Abstract
Anaerobic benzene oxidation coupled to the reduction of Fe(III) was studied in Ferroglobus placidus in order to learn more about how such a stable molecule could be metabolized under strict anaerobic conditions. F. placidus conserved energy to support growth at 85°C in a medium with benzene provided as the sole electron donor and Fe(III) as the sole electron acceptor. The stoichiometry of benzene loss and Fe(III) reduction, as well as the conversion of [14C]benzene to [14C]carbon dioxide, was consistent with complete oxidation of benzene to carbon dioxide with electron transfer to Fe(III). Benzoate, but not phenol or toluene, accumulated at low levels during benzene metabolism, and [14C]benzoate was produced from [14C]benzene. Analysis of gene transcript levels revealed increased expression of genes encoding enzymes for anaerobic benzoate degradation during growth on benzene versus growth on acetate, but genes involved in phenol degradation were not upregulated during growth on benzene. A gene for a putative carboxylase that was more highly expressed in benzene- than in benzoate-grown cells was identified. These results suggest that benzene is carboxylated to benzoate and that phenol is not an important intermediate in the benzene metabolism of F. placidus. This is the first demonstration of a microorganism in pure culture that can grow on benzene under strict anaerobic conditions and for which there is strong evidence for degradation of benzene via clearly defined anaerobic metabolic pathways. Thus, F. placidus provides a much-needed pure culture model for further studies on the anaerobic activation of benzene in microorganisms.
INTRODUCTION
The anaerobic degradation of benzene is of interest because of its environmental significance (25, 38, 63) and because the mechanisms for the initial metabolic attack on such a stable molecule are expected to be novel (14, 22, 63, 66). The lack of pure cultures in which it can definitely be stated that benzene is metabolized without molecular oxygen has greatly stymied the study of this process.
The only previously described pure cultures suggested to anaerobically degrade benzene have used nitrate or chlorate as an electron acceptor (21, 32, 62). Anaerobic benzene degradation has been studied in detail in one of these organisms, Dechloromonas aromatica (15, 16, 21, 55). One of the most surprising findings in the study of D. aromatica is that it lacks all of the otherwise highly conserved genes involved in the anaerobic degradation of monoaromatic compounds (55), such as benzoate and phenol, which are otherwise found in all Bacteria (13) and Archaea (30) that can metabolize these compounds. In addition, it appears that oxygen incorporated into the benzene ring does not come from water as would be expected for anaerobic benzene degradation (15). These results have lead to the suggestion that D. aromatica might activate benzene with oxygen produced intracellularly during growth on nitrate (55, 63).
However, benzene is clearly degraded under anoxic conditions in natural communities and enrichment cultures. Benzene was anaerobically degraded in sediments coupled to the reduction of Fe(III) (4, 10, 43, 44, 53), Mn(IV) (61), sulfate (3, 20, 23, 27, 34, 35, 40, 64), carbon dioxide (17, 65), and graphite electrodes (67). Enrichment cultures capable of anaerobic oxidation of benzene with either sulfate (1, 7, 18, 19, 49–51), carbon dioxide (26, 54, 59), or Fe(III) (11, 31, 37, 53) as the electron acceptor have been described. A number of different species appear to be involved in benzene degradation in these enrichment cultures. For example, different species of Gram-positive bacteria and Delta- and/or Epsilonproteobacteria have been associated with benzene degradation in sulfate-reducing (1, 52), Fe(III)-reducing (37), and methanogenic (60) enrichments. Archaeal species have also been detected in sulfate-reducing enrichments (7).
Multiple pathways for anaerobic benzene degradation have been proposed, and different microorganisms may employ alternate pathways (22, 46). In early studies with methanogenic benzene-degrading enrichment cultures, phenol and cyclohexanone were detected as intermediates, leading to the suggestion that first benzene was hydroxylated to phenol and then the ring was reduced to produce cyclohexanone (26). Detection of phenol in sulfate-reducing enrichments also supported the hydroxylation pathway (12). However, the potential for abiotic production of phenol from benzene during sampling has demonstrated that evidence beyond mere detection of phenol may be required before phenol can be designated an intermediate (1, 36). Production of [13C]toluene and [13C]benzoate in methanogenic and nitrate-reducing enrichments metabolizing [13C]benzene suggested that benzene was initially methylated, followed by transformation of toluene to benzoate (59). It has also been proposed that benzene may be directly carboxylated to benzoate, based on the production of benzoate in enrichment cultures (1, 36), and a putative carboxylase was detected in protein extracts from an Fe(III)-reducing enrichment culture during growth on benzene but not phenol or benzoate (2).
The vast majority of studies on the anaerobic degradation of aromatic compounds have focused on mesophilic bacteria. However, it was previously demonstrated that the hyperthermophilic archaeon Ferroglobus placidus is able to anaerobically oxidize benzoate and phenol with Fe(III) as the electron acceptor (30, 58). The F. placidus genes for benzoate and phenol metabolism are homologous to those found in mesophilic bacteria, many of the genes are arranged in clusters similar to those found in bacteria, and the expression of the appropriate genes is specifically increased during growth on benzoate or phenol (30). A unique feature of benzoate metabolism in F. placidus is that it appears to use a ATP-consuming class I benzoyl coenzyme A (benzoyl-CoA) reductase, similar to those found in facultative bacteria, rather than the ATP-independent class II benzoyl-CoA reductase found in all other strict anaerobes (30).
Here we report that F. placidus is capable of anaerobic growth on benzene with Fe(III) serving as the electron acceptor. Gene expression patterns and metabolite data suggest that benzene is converted directly to benzoate and that phenol is not an important intermediate. This study also identifies a putative carboxylase protein involved in benzene activation that is homologous to a protein identified by proteomic and genomic analysis of benzene degradation in an Fe(III)-reducing enrichment culture (2).
