Abstract
A diverse set of 24 novel phages infecting the fire blight pathogen Erwinia amylovora was isolated from fruit production environments in Switzerland. Based on initial screening, four phages (L1, M7, S6, and Y2) with broad host ranges were selected for detailed characterization and genome sequencing. Phage L1 is a member of the Podoviridae, with a 39.3-kbp genome featuring invariable genome ends with direct terminal repeats. Phage S6, another podovirus, was also found to possess direct terminal repeats but has a larger genome (74.7 kbp), and the virus particle exhibits a complex tail fiber structure. Phages M7 and Y2 both belong to the Myoviridae family and feature long, contractile tails and genomes of 84.7 kbp (M7) and 56.6 kbp (Y2), respectively, with direct terminal repeats. The architecture of all four phage genomes is typical for tailed phages, i.e., organized into function-specific gene clusters. All four phages completely lack genes or functions associated with lysogeny control, which correlates well with their broad host ranges and indicates strictly lytic (virulent) lifestyles without the possibility for host lysogenization. Comparative genomics revealed that M7 is similar to E. amylovora virus ΦEa21-4, whereas L1, S6, and Y2 are unrelated to any other E. amylovora phage. Instead, they feature similarities to enterobacterial viruses T7, N4, and ΦEcoM-GJ1. In a series of laboratory experiments, we provide proof of concept that specific two-phage cocktails offer the potential for biocontrol of the pathogen.
INTRODUCTION
Fire blight is a devastating plant disease caused by the Gram-negative bacterium Erwinia amylovora. The disease affects species of the Rosaceae family, mainly plants belonging to the pome fruit subfamily Maloideae, e.g., apple, pear, and quince (39, 56). After infection (e.g., through the nectarthodes or small lesions), bacterial multiplication leads to the collapse of the parenchyma and to the migration of the bacteria into the plant tissue, causing the typical symptoms of fire blight: necrosis, wilting, and ooze production (58). Depending upon the climatic conditions during the flowering period, fire blight outbreaks can destroy entire trees and orchards within a single season. Since its first observation in the 1780s, fire blight has spread within North America, Central Europe, the Middle East, and New Zealand. In affected countries, it is the major threat to sustainable pome fruit production, resulting in severe economic losses (8) through reduced yield, trade barriers, tree and orchard decimation, sanitation labor, and eradication regulations (16). Since the late 1950s until today, the primary and most reliable control option against fire blight is the application of antibiotics (e.g., streptomycin, oxytetracycline, and gentamicin). However, antibiotic use involves practical drawbacks in terms of pathogen resistance development (28) and regulatory restrictions based upon public health risks (46). Therefore, alternatives, like biological control methods, are in urgent demand from conventional, integrated, and organic growers (26, 27).
Biological control with bacteriophages (phages) features several advantages. Phages are considered natural and ubiquitously distributed, and they represent the most abundant group of biological entities in our environment (9). Their isolation, production, and storage are relatively simple and inexpensive. In general, phages infect only a specific host group, leaving other resident bacteria unharmed. The strictly virulent, nontransducing phages are regarded as environmentally safe tools for the control of bacterial pathogens (22). The potential of phages for the biocontrol of E. amylovora and other plant pathogens (e.g., Pectobacterium carotovora, Xanthomonas campestris pv. vesicatoria, and Agrobacterium tumefaciens) has been demonstrated (4, 6, 20, 21, 45, 49, 53). Balogh et al. (5) offer a thorough review of the history, success, and unique considerations for phage therapy in plant agriculture.
The first E. amylovora phages were described in the 1970s (18, 47). Later on, several E. amylovora-infecting phages were isolated and classified into different subtypes based on plaque morphology, host range analysis, electron microscopy, restriction fragment length polymorphism (RFLP), and PCR (21, 50). The current sequence information is limited, as the genome sequences of phages of only two types are available: the podovirus ERA103 (NC_009014) and its relatives ΦEa1h and ΦEa100 (40) and the Felix O1-like myovirus ΦEa21-4 (35) and its close relative ΦEa104 (40).
Major challenges in phage therapy for plant disease control are the emergence of resistant strains and environmental factors that would rapidly reduce the number of infective phages, particularly desiccation and UV destruction (5). The phage persistence in the phyllosphere can be improved by appropriate formulations (6) and by avoiding exposure to sunlight. The probability of the development of phage-resistant strains can be reduced by the application of cocktails consisting of phages featuring different replication pathways. Therefore, a thorough characterization of the phages, including sequencing of the complete genomes, is required if they are intended to be used as biocontrol agents.
We report here the isolation of a set of E. amylovora-infecting phages and the complete sequencing and molecular analysis of the phages L1, M7, S6, and Y2. Moreover, we demonstrate that specific two-phage cocktails are well suited to control E. amylovora in vitro. The data presented yield novel insights regarding the molecular biology of E. amylovora phages and provide a sound basis for their development as fire blight biocontrol agents.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strains of Erwinia spp. and Pantoea spp. were grown in Luria Bertani (LB) broth at 30°C or at 27°C when grown on plates. Escherichia coli strain XL1-Blue MRF′ (Agilent Technologies, Santa Clara, CA) was cultivated in LB at 37°C.
Phage isolation and propagation.
Soil samples were collected from 32 locations throughout Switzerland between the years 2007 and 2009. Locations that had orchard and old-growth landscape apple or pear trees and that had a recorded recent, a historical, or no fire blight outbreak were chosen. Phages were isolated and enriched by a standard method. Briefly, 190 ml of SM-buffered (50 mM Tris, 100 mM NaCl, 8 mM MgSO4, pH 7.4) LB broth was inoculated with 10 ml of an overnight culture of one of several different E. amylovora host strains. The use of various bacterial strains increases the probability of the isolation of diverse phage types (21). Ten grams of soil was added and incubated under constant agitation at 30°C. After 18 h, soil and bacterial host cells were pelletized by centrifugation (1 min, 10,000 × g), and the supernatant was filtered (0.2-μm-pore-size filter; Sarstedt, Nümbrecht, Germany). Phage presence was determined using the soft-agar overlay method (2) by spotting 10 μl of the supernatant onto LB+ soft agar (LB containing 1% [wt/vol] glucose, 2 mM MgSO4, and 0.4% [wt/vol] agar) inoculated with freshly growing E. amylovora. Phages were isolated by picking individual plaques using a Pasteur pipette and resuspended in 250 μl SM buffer. This step was repeated at least twice to ensure the purity of the phage isolate. To propagate each phage, 40 soft-agar overlay plates with semiconfluent phage lysis were produced. Phages were recovered from the plates by the addition of SM buffer to the top surface (5 ml per plate). Then, the plates were gently agitated at room temperature (RT) (20 rpm, 6 h; Heidolph Polymax 1040), and the buffer liquid was harvested. Phages were precipitated using polyethylene glycol (10% [wt/vol] PEG 8000, 1 M NaCl; 4°C, overnight) and pelletized (10 min, 10,000 × g), and the pellet was resuspended in 5 ml SM buffer. Phages were purified using CsCl gradient centrifugation (48) for subsequent electron microscopic analysis and DNA extraction.
