made by this second model are similar to those of the first model, because if all origins share the same Cdc7 requirement, both early and late origins would be expected to be activated to the same extent in response to a brief pulse of Cdc7 activity. As discussed above for the first model, however, the data are not consistent with this as the sole mode of action of Cdc7 kinase.
The third model also has Cdc7 acting separately on individual replication origins, but in this case, early- and late-activated origins are proposed to have either temporally or quantitatively distinct Cdc7 requirements. If this model were correct, then it should be possible to dissect the different Cdc7 requirements of early and late replication origins by subjecting the cells to limiting doses of the kinase activity. The data presented in this paper are more consistent with this third possibility than with either of the first two, as is most clearly shown by the plasmid replication data. As illustrated by Figure 4, plasmids are replicated to ∼80% during a “normal” S phase at 23°C. A short exposure to a limiting quantity of Cdc7 activity at the beginning of S phase causes activation of just over 70% of early p306.10 plasmid replication origins in the culture, but an identical pulse causes only 20% of late p12 plasmid origins to fire (Fig. 5). Additionally, our data indicate that under the same conditions, only early chromosomal origins fire, whereas late regions are passively duplicated. Therefore, the results presented here are best explained by proposing that at least one role of Cdc7 kinase is to promote initiation at individual replication origins, and that the requirements of early and late replication origins for Cdc7 kinase are either quantitatively or temporally different.
Cdc7 kinase is required throughout S phase for activation of replication origins
The results of the experiments in which cells were released briefly from a cdc7-1 block suggest that early- and late-activated origins differ in their Cdc7 requirements. We cannot, however, deduce from these experiments whether the difference is quantitative or temporal. For example, both early and late origins could require Cdc7 activity only at the start of S phase, with late origins requiring significantly higher levels for their eventual activation (a quantitative distinction). The brief interval at 23°C used to release from the cdc7-1 block may not have allowed kinase activity to build up to the levels required for late origin activation. An alternative possibility is that Cdc7 executes its function at each replication origin at the moment of activation, acting early during S phase on early-activated origins but at a later time on those origins that fire late. If this explanation were true, then the distinction in Cdc7 requirement between early and late origins would be temporal.
We addressed this issue by examining origin activation in asynchronous cultures grown at semi-permissive temperature for the cdc7-1 allele. Under such conditions, the available Cdc7 activity is assumed to be at a constantly limiting level throughout S phase; if late replication origins required higher levels of Cdc7 kinase for activation, the effects would be more severe for late origins. The data obtained from both two-dimensional gels and plasmid loss rate analysis gave no indication that semi-permissive temperature decreases firing of late origins more than early origins—both were reduced by increased temperatures, suggesting that the difference in requirement for Cdc7 is not quantitative. We infer that the distinction in the requirement for Cdc7 activity at early and late replication origins is likely to be temporal.
Data in the accompanying paper by Bousset and Diffley (1998) also favor a later requirement for Cdc7 activity at late origins. Their experiments make use of a hydroxyurea block to interrupt S phase, followed by release under restrictive conditions for cdc7. During this experiment, cells appear able to fire early origins but not late ones. Because cells presumably enter the hydroxyurea block with normal levels of Cdc7, these data imply that even normal levels of Cdc7 at the start of S phase are not adequate for late origin firing. Instead, these data concur with ours in suggesting that Cdc7 activity must also be present later in S phase (after release from the hydroxyurea block) for late origin activation.
The finding that Cdc7 is required later in S phase for late origin activation seems initially inconsistent with the original determination of an execution point for CDC7 at the start of S phase. In light of the results presented here, it seems possible that late replication origins were not activated as usual after the temperature shift in Hartwell’s experiment (Fig. 1B)—this possibility would be consistent with the somewhat slowed incorporation of radioactivity observed in his culture after shift to restrictive temperature (Hartwell 1973). The fact that Cdc7 was inferred to act only to initiate S phase highlights a complexity in interpreting the meaning of the “execution point” of a gene. In the case of CDC7, the gene product has completed its essential function soon after the start of S phase, at the time when enough replication origins have fired to allow the eventual completion of chromosome replication. Although its essential role is complete, Cdc7 is still used later during the normal S phase to fire late origins. The fact that Cdc7 is not essential throughout S phase presumably reflects the fact that firing of all the late origins is not absolutely required for completion of S phase.