MATERIALS AND METHODS
Growth of Ferroglobus placidus.
Ferroglobus placidus strain AEDII12DO DSM 10642 was obtained from the type culture collection of the Deutsche Sammlung von Mikroorganismen and Zelkuturen (DSMZ), Braunschweig, Germany. Strict anaerobic culturing and sampling techniques were used throughout (6, 48). The growth medium was prepared as previously described (30, 57) with slight alterations that included additional selenate and tungstate (Na2SeO4 was 30 μg/liter and Na2WO4 was 40 μg/liter), 1 g/liter MgCl2, and 0.23 g/liter CaCl2. Cells were grown with either Fe(III) citrate (56 mM) or amorphous Fe(III) oxyhydroxide (100 mmol/liter) as an electron acceptor, and electron donors were supplied from stock solutions to provide a final concentration of 10 mM acetate, 0.5 mM phenol, 1 mM benzoate, or 2 mM benzene. All incubations were under N2-CO2 (80:20) at 85°C in the dark.
In order to adapt F. placidus for growth on benzene, it was initially grown in the presence of benzene with hydrogen added to provide an initial partial pressure of 101 kPa. Following growth in this medium, the culture was propagated in medium without hydrogen.
Analytical techniques.
Cell numbers were determined with epifluorescence microscopy as previously described (28, 41). Culture samples were anaerobically removed and fixed with glutaraldehyde (final concentration, 2.5%). The iron minerals were dissolved with an acidic oxalate solution (28 g/liter ammonium oxalate and 15 g/liter oxalic acid) for 1 h. The cells were treated with an acridine orange solution (final concentration of 0.01%) and collected on a Nuclepore membrane filter (0.2-μm pore diameter, GTBP; Millipore). These filters were then observed under oil immersion with a Nikon Eclipse E600 epifluorescence microscope.
Fe(III) reduction was monitored by measuring the formation of Fe(II) in the medium over time as previously described (41, 42).
Benzene and toluene concentrations were quantified with a gas chromatograph (Perkin-Elmer Clarus 600 gas chromatograph) equipped with a flame ionization detector. The hydrocarbons were separated on a Supelco Vocol fused-silica capillary column (no. 24154) held at 50°C for 0.5 min, followed by an increase to 200°C at a rate of 10°C/min. The concentration of benzene in the aqueous phase was calculated with Henry's law constants for 85°C.
Cultures of F. placidus with insoluble Fe(III) as an electron acceptor were amended with 1 μCi of [ring-UI-14C]benzene (75 mCi/mmol; Moravek Biochemicals, Brea, CA). Aqueous stock solutions of [14C]benzene were prepared as previously described (4). Production of 14CO2 in the headspace was monitored using gas chromatography coupled with a gas proportional counter detector (39).
Concentrations of benzoate were determined with high-performance liquid chromatography (HPLC) (Agilent 1100 HPLC series) with an Altima HP C18 HL column. The eluent consisted of methanol (MeOH)-H2O (60:40) and 0.1% H2PO4, and the compounds were detected at an absorbance of 280 nm. The detection limit for benzoate with this system was 0.5 μM.
For thin-layer chromatography, cultures grown with insoluble Fe(III) as an electron donor were amended with 1 μCi of [ring-UI-14C]benzene, as described above. After 27 days of incubation, when benzene was being actively metabolized, 0.25-ml aliquots were extracted and centrifuged at 16,100 × g for 5 min to separate the solid Fe(III). The supernatant was filtered with a 0.2-μm filter, and 30 μl of the supernatant was loaded on Whatman flexible plates (250-μm layer Al/Sil G/UV), 1 μl at a time, with drying in between applications. The solvent mix was benzene-dioxane-acetic acid (8:1:1) (9), and 3 μl of 7.68-μCi/ml stock [14C]phenol and 8.6-μCi/ml stock [14C]benzoate were used as standards. The plates were exposed to a phosphor screen (Molecular Dynamics) for 3 days and digitized with a Typhoon 9210 scanner (Amersham).
Extraction of RNA from samples.
RNA was extracted from triplicate cultures during exponential growth. Cells were pelleted by centrifugation at 3,000 × g for 15 min at 4°C. Pellets were then immediately frozen in liquid nitrogen and stored at −80°C. RNA was extracted from these pellets as previously described (30). All extracted RNA samples had A260/A280 ratios of 1.8 to 2.0, indicating that they were of high purity (5). In order to ensure that RNA samples were not contaminated with DNA, PCR amplification with primers targeting the 16S rRNA gene was conducted on RNA samples that had not undergone reverse transcription.
Microarray analysis.
Whole-genome microarray hybridizations were carried out by Roche NimbleGen, Inc. RNA was amplified with the TransPlex whole-transcriptome amplification kit (Sigma) prior to transcriptomic analyses. Triplicate biological and technical replicates were conducted for all microarray analyses. All cDNA samples were chemically labeled with Cy3 and hybridized to oligonucleotide microarrays that were designed based on the first draft of the F. placidus genome sequence (accession number NC_013849) obtained from the DOE Joint Genome Institute (JGI) website (www.jgi.doe.gov).
Results from microarray hybridizations were analyzed with the software Array 4 Star (DNASTAR). P values were determined with Student t test analysis. A gene was considered differentially expressed only if the P value was less than or equal to 0.01.
Testing and design of primers for qRT-PCR.
Genome sequence data obtained from the DOE Joint Genome Institute (JGI) website (www.jgi.doe.gov) were used to design quantitative reverse transcription-PCR (qRT-PCR) primers. All qRT-PCR primers were designed according to the manufacturer's specifications (amplicon size, 100 to 200 bp), and representative products from each of these primer sets were verified by sequencing clone libraries. All of the primer pairs used in this study were previously described elsewhere (30).