Phage nomenclature.
For the submission of the genome sequences to GenBank, we named the phages according to the current bacterial virus nomenclature to ensure a clear identification (e.g., L1 is vB_EamP-L1, with vB as the virus of bacteria; Eam as E. amylovora, the host species; P as Podoviridae, the virus family; followed by L1, the laboratory name) (33). For reasons of simplicity, only the laboratory names are used in this paper.
Host range analysis.
Using the soft-agar overlay method (2), all phage isolates were tested for their abilities to infect a collection of E. amylovora strains and closely related species. The occurrence of single plaques in the soft-agar overlay was determined after overnight incubation at 27°C.
Phage DNA extraction and analysis.
CsCl-purified virus particles were dialyzed against a 1,000-fold excess of SM buffer (6 h, RT) and afterwards treated for 1 h at 56°C with EDTA (pH 8.0, 20 mM), proteinase K (50 μg/ml), and SDS (0.5% [wt/vol]). DNA was isolated and purified using a phenol-chloroform extraction procedure as described previously (32, 48). The phage whole-genome sizes were determined by pulsed-field gel electrophoresis (PFGE) as described earlier (30) using a Chef DR III apparatus (Bio-Rad, Reinach, Switzerland).
TEM.
For transmission electron microscopy (TEM) imaging, CsCl-purified phages were negatively stained with 2% (wt/vol) uranyl acetate, 2% (wt/vol) ammonium molybdate, or 2% (wt/vol) Na-phosphotungstic acid and then analyzed as previously described (32). Actual magnifications were determined using cross grating replica or catalase crystals (Electron Microscopy Sciences, Hatfield, PA) (60).
Genome sequencing and bioinformatic analysis.
The genomes of L1, S6, and Y2 were sequenced using a shotgun approach. Genomic libraries of L1 were constructed by cloning TasI- and MspI-digested DNA into EcoRI- and ClaI-linearized cloning vector pBluescript II SK(−) (Agilent Technologies), respectively. A third library was generated by sonication. DNA was sheared, followed by gel extraction of fragments of 1.0 to 2.5 kbp in size. The fragments were end repaired (End-It DNA end repair kit; Epicentre Biotechnologies, Madison, WI) and inserted into EcoRV-linearized pBluescript II SK(−). The libraries of S6 and Y2 genomic DNA were generated after sonication only. The vectors were electroporated (2.5 kV, 25 μF, 200 Ω; GenePulser; Bio-Rad, Hercules, CA) into electrocompetent E. coli XL1-Blue MRF′ cells and incubated on LB plates containing ampicillin (100 μg/ml) and tetracycline (10 μg/ml) at 37°C. The addition of 80 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and 20 mM IPTG (isopropyl-β-d-1-thiogalactopyranoside) to the plates enabled the identification of insert-bearing clones by blue/white screening. Constructs containing inserts of 1.0 to 2.5 kbp in size were selected for sequencing using the ABI 3730xl DNA analyzer (Sanger Technology). Sequences were assembled using the ContigExpress module of Vector NTI Advance 10 (Invitrogen, Paisley, United Kingdom) with default parameters and CLC Main Workbench 5 (CLC Bio, Aarhus, Denmark). Gaps were closed by primer walking using phage DNA as the template. Every position was sequenced at least twice. Phage M7 was sequenced using the Roche GS FLX system with Titanium reagents and the Roche GS Assembler 2.3 software (default settings) for sequence assembly.
Open reading frames (ORFs) of all phage genomes were predicted using CLC Main Workbench 5 and Vector NTI software; the minimal ORF length was set to 30 codons, with ATG, GTG, or TTG as start codons and TAA, TGA, or TAG as stop codons. Putative genes were annotated based on similarities to database entries and the presence of a putative Shine-Dalgarno sequence (ribosome binding site) similar to the consensus sequence of E. coli GGA GGT (51). Translated amino acid sequences were then compared to the nonredundant GenBank protein database, using BlastP (3). Pairwise alignments were performed with CLC Main Workbench 5.
Peptide mass fingerprinting.
Structural proteins were separated by horizontal SDS-PAGE as outlined previously (15). Distinct bands were excised from the gels, and the polypeptides were analyzed using liquid chromatography/tandem mass spectrometry (LC/MS-MS) as described earlier (32, 62). The identified proteins were compared to all possible ORFs >60 nucleotides (nt) in size and summarized using Scaffold 2 Proteome Software (Proteome Software Inc., Portland, OR).
Determination of genome end structures.
Physical structures of the genome ends were determined as described elsewhere (32, 37). Briefly, phage DNA was treated with BAL31 nuclease (NEB, Ipswich, MA) at 30°C for different incubation times prior to digestions with selected restriction enzymes. For the identification of potential cohesive ends (cos), half of the restriction-digested phage DNA was incubated at 62°C for 10 min to separate potentially ligated cos sites and immediately stored on ice. The RFLP patterns were then analyzed for changes. Based on RFLP and BAL31 experiments, the genome ends could roughly be localized. Primers were designed that annealed approximately 300 bp apart from the expected ends to initiate sequencing toward the putative ends. An abrupt drop-off in the sequencing signal intensity indicated the physical end of the phage genomic DNA molecule. The experimental data were verified by phylogenetic analysis of the large terminase proteins (11). A tree was constructed by applying the neighbor-joining method (1,000 bootstrap replicates) using alignments generated with the following parameters: gap open cost, 10; gap extension cost, 1; end gap cost, free (CLC Main Workbench 5).
In vitro activity.
The efficacies of the strictly virulent phages L1, M7, S6, and Y2 to control E. amylovora were tested in vitro. Strain CFBP 1430 (105 CFU/ml) was infected with 108 PFU/ml in 4 ml LB broth. The efficacies of two-phage cocktails were determined by mixing equal amounts of individual phages to reach a total concentration of 108 PFU/ml (i.e., 5 × 107 PFU/ml of each phage). The phage-bacterium mixtures were incubated at 30°C under constant shaking. Suspensions of bacteria and phage were dilution plated immediately after phage addition (0 h) and after 1, 2, 4, 6, 8, and 24 h of incubation to determine the CFU/ml of surviving bacteria. The plates were incubated at 27°C until quantification of viable bacteria. Each experiment consisted of three replicates per treatment and was carried out at least three times.
Nucleotide sequence accession numbers.
Complete nucleotide sequences of L1 (HQ728265), M7 (HQ728263), S6 (HQ728266), and Y2 (HQ728264) were deposited in GenBank under the accession numbers listed in parentheses.
RESULTS
Novel E. amylovora phages feature unique characteristics.