The identity of the substrate phosphorylated by Cdc7 to cause origin activation remains unknown. However, a single amino acid change in the Cdc46/Mcm5 protein, can bypass the effects of cdc7 or dbf4 null mutations. This intriguing observation has led to the suggestion that the essential role of Cdc7 is to regulate Cdc46/Mcm5 function; one possibility is that Cdc46/Mcm5 poses a block to replication that is normally removed by Cdc7 kinase (Hardy et al. 1997). Cdc46/Mcm5 is a member of the Mcm/P1 family of six essential proteins (for review, see Chong et al. 1996; Kearsey et al. 1996; see also Hardy et al. 1997) that mediate the licensing of DNA necessary for its replication (Madine et al. 1995; Kubota et al. 1997; Thömmes et al. 1997). MCM family proteins appear to be present in all eukaryotes. Also, genes homologous to S. cerevisiae CDC7 have been identified in Schizosaccharomyces pombe, Xenopus, and human (Masai et al. 1995; Sato et al. 1997). The ubiquitous occurrence of Cdc7-related proteins, and of homologs of the CDC46 gene known to interact with CDC7, suggests that activities homologous to that of Cdc7 kinase may be generally important in controlling the S phase of eukaryotic cells.
Replication origins in Saccharomyces cerevisiae are programmed during the G1 phase of the cell cycle to be fired either early or late in the following S phase (Raghuraman et al. 1997). Origins so programmed for early or late activation must have molecular features that distinguish them. Whether Cdc7 kinase activates replication origins by regulating Cdc46/Mcm5 function or by another mechanism, the results presented here show that Cdc7–Dbf4 executes its function at different times during S phase, first at early origins and subsequently at late origins. We conclude that the Cdc7–Dbf4 kinase can directly or indirectly detect the molecular distinction between early and late origins, and bring about their activation at the appropriate time.
Materials and methods
Strains and plasmids
All strains were constructed from RM14-3a (MATa cdc7-1 bar1 ura3-52 trp1-289 leu2-3,112 his6). A CDC7 derivative of RM14-3a was made by transformation with a 2489-bp EcoRV fragment containing wild-type CDC7 sequence [obtained by digesting pRS277 (Sclafani and Fangman 1986), a gift from Robert Sclafani], followed by selection for growth at 37°C (restrictive temperature for cdc7-1).
Both p12 and p306.10 are URA3 CEN5 plasmids containing inserts at the BamHI site of vector YIp5-5 (Ferguson et al. 1991). Plasmid p12 (a gift from John Diller) has been described (Friedman et al. 1996). Plasmid p306.10 was obtained by screening a library of yeast inserts (Ferguson et al. 1991) with the 1391-bp EcoRI subfragment of the chromosome III BamHI fragment C1G (Reynolds et al. 1989). p306.10 contains a 13.3-kb insert that extends from nucleotide number 69,586 to 82,849 of the chromosome III sequence as given in the S. cerevisiae Genome Database, oriented with the 82,849 end of the insert lying closer to the YIp5-5 centromere. ARS306 lies between nucleotides 74,102 and 74,642 of the chromosome III sequence (Zhu et al. 1992).
Cells were grown in standard synthetic complete medium or selective medium as appropriate.
Temperature shifts
The temperature of cultures was shifted rapidly from 37°C to 23°C or vice versa either by swirling the flask in an iced or 55°C water bath, or, for larger culture volumes, by addition of a predetermined volume of warmed or chilled medium. Attainment of the target temperature generally took between 1 and 2 min. The time of transfer was taken to be at the beginning of the shift procedure.
Two-dimensional agarose gel electrophoresis
DNA was prepared from either pooled S-phase cells (Fig. 2) or asynchronously growing cultures (Fig. 6) as described previously (Huberman et al. 1987; Brewer et al. 1992). Two-dimensional gels were run as described (Friedman and Brewer 1995). First dimension gels were 0.4% agarose. Second dimension gels were either 1% or 1.1% agarose, depending on the size of the fragment to be examined. Fragments probed were ARS306, 5.405-kb NcoI; ARS1412, 2.574-kb MscI; 3LT, 4.877-kb NcoI; ARS607, 2.764-kb BclI; ARS603, 2.793-kb PstI (Fig. 2 and data not shown) and ARS306, 4.223-kb ClaI; ARS1412, 2.574-kb MscI (Fig. 6). All restriction digests (including digests using BclI) were carried out at 37°C. Probe fragments were prepared by appropriate digestion and gel purification of fragments from plasmids p306.10, p12, or p78-4.4 (McCarroll and Fangman 1988) or were PCR-amplified fragments [gifts from Katherine Friedman (Friedman et al. 1997)].