PCR amplification parameters and clone library construction.
A DuraScript enhanced avian RT single-strand synthesis kit (Sigma) was used to generate cDNA as previously described (29). For clone library construction, PCR products were purified with a gel extraction kit (Qiagen), and clone libraries were constructed with a TOPO TA cloning kit, version M (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. One hundred plasmid inserts from each clone library were sequenced with the M13F primer at the University of Massachusetts Sequencing Facility.
Quantification of gene expression by quantitative RT-PCR.
Once the appropriate cDNA fragments were generated by RT-PCR, quantitative PCR amplification and detection were performed with the 7500 real-time PCR system (Applied Biosystems). Optimal quantitative RT-PCR conditions were determined using the manufacturer's guidelines. Each PCR mixture consisted of a total volume of 25 μl and contained 1.5 μl of the appropriate primers (stock concentrations, 15 μM) and 12.5 μl of Power SYBR green PCR master mix (Applied Biosystems). Standard curves covering 8 orders of magnitude were constructed with serial dilutions of known amounts of purified cDNA quantified with a NanoDrop ND-1000 spectrophotometer at an absorbance of 260 nm.
The housekeeping gene gyrA, which codes for the alpha subunit of DNA topoisomerase, was used as an external control. This gene was selected because it appears to be constitutively expressed by F. placidus species under a variety of growth conditions (30), and it was not differentially expressed in any of the microarray experiments. This gene has also been used as an external control for studies with other organisms (56).
Microarray data accession numbers.
A complete record of all oligonucleotide sequences used and raw and statistically treated data files is available in the NCBI Gene Expression Omnibus database (GEO data series numbers GSE30798 [benzene versus acetate], GSE30801 [benzene versus benzoate], and GSE30799 [benzene versus phenol]).
RESULTS AND DISCUSSION
Growth and stoichiometry of benzene degradation.
Cultures of F. placidus that had been propagated for more than a year through 15 sequential transfers with benzene as the sole electron donor grew without a detectable lag phase in medium with benzene provided as the sole electron donor and Fe(III) oxide as the sole electron acceptor (Fig. 1). It was previously reported that F. placidus could not grow on benzene (58). The success in growing F. placidus on benzene in the studies reported here may be attributed to slight changes in the medium composition and/or the initial culturing of F. placidus in the presence of hydrogen and benzene, with hydrogen potentially providing energy to permit adaptation for growth on benzene. Additional nutritional studies might indicate whether any of the medium modifications were significant, which could provide further insight into the biochemistry of benzene metabolism.
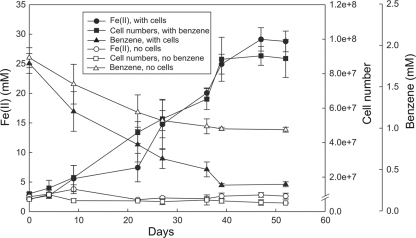
Fig. 1.
Growth of and Fe(III) reduction by Ferroglobus placidus under anaerobic conditions with benzene as the sole electron donor and Fe(III) oxide as the sole electron acceptor. A relatively high concentration of benzene (ca. 2 mM) was provided to permit maximum cell growth with the Fe(III) available. Results are the means and standard deviations from three replicate cultures.
During growth on benzene, the increase in cell numbers tracked well with the accumulation of Fe(II) from Fe(III) reduction (Fig. 1). There was no increase in Fe(II) or cell numbers in controls without benzene. There was some loss of benzene over time in controls without cells, which was attributed to adsorption of benzene in the butyl rubber stoppers that were used to seal the tubes. However, there was substantially more benzene loss in the presence of cells (Fig. 1). Given that most of the carbon and electron flow during the oxidation of organic compounds coupled to Fe(III) reduction goes toward respiration (45), the additional loss of benzene in the presence of cells (0.66 ± 0.03 mM) and the accumulation of Fe(II) (23.33 ± 1.62 mM) over time were close to those expected for the stoichiometry of the reaction: C6H6 + 30Fe3+ + 12H2O → 6CO2 + 30Fe2+ + 30H+.
Benzene metabolism stopped after Fe(III) was depleted as an electron acceptor (Fig. 1). Although 100 mmol/liter of Fe(III) oxide was initially provided, typically only one-third of this Fe(III) is available for reduction because the Fe(II) produced combines with the remaining Fe(III) to produce magnetite, which is not a suitable electron acceptor (33, 42). In order to further evaluate anaerobic benzene oxidation, the cells were cultured with lower concentrations of benzene to favor a more complete utilization of the added benzene and [14C]benzene was added to facilitate direct monitoring of benzene oxidation (Fig. 2).
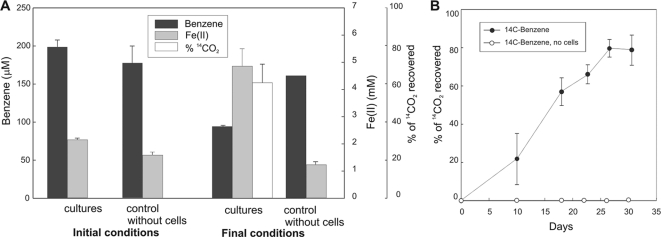
Fig. 2.
Metabolism of benzene at low benzene concentrations. (A) Initial and final (29 days of incubation) concentrations of benzene, Fe(II), and 14CO2 in benzene-Fe(III) oxide cultures amended with [14C]benzene. (B) Conversion of [14C]benzene to 14CO2 in cultures with a relatively low concentration of benzene (200 μM) and a high electron acceptor concentration [56 mM Fe(III) citrate] to promote maximum benzene utilization. The data points represent the means and standard deviations from three replicate cultures.