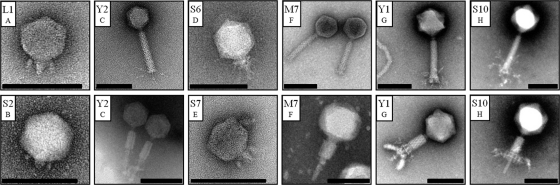
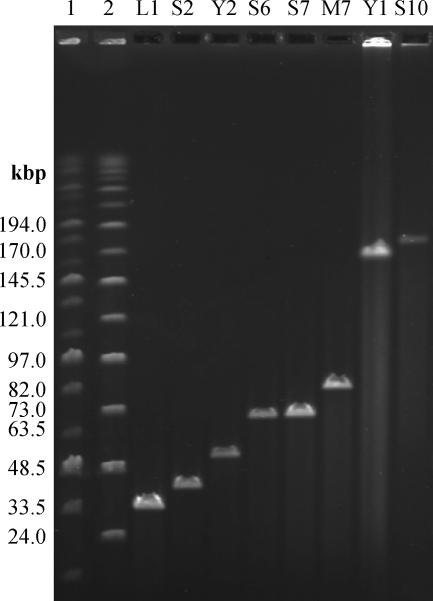
Preliminary results from plaque morphology, PFGE, RFLP, and host range analysis indicated that the 24 phage isolates can be grouped into eight subtypes (A to H). From each type, a single representative was selected for further examination by electron microscopy. They belong to the Myoviridae or Podoviridae family, having either A1 or C1 morphotypes, respectively (1). Their capsid diameters range from 58 to 95 nm (Fig. 1). PFGE indicated phage genomes of 39 to 180 kbp in size (Fig. 2). Most phages isolated in this study are novel representatives of phages infecting E. amylovora and feature unique characteristics. Only two subgroups are related to the previously described North American isolates (18, 21, 47, 50). Group F phages seem to be similar to group 1 viruses described by Gill et al. (21). This could later be confirmed by sequencing of phage M7 (see below). Basic characterization of group B phages indicated that they resemble phage ΦEa1 (group 3) (21, 47, 50).
Fig. 1.
Electron micrographs of negatively stained Erwinia amylovora phages. Phages were classified into the Podoviridae (L1, S2, S6, and S7) and Myoviridae (Y2, M7, Y1, and S10) families. From the latter, virions with both contracted and uncontracted tails are shown. Letters A to H indicate phage subtypes. Scale bars represent 100 nm.
Fig. 2.
Pulsed-field gel electrophoresis analysis of full-length Erwinia amylovora phage genomes (as indicated). Lanes 1 and 2 include MidRange PFGE markers I and II (NEB), respectively. Sizes are as indicated.
Broad host range is a common feature of E. amylovora phages.
Most phages investigated here were able to infect a majority of tested hosts (Table 1). Phages Y2 and M7 successfully infected all E. amylovora strains. Phage Y2 was highly specific toward E. amylovora, while other phages also lysed closely related bacteria such as Pantoea agglomerans or Erwinia billingiae (Table 1).
Table 1.
Host ranges of eight Erwinia amylovora phages isolated on different hosts
| Species | Host strain | Infectivity of phage (phage group)a: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| L1 (A) | S2 (B) | Y2 (C) | S6 (D) | S7 (E) | M7 (F) | Y1 (G) | S10 (H) | ||
| E. amylovora | CFBP1430 | +* | +* | + | + | + | + | − | +* |
| CFBP1232 | + | + | + | + | + | + | − | + | |
| Ea153 | + | + | + | + | + | + | − | + | |
| Ea1/74 | + | + | + | + | + | + | − | + | |
| Ea1/79 | + | + | + | + | + | + | − | + | |
| ACW38899 | + | + | + | + | + | + | − | + | |
| ACW56400 | + | + | + | +* | +* | + | + | + | |
| ACW44274 | + | + | + | + | + | + | − | + | |
| ACW42912 | + | + | + | + | + | + | − | + | |
| ACW30560 | + | + | + | + | + | + | − | + | |
| Ea Rac 3075 | + | + | + | + | + | + | − | + | |
| ACW55500 | + | + | + | + | + | + | − | + | |
| ACW55835 | + | + | + | + | + | + | − | − | |
| ACW55955 | + | + | + | + | + | + | − | + | |
| Ea4/82 | − | − | +* | + | + | + | − | − | |
| IPV 1077/7 | + | + | + | + | + | + | + | + | |
| IPV-BD 5357 | + | + | + | + | + | + | +* | + | |
| 01SFR-BO | + | − | + | + | + | + | − | − | |
| OMPBO 691.2 | + | + | + | + | + | + | + | − | |
| LA 469 | − | + | + | + | + | + | − | − | |
| LA 071 | − | − | + | + | + | + | − | − | |
| LA 411 | + | + | + | + | + | + | − | + | |
| LA 468 | + | + | + | + | + | + | − | − | |
| LA 477 | − | − | + | − | − | +* | − | − | |
| Erwinia persicina | ACW40943 | − | − | − | − | − | + | − | − |
| ACW40560x | − | − | − | − | − | − | − | − | |
| ACW41072 | − | − | − | − | − | − | − | − | |
| Erwinia billingiae | B90 | − | − | − | − | + | + | − | − |
| Pantoea agglomerans | Eh 42 | − | − | − | − | − | + | − | − |
| Em 406 | − | − | − | − | + | − | − | − | |
| Em 283 | + | + | − | − | + | − | − | − | |
| Pantoea vagans | C9-1 | − | − | − | − | + | − | − | − |
| Pantoea ananatis | 351 Lys | + | + | − | − | + | − | − | + |
+, sensitive; −, no infection. Isolation strains are highlighted with asterisks. Phage groups are given in parentheses.
Genome analyses indicated relatedness to Enterobacteriaceae phages.
Four broad-host-range phages (L1, S6, Y2, and M7) were chosen for complete genome sequencing. L1 formed distinct plaques surrounded by secondary haloes in soft-agar overlays, suggesting a diffusible enzyme activity. Y2 was selected because of its species specificity, M7 because it infected every E. amylovora strain as well as strains of closely related species, and S6 because of its different morphological properties.
The average sequence coverages of the genomes were 4.8-fold (L1), 3.3-fold (S6), 4.7-fold (Y2), and 130-fold (M7). The assemblies of the phage genomes from individual reads yielded unique unit genomes for all four phages. In silico predictions were confirmed by restriction enzyme analysis. Table 2 summarizes the major characteristics for the phages sequenced in this study, including data for ΦEa21-4 and ERA103. Additional information on features and database matches of predicted proteins encoded by L1, M7, S6, and Y2 can be found in the supplemental material.
Table 2.
Major characteristics of Erwinia amylovora phages
| Characteristic | L1 | S6 | Y2 | M7 | ΦEa21-4 | ERA103 |
|---|---|---|---|---|---|---|
| Virus familya | C | C | A | A | A | C |
| Lifestyle | Virulent | Virulent | Virulent | Virulent | Virulent | Virulent |
| Capsid diameter (nm) | 58 | 66 | 67 | 77 | 60 | NDc |
| Tail length (nm) | 124 | 116 | 90 | |||
| Genome size (kbp) | 39.3 | 74.7 | 56.6 | 84.7 | 84.6 | 45.4 |
| Genome structureb | DTR | DTR | DTR | DTR | DTR | DTR |
| G+C content (mol%) | 51.9 | 52.1 | 44.2 | 43.4 | 43.8 | 49.8 |
| No. of predicted ORFs/no. of ORFs with function assigned | 49/27 | 115/18 | 90/14 | 117/30 | 117/23 | 53/29 |
| Reference | This study | This study | This study | This study | 35 | NC_009014 |
A, Myoviridae; C, Podoviridae (according to Ackermann [1]).
