Abstract
A genome mining study in the plant pathogenic bacterium Ralstonia solanacearum GMI1000 unveiled a polyketide synthase/nonribosomal peptide synthetase gene cluster putatively involved in siderophore biosynthesis. Insertional mutagenesis confirmed the respective locus to be operational under iron-deficient conditions and spurred the isolation of the associated natural product. Bioinformatic analyses of the gene cluster facilitated the structural characterization of this compound, which was subsequently identified as the antimycoplasma agent micacocidin. The metal-chelating properties of micacocidin were evaluated in competition experiments, and the cellular uptake of gallium-micacocidin complexes was demonstrated in R. solanacearum GMI1000, indicating a possible siderophore role. Comparative genomics revealed a conservation of the micacocidin gene cluster in defined, but globally dispersed phylotypes of R. solanacearum.
INTRODUCTION
Ralstonia solanacearum is an economically important bacterial plant pathogen, causing a lethal wilt disease that affects over 200 mono- and dicotyledonous plant species (9, 44). The soilborne pest infects plants through their root system, using wounded root tissue or cracks where secondary roots emerge as entry points (18). Following a successful invasion, the bacterium preferentially colonizes the large xylem vessels, from where it spreads systemically. Through the secretion of highly polymerized polysaccharides, R. solanacearum avoids recognition of its surface structures by plant defense mechanisms (1). This camouflage technique is concomitant with an interference of the host vascular system, which eventually leads to clogging of the xylem vessels and wilting of the plant (19). Infections by R. solanacearum cause great losses in agriculture due to the global occurrence of the pathogen, the susceptibility of many crop plants, and the limited means of protection currently available (14). Notwithstanding its significance as a pathogen, we know surprisingly little about the secondary metabolome of this bacterium and the ensuing implications for virulence. Recent studies have shown that R. solanacearum is capable of natural product biosynthesis and that the expression of the required biosynthesis genes responds to virulence-associated regulators (8, 37), assuming a role of secondary metabolites in host adaptation and/or pathogenesis.
The importance of scavenging iron from plant host tissues has been reported for the enterobacterial phytopathogens Erwinia chrysanthemi and E. amylovora (16). Both pests secrete siderophores in planta and mutants that are deficient in the biosynthesis of these molecules exhibit reduced virulence on their plant hosts (16). R. solanacearum secretes the natural product staphyloferrin B when exposed to low-iron conditions (3). The metabolite was proposed to function as a siderophore based upon its activity in the CAS assay; however, according to a gene inactivation study, staphyloferrin B production appears to be dispensable for the ability of the bacterium to infect tomato plants (3). Bioinformatic studies now have revealed the existence of a second putative siderophore gene cluster (mic) on the genome of R. solanacearum strain GMI1000, which is not only conserved among different phylotypes of the R. solanacearum species complex, but also found, albeit rearranged, on a plasmid of the phytopathogenic Burkholderia gladioli strain BSR3 (Fig. 1 and Table 1). The mic locus includes three genes encoding polyketide synthases (PKS) and/or nonribosomal peptide synthetases (NRPS), the domain architecture of which was suggestive of a yersiniabactin-like biosynthesis (29). The siderophore yersiniabactin is an important virulence factor for many human pathogenic enterobacteria, e.g., Yersinia pestis and Klebsiella pneumoniae, mediating bacterial iron uptake during infection and limiting the availability of iron to cells of the innate immune system (31). Gene clusters encoding compounds in the yersiniabactin family have been increasingly detected in whole-genome sequencing projects (41, 43); however, in only a few cases are the associated metabolites known. To identify the product of the corresponding gene cluster from the R. solanacearum GMI1000 genome, we disrupted one of the key biosynthetic genes by means of insertional mutagenesis and subsequently compared the metabolite profiles of mutant and wild-type strain under iron deficiency. This approach resulted in the isolation and structural characterization of micacocidin (compound 1 [ Fig. 2 ]), a metal-complexing antibiotic first isolated from a Pseudomonas sp. (24, 25).
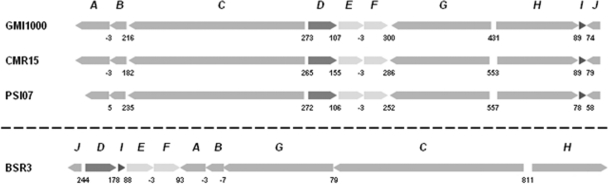
Fig. 1.
Organization of the micacocidin biosynthesis (mic) gene cluster in R. solanacearum GMI1000 and conserved loci in the phytopathogenic strains CMR15 and PSI07 from the R. solanacearum species complex. A rearranged mic cluster was found on plasmid 4 of the Burkholderia gladioli BSR3 genome.
Table 1.