In stoichiometry studies (Fig. 2A) only initial and final time points were used to prevent loss of benzene from repeated headspace sampling. The consumption of benzene in the cultures beyond that lost in the controls without cells (103.7 ± 10.35 μM [mean ± standard deviation; n = 3]) and the production of Fe(II) (2.7 ± 1.1 mM) compared favorably with the 3.1 mM Fe(II) expected from complete benzene oxidation. A total of 60.83% ± 11.76% of the added [14C]benzene was recovered as 14CO2, which was consistent with loss of 52.12% ± 5.39% of the initial benzene that could be attributed to microbial activity in the culture. In a study designed to track benzene oxidation over time, there was a steady production of 14CO2 from [14C]benzene over time and a maximum of 81.2% ± 14.0% of the added [14C]benzene was recovered as 14CO2 (Fig. 2B), providing further proof that F. placidus effectively oxidized benzene to carbon dioxide.
Production of benzoate from benzene.
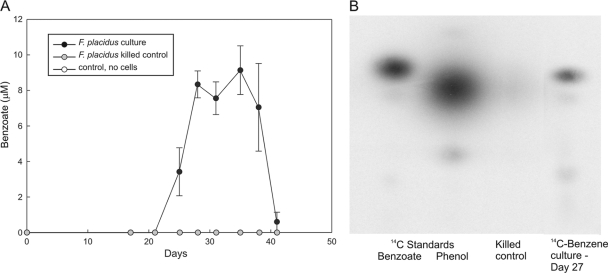
Benzoate, but not phenol, was detected with high-performance liquid chromatography while F. placidus was growing with benzene as the sole electron donor (Fig. 3A). Up to 10.5 μM benzoate was detected in F. placidus cultures grown with benzene, whereas benzoate was below the detection limit in sterile and killed controls. No toluene was detected with gas chromatography (detection limit, approximately 10 ppb). Thin-layer chromatography analysis of filtrates from cultures actively metabolizing [14C]benzene also detected [14C]benzoate (Fig. 3B).
Fig. 3.
Detection of benzoate during growth on benzene with Fe(III) oxide as the electron acceptor. (A) High-pressure liquid chromatography analysis. Benzoate levels were below the HPLC detection limit in killed controls or controls with no cells. Results are the means and standard deviations from triplicate cultures. (B) Production of [14C]benzoate from [14C]benzene detected with thin-layer chromatography.
Transcriptomic analysis indicative of benzene metabolism to benzoate.
Three different whole-genome DNA microarray studies were analyzed to gain insight into benzene metabolism by F. placidus: benzene versus acetate, benzene versus benzoate, and benzene versus phenol (see Tables S1 to S3 in the supplemental material). The number of genes that exhibited at least a 2-fold change in expression (P value cutoff = 0.01) was relatively high for all comparisons (benzene versus acetate, 209 upregulated and 188 downregulated; benzene versus benzoate, 77 upregulated and 977 downregulated; benzene versus phenol, 151 upregulated and 138 downregulated). Unfortunately, it was not possible to grow F. placidus in chemostats to control for factors such as different growth rates on the various substrates in batch culture, and it is expected that many of the changes in gene expression were related to the use of batch cultures. Therefore, analysis of the microarray results focused on genes thought to be involved in the anaerobic degradation of aromatic compounds.
The microarray analysis of gene transcript levels in cells grown on benzene versus cells grown on acetate (Table 1; see Table S1 in the supplemental material) demonstrated a widespread increase in transcript abundance for genes that are homologous to genes that are known to encode proteins involved in anaerobic benzoate degradation in other microorganisms and were previously reported to be specifically upregulated in F. placidus cells metabolizing benzoate (30). More quantitative analysis of transcript levels confirmed this finding (Fig. 4).
Table 1.
Genes encoding proteins from the benzoate-CoA ligation pathway that were upregulated according to microarray and qRT-PCR analyses during growth on 1 mM benzene (experimental condition) and 10 mM acetate or 1 mM benzoate (control conditions) provided as the electron donor and Fe(III) citrate (55 mM) provided as the electron acceptor
| Gene ID | Gene annotation | Gene name | Fold change (mean ± SD)a |
|||
|---|---|---|---|---|---|---|
| Benzene vs acetate |
Benzene vs benzoate |
|||||
| Microarray | qRT-PCR | Microarray | qRT-PCR | |||
| Ferp_1031 | Didehydro-pimeloyl-CoA hydratase | dph | 3.72 | 6.80 ± 2.35 | ND | 1.22 ± 0.32 |
| Ferp_1035 | Cyclohex-1-ene-1-carboxyl-CoA hydratase | badK1 | ND | 2.11 ± 0.98 | ND | 1.35 ± 0.55 |
| Ferp_1040 | 2-Ketocyclohexanecarboxyl-CoA hydrolase | badI | ND | 2.75 ± 1.35 | ND | 1.24 ± 0.64 |
| Ferp_1041 | 2-Hydroxy-glutaryl-CoA reductase, component A | hgdC | 2.38 | 4.15 ± 1.65 | −2.61 | −3.17 ± 1.53 |