DTR, direct terminal repeats.
ND, not determined.
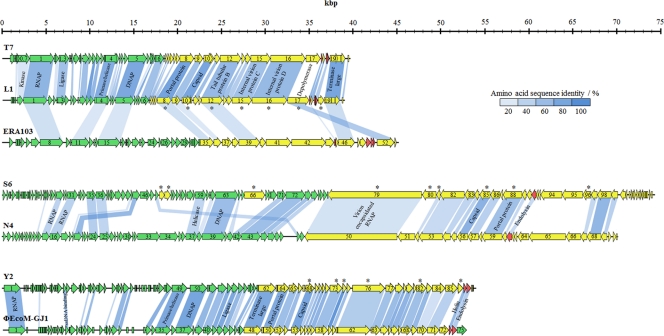
The genome of phage L1 has a length of 39,282 bp with direct terminal repeats of 172 bp. Forty-nine putative open reading frames were annotated based on similarities to proteins in the database as well as on the presence of a potential ribosome binding site preceding the start codon. The putative functions of the gene products (gp) could be grouped into early and late gene clusters (Fig. 3). The early gene cluster comprises genes that are involved at the beginning of the infection process in host takeover, DNA replication, modification, and translation. The late gene cluster is composed of structural, DNA assembly, and cell lysis proteins. The lack of a lysogeny control region indicates that it also is a virulent phage. L1 is a member of the T7-like phages: the genomes of L1 and T7 share a 61.2% nucleotide identity over their full lengths and feature the same architectures, with function-specific clusters and transcription exclusively from left to right. Out of 49 putative gene products, 24 share overall sequence identities of more than 50% with T7 proteins (17). Only 10 gene products have identities below 20% (Fig. 3). Further common features are the putative internal in-frame start in orf04 (primase/helicase) at nucleotide position 10676 of L1 producing gp4B (helicase domain) and a potential frameshift in the gene encoding the major capsid protein of L1 (gp10A). A putative frameshift site was identified at nucleotide positions 21483 to 21488 (GUU UUC), where the phenylalanine-tRNA can also pair in the −1 frame. The stop codon of the −1 frame is located 145 nt downstream of the frameshift site and extends gp10A by an additional 43 amino acids, generating the minor capsid protein (gp10B). In T7, the frameshift also consists of overlapping phenylalanine codons (13, 38). A potential transcriptional terminator is located approximately 200 nt downstream of the frameshift site in both phages.
Fig. 3.
Genome maps of phages L1, S6, and Y2 and alignment with related phages. Genes were classified into functional clusters based on database comparisons, peptide fingerprinting, and their locations on the genome. They are represented in green (early genes), yellow (late genes), and red (lysis genes). Shading indicates the degree of amino acid sequence identity of gene products with an identity of >20%. Putative functions of selected genes are indicated. Gene products that were identified using mass spectrometry are marked with an asterisk.
Phage S6 has a genome size of 74,669 bp, including direct terminal repeats of 397 bp. A function was assigned to 18 out of 115 putative gene products (Table 2). In contrast to phage L1, the early and late genes are oriented in opposite directions (Fig. 3). The first gene in the late gene cluster encodes a putative virion-encapsidated RNA polymerase (vRNAP). This genome organization resembles those of E. coli phage N4 (19, 29) and other N4-like phages infecting marine roseobacters (DSS3Φ2, EE36Φ1) (61) and Pseudomonas aeruginosa (LUZ7, LIT1, PEV2) (12). A major hallmark of N4-like phages is the use of three different DNA-dependent RNA polymerases during their growth cycles (29). Unlike other phages, N4 uses the vRNAP for the transcription of the early genes. The vRNAP is functionally present in the capsid and injected into the host cell during infection (19, 29). Phages S6 and N4 display significant similarities across their entire genomes, sharing an overall nucleotide sequence identity of 54.4%, with 29 gene products having greater than 20% sequence identity (Fig. 3). However, putative functions could not be assigned to each of them.
Phage Y2 features a 56,621-bp genome with invariable genome ends and long direct terminal repeats of 2,608 bp. Ninety ORFs were annotated (Table 2). Similarly to L1 and S6, phage Y2 is strictly virulent since it does not possess a lysogeny control region (Fig. 3). Transcription of the ORFs is exclusively from left to right. Gene products involved in host takeover and DNA metabolism are located at the left end of the genome in the early gene cluster. The late gene cluster and the cell lysis cassette are located at the right end. Y2 shows high sequence identities only to ΦEcoM-GJ1, a phage infecting porcine enterotoxigenic E. coli strains (25). They share a 61.0% overall nucleotide sequence identity and 43 gene products with sequence identities above 20%.
Sequencing of phage M7 yielded an 84,694-bp genome, with direct terminal repeats of 444 bp. A putative function of the 117 annotated ORFs was found for 30 gene products (Table 2). The densely distributed ORFs are interrupted by an ∼3.5-kbp region, including a series of tRNAs. No lysogeny control region was identified, confirming the strictly lytic life cycle of M7. Comparative genomics revealed its close relationship to the Felix O1-like phage ΦEa21-4 (35). The overall nucleotide sequence identity between these two phages is 87.4%, and their genome architectures are virtually identical. Thus, M7 was considered a close relative of ΦEa21-4 and was not studied further (except for its genome structure [see below]).
Proteomes of E. amylovora phages.
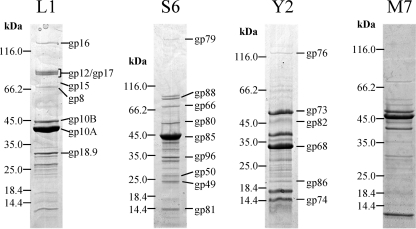
Peptide mass fingerprinting identified eight structural proteins of L1, nine of S6, and six of Y2 (M7 was not analyzed); corresponding genes are marked with asterisks in Fig. 3. Putative capsid components are the most prominent bands on the SDS gels (Fig. 4). The minor capsid protein of L1 (gp10B) was identified on the gel with a molecular mass approximately 5 kDa greater than that of the major capsid protein gp10A. Both gp10A and gp10B were found to be encoded by the same gene. This confirms the frameshift at nucleotide positions 21483 to 21488 (see above). The differing intensities of the protein bands allowed for an estimated quantification of the number of frameshift events. The band of gp10A is roughly 10-fold more intense than the band of gp10B, indicating that the frameshift occurs in 10% of the recoding events. This is similar to T7 (13). Other proteins of L1 identified as structural components are the putative portal protein (gp8), the tail tubular protein B (gp12), internal virion proteins C and D (gp15, gp16), an exopolysaccharide (EPS) depolymerase (gp17), and an unknown structural protein (gp18.9).
Fig. 4.
Analysis of virion proteins of phages L1, S6, Y2, and M7 by SDS-PAGE. Molecular mass markers are indicated on the left of each gel segment. Identification of individual protein bands was performed by peptide fingerprinting and mass spectrometry. The corresponding gene product names are indicated on the right (except for phage M7).