Distribution and putative function of micacocidin biosynthesis genesa
| Code |
R. solanacearum GMI1000 |
R. solanacearum CMR15 |
R. solanacearum PSI07 |
B. gladioli BSR3 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF | Putative function | ORF | % Id | % Pos | ORF | % Id | % Pos | ORF | % Id | % Pos | |
| A | 1804 | Type II thioesterase | 11809 | 93 | 95 | 1874 | 88 | 91 | 4p_2100 | 61 | 70 |
| B | 1805 | Thiazolinyl imide reductase | 11810 | 98 | 99 | 1873 | 91 | 94 | 4p_2110 | 69 | 76 |
| C | 1806 | PKS/NRPS | 11812 | 96 | 97 | 1872 | 90 | 93 | 4p_2130 | 66 | 75 |
| D | 1807 | TonB-dependent receptor | 11813 | 98 | 99 | 1871 | 94 | 97 | 4p_2060 | 36 | 50 |
| E | 1808 | ABC transporter | 11814 | 98 | 99 | 1870 | 94 | 97 | 4p_2080 | 38 | 58 |
| F | 1809 | ABC transporter | 11815 | 98 | 99 | 1869 | 94 | 97 | 4p_2090 | 35 | 51 |
| G | 1810 | PKS | 11816 | 94 | 96 | 1868 | 88 | 91 | 4p_2120 | 60 | 69 |
| H | 1811 | NRPS | 11817 | 96 | 97 | 1867 | 89 | 92 | 4p_2140 | 61 | 70 |
| I | 1812 | Unknown | 11818 | 99 | 99 | 1866 | 94 | 96 | 4p_2070 | 70 | 82 |
| J | 1813 | AraC-like regulator | 11819 | 97 | 98 | 1865 | 92 | 95 | 4p_2050 | 43 | 62 |
Similarities at the amino acid level for corresponding enzymes in R. solanacearum strains CMR15 and PSI07, as well as in B. gladioli strain BSR3, are indicated. Abbreviations: % Id, percent identical amino acids; % Pos, percent similar amino acids.
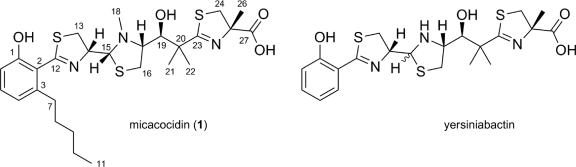
Fig. 2.
Chemical structures of micacocidin (compound 1) and yersiniabactin, metal-chelating agents produced by R. solanacearum GMI1000 and Y. pestis, respectively.
We describe here the previously unrecognized micacocidin biosynthesis gene cluster, as well as the isolation, the structural characterization, and the siderophore properties of its product.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in the present study are described in Table 2. R. solanacearum GMI1000 and the mutant strain RS6 were grown at 30°C in liquid 1/4× M63 minimal medium lacking FeSO4 but supplemented with 0.2% glucose and 0.02% sodium acetate. The influence of Fe3+ on micacocidin production was tested in the same medium containing 4 μM FeCl3. The mutant strain RS6 was continuously kept under antibiotic selection pressure (25 μg of kanamycin/ml). Escherichia coli strains were cultured in liquid or solidified LB medium. When required, antibiotics were added at the following concentrations: 100 μg of ampicillin/ml, 25 μg of chloramphenicol/ml, and 50 μg of kanamycin/ml.
Table 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| Strains | ||
| R. solanacearum | ||
| GMI1000 | Wild-type strain | 33 |
| RS6 | RSc1806::pHiK002 in GMI1000 | This study |
| E. coli | ||
| TOP10 | F−mcrA Δ(mrr–hsdRMS–mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| BW25113 | K-12 derivative: ΔaraBAD ΔrhaBAD | 6 |
| BW25113-2 | BW21533 harboring pIJ790 and pHiK001 | This study |
| Plasmids | ||
| pJET1.2 | Cloning vector; Ampr | Fermentas |
| pIJ790 | Lambda-Red (gam, beta, exo), araC, rep101(Ts); Cmr | 22 |
| pAphA-3 | Kanamycin resistance cassette; Kmr | 27 |
| pHiK001 | pJET1.2 carrying 1.7-kb RSc1806 fragment | This study |
| pHiK002 | pHiK001 carrying RSc1806::Kmr | This study |
Abbreviations: Ampr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance.
Sequence analyses.
The genome of R. solanacearum GMI1000 consists of two circular replicons (GenBank accession numbers NC_003295 and NC_003296, respectively), which were screened for genes putatively involved in secondary metabolism using homology-based alignments against a library of conserved PKS and NRPS domains (41). To identify potential biosynthetic gene clusters, initial hits were grouped by physical proximity and mapped onto the genome using Vector NTI (Invitrogen). Identified loci were further analyzed by using web-based bioinformatic software, as well as natural product-specific databases such as ASMPKS and NORINE (4, 40).
General DNA methods.
Genomic DNA from R. solanacearum GMI1000 was isolated according to previously established methods (6). Plasmid isolation from E. coli strains was accomplished with a commercial plasmid isolation kit (Roti-Prep MINI; Roth). DNA fragments from agarose electrophoresis gels were extracted with a QIAquick gel extraction kit (Qiagen). For the preparation of competent cells, E. coli BW25113 and E. coli BW25113-2 were grown in LB medium at 30°C to an optical density at 600 nm of 0.4. The cultures were thoroughly washed with ice-cold 10% glycerol and resuspended in the remaining drop of 10% glycerol after centrifugation. Competent cells (40 μl) were electrotransformed with 100 ng of purified DNA in 0.2-cm gapped ice-cold electroporation chambers using a Bio-Rad GenePulser II set to 200 Ω, 25 μF, and 2.5 kV.
Construction of the RSc1806 disruption plasmid.