| Ferp_1042 | 2-Hydroxy-glutaryl-CoA reductase, component D | hgdA | 2.42 | 9.30 ± 3.27 | ND | 1.01 ± 0.80 |
| Ferp_1043 | 2-Hydroxy-glutaryl-CoA reductase, component D | hgdB | ND | 2.25 ± 0.76 | ND | 1.26 ± 0.88 |
| Ferp_1044 | Benzoate-CoA ligase | bcl1 | 3.01 | 7.32 ± 2.16 | ND | 1.99 ± 0.23 |
| Ferp_1180 | Ferredoxin | fdx | 3.54 | 11.80 ± 2.34 | ND | 1.75 ± 0.57 |
| Ferp_1184 | Benzoyl-CoA reductase, gamma subunit | bzdN | 3.83 | 5.32 ± 3.12 | ND | 1.44 ± 0.49 |
| Ferp_1185 | Benzoyl-CoA reductase, beta subunit | bzdO | 2.01 | 5.23 ± 2.96 | ND | 1.23 ± 0.21 |
| Ferp_1186 | Benzoyl-CoA reductase, delta subunit | bzdP | 3.62 | 9.81 ± 3.75 | ND | 1.96 ± 0.35 |
| Ferp_1187 | Benzoyl-CoA reductase, alpha subunit | bzdQ | ND | 2.27 ± 1.18 | ND | 1.89 ± 0.77 |
| Ferp_1233 | 2-Hydroxycyclohexanecarboxyl-CoA dehydrogenase | badH | 2.62 | 5.65 ± 2.33 | ND | 1.76 ± 0.50 |
| Ferp_1543 | Carboxylase-like protein | ppcX1 | ND | 0.96 ± 0.32 | ND | 1.24 ± 0.45 |
| Ferp_1566 | Pimeloyl-CoA dehydrogenase (large subunit) | pimC | ND | 2.41 ± 1.31 | ND | 1.11 ± 0.62 |
| Ferp_1579 | Pimeloyl-CoA dehydrogenase (small subunit) | pimD | ND | 2.25 ± 1.77 | −2.14 | −4.77 ± 2.11 |
| Ferp_1630 | Carboxylase UbiD-like protein | ppcX2 | 2.98 | 5.63 ± 1.46 | 2.53 | 5.65 ± 2.15 |
Results were obtained from triplicate biological and technical replicates. A negative sign represents a negative fold change in expression. ND, no difference in gene expression.
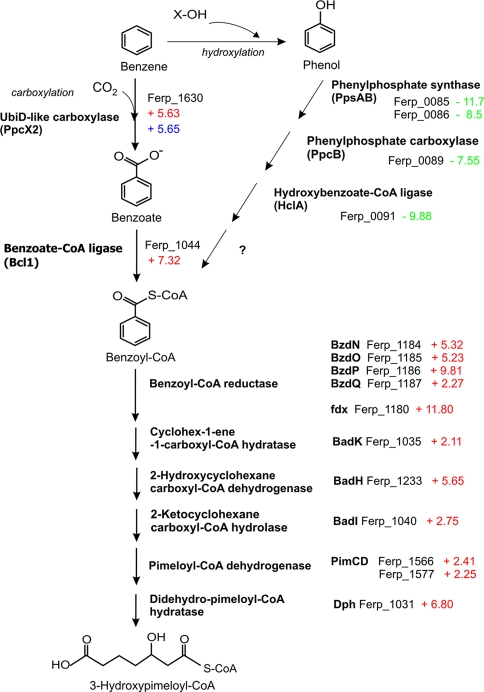
Fig. 4.
Transcriptomic analysis of benzene degradation in F. placidus. The numbers next to the gene designations represent the fold change in transcript abundance determined with quantitative RT-PCR analyses in benzene- versus acetate-grown cells (red), benzene- versus phenol-grown cells (green), or benzene- versus benzoate-grown cells (blue). The results are the means of triplicate determinations from each of three replicate cultures.
For example, the gene encoding benzoate-CoA ligase, the enzyme responsible for benzoate activation to benzoyl-CoA, was significantly upregulated during growth on benzene compared to growth on acetate. There are 12 genes in the F. placidus genome annotated as AMP-dependent synthetase/ligase genes (30), and two of these genes, Ferp_1044 and Ferp_1228, were upregulated 3- and 2.6-fold, respectively, during growth on benzene compared to growth on acetate (Table 1; see Table S1 in the supplemental material). These same genes were significantly upregulated in previous transcriptomic studies comparing benzoate- and acetate-grown cells (30). Ferp_1044 is the most likely candidate for benzoate-CoA ligase because it is clustered with other genes associated with benzoate metabolism and was the most highly upregulated candidate gene during growth on benzoate (30).
Other genes further down the benzoate pathway were also significantly upregulated in benzene- versus acetate-grown cells. For example, all of the genes encoding subunits of benzoyl-CoA reductase (BzdNOPQ), the enzyme responsible for the ring reduction of benzoate, as well as enzymes involved in the formation of 3-hydroxypimeloyl-CoA, were upregulated in benzene-grown cells compared to acetate-grown cells (Table 1 and Fig. 4).
These results indicated that benzoate was an intermediate in benzene metabolism. Transcript abundances for genes involved in benzoate metabolism are comparable in benzene- and benzoate-grown cells (Table 1; see Table S2 in the supplemental material), further supporting this conclusion.
In contrast, genes that have high homology to genes known to be involved in anaerobic degradation of phenol in other microorganisms and that are specifically upregulated during growth of F. placidus on phenol (30) did not have increased transcript abundance in benzene- versus acetate-grown cells (Fig. 4; see Table S1 in the supplemental material). The transcript abundance of genes from the phenylphosphate/phenylcarboxylase pathway, which previous studies have shown to be involved in anaerobic phenol degradation by F. placidus (30), was much lower in benzene- than in phenol-grown cells (Fig. 4; see Table S3 in the supplemental material). For example, genes from a phenol-specific cluster coding for phenylphosphate synthase (ppsAB; Ferp_0085-86), phenylphosphate carboxylase (ppcB; Ferp_0089), and 4-hydroxybenzoate-CoA ligase (hclA; Ferp_0091) were all significantly downregulated in benzene-grown cells compared to phenol-grown cells (Table 2, Fig. 4). These results indicated that phenol is not an intermediate in benzene metabolism in F. placidus.