Phage S6 possesses a component (gp79) which is significantly larger than its other structural proteins. It was identified as the putative vRNAP. A set of three proteins (gp49, gp50, and gp66) encoded in the early gene region (i.e., left of the putative vRNAP gene) was also found to be present in the mature virion. Unfortunately, functional predictions could not be made, as there were no closely related proteins in the database. Other structural proteins of S6 are the major capsid protein (gp85), the portal protein (gp88), and three unknown proteins (gp80, gp81, and gp96).
As phage Y2 is relatively novel, there is very limited information available on similar phages. Besides the major capsid protein (gp68), a putative tail fiber (gp76) was identified. The functions of other structural components, such as gp73, gp74, gp82, and gp86, are unknown.
Phage genomes L1, M7, S6, and Y2 feature direct terminal repeats of different lengths.
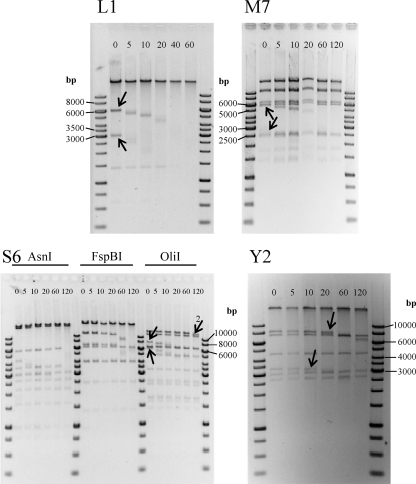
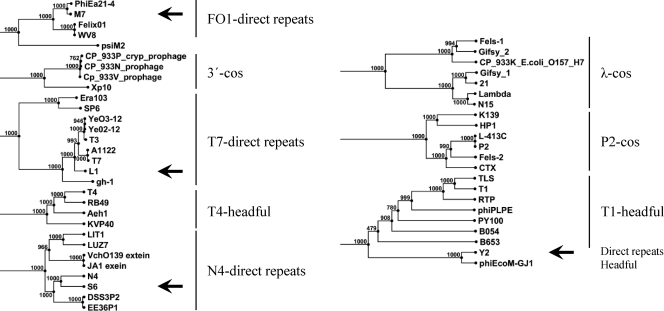
Our data showed that all four phages contain invariable genome ends. Restriction digestion analysis after BAL31 nuclease treatment of phage DNAs revealed that the two fragments located at the distal portions of the phage DNA molecules were progressively shortened by the enzyme, until they completely disappeared (Fig. 5). In some cases (e.g., AsnI- or FspBI-treated S6-DNA), the disappearing fragments were not visible because of their relatively small sizes (<1,000 nt). In the case of terminally redundant, circular permuted genomes, all fragments would have been degraded simultaneously by BAL31 (37). The phage genomes also lack cohesive ends, since heat treatment prior to electrophoresis did not change the restriction patterns (10) (data not shown). To confirm the genome end structures, sequencing primers were designed that bind in a suitable distance to the putative ends to initiate synthesis toward the physical end of the genome. When linear phage DNA or isolated terminal fragments were used as templates, typical sudden drop-offs of the Sanger sequencing signals were observed. It was found that the sequences of the two individual ends of phages L1, M7, S6, and Y2 feature direct terminal repeats of 172 bp, 444 bp, 397 bp, and 2,608 bp, respectively. Phylogenetic analysis of the large terminase proteins confirmed the experimental data for phages L1, M7, and S6. The terminases cluster together with terminases of phages having similar packaging strategies (Fig. 6). The Y2 terminase was found to be similar to terminases of phages with a T1-like headful packaging mechanism.
Fig. 5.
Analysis of phage genome physical structures. Shown are fragment patterns of phage DNA, following time-limited BAL31 treatment and subsequent digestion with different endonucleases. Phage L1 was digested with MfeI, M7 with FspBI, S6 with three different restriction endonucleases, and Y2 with OliI. Arrows indicate terminal fragments, which are progressively shortened by the BAL31 nuclease. The numbered arrow in the lower left panel (S6) indicates the second-to-last fragment, which is shortened only after the terminal fragment is completely degraded. Incubation times with BAL31 in minutes are indicated at the top of each gel.
Fig. 6.
Phylogenetic analysis of large terminase subunits of phages L1, M7, S6, and Y2 (indicated by arrows) and comparison to other phages with known packaging mechanisms (11, 30) as indicated. The tree was generated from an alignment (gap open cost, 10; gap extension cost, 1; end gap cost, free) using the neighbor-joining method with 1,000 bootstrap replicates (CLC Main Workbench 5).
Phage cocktails efficiently control E. amylovora.
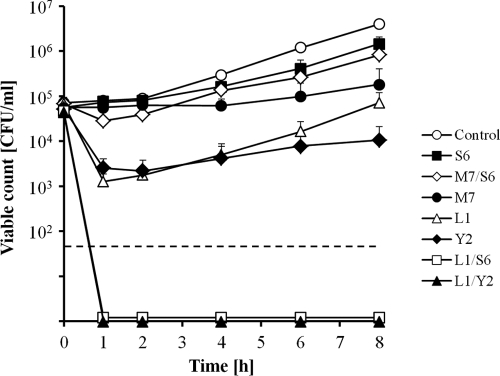
The application of a single virulent phage did not result in a complete eradication of E. amylovora. However, specific combinations of two different phages (L1/Y2 and L1/S6) very efficiently reduced the viable counts below the detection limit (50 CFU/ml) after 1 h of incubation (Fig. 7). Other phage combinations had no enhanced efficacy compared to that of the single phages (e.g., M7/S6 and others; data not shown). After 24 h, all bacterial cultures exposed to phage showed regrowth. Nevertheless, the remaining cell counts were significantly reduced in all samples compared to that of the uninfected control. Using single phages, reductions of 0.5 (L1), 2.5 (M7), 2.0 (S6), and 2.9 (Y2) logs (CFU/ml) were achieved. Applying two-phage cocktails, final viable counts were reduced by 3.1 (L1/S6), 3.3 (L1/Y2), and 1.6 (M7/S6) logs (CFU/ml). The observed effects could be reproduced using other E. amylovora strains (i.e., ACW56400, ACW55500, and LA411).
Fig. 7.
Viable counts of E. amylovora CFBP1430 after infection with single phages or two-phage cocktails. The values indicate the means of results from three independent trials, including standard deviations. The dashed line indicates the detection limit of 50 CFU/ml.