A gene fragment of RSc1806 (accession number NC_003295) was amplified by PCR from R. solanacearum GMI1000 genomic DNA by using the primers P1-RSc1806 (5′-AACCCGATCGGCACCTTC-3′) and P2-RSc1806 (5′-TAGGC GTCGAACATGGCATC-3′). The PCR product was blunt end cloned into pJET1.2 to give pHiK001, which was subsequently used to transform E. coli BW25113 harboring pIJ790 (7, 22). Insertion of a kanamycin resistance gene into the 1.7-kb gene fragment of RSc1806 was achieved by lambda red-mediated recombination. To this end, the antibiotic resistance cassette was amplified from pAphA-3 by PCR with the primer pair P3-kan (5′-AGCACGCCCCCCAGGGCGCCGTGCCCGCACCGGCGGCAGCCGAGCCCATCGTGCATGCGCCGGACCACGCGCATCTGCCCTGACTAACTAGGAGGAATAA-3′) and P4-kan (5′-AAGGCGCGGTGCTGCAGGTGCTCGAGCACGGCCCCGTTGACGGCGGCGGCGCGCTGG CCGAAGGGCAGGGGCGCGGCGCATATTCCCTCCAGGTACTAAA-3′), generating extensions homologous to the target RSc1806 sequence at either end of the marker. E. coli BW25113-2 was electrotransformed with the PCR product following induction of lambda red genes with 10 mM l-arabinose at 30°C for 3 h. After recovery in SOC medium (2% tryptone, 0.5% yeast extract, 20 mM glucose, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2) (37°C, 1 h), the transformants were spread onto LB agar plates containing 50 μg of kanamycin/ml, followed by incubation overnight at 37°C. Integration of the marker in the plasmid DNA by homologous recombination yielded the RSc1806 disruption plasmid pHiK002, which was isolated and verified by PCR analysis with the primers P5 (5′-CGACTCACTATAGGGAGAGCGGC-3′) and P6 (5′-AAGAACATCGATTTTCCATGGCAG-3′).
Construction of the RSc1806 mutant.
For electroporation, R. solanacearum GMI1000 cells were grown in 1/4× M63 medium supplemented with 0.2% glucose and 4 ml of glycerol/liter at 30°C to an optical density at 580 nm of 0.5. After centrifugation, the harvested cells were successively washed with sterile Milli-Q water and ice-cold 10% glycerol. The ensuing competent cells were electrotransformed with pHiK002. Electroporation was carried out in a 0.2-cm gap cuvette using a Bio-Rad GenePulser II set to 200 Ω, 25 μF, and 2.5 kV. SOC medium was immediately added to the shocked cells, which were then incubated at 30°C for 3 h. The resulting culture was spread onto B agar containing 25 μg of kanamycin/ml, followed by incubation at 30°C. The colonies were checked by PCR with the primers P7 (5′-TTTACCTCGTCGCCGGCGCTTA-3′), P8 (5′-TCAATTCCGGTGATATTCTCATTTTAGC-3′), P9 (5′-CTTACTGGGGATCAAGCCTGATTG-3′), and P10 (5′-GCGAAGCTGCCGAAGAGATCGTAGA-3′) to verify the mutation.
Analytical methods.
High-pressure liquid chromatography (HPLC) was performed on a Shimadzu UFLC liquid chromatography system equipped with a Nucleodur Sphinx RP column (250 by 10 mm, 5-μm pore diameter). Micacocidin was isolated by using a linear gradient of acetonitrile in water plus 0.1% trifluoroacetic acid (10% → 90% acetonitrile within 20 min; flow rate, 3 ml/min) with wavelength monitoring at 254 nm. HPLC-mass spectrometry (MS) analyses were conducted on an Agilent 1100 series HPLC-DAD system coupled with an MSD trap (Agilent) operating in alternating ionization mode and an Antek 8060 HPLC-CLN-detector (Antek Instruments GmbH) using a C8 column (Zorbax Eclipse XDB C8, 150 by 4.6 mm; 5-μm pore diameter). A linear gradient of methanol in water plus 0.1% trifluoroacetic acid (10% → 90% methanol within 15 min; flow rate, 1 ml/min) was used for metabolic profiling and competition experiments. Radio HPLC was carried out on a system from Sykam GmbH equipped with a Smartline UV detector 2600 and a scintillation radio detector (γ-sensorPE; Scintomics). The system was operated with a reversed-phase column (Nucleosil C4; 125 by 4 mm, 5-μm pore diameter) using a linear gradient of acetonitrile in water plus 0.05% trifluoroacetic acid (10% → 90% acetonitrile within 10 min; flow rate, 1 ml/min). The retention time of the 68Ga-labeled micacocidin was 8.5 min (see Fig. SA2 in the supplemental material). High purity and labeling yield was proven additionally by radio instant thin-layer chromatography (ITLC) using Biodex strips (TEC control chromatography strips 150 to 771) and citric acid (pH 4.9) as a buffer system. The 68Ga-chelated micacocidin showed a strong peak at the starting point. A signal corresponding to free 68Ga was not observed (see Fig. SA3 in the supplemental material). High resolution mass determination was carried out using an Exactive mass spectrometer (Thermo-Scientific). For the detection of staphyloferrin B, supernatants from iron-deficient cultures of R. solanacearum GMI1000 and RS6 were lyophilized and redissolved in 50% methanol. Samples (5 μl) were loop injected into the mass spectrometer, and mass spectra were acquired in the negative ionization mode. Nuclear magnetic resonance (NMR) spectra were recorded at 300 K on a Bruker Avance III 600 MHz spectrometer using methanol-d4 as a solvent and internal standard (δH = 3.31 ppm; δC = 49.0 ppm).
Siderophore screening.
Chrome azurol S (CAS) plates were prepared as previously reported (28, 38). Half of each CAS agar layer was cut out, and the gap was filled with 1/4× M63 minimal medium agar. R. solanacearum wild-type and mutant strains were plated on the M63 half of the plates. The secretion of iron-scavenging molecules was detected by a color change from blue to orange after incubation at 30°C for 3 days.
Competition experiments for metal binding.
Micacocidin (10 mM) was added to an aqueous solution containing 10 mM FeCl3 and 10 mM NiCl2. The formation of complexes was determined after 24 h by using HPLC-MS according to a previously described method (10). Displacement of already existing complexes was measured in 10 mM aqueous solutions of ferric micacocidin and nickel micacocidin. Both complexes were incubated for 24 h with a 10 mM concentration of the opposing metal ion and analyzed by HPLC-MS.