Dense isotope transfer experiments
Dense isotope transfer experiments were carried out using an adaptation of the synchronous density transfer procedure described previously (McCarroll and Fangman 1988). Cells were grown for at least seven generations in minimal medium containing 0.1% [13C]glucose and 0.01% [15N]ammonium sulfate and arrested by incubation with 200 nm α-factor. Cultures were shifted to 37°C, and pronase was added to degrade the α-factor, allowing cells to progress to the cdc7 block. Cultures were transferred rapidly to 23°C (see above) and held at 23°C for between 3 and 7.5 min before rapid transfer to 37°C. Samples were collected at appropriate times after the initial transfer to 23°C, and processed for CsCl density gradient centrifugation (McCarroll and Fangman 1988). Chromosomal EcoRI fragments probed were as described previously (McCarroll and Fangman 1988; Friedman et al. 1996). Replication levels of plasmids (Fig. 5; Table 2) were determined using a probe unique to the vector sequences. Replication levels were quantitated by analysis on an InstantImager (Packard).
Determination of plasmid loss rates
Loss rates of plasmids p306.10 and p12 were measured by a comparative Southern blot method (Brewer and Fangman 1994). Log phase cultures containing either plasmid were transferred from selective to non-selective medium and grown for an additional 24 hr. A sample of the culture was harvested for DNA isolation. The remaining culture was split and diluted 200×. Half the culture was maintained at 23°C, whereas the other half was transferred to 26.5°C. DNA was isolated from samples of each culture taken at approximately seven doublings after the temperature split. Growth rate was followed by measuring cell density, and DNA was prepared by the smash and grab procedure of Rose et al. (1990). Aliquots of DNA were digested with NcoI, and subjected to Southern blot analysis. Relative maintenance levels of the plasmids after transfer to nonselective conditions were estimated by comparing changes in relative intensity of Southern blot signal from the plasmid with the signals produced by restriction fragments on chromosome XIV or at the rDNA locus. Southern blots were quantitated using an InstantImager (Packard).
Acknowledgments
We thank Kristine Bousset and John Diffley for sharing data before publication. Thanks to John Diller, Carol Newlon, and M.K. Raghuraman for helpful discussions, and to Léon Dirick, R. Scott Hansen, Katherine Kolor, and M.K. Raghuraman for comments on the manuscript. Katherine Friedman and John Diller generously allowed us to show previously published data. We acknowledge gifts of plasmids and probes from John Diller, Katherine Friedman, and Robert Sclafani. A.D.D. was the recipient of an SERC/NATO postdoctoral fellowship. This work was supported by National Institutes of General Medical Sciences grant 18926 to W.L.F. and B.J.B.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL bbrewer@genetics.washington.edu; FAX (206) 543-0754.