DISCUSSION
A set of 24 novel phage isolates infecting E. amylovora was isolated. Based on host range, virion morphology, genome size, and DNA restriction patterns, they could be grouped into eight subtypes. Only two subtypes appeared related to previously described phages from samples collected in North America (21). Sequencing of phages L1, M7, S6, and Y2 revealed relationships to phages infecting other members of the Enterobacteriaceae. Phages L1, S6, and Y2 are novel among E. amylovora phages, with little relatedness to other known phages infecting this species. It is possible that they have evolved more or less recently, during a process of adaptation to E. amylovora in the second half of the last century, when the pathogen arrived in Central Europe (8). The close relationship of M7 and the Felix O1-like virus ΦEa21-4 (35) and the relatedness between group B and ΦEa1-like phages (21) could indicate that E. amylovora phages also spread from North America to Central Europe. It is unclear if the different phage isolates are just a result of diverse strains used for the isolation or if the phage collections represent the local phage populations. The possible influence of isolation strains on the phage variety was reported earlier (21, 50). Using E. amylovora strain Ea110 as a host, a distinct phage similar to ΦEa1 was repeatedly isolated (50). In our study, the variety of phages did not increase proportionally to the total number of phage isolates, and a plateau may have been reached due to the use of a particular set of propagation strains.
The broad host ranges observed for most of the phages suggest obligately lytic life cycles, which is an important consideration regarding application of phages against target bacteria (22). True virulent lifestyles could be confirmed for phages L1, M7, S6, and Y2 through sequencing of their genomes. Nevertheless, some E. amylovora strains tested were insensitive to phages L1 and S6. A known factor influencing the sensitivity or resistance of E. amylovora to phages is the production of a capsule (7). Our observations support this theory. For example, the L1-like small podoviruses (group A), isolated on strain CFBP 1430, were not able to infect strain 4/82, which produces smaller amounts of EPS.
In addition to a virulent lifestyle and a broad host range, host specificity is of major interest (22). Our analyses showed that several of the 24 phages were able to infect species closely related to E. amylovora, which may potentially serve for phage replication in the phyllosphere (5, 34). In contrast, phage Y2 is strictly species specific and therefore unlikely to affect the nonpathogenic resident microflora when applied as a biocontrol agent, even in large quantities.
The inability to transduce nonphage DNA is another desirable property of phages designated for biocontrol purposes. The enterobacterial relatives of the phages described here are not known as generalized transducers (55), which can be explained by their physical genome structures. Their terminase enzymes recognize specific sequence motifs for the internalization of the viral DNA molecules into the newly assembled capsids. This lowers the probability that nonphage DNA is accidentally incorporated into the virus particle (11). Terminases of transducing phages generally feature less stringent sequence specificity. They possess circularly permuted and terminally redundant genomes and package their DNA by a headful mechanism (11). L1, M7, S6, and Y2 feature fixed genome ends and are therefore unlikely to be able to transduce host DNA. Phylogenetic analysis of the large terminase subunits indicated that the L1 terminase is related to terminases of phages T7, T3, and SP6, all of which feature direct terminal repeats of variable lengths (11, 14, 17, 38, 43). Phage S6 clusters together with N4 and other phages that use an N4-like packaging mechanism. Phage N4 has short direct terminal repeats of variable lengths (390 to 440 nt) containing 3′-single-stranded extensions (42). Phylogenetic analysis of the M7 terminase indicated its relatedness to Felix O1-type terminases. Felix O1 and ΦEa21-4 were proposed to have invariable ends with short direct terminal repeats (35, 59), in line with our findings.
The striking homologies between phages Y2 and ΦEcoM-GJ1 (including their terminases) could suggest a circularly permuted genome for Y2, containing a pac site. However, attempts to obtain experimental support for this hypothesis failed, i.e., restriction patterns did not yield the characteristic submolar fragment(s) occurring from digestion of phage DNA molecules originating from pac-dependent headful packaging (10). Moreover, BAL31 nuclease simultaneously degraded two fragments, and digestions with various restriction enzymes with n recognition sites always yielded n + 1 fragments. Therefore, we conclude that Y2 does not use headful packaging but that all phage genome molecules are identical and consist of a 54,013-bp information genome and a 3′ terminal repeat of 2,608 bp, yielding a 56,621-bp physical genome.
The clustered organization of the E. amylovora phage genomes into transcriptional units is typical for tailed phages, and the architecture is concordant with the model of phage genomes being modular built genetic mosaics (9, 24). This proposed mosaicism likely served as a driving force for the rapid adaptation to new host bacteria. Genetic modules, genes, or entire transcriptional units can be exchanged via a single recombination event. The similarity between two related phages abruptly ceases at the boundaries of genetic material exchanged by horizontal transfer (9). Analogously, L1, M7, S6, and Y2 and their enterobacterial relatives share significant homologies among most late genes, interrupted by gaps with apparently lower sequence identities. These gaps are located mainly in the region encoding tail-associated proteins. In general, host specificity seems to be related to sequence diversities in these tail-associated proteins. In the case of L1, the similarity to T7 is interrupted in gp17, whereas the genome of ERA103 shares stronger similarity to L1 in this particular region. The N-terminal domains of gp17, which link the tail fiber to the tail tube (52), are similar in L1 and T7. The major C-terminal part of L1-gp17 appears to have been replaced by a depolymerase (23, 31, 57), leading to a different function of the tail fiber and, thus, to nonoverlapping host ranges. Genes encoding tail components such as fibers or spikes are known hot spots for recombination. For example, K1-specific E. coli phages originating from different ancestors gained their ability to infect encapsulated bacteria via horizontal uptake of an endosialidase gene (54). Although these phages have different ancestors, their host ranges overlap due to the shared tail spikes. Accordingly, the host ranges of L1 and ERA103 overlap due to the presence of tail fibers with a depolymerase activity, even if they likely have different ancestors. Similar observations were made when comparing M7, S6, and Y2 to related enterobacterial phages. The ORFs encoding putative tail structures of M7 (orf40 to orf42) and Felix O1 (orf77) are unrelated, while the genes upstream and downstream are homologous. The same findings hold true for S6 and N4. There is an exceptional region of low similarity between S6 and N4 comprising orf94 and orf95 of S6. These gene products have putative glycoside hydrolase activity which may facilitate degradation of the host cell capsule during infection, similar to the depolymerase from phage L1. Finally, phage Y2 and ΦEcoM-GJ1 share significant similarities throughout their late gene cluster, with the exception of orf86 of Y2 and orf72 of ΦEcoM-GJ1. Again, both regions encode apparently unrelated putative tail fiber components (25).
In vitro experiments indicated a significant reduction of viable host cells by phage infection but also revealed the development of insensitive bacteria following exposure to phage preparations. Earlier studies, including E. coli, Campylobacter jejuni, and X. campestris pv. pruni, also reported the development of temporarily resistant bacteria after phage treatment. It is interesting to note that these resistant bacteria were highly attenuated in virulence and reverted to a phage-sensitive wild type in the absence of the selective pressure (36, 41, 44). Erskine (18) also observed delayed symptom development when pear slices were infected with a phage-resistant variant of E. amylovora which was not lysogenic. Therefore, the survival of insensitive host cells might not have a negative impact on the efficacy of phage application in planta.
Phage challenge and infection experiments demonstrated that selected two-phage cocktails can feature a much enhanced control efficacy on the growth of E. amylovora. Enhanced efficacy through the combined action of different types of phages with complementary host ranges represents a useful strategy and has been suggested for E. amylovora (49). Synergism of two or more different phages applied as a cocktail is likely based upon recognition of two different phage receptor molecules at the host cell surface and employment of different mechanisms for initial host cell takeover and virus multiplication. The exact reason for the apparently synergistic killing effect observed in our work is currently being investigated; preliminary data indicate a role of enzymes involved in depolymerizing the bacterial capsular material.