68Ga3+ uptake assay in R. solanacearum GMI1000 and RS6.
A 50-ml aliquot of a 4-day-old culture of R. solanacearum was centrifuged at 5,700 × g for 5 min to separate the cell pellet from the supernatant. The cell pellet was resuspended in 1 ml of the modified 1/4× M63 medium and mixed with either a 68Ga-micacocidin incubation solution or 8 nmol of 68Ga3+ solely. The incubation solution contained 8 nmol of micacocidin in 1.1 M sodium acetate buffer and freshly eluted 68Ga-HCl (set to final pH 5.3) from a 68Ge/68Ga-generator system (Cyclotron Co., Ltd.) with a radioactivity of about 10 MBq. The formation of the 68Ga-micacocidin complex was monitored by radio HPLC and further proven by radio ITLC. After 1 h of shaking at 30°C, the cells were separated from the supernatant by centrifugation at 5,700 × g for 5 min, and the radioactivity was measured using an Isomed 2000 activimeter (MED Nuklear-Medizintechnik GmbH). Subsequently, the cells were resuspended in 1 ml of 1.1 M sodium acetate (pH 8.4), vortexed, and centrifuged. This washing procedure was repeated three times. The leftover radioactivity of the cell pellet and the supernatant was measured after each washing step. Radioactivity values were obtained by using the mean of three independently collected data sets, including the standard deviation.
RESULTS
Architecture and features of the PKS/NRPS gene cluster encoded on the GMI1000 chromosome.
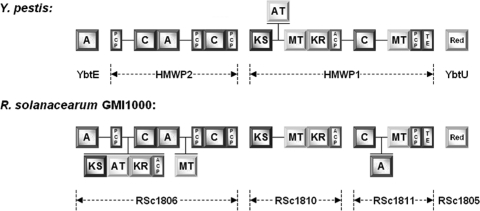
Bioinformatic studies revealed the presence of a discrete locus in the 52.3- to 53.3-centisome region of the R. solanacearum GMI1000 chromosome. The mic locus covers 38.1 kb of contiguous DNA and includes three genes for enzymes with a predicted function as PKS and/or NRPS (Fig. 1). The colocalization with a TonB-dependent receptor (RSc1807), as well as two iron-siderophore ABC transporters (RSc1808 and RSc1809), suggested an involvement of the gene cluster in siderophore biosynthesis. A more thorough analysis unveiled a close relationship to the yersiniabactin locus of the plague bacterium Yersinia pestis (29). Although the corresponding genes lack direct sequence similarity, the general domain architecture of the PKS and NRPS biosynthesis enzymes is largely conserved in both gene clusters (Fig. 3). While the C terminus of RSc1806 is organized similarly to the HMWP2 subunit of the yersiniabactin synthetase (HMWP, high-molecular-weight protein), RSc1810 and RSc1811 correspond to the nine-domain HMWP1 subunit. On the other hand, the R. solanacearum assembly line features also some deviations from the known motif, including an additional polyketide synthase module and an extra methyltransferase domain in RSc1806 that are missing in HMWP2. Comparison of the domains YbtE and RSc1806-A1, which are assumed to specify the respective biosynthetic starter unit, revealed another putative inconsistency between the two assembly lines. The stand-alone AMP-dependent ligase YbtE from the yersiniabactin gene cluster has a strong preference for salicylic acid (17), and the 10-amino-acid “nonribosomal code” signature of its binding pocket (PLPAQGVLCK) is consistent with the priming of this aryl carboxylic acid (39), whereas RSc1806-A1, the YbtE counterpart in the GMI1000 cluster, exhibits a distinct motif (DLMPMAALLK). Taken together, the data strongly suggested (i) that the natural product encoded by the mic gene cluster in GMI1000 is structurally related, but not identical to yersiniabactin, and (ii) that the most important deviation would likely involve the salicylate-derived phenolic group of yersiniabactin, which was shown to be crucial for the coordination of ferric iron and thus the siderophore properties of yersiniabactin (30).
Fig. 3.
Domain architecture of modular biosynthesis enzymes from gene clusters of Y. pestis and R. solanacearum GMI1000. The former system encodes the known siderophore and virulence factor yersiniabactin. Domain notation: A, adenylation; PCP, peptidyl carrier protein; C, condensation; KS, ketosynthase; AT, acyltransferase; MT, SAM-dependent methyltransferase; KR, ketoreductase; TE, thioesterase; Red, thiazolinyl imide reductase. Domains that are unique to one assembly line are drawn on different planes.
Other available R. solanacearum genomes were subsequently searched for orthologous genes (20, 32, 33). The mic locus was found, along with GMI1000, in strains CMR15 and PSI07 as well, whereas the genomes of strains IPO1609/UW551, Molk2, and CFBP2957 did not include such a cluster of genes.
Metabolic profiling of GMI1000 and the mutant strain RS6.
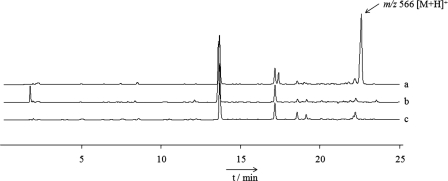
Based on the assumption that the identified PKS/NRPS gene cluster encodes a siderophore, we grew the GMI1000 strain under low-iron conditions to induce its expression. The ethyl acetate extract from this culture was subsequently subjected to LC-MS profiling. From the comparison with a culture grown in the same medium, but containing 4 μM FeCl3, it was evident that GMI1000 produces at least one additional metabolite at low ferric iron levels (Fig. 4). The detected pseudomolecular ion of this compound at m/z 566 [M+H]+ did not match the previously described staphyloferrin B, which has a mass of 448 (3, 12). To examine a potential role of the identified PKS/NRPS gene cluster in the production of the discovered metabolite, we disrupted the N-terminal C domain of RSc1806. The latter was predicted to code for an essential component of the hybrid synthase catalyzing the formation of a thiazoline ring in analogy to HMWP2 (29). As shown in Fig. 4, profile c, the peak with the target ion at m/z 566 [M+H]+ was completely absent in the mutant strain RS6, thus confirming the involvement of RSc1806 in the biosynthesis of the putative siderophore. It is important to note that the mutant retained iron-scavenging activity, as evidenced by the color change on CAS indicator plates. By means of MS analyses, this activity could be traced back to the presence of staphyloferrin B (see the supplemental material).