Table 2.
Genes encoding proteins from the phenol degradation pathway that were downregulated according to microarray and qRT-PCR analyses during growth on 1 mM benzene (experimental condition) or 0.5 mM phenol (control condition) provided as the electron donor and Fe(III) citrate (55 mM) provided as the electron acceptor
| Gene ID | Gene annotation | Gene name | Fold change (mean ± SD) in benzene vs phenola |
|
|---|---|---|---|---|
| Microarray | qRT-PCR | |||
| Ferp_0085 | Phenylphosphate synthase subunit B | ppsB | −3.90 | −11.7 ± 3.26 |
| Ferp_0086 | Phenylphosphate synthase subunit A | ppsA | −9.47 | −8.5 ± 3.33 |
| Ferp_0089 | Phenylphosphate carboxylase subunit beta | ppcB | −6.17 | −7.55 ± 2.11 |
| Ferp_0090 | Dehalogenase domain protein hydrolase | −9.68 | −11.31 ± 1.32 | |
| Ferp_0091 | 4-Hydroxybenzoate-CoA ligase | hclA | −7.74 | −9.88 ± 2.25 |
| Ferp_0092 | Carboxylase-like ORF 8 | ppcY | −2.36 | −6.35 ± 1.45 |
| Ferp_0093 | HAD superfamily hydrolase | −5.17 | −10.32 ± 3.28 | |
| Ferp_0094 | Molybdopterin oxidoreductase | ND | 0.97 ± 0.12 | |
Results were obtained from triplicate biological and technical replicates. A negative sign represents a negative fold change in expression. ND, no difference in gene expression.
Putative carboxylase protein potentially involved in the initial activation of benzene.
A protein from a Clostridium species (ORF 138) was proposed to be involved in the carboxylation of benzene in an anaerobic benzene-degrading, Fe(III)-reducing enrichment culture based on the increased expression of this protein in benzene-grown but not in phenol- or benzoate-grown cultures (2). F. placidus has two genes with homology to ORF 138, Ferp_0089 and Ferp_1630. Previous studies have demonstrated that Ferp_0089 (32% identity and 48% similarity to ORF 138) codes for PpcB, a phenolphosphate carboxylase involved in phenol degradation (30). Furthermore, expression of this gene was not upregulated in benzene- versus benzoate- or acetate-grown cells, and its expression was downregulated in benzene- versus phenol-grown cells (Fig. 4 and Table 1; see Table S3 in the supplemental material). These results suggested that Ferp_0089 is not involved in benzene metabolism.
In contrast, Ferp_1630, the other ORF 138 homolog (31% identity and 47% similarity), had increased transcript abundance in benzene- versus acetate- or benzoate-grown cells (Fig. 4 and Table 1). The predicted protein encoded by Ferp_1630 shows high homology to UbiD from Escherichia coli (44% identical and 63% similar), which catalyzes the decarboxylation of 3-octaprenyl-4-hydroxybenzoic acid into 2-octaprenylphenol during the biosynthesis of ubiquinone. It is unlikely that the protein encoded by Ferp_1630 is involved in ubiquinone biosynthesis in F. placidus because none of the other genes that would be required for ubiquinone biosynthesis are present in the genome. Transcript abundance for Ferp_1630 was higher in benzene-grown cells than in cells grown on acetate or, more importantly, benzoate. These results suggest that Ferp_1630 may encode the enzyme responsible for carboxylation of benzene to benzoate.
The gene coding for ORF 138 from the benzene-degrading, Fe(III)-reducing enrichment culture was located within a cluster of genes containing three other potential carboxylase genes and a benzoate-CoA ligase gene (2). However, none of the genes in the proximity of Ferp_1630 are likely to code for carboxylase proteins, benzoate-CoA ligase proteins, or any other proteins involved in the metabolism of aromatic compounds. This is in contrast to most of the other genes involved in anaerobic aromatic degradation in F. placidus, which are localized within gene clusters. Additional analyses with either gene deletions or biochemical assays of the purified protein are warranted to further evaluate the role of the enzyme encoded by Ferp_1630 in benzene metabolism. However, as of now a genetic system for F. placidus has not been developed. Biochemical analysis will require strategies to generate much more biomass of benzene-grown F. placidus than is presently feasible or successful expression of the enzyme in another host.
Implications.
F. placidus is the first microorganism reported to be able to grow on benzene under strict anoxic conditions in pure culture and for which there is strong evidence for degradation of benzene via clearly defined anaerobic metabolic pathways. The accumulation of Fe(II) produced during Fe(III) reduction ensured removal of any trace oxygen contamination, and unlike with nitrate or chlorate, there is no conceivable mechanism to produce molecular oxygen from Fe(III) or Fe(II). Furthermore, F. placidus does not possess genes for the monooxygenases necessary to catalyze an oxidative attack on benzene.
The available evidence is consistent with carboxylation of benzene to benzoate. The upregulation of genes encoding enzymes catalyzing anaerobic benzoate degradation, as well as the accumulation of small amounts of benzoate during benzene metabolism, suggests that benzoate is a key intermediate. This evidence alone does not rule out other possible intermediates as benzoate precursors. However, phenol is not likely to be an important intermediate because during growth on benzene there was no upregulation of phenol degradation genes whose expression is induced when F. placidus is metabolizing phenol. Furthermore, phenol was not detected as an intermediate during growth on benzene. Toluene, a third alternative, was also not detected during benzene metabolism.