In conclusion, the results presented in this study demonstrate that the phages L1, M7, S6, and Y2 offer potential for phage-based biocontrol of fire blight. All four phages feature broad host ranges and virulent lifestyles. Detailed sequence analyses provided information to circumvent undesired transfer of host genetic material and facilitated selection of complementary phages for pooled applications.
Supplementary Material
ACKNOWLEDGMENTS
We thank Antonet Svircev for providing phage RFLP patterns, Jochen Klumpp for help with genome end analysis, Silvio Peng and Martin Zgraggen for assistance with phage isolation and characterization, and Jenna Denyes for helpful discussions.
This work was funded by the Swiss Federal Office for Agriculture (BLW Fire Blight Project—Biocontrol) and was conducted within the Swiss ProfiCrops and European Science Foundation COST Action 864 research networks.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Ackermann H.-W. 1996. Frequency of morphological phage descriptions in 1995. Arch. Virol. 141:209–218 [DOI] [PubMed] [Google Scholar]
- 2. Adams M. H. 1959. Bacteriophages. Interscience Publishers Inc., New York, NY [Google Scholar]
- 3. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 4. Balogh B., Canteros B. I., Stall R. E., Jones J. B. 2008. Control of citrus canker and citrus bacterial spot with bacteriophages. Plant Dis. 92:1048–1052 [DOI] [PubMed] [Google Scholar]
- 5. Balogh B., Jones J. B., Iriarte F. B., Momol M. T. 2010. Phage therapy for plant disease control. Curr. Pharm. Biotechnol. 11:48–57 [DOI] [PubMed] [Google Scholar]
- 6. Balogh B., et al. 2003. Improved efficacy of newly formulated bacteriophages for management of bacterial spot on tomato. Plant Dis. 87:949–954 [DOI] [PubMed] [Google Scholar]
- 7. Billing E. 1960. An association between capsulation and phage sensitivity in Erwinia amylovora. Nature 186:819–820 [DOI] [PubMed] [Google Scholar]
- 8. Bonn W. G., van der Zwet T. 2000. Distribution and economic importance of fire blight, p. 37–53 In Vanneste J. L. (ed.), Fire blight: the disease and its causative agent, Erwinia amylovora. CAB International, New York, NY [Google Scholar]
- 9. Casjens S. R. 2005. Comparative genomics and evolution of the tailed-bacteriophages. Curr. Opin. Microbiol. 8:451–458 [DOI] [PubMed] [Google Scholar]
- 10. Casjens S. R., Gilcrease E. B. 2009. Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions, p. 91–111 In Clokie M. R. J., Kropinski A. M. (ed.), Bacteriophages: methods and protocols, vol. 2 Molecular and applied aspects. Humana Press, New York, NY: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Casjens S. R., et al. 2005. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J. Bacteriol. 187:1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ceyssens P.-J., et al. 2010. Molecular and physiological analysis of three Pseudomonas aeruginosa phages belonging to the “N4-like viruses.” Virology 405:26–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Condron B. G., Atkins J. F., Gesteland R. F. 1991. Frameshifting in gene 10 of bacteriophage T7. J. Bacteriol. 173:6998–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dobbins A. T., et al. 2004. Complete genomic sequence of the virulent Salmonella bacteriophage SP6. J. Bacteriol. 186:1933–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dorscht J., et al. 2009. Comparative genome analysis of Listeria bacteriophages reveals extensive mosaicism, programmed translational frameshifting, and a novel prophage insertion site. J. Bacteriol. 191:7206–7215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duffy B., Schärer H.-J., Bünter M., Klay A., Holliger E. 2005. Regulatory measures against Erwinia amylovora in Switzerland. Bull. OEPP 35:239–244 [Google Scholar]
- 17. Dunn J. J., Studier F. W. 1983. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 166:477–535 [DOI] [PubMed] [Google Scholar]
- 18. Erskine J. M. 1973. Characteristics of Erwinia amylovora bacteriophage and its possible role in the epidemiology of fire blight. Can. J. Microbiol. 19:837–845 [DOI] [PubMed] [Google Scholar]
- 19. Falco S. C., Vander Laan K., Rothman-Denes L. B. 1977. Virion-associated RNA polymerase required for bacteriophage N4 development. Proc. Natl. Acad. Sci. U. S. A. 74:520–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Flaherty J. E., Jones J. B., Harbaugh B. K., Somodi G. C., Jackson L. E. 2000. Control of bacterial spot on tomato in the greenhouse and field with h-mutant bacteriophages. HortScience 35:882–884 [Google Scholar]
- 21. Gill J. J., Svircev A. M., Smith R., Castle A. J. 2003. Bacteriophages of Erwinia amylovora. Appl. Environ. Microbiol. 69:2133–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hagens S., Loessner M. J. 2010. Bacteriophage for biocontrol of foodborne pathogens: calculations and considerations. Curr. Pharm. Biotechnol. 11:58–68 [DOI] [PubMed] [Google Scholar]
- 23. Hartung J. S., Fulbright D. W., Klos E. J. 1988. Cloning of a bacteriophage polysaccharide depolymerase gene and its expression in Erwinia amylovora. Mol. Plant Microbe Interact. 1:87–93 [Google Scholar]
- 24. Hatfull G. F. 2008. Bacteriophage genomics. Curr. Opin. Microbiol. 11:447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jamalludeen N., et al. 2008. Complete genomic sequence of bacteriophage ΦEcoM-GJ1, a novel phage that has myovirus morphology and a podovirus-like RNA polymerase. Appl. Environ. Microbiol. 74:516–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johnson K. B., Stockwell V. O. 2000. Biological control of fire blight, p. 319–337 In Vanneste J. L. (ed.), Fire blight: the disease and its causative agent, Erwinia amylovora. CAB International, New York, NY [Google Scholar]
- 27. Johnson K. B., Stockwell V. O. 1998. Management of fire blight: a case study in microbial ecology. Annu. Rev. Phytopathol. 36:227–248 [DOI] [PubMed] [Google Scholar]
- 28. Jones A. L., Schnabel E. L. 2000. The development of streptomycin-resistant strains of Erwinia amylovora, p. 235–251 In Vanneste J. L. (ed.), Fire blight: the disease and its causative agent, Erwinia amylovora. CAB International, New York, NY [Google Scholar]
- 29. Kazmierczak K. M., Rothman-Denes L. B. 2006. Bacteriophage N4, p. 302–314 In Calendar R. (ed.), The bacteriophages, 2nd ed Oxford University Press, New York, NY [Google Scholar]
- 30. Kilcher S., Loessner M. J., Klumpp J. 2010. Brochothrix thermosphacta bacteriophages feature heterogeneous and highly mosaic genomes and utilize unique prophage insertion sites. J. Bacteriol. 192:5441–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim W.-S., Geider K. 2000. Characterization of a viral EPS-depolymerase, a potential tool for control of fire blight. Phytopathology 90:1263–1268 [DOI] [PubMed] [Google Scholar]