Fig. 4.
Metabolic profiles of crude culture extracts from GMI1000 grown under iron-deficient conditions (profile a) and in the presence of 4 μM FeCl3 (profile b), as well as from the mutant strain RS6 at reduced iron levels (profile c). The peak with m/z 566 [M+H]+ is only produced by the wild-type strain under iron-limited conditions.
Isolation and structure elucidation of micacocidin from GMI1000.
For the isolation of the metabolite that derived from the mic gene cluster, R. solanacearum GMI1000 was grown in a 3-liter scale under iron-limiting conditions. The culture broth was extracted twice with ethyl acetate, and the combined organic layers were concentrated in vacuo. The resulting crude extract was dissolved in a small volume of acetonitrile and purified by using reversed-phase HPLC to yield 11.2 mg of a white compound (i.e., compound 1) that gave a positive response in the CAS assay (38).
High-resolution electrospray ionization (ESI)-MS measurements of compound 1 showed pseudomolecular ions at 566.2175 [M+H]+ and 564.2022 [M−H]−, respectively, which were consistent with an empirical formula of C27H39O4N3S3. Structure elucidation was accomplished using one- and two-dimensional NMR experiments after preparation of the gallium complex of compound 1 to suppress epimerization (11, 36). Most of the detected 13C and 1H resonances correlated well with published values of yersiniabactin (11), allowing for the rapid identification of the thiazolidine-thiazoline framework in compound 1 (see Table SA1 in the supplemental material). In accordance with the genomic data, this part of the molecule was found to be almost identical to yersiniabactin except for the presence of an N-bound methyl group at the thiazolidine ring, which can likely be attributed to the additional methyltransferase domain in RSc1806. Major differences between yersiniabactin and compound 1 were related to the expected replacement of the salicylate-derived moiety. The 1H NMR spectrum of the GMI1000-derived metabolite displayed only three resonances in the aromatic region, which occur as an isolated AMX spectrum (δA = 6.75, δM = 7.23, δX = 6.64, JAM = 8.4 Hz, JMX = 7.4 Hz, JAX = 1.3 Hz) characteristic of a 1,2,3-trisubstituted benzene derivative. The substituent at C-1 of the benzoid system was identified as a phenolate function due to the downfield resonance of its adjacent carbon atom (δC = 168.1 ppm) and the bathochromic shift observed in the UV spectrum of the nonchelated compound 1 upon addition of methanolic NaOH. Five signals in the upfield region that were not present in yersiniabactin could be assigned to a pentyl side chain by interpretation of COSY data. 1H,13C long-range correlations allocated this moiety at C-3 of the benzene ring in ortho position to the proton resonating at a δH of 6.64 ppm. Weak heteronuclear four-bond correlations from the latter and H-6 to C-12 allowed the linkage of the resolved 1-hydroxy-3-pentyl-benzene moiety with the thiazolidine-thiazoline partial structure via C-2, thus establishing the planar structure of compound 1. The relative configuration of the chiral carbon atoms at positions 14, 15, 17, and 19 was determined by nuclear Overhauser enhancement (NOE) spectroscopy. Strong NOE interactions were detected from H-14 to H-13a and H-16b on the one hand and from H-15 to H-13b and H3-18, as well as from H-17 to H-16a, H3-18, and H-19, on the other. Consolidating this information, the stereochemical arrangement was deduced as 14R*, 15R*, 17R*, and 19S*. A subsequent database search identified compound 1 as the known antimycoplasma agent micacocidin (Fig. 2) from Pseudomonas sp. (24, 26).
Competition experiments for metal binding.
Micacocidin was previously reported to coordinate different metal ions, including Zn2+, Cu2+, and Fe3+ (24). To probe the discrimination of compound 1 between divalent and trivalent cations, we conducted metal competition assays with Ni2+ and Fe3+, both of which form stable, monomeric 1:1 complexes with micacocidin that can be easily distinguished in LC-MS due to their different retention times and isotopic patterns, respectively. When micacocidin was incubated with equimolar amounts of Ni2+ and Fe3+, it was preferentially converted into its ferric complex and showed only poor affinity toward Ni2+ (see Fig. SA1 in the supplemental material). Surprisingly, however, Fe3+ did not displace the metal ligand from an existing complex of micacocidin with a divalent cation, even after incubation for 24 h.
Gallium uptake assay.