Activation of benzene via carboxylation has been previously proposed for a benzene-oxidizing, Fe(III)-reducing enrichment culture (2, 36). The high similarity of the putative carboxylase encoded by the gene Ferp_1630 in F. placidus and the putative carboxylase proposed to be responsible for benzene carboxylation in the enrichment culture suggests a similar enzymatic strategy. This may not be surprising because even though F. placidus is a hyperthermophile and phylogenetically distant, its other genes for the metabolism of aromatic compounds are closely related to those of mesophilic Bacteria (30). The increased expression of Ferp_1630 during growth on benzene versus growth on benzoate strongly suggests that it is specifically involved in benzene metabolism. Further study of the enzyme that Ferp_1630 encodes is warranted.
These studies extend the known metabolic potential of hyperthermophilic microorganisms. Aromatic compounds such as benzene are present in modern hot environments such as the hydrothermal vent in Vulcano, where F. placidus was first isolated (47), and could have been abundant components of organic matter on a hot, early Earth (8, 24). To date, F. placidus is the only hyperthermophile known to anaerobically oxidize aromatic compounds. However, genes from this metabolic pathway may be used as molecular markers to identify other hyperthermophiles involved in anaerobic degradation of benzene and other aromatic compounds in situ.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Office of Science (BER), U.S. Department of Energy, Cooperative Agreement no. DE-FC02-02ER63446.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 8 July 2011.
REFERENCES
- 1. Abu Laban N., Selesi D., Jobelius C., Meckenstock R. U. 2009. Anaerobic benzene degradation by Gram-positive sulfate-reducing bacteria. FEMS Microbiol. Ecol. 68:300–311 [DOI] [PubMed] [Google Scholar]
- 2. Abu Laban N., Selesi D., Rattei T., Tischler P., Meckenstock R. U. 2010. Identification of enzymes involved in anaerobic benzene degradation by a strictly anaerobic iron-reducing enrichment culture. Environ. Microbiol. 12:2783–2796 [DOI] [PubMed] [Google Scholar]
- 3. Anderson R. T., Lovley D. R. 2000. Anaerobic bioremediation of benzene under sulfate-reducing conditions in a petroleum-contaminated aquifer. Environ. Sci. Technol. 34:2261–2266 [Google Scholar]
- 4. Anderson R. T., Rooney-Varga J. N., Gaw C. V., Lovley D. R. 1998. Anaerobic benzene oxidation in the Fe(III) reduction zone of petroleum contaminated aquifers. Environ. Sci. Technol. 32:1222–1229 [Google Scholar]
- 5. Ausubel F. M., et al. 1997. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, NY [Google Scholar]
- 6. Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Berlendis S., Lascourreges J. F., Schraauwers B., Sivadon P., Magot M. 2010. Anaerobic biodegradation of BTEX by original bacterial communities from an underground gas storage aquifer. Environ. Sci. Technol. 44:3621–3628 [DOI] [PubMed] [Google Scholar]
- 8. Bernstein M. P., et al. 1999. UV irradiation of polycyclic aromatic hydrocarbons in ices: production of alcohols, quinones, and ethers. Science 283:1135–1138 [DOI] [PubMed] [Google Scholar]
- 9. Biegert T., Fuchs G., Heider J. 1996. Evidence that anaerobic oxidation of toluene in the denitrifying bacterium Thauera aromatica is initiated by formation of benzylsuccinate from toluene and fumarate. Eur. J. Biochem. 238:661–668 [DOI] [PubMed] [Google Scholar]
- 10. Botton S., Parsons J. R. 2006. Degradation of btex compounds under iron-reducing conditions in contaminated aquifer microcosms. Environ. Toxicol. Chem. 25:2630–2638 [DOI] [PubMed] [Google Scholar]
- 11. Botton S., Parsons J. R. 2007. Degradation of BTX by dissimilatory iron-reducing cultures. Biodegradation 18:371–381 [DOI] [PubMed] [Google Scholar]
- 12. Caldwell M. E., Suflita J. M. 2000. Detection of phenol and benzoate as intermediates of anaerobic benzene biodegradation under different terminal electron-accepting conditions. Environ. Sci. Technol. 34:1216–1220 [Google Scholar]
- 13. Carmona M., et al. 2009. Anaerobic catabolism of aromatic compounds: a genetic and genomic view. Microbiol. Mol. Biol. Rev. 73:71–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chakraborty R., Coates J. D. 2004. Anaerobic degradation of monoaromatic hydrocarbons. Appl. Microbiol. Biotechnol. 64:437–446 [DOI] [PubMed] [Google Scholar]
- 15. Chakraborty R., Coates J. D. 2005. Hydroxylation and carboxylation: two crucial steps of anaerobic benzene degradation by Dechloromonas strain RCB. Appl. Environ. Microbiol. 71:5427–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chakraborty R., O'Connor S. M., Chan E., Coates J. D. 2005. Anaerobic degradation of benzene, toluene, ethylbenzene, and xylene compounds by Dechloromonas strain RCB. Appl. Environ. Microbiol. 71:8649–8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chang W., Um Y., Pulliam Holoman T. R. 2005. Molecular characterization of anaerobic microbial communities from benzene-degrading sediments under methanogenic conditions. Biotechnol. Prog. 21:1789–1794 [DOI] [PubMed] [Google Scholar]
- 18. Chaudhuri B. K., Wiesmann U. 1995. Enhanced anaerobic degradation of benzene by enrichment of mixed microbial culture and optimization of the culture medium. Appl. Microbiol. Biotechnol. 43:178–187 [DOI] [PubMed] [Google Scholar]
- 19. Chen C. I., Taylor R. T. 1997. Thermophilic biodegradation of BTEX by two consortia of anaerobic bacteria. Appl. Microbiol. Biotechnol. 48:121–128 [DOI] [PubMed] [Google Scholar]