- 32. Klumpp J., et al. 2008. The terminally redundant, nonpermuted genome of Listeria bacteriophage A511: a model for the SPO1-like myoviruses of Gram-positive bacteria. J. Bacteriol. 190:5753–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kropinski A. M., Prangishvili D., Lavigne R. 2009. Position paper: the creation of a rational scheme for the nomenclature of viruses of Bacteria and Archaea. Environ. Microbiol. 11:2775–2777 [DOI] [PubMed] [Google Scholar]
- 34. Lehman S. M. 2007. Ph.D. thesis. Development of a bacteriophage-based biopesticide for fire blight. Brock University, St. Catharines, Ontario, Canada [Google Scholar]
- 35. Lehman S. M., Kropinski A. M., Castle A. J., Svircev A. M. 2009. Complete genome of the broad-host-range Erwinia amylovora phage ΦEa21-4 and its relationship to Salmonella phage Felix O1. Appl. Environ. Microbiol. 75:2139–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Loc Carrillo C., et al. 2005. Bacteriophage therapy to reduce Campylobacter jejuni colonization of broiler chickens. Appl. Environ. Microbiol. 71:6554–6563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Loessner M. J., Inman R. B., Lauer P., Calendar R. 2000. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol. Microbiol. 35:324–340 [DOI] [PubMed] [Google Scholar]
- 38. Molineux I. J. 2006. The T7 group, p. 277–301 In Calendar R. (ed.), The bacteriophages, 2nd ed Oxford University Press, New York, NY [Google Scholar]
- 39. Momol M. T., Aldwinckle H. S. 2000. Genetic diversity and host range of Erwinia amylovora, p. 55–72 In Vanneste J. L. (ed.), Fire blight: the disease and its causative agent, Erwinia amylovora. CAB International, New York, NY [Google Scholar]
- 40. Müller I., Kube M., Reinhardt R., Jelkmann W., Geider K. 2011. Complete genome sequences of three Erwinia amylovora phages isolated in North America and a bacteriophage induced from an Erwinia tasmaniensis strain. J. Bacteriol. 193:795–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. O'Flynn G., Ross R. P., Fitzgerald G. F., Coffey A. 2004. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl. Environ. Microbiol. 70:3417–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ohmori H., Haynes L. L., Rothman-Denes L. B. 1988. Structure of the ends of the coliphage N4 genome. J. Mol. Biol. 202:1–10 [DOI] [PubMed] [Google Scholar]
- 43. Pajunen M. I., Elizondo M. R., Skurnik M., Kieleczawa J., Molineux I. J. 2002. Complete nucleotide sequence and likely recombinatorial origin of bacteriophage T3. J. Mol. Biol. 319:1115–1132 [DOI] [PubMed] [Google Scholar]
- 44. Randhawa P. S., Civerolo E. L. 1986. Interaction of Xanthomonas campestris pv. pruni with pruniphage and epiphytic bacteria on detached peach leaves. Phytopathology 76:549–553 [Google Scholar]
- 45. Ravensdale M., Blom T. J., Gracia-Garza J. A., Svircev A. M., Smith R. J. 2007. Bacteriophages and the control of Erwinia carotovora subsp. carotovora. Can. J. Plant Pathol. 29:121–130 [Google Scholar]
- 46. Rezzonico F., Stockwell V. O., Duffy B. 2009. Plant agricultural streptomycin formulations do not carry antibiotic resistance genes. Antimicrob. Agents Chemother. 53:3173–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ritchie D. F., Klos E. J. 1979. Some properties of Erwinia amylovora bacteriophages. Phytopathology 69:1078–1083 [Google Scholar]
- 48. Sambrook J., Russell D. W. 2001. Molecular cloning, 3rd ed, vol. 1 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 49. Schnabel E. L., Fernando W. G. D., Meyer M. P., Jones A. L., Jackson L. E. 1999. Bacteriophage of Erwinia amylovora and their potential for biocontrol, p. 649–653 In Momol M. T., Saygili H. (ed.), Proceedings of the 8th international workshop on fire blight, vol. 489 ISHS Acta Hortic., Leuven, Belgium [Google Scholar]
- 50. Schnabel E. L., Jones A. L. 2001. Isolation and characterization of five Erwinia amylovora bacteriophages and assessment of phage resistance in strains of Erwinia amylovora. Appl. Environ. Microbiol. 67:59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shine J., Dalgarno L. 1974. The 3′-terminal sequence of Escherichia coli 16S rRNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. U. S. A. 71:1342–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Steven A. C., et al. 1988. Molecular substructure of a viral receptor-recognition protein—the gp17 tail-fiber of bacteriophage T7. J. Mol. Biol. 200:351–365 [DOI] [PubMed] [Google Scholar]
- 53. Stonier T., McSharry J., Speitel T. 1967. Agrobacterium tumefaciens Conn IV. Bacteriophage PB2 1 and its inhibitory effect on tumor induction. J. Virol. 1:268–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stummeyer K., et al. 2006. Evolution of bacteriophages infecting encapsulated bacteria: lessons from Escherichia coli K1-specific phages. Mol. Microbiol. 60:1123–1135 [DOI] [PubMed] [Google Scholar]
- 55. Thierauf A., Perez G., Maloy S. 2009. Generalized transduction, p. 267–286 In Clokie M. R. J., Kropinski A. M. (ed.), Bacteriophages: methods and protocols, vol. 1: isolation, characterization, and interactions. Humana Press, New York, NY [Google Scholar]
- 56. Thomson S. V. 2000. Epidemiology of fire blight, p. 9–36 In Vanneste J. L. (ed.), Fire blight: the disease and its causative agent, Erwinia amylovora. CAB International, New York, NY [Google Scholar]
- 57. Vandenbergh P. A., Wright A. M., Vidaver A. K. 1985. Partial purification and characterization of a polysaccharide depolymerase associated with phage-infected Erwinia amylovora. Appl. Environ. Microbiol. 49:994–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vanneste J. L., Eden-Green S. 2000. Migration of Erwinia amylovora in host plant tissues, p. 73–83 In Vanneste J. L. (ed.), Fire blight: the disease and its causative agent, Erwinia amylovora. CAB International, New York, NY [Google Scholar]
- 59. Whichard J. M., et al. 2010. Complete genomic sequence of bacteriophage Felix O1. Viruses 2:710–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wrigley N. G. 1968. Lattice spacing of crystalline catalase as an internal standard of length in electron microscopy. J. Ultrastruct. Res. 24:454–464 [DOI] [PubMed] [Google Scholar]
- 61. Zhao Y., Wang K., Jiao N., Chen F. 2009. Genome sequences of two novel phages infecting marine roseobacters. Environ. Microbiol. 11:2055–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zimmer M., Sattelberger E., Inman R. B., Calendar R., Loessner M. J. 2003. Genome and proteome of Listeria monocytogenes phage PSA: an unusual case for programmed +1 translational frameshifting in structural protein synthesis. Mol. Microbiol. 50:303–317 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.