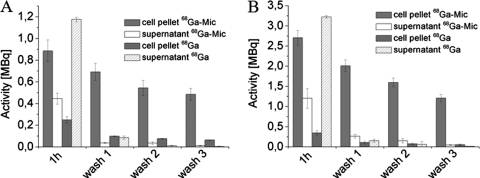
To investigate the cellular uptake of micacocidin and thus evaluate possible siderophore properties, the compound was chelated with 68Ga3+ as a surrogate for Fe3+ (2, 13), and the resulting complex was added to a culture of R. solanacearum GMI1000. As a control experiment and to consider a potential effect of endogenously produced staphyloferrin B in the internalization of 68Ga3+, R. solanacearum GMI1000 was solely treated with 68Ga3+. In both preparations, the radioactivity of cells and supernatant was determined after 1 h of incubation. Subsequent washing steps with 1.1 mM sodium acetate removed residual 68Ga3+ (in the control experiment) or 68Ga3+-micacocidin as monitored by radioactivity measurements (Fig. 5 A). The data clearly showed that the radioactivity level was higher in the cell pellet than in the supernatant for the micacocidin-containing samples. The control samples that contained no micacocidin exhibited a higher concentration of radioactivity in the supernatant, indicating that fewer 68Ga3+ had been taken up by the cells. After three successive washing steps, the amount of radioactivity decreased to nearly zero in the wash fractions, whereas the cell pellets retained high radioactivity levels. Noteworthy, the cell pellets of the micacocidin samples had a leftover radioactivity of 0.485 ± 0.054 MBq following the washing steps, while the corresponding value from the control samples was significantly lower with 0.063 ± 0.003 MBq. These results imply that micacocidin serves as a carrier for Ga3+, shuttling the trivalent metal into cells of GMI1000, and corroborate a potential siderophore role for this natural product. The low leftover radioactivity in the control experiment also suggests that the presence of endogenously produced siderophores played only a negligible role in the 68Ga3+ uptake assay. Subsequent testing of the mutant strain RS6, which was impaired in micacocidin biosynthesis, but not in its iron-siderophore ABC transporter genes, yielded consistent incorporation data for the 68Ga- micacocidin complex and 68Ga3+, respectively (Fig. 5B).
Fig. 5.
68Ga3+ uptake of R. solanacearum GMI1000 (A) and the mutant strain RS6 (B) after a 1-h incubation with the 68Ga-micacocidin complex (68Ga-Mic) and three successive washing steps compared to a micacocidin-free control sample (68Ga). Radioactivity is retained in the cell pellet, while the washing fractions are nearly free of activity after two washing steps.
DISCUSSION
An important group of virulence factors produced by bacteria and fungi that invade vertebrate hosts are siderophores (35). Siderophore-mediated iron uptake counters the growth limiting effects imposed by iron-binding proteins in animals and humans and is often a prerequisite for effective host colonization (23). In contrast, the role of siderophores in plant pathogenesis is less clear, and various scenarios are conceivable that provoke their biosynthesis. The vascular and apoplastic fluids of plants are a hostile environment for bacterial intruders, where siderophore release may weaken host defense reactions and guarantee a sufficient iron supply for the pathogen on the one hand or represent a resistance mechanism under conditions of elevated metal concentrations on the other (15). To explore possible modes of iron competition in such biological systems, profound knowledge of the involved siderophores is required. A previous study on the economically relevant pest R. solanacearum has shown that it produces the supposed siderophore staphyloferrin B under iron deficiency and that a mutant defective in the biosynthesis of this metal chelator remained fully virulent on tomato plants (3). Noteworthy, the iron-scavenging activity of the mutant was not completely abolished (3), suggesting that the loss of staphyloferrin B might have been partially compensated for by the bacterium.
In the present study we demonstrate that some R. solanacearum strains harbor a putative siderophore gene cluster on their chromosomes (Fig. 1) in addition to the plasmid-borne staphyloferrin B locus, which appears to be conserved in the entire species complex according to genomic data (20, 32, 33). Bioinformatic analysis of the newly identified cluster pointed to the biosynthesis of a thiazoline-containing natural product but did not allow an accurate structural prediction. Using the same cultivation conditions as described for ralfuranone production (37), while omitting the iron source, enabled the isolation of the metal-chelating metabolite micacocidin from R. solanacearum GMI1000. Targeted gene inactivation linked the biosynthesis of micacocidin with the aforementioned cluster and confirmed that micacocidin production is not vital under iron-limited conditions. Either the trace amounts of iron still present in the medium were sufficient for growth of the bacterium or GMI1000 possesses an auxiliary system for iron acquisition. Since the staphyloferrin B biosynthesis genes were not impaired and the micacocidin-negative mutant RS6 was still producing halos on CAS agar plates, we rather tend to the latter explanation, which is also supported by MS data. Subsequent analyses revealed that micacocidin and staphyloferrin B are produced simultaneously in the wild-type strain when iron becomes scarce (see the supplemental material).
Micacocidin was shown to coordinate diverse metal ions (including Fe3+), which provides an explanation for its activity in the CAS assay. Even though the coordination of ferric iron is an essential property, it is not sufficient to identify a compound as a siderophore. The addition of FeCl3 in a concentration of 4 μM repressed the production of micacocidin in the GMI1000 wild-type strain, which is typical for a siderophore, since the unregulated uptake of iron can lead to unfavorable intracellular chemistry such as Fenton or Haber-Weiss reactions ending in the formation of reactive oxygen species (34). Furthermore, we demonstrated that micacocidin prefers ferric iron over Ni2+ as a ligand and that it increases the uptake of 68Ga3+ (as a surrogate for Fe3+) in R. solanacearum cells compared to control experiments. Taken together, these data strongly support a siderophore function for micacocidin; however, further testing is required to confirm an active transport mechanism for the iron-loaded metabolite.