References
- Bahman M, Buck V, White A, Rosamond J. Characterisation of the CDC7 gene product of Saccharomyces cerevisiae as a protein kinase needed for the initiation of mitotic DNA synthesis. Biochim Biophys Acta. 1988;951:335–343. doi: 10.1016/0167-4781(88)90104-2. [DOI] [PubMed] [Google Scholar]
- Bousset, K. and J.F.X. Diffley. 1998. The Cdc7 protein kinase is required for origin firing during S phase. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- ————— Mapping replication origins in yeast chromosomes. BioEssays. 1991;13:317–322. doi: 10.1002/bies.950130702. [DOI] [PubMed] [Google Scholar]
- ————— Initiation preference at a yeast origin of replication. Proc Natl Acad Sci. 1994;91:3418–3422. doi: 10.1073/pnas.91.8.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Lockshon D, Fangman WL. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992;71:267–276. doi: 10.1016/0092-8674(92)90355-g. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Diller JD, Friedman KL, Kolor KM, Raghuraman MK, Fangman WL. The topography of chromosome replication in yeast. Cold Spring Harbor Symp Quant Biol. 1993;58:425–344. doi: 10.1101/sqb.1993.058.01.049. [DOI] [PubMed] [Google Scholar]
- Chapman JW, Johnston LH. The yeast gene, DBF4, essential for entry into S phase is cell cycle regulated. Exp Cell Res. 1989;180:419–428. doi: 10.1016/0014-4827(89)90068-2. [DOI] [PubMed] [Google Scholar]
- Chong JP, Thömmes P, Blow JJ. The role of MCM/P1 proteins in the licensing of DNA replication. Trends Biochem Sci. 1996;21:102–106. [PubMed] [Google Scholar]
- Culotti J, Hartwell LH. Genetic control of the cell division cycle in yeast. III. Seven genes controlling nuclear division. Exp Cell Res. 1971;67:389–401. doi: 10.1016/0014-4827(71)90424-1. [DOI] [PubMed] [Google Scholar]
- Dowell SJ, Romanowski P, Diffley JF. Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science. 1994;265:1243–1246. doi: 10.1126/science.8066465. [DOI] [PubMed] [Google Scholar]
- Ferguson BM, Brewer BJ, Reynolds AE, Fangman WL. A yeast origin of replication is activated late in S phase. Cell. 1991;65:507–515. doi: 10.1016/0092-8674(91)90468-e. [DOI] [PubMed] [Google Scholar]
- Friedman KL, Brewer BJ. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- Friedman KL, Diller JD, Ferguson BM, Nyland SVM, Brewer BJ, Fangman WL. Multiple determinants controlling activation of yeast replication origins late in S phase. Genes & Dev. 1996;10:1595–1607. doi: 10.1101/gad.10.13.1595. [DOI] [PubMed] [Google Scholar]
- Friedman KL, Brewer BJ, Fangman WL. Replication profile of Saccharomyces cerevisiae chromosome VI. Genes to Cells. 1997;2:667–678. doi: 10.1046/j.1365-2443.1997.1520350.x. [DOI] [PubMed] [Google Scholar]
- Hardy CF, Dryga O, Seematter S, Pahl PM, Sclafani RA. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH. Three additional genes required for deoxyribonucleic acid synthesis in Saccharomyces cerevisiae. J Bacteriol. 1973;115:966–974. doi: 10.1128/jb.115.3.966-974.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J Mol Biol. 1976;104:803–817. doi: 10.1016/0022-2836(76)90183-2. [DOI] [PubMed] [Google Scholar]
- Hollingsworth REJ, Sclafani RA. DNA metabolism gene CDC7 from yeast encodes a serine (threonine) protein kinase. Proc Natl Acad Sci. 1990;87:6272–6276. doi: 10.1073/pnas.87.16.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao CL, Carbon J. High-frequency transformation of yeast by plasmids containing the cloned yeast ARG4 gene. Proc Natl Acad Sci. 1979;76:3829–3833. doi: 10.1073/pnas.76.8.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman JA, Spotila LD, Nawotka KA, El-Assouli SM, Davis LR. The in vivo replication origin of the yeast 2 micron plasmid. Cell. 1987;51:473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- Huberman JA, Zhu JG, Davis LR, Newlon CS. Close association of a DNA replication origin and an ARS element on chromosome III of the yeast, Saccharomyces cerevisiae. Nucleic Acids Res. 1988;16:6373–6384. doi: 10.1093/nar/16.14.6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Pahl PM, Harrison K, Rosamond J, Sclafani RA. Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol Cell Biol. 1993;13:2899–2908. doi: 10.1128/mcb.13.5.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey SE, Maiorano D, Holmes EC, Todorov IT. The role of MCM proteins in the cell cycle control of genome duplication. BioEssays. 1996;18:183–190. doi: 10.1002/bies.950180305. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Mimura S, Nishimoto S, Masuda T, Nojima H, Takisawa H. Licensing of DNA replication by a multi-protein complex of MCM/P1 proteins in Xenopus eggs. EMBO J. 1997;16:3320–3331. doi: 10.1093/emboj/16.11.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madine MA, Khoo CY, Mills AD, Musahl C, Laskey RA. The nuclear envelope prevents reinitiation of replication by regulating the binding of MCM3 to chromatin in Xenopus egg extracts. Curr Biol. 1995;5:1270–1279. doi: 10.1016/s0960-9822(95)00253-3. [DOI] [PubMed] [Google Scholar]