The identification of a second assembly line in GMI1000, which is involved in the biosynthesis of a metal-chelating molecule, provides the basis for future studies that aim at clarifying the role of iron in plant host-pathogen interactions. Further investigations may also emerge from comparative genomics. Those strains (GMI1000, CMR15, and PSI07) that were found to contain both the micacocidin and the staphyloferrin B gene cluster correspond to the phylotypes I, III, and IV of the R. solanacearum species complex, whereas strains that lack the genetic capacity for micacocidin production, namely, IPO1609/UW551, Molk2, and CFBP2957, exclusively belong to phylotype II (32). The four phylotypes or phylogenetic groups of R. solanacearum largely correlate with the geographical distribution of the strains (21, 32). Even though only a limited number of R. solanacearum genomes is publicly available which, therefore, may not sufficiently reflect the diversity within the species, it is still tempting to speculate that the acquisition (or loss) of micacocidin biosynthesis genes reflects their evolutionary divergence to some extent. Analysis of the corresponding open reading frames (ORFs) in GMI1000 did not reveal any significant deviations in GC content or codon adaptation index values from the rest of the genome, which makes a recent acquisition of the mic cluster by horizontal gene transfer in this strain rather unlikely. It should be noted, however, that a region with alternative codon usage and below-average GC content is located immediately downstream of the mic locus in GMI1000.
From a biosynthetic perspective, micacocidin represents a new and discrete natural product class made by R. solanacearum and expands the known metabolome of this species, which already includes staphyloferrin B (3), the HrpB-dependent factor HDF (8), and the ralfuranone family of compounds (37). It is worth mentioning that the availability of the genome sequence of GMI1000 (33) not only was instrumental in the discovery of these metabolites by promoting the development of specific isolation strategies but also granted insights into their biosyntheses (8, 42). While micacocidin and the ralfuranones are synthesized using thiotemplate-based assembly line enzymology (42), the condensation of amine and carboxylate precursors in staphyloferrin B biosynthesis is mechanistically distinct, involving NRPS-independent siderophore synthetases, as shown in Staphylococcus aureus (5). Finally, the isatin derivative HDF derives via a shunt in tryptophan catabolism (8). The biosynthetic versatility of R. solanacearum is intriguing and certainly warrants additional studies.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully thank C. Allen, University of Wisconsin-Madison, Madison, WI, for expert technical advice, A. Perner for HR-ESI-MS measurements, and M. Grigsby for proofreading of the manuscript. H.K. thanks the Jena School for Microbial Communication for a postdoctoral research fellowship.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 1 July 2011.
REFERENCES
- 1. Araud-Razou I., Vasse J., Montrozier H., Etchebar C., Trigalet A. 1998. Detection and visualization of the major acidic exopolysaccharide of Ralstonia solanacearum and its role in tomato root infection and vascular colonization. Eur. J. Plant Pathol. 104:795–809 [Google Scholar]
- 2. Ardon O., et al. 2001. Identification of a Candida albicans ferrichrome transporter and its characterization by expression in Saccharomyces cerevisiae. J. Biol. Chem. 276:43049–43055 [DOI] [PubMed] [Google Scholar]
- 3. Bhatt G., Denny T. P. 2004. Ralstonia solanacearum iron scavenging by the siderophore staphyloferrin B is controlled by PhcA, the global virulence regulator. J. Bacteriol. 186:7896–7904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caboche S., et al. 2008. NORINE: a database of nonribosomal peptides. Nucleic Acids Res. 36:D326–D331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheung J., Beasley F. C., Liu S., Lajoie G. A., Heinrichs D. E. 2009. Molecular characterization of staphyloferrin B biosynthesis in Staphylococcus aureus. Mol. Microbiol. 74:594–608 [DOI] [PubMed] [Google Scholar]
- 6. Coupat B., et al. 2008. Natural transformation in the Ralstonia solanacearum species complex: number and size of DNA that can be transferred. FEMS Microbiol. Ecol. 66:14–24 [DOI] [PubMed] [Google Scholar]
- 7. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Delaspre F., et al. 2007. The Ralstonia solanacearum pathogenicity regulator HrpB induces 3-hydroxy-oxindole synthesis. Proc. Natl. Acad. Sci. U. S. A. 104:15870–15875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Denny T. P. 2006. Plant pathogenic Ralstonia species, p. 573–644 In Gnanamanickam S. S. (ed.), Plant-associated bacteria. Springer, Dordrecht, Netherlands [Google Scholar]
- 10. Dimkpa C. O., Merten D., Svatoš A., Büchel G., Kothe E. 2009. Metal-induced oxidative stress impacting plant growth in contaminated soil is alleviated by microbial siderophores. Soil Biol. Biochem. 41:154–162 [Google Scholar]
- 11. Drechsel H., et al. 1995. Structure elucidation of yersiniabactin, a siderophore from highly virulent Yersinia strains. Liebigs Ann. 1995:1727–1733 [Google Scholar]
- 12. Drechsel H., et al. 1993. Purification and chemical characterization of staphyloferrin B, a hydrophilic siderophore from staphylococci. Biometals 6:185–192 [DOI] [PubMed] [Google Scholar]
- 13. Emery T. 1971. Role of ferrichrome as a ferric ionophore in Ustilago sphaerogena. Biochemistry 10:1483–1488 [DOI] [PubMed] [Google Scholar]
- 14. Ephinstone J. G. 2005. The current bacterial wilt situation: a global overview, p. 9–28 In Allen C., Prior P., Hayward A. C. (ed.), Bacterial wilt: the disease and the Ralstonia solanacearum species complex. American Phytopathological Society, St. Paul, MN [Google Scholar]
- 15. Expert D., Enard C., Masclaux C. 1996. The role of iron in plant host-pathogen interactions. Trends Microbiol. 4:232–237 [DOI] [PubMed] [Google Scholar]
- 16. Expert D. 1999. Withholding and exchanging iron: interactions between Erwinia spp. and their plant hosts. Annu. Rev. Phytopathol. 37:307–334 [DOI] [PubMed] [Google Scholar]
- 17. Gehring A. M., Mori I., Perry R. D., Walsh C. T. 1998. The nonribosomal peptide synthetase HMWP2 forms a thiazoline ring during biogenesis of yersiniabactin, an iron-chelating virulence factor of Yersinia pestis. Biochemistry 37:11637–11650 [DOI] [PubMed] [Google Scholar]
- 18. Genin S., Boucher G. 2002. Ralstonia solanacearum: secrets of a major pathogen unveiled by analysis of its genome. Mol. Plant Pathol. 3:111–118 [DOI] [PubMed] [Google Scholar]
- 19. Genin S. 2010. Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum. New Phytologist 187:920–928 [DOI] [PubMed] [Google Scholar]
- 20. Guidot A., et al. 2009. Specific genes from the potato brown rot strains of Ralstonia solanacearum and their potential use for strain detection. Phytopathology 99:1105–1112 [DOI] [PubMed] [Google Scholar]
- 21. Guidot A., et al. 2007. Genomic structure and phylogeny of the plant pathogen Ralstonia solanacearum inferred from gene distribution analysis. J. Bacteriol. 189:377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gust B., Challis G. L., Fowler K., Kieser T., Chater K. F. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hider R. C., Kong X. 2010. Chemistry and biology of siderophores. Nat. Prod. Rep. 27:637–657 [DOI] [PubMed] [Google Scholar]
- 24. Kobayashi S., et al. 1998. Micacocidin A, B and C, novel antimycoplasma agents from Pseudomonas sp. II. Structure elucidation. J. Antibiot. 51:328–332 [DOI] [PubMed] [Google Scholar]
- 25. Kobayashi S., Hidaka S., Kawamura Y., Ozaki M., Hayase Y. 1998. Micacocidin A, B and C, novel antimycoplasma agents from Pseudomonas sp. I. Taxonomy, fermentation, isolation, physico-chemical properties, and biological activities. J. Antibiot. 51:323–327 [DOI] [PubMed] [Google Scholar]
- 26. Laatsch H. 2008. AntiBase: the natural compound identifier. Wiley-VCH, Weinheim, Germany [Google Scholar]
- 27. Ménard R., Sansonetti P. J., Parsot C. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899–5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Milagres A. M., Machuca A., Napoleão D. 1999. Detection of siderophore production from several fungi and bacteria by a modification of chrome azurol S (CAS) agar plate assay. J. Microbiol. Methods 37:1–6 [DOI] [PubMed] [Google Scholar]
- 29. Miller D. A., Luo L. S., Hillson N., Keating T. A., Walsh C. T. 2002. Yersiniabactin synthetase: a four-protein assembly line producing the nonribosomal peptide/polyketide hybrid siderophore of Yersinia pestis. Chem. Biol. 9:333–344 [DOI] [PubMed] [Google Scholar]
- 30. Miller M. C., Parkin S., Fetherstone J. D., Perry R. D., DeMoll E. 2006. Crystal structure of ferric-yersiniabactin, a virulence factor of Yersinia pestis. J. Inorg. Biochem. 100:1495–1500 [DOI] [PubMed] [Google Scholar]
- 31. Paauw A., Leverstein-van Hall M., van Kessel K. P. M., Verhoef J., Fluit A. C. 2009. Yersiniabactin reduces the respiratory oxidative stress response of innate immune cells. PLoS One 4:e8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Remenant B., et al. 2010. Genomes of three tomato pathogens within the Ralstonia solanacearum species complex reveal significant evolutionary divergence. BMC Genomics 11:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Salanoubat M., et al. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497–502 [DOI] [PubMed] [Google Scholar]
- 34. Sandy M., Butler A. 2009. Microbial iron acquisition: marine and terrestrial siderophores. Chem. Rev. 109:4580–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schaible U. E., Kaufmann S. H. E. 2004. Iron and microbial infection. Nat. Rev. Microbiol. 2:946–953 [DOI] [PubMed] [Google Scholar]
- 36. Schlegel K., Taraz K., Budzikiewicz H. 2004. The stereoisomers of pyochelin, a siderophore of Pseudomonas aeruginosa. Biometals 17:409–414 [DOI] [PubMed] [Google Scholar]
- 37. Schneider P., et al. 2009. The global virulence regulators VsrAD and PhcA control secondary metabolism in the plant pathogen Ralstonia solanacearum. ChemBioChem 10:2730–2732 [DOI] [PubMed] [Google Scholar]
- 38. Schwyn B., Neilands J. B. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47–56 [DOI] [PubMed] [Google Scholar]
- 39. Stachelhaus T., Mootz H. D., Marahiel M. A. 1999. The selectivity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493–505 [DOI] [PubMed] [Google Scholar]
- 40. Tae H., Kong E. B., Park K. 2007. ASMPKS: an analysis system for modular polyketide synthases. BMC Bioinformatics. 8:327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Udwary D. W., et al. 2007. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. U. S. A. 104:10376–10381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wackler B., et al. 2011. Ralfuranone biosynthesis in Ralstonia solanacearum suggests functional divergence in the quinone synthetase family of enzymes. Chem. Biol. 18:354–360 [DOI] [PubMed] [Google Scholar]
- 43. Waterfield N. R., et al. 2008. Rapid virulence annotation (RVA): identification of virulence factors using a bacterial genome library and multiple invertebrate hosts. Proc. Natl. Acad. Sci. U. S. A. 105:15967–15972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yabuuchi E., Kawamura Y., Ezaki T. 2005. Genus Ralstonia Yabuuchi, Kosako, Yano, Hotta, and Nishiuchi 1996, p. 609–620 In Garrity G. M., Brenner D. J., Krieg N. R., Staley J. T. (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2 Springer, New York, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.