- Masai H, Miyake T, Arai K. hsk1+, a Schizosaccharomyces pombe gene related to Saccharomyces cerevisiae CDC7, is required for chromosomal replication. EMBO J. 1995;14:3094–3104. doi: 10.1002/j.1460-2075.1995.tb07312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll RM, Fangman WL. Time of replication of yeast centromeres and telomeres. Cell. 1988;54:505–513. doi: 10.1016/0092-8674(88)90072-4. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Viewpoint: Putting the cell cycle in order. Science. 1996;274:1643–1645. doi: 10.1126/science.274.5293.1643. [DOI] [PubMed] [Google Scholar]
- Newlon CS, Theis JF. The structure and function of yeast ARS elements. Curr Opin Genet Dev. 1993;3:752–758. doi: 10.1016/s0959-437x(05)80094-2. [DOI] [PubMed] [Google Scholar]
- Newlon CS, Collins I, Dershowitz A, Deshpande AM, Greenfeder SA, Ong LY, Theis JF. Analysis of replication origin function on chromosome III of Saccharomyces cerevisiae. Cold Spring Harbor Symp Quant Biol. 1993;58:415–423. doi: 10.1101/sqb.1993.058.01.048. [DOI] [PubMed] [Google Scholar]
- Raghuraman MK, Brewer BJ, Fangman WL. Cell cycle-dependent establishment of a late replication program. Science. 1997;276:806–809. doi: 10.1126/science.276.5313.806. [DOI] [PubMed] [Google Scholar]
- Reynolds AE, McCarroll RM, Newlon CS, Fangman WL. Time of replication of ARS elements along yeast chromosome III. Mol Cell Biol. 1989;9:4488–4494. doi: 10.1128/mcb.9.10.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics: A laboratory course manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Sato N, Arai K, Masai H. Human and Xenopus cDNAs encoding budding yeast Cdc7-related kinases: In vitro phosphorylation of MCM subunits by a putative human homologue of Cdc7. EMBO J. 1997;16:4340–4351. doi: 10.1093/emboj/16.14.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani RA, Fangman WL. Thymidine utilization by tut mutations and facile cloning of mutant alleles by plasmid conversion in S. cerevisiae. Genetics. 1986;114:753–767. doi: 10.1093/genetics/114.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani RA, Jackson AL. Cdc7 protein kinase for DNA metabolism comes of age. Mol Microbiol. 1994;11:805–810. doi: 10.1111/j.1365-2958.1994.tb00358.x. [DOI] [PubMed] [Google Scholar]
- Sclafani RA, Patterson M, Rosamond J, Fangman WL. Differential regulation of the yeast CDC7 gene during mitosis and meiosis. Mol Cell Biol. 1988;8:293–300. doi: 10.1128/mcb.8.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb DT, Struhl K, Davis RW. Isolation and characterization of a yeast chromosomal replicator. Nature. 1979;282:39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- Thömmes P, Kubota Y, Takisawa H, Blow JJ. The RLF-M component of the replication licensing system forms complexes containing all six MCM/P1 polypeptides. EMBO J. 1997;16:3312–3319. doi: 10.1093/emboj/16.11.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Hori Y, Shinomiya T, Obuse C, Tsurimoto T, Yoshikawa H, Shirahige K. The efficiency and timing of initiation of replication of multiple replicons of the Saccharomyces cerevisiae chromosome VI. Genes Cells. 1997;2:655–666. doi: 10.1046/j.1365-2443.1997.1530351.x. [DOI] [PubMed] [Google Scholar]
- Yoon HJ, Campbell JL. The CDC7 protein of Saccharomyces cerevisiae is a phosphoprotein that contains protein kinase activity. Proc Natl Acad Sci. 1991;88:3574–3578. doi: 10.1073/pnas.88.9.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Newlon CS, Huberman JA. Localization of a DNA replication origin and termination zone on chromosome III of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4733–4741. doi: 10.1128/mcb.12.10.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]