- 20. Coates J. D., Anderson R. T., Woodward J. C., Phillips E. J. P., Lovley D. R. 1996. Anaerobic hydrocarbon degradation in petroleum-contaminated harbor sediments under sulfate-reducing and artificially imposed iron-reducing conditions. Environ. Sci. Tech. 30:2784–2789 [Google Scholar]
- 21. Coates J. D., et al. 2001. Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature 411:1039–1043 [DOI] [PubMed] [Google Scholar]
- 22. Coates J. D., Chakraborty R., McInerney M. J. 2002. Anaerobic benzene biodegradation—a new era. Res. Microbiol. 153:621–628 [DOI] [PubMed] [Google Scholar]
- 23. Edwards E. A., Grbic-Galic D. 1992. Complete mineralization of benzene by aquifer microorganisms under strictly anaerobic conditions. Appl. Environ. Microbiol. 58:2663–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ehrenfreund P. 1999. Molecules on a space odyssey. Science 283:1123–1124 [DOI] [PubMed] [Google Scholar]
- 25. Farhadian M., Vachelard C., Duchez D., Larroche C. 2008. In situ bioremediation of monoaromatic pollutants in groundwater: a review. Bioresour. Technol. 99:5296–5308 [DOI] [PubMed] [Google Scholar]
- 26. Grbic-Galic D., Vogel T. M. 1987. Transformation of toluene and benzene by mixed methanogenic cultures. Appl. Environ. Microbiol. 53:254–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Herrmann S., et al. 2010. Functional characterization of an anaerobic benzene-degrading enrichment culture by DNA stable isotope probing. Environ. Microbiol. 12:401–411 [DOI] [PubMed] [Google Scholar]
- 28. Hobbie J. E., Daley R. J., Jasper S. 1977. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Holmes D. E., Nevin K. P., Lovley D. R. 2004. In situ expression of nifD in Geobacteraceae in subsurface sediments. Appl. Environ. Microbiol. 70:7251–7259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Holmes D. E., Risso C., Smith J., Lovley D. R. 2011. Genome-scale analysis of anaerobic benzoate and phenol metabolism in the hyperthermophilic archaeon Ferroglobus placidus. ISME J. [Epub ahead of print.] doi:10.1038/ismej.2011.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jahn M. K., Haderlein S. B., Meckenstock R. U. 2005. Anaerobic degradation of benzene, toluene, ethylbenzene, and o-xylene in sediment-free iron-reducing enrichment cultures. Appl. Environ. Microbiol. 71:3355–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kasai Y., Takahata Y., Manefield M., Watanabe K. 2006. RNA-based stable isotope probing and isolation of anaerobic benzene-degrading bacteria from gasoline-contaminated groundwater. Appl. Environ. Microbiol. 72:3586–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kashefi K., Lovley D. R. 2000. Reduction of Fe(III), Mn(IV), and toxic metals at 100 degrees C by Pyrobaculum islandicum. Appl. Environ. Microbiol. 66:1050–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kazumi J., Caldwell M. E., Suflita J. M., Lovley D. R., Young L. Y. 1997. Anaerobic degradation of benzene in diverse anoxic environments. Environ. Sci. Technol. 31:813–818 [Google Scholar]
- 35. Kleinsteuber S., et al. 2008. Molecular characterization of bacterial communities mineralizing benzene under sulfate-reducing conditions. FEMS Microbiol. Ecol. 66:143–157 [DOI] [PubMed] [Google Scholar]
- 36. Kunapuli U., Griebler C., Beller H. R., Meckenstock R. U. 2008. Identification of intermediates formed during anaerobic benzene degradation by an iron-reducing enrichment culture. Environ. Microbiol. 10:1703–1712 [DOI] [PubMed] [Google Scholar]
- 37. Kunapuli U., Lueders T., Meckenstock R. U. 2007. The use of stable isotope probing to identify key iron-reducing microorganisms involved in anaerobic benzene degradation. ISME J. 1:643–653 [DOI] [PubMed] [Google Scholar]
- 38. Lovley D. R. 1997. Potential for anaerobic bioremediation of BTEX in petroleum-contaminated aquifers. J. Ind. Microbiol. 18:75–81 [Google Scholar]
- 39. Lovley D. R., Chapelle F. H., Woodward J. C. 1994. Use of dissolved H2 concentrations to determine the distribution of microbially catalyzed redox reactions in anoxic groundwater. Environ. Sci. Technol. 28:1992–1992 [DOI] [PubMed] [Google Scholar]
- 40. Lovley D. R., Coates J. D., Woodward J. C., Phillips E. J. P. 1995. Benzene oxidation coupled to sulfate reduction. Appl. Environ. Microbiol. 61:953–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lovley D. R., Phillips E. J. P. 1988. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54:1472–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lovley D. R., Stolz J. F., Nord G. L., Phillips E. J. P. 1987. Anaerobic production of magnetite by a dissimilatory iron-reducing microorganism. Nature 330:252–254 [Google Scholar]
- 43. Lovley D. R., Woodward J. C., Chapelle F. H. 1994. Enhancement of aromatic hydrocarbon degradation in the suboxic zone of a contaminated aquifer. Abstr. Papers Am. Chem. Soc. 207:124–GEOC [Google Scholar]
- 44. Lovley D. R., Woodward J. C., Chapelle F. H. 1996. Rapid anaerobic benzene oxidation with a variety of chelated Fe(III) forms. Appl. Environ. Microbiol. 62:288–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mahadevan R., et al. 2006. Characterization of metabolism in the Fe(III) reducing organism, Geobacter sulfurreducens, by constraint-based modeling. Appl. Environ. Microbiol. 72:1558–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mancini S. A., et al. 2008. Isotopic evidence suggests different initial reaction mechanisms for anaerobic benzene biodegradation. Environ. Sci. Technol. 42:8290–8296 [DOI] [PubMed] [Google Scholar]
- 47. Mangani F., Cappiello A., Capaccioni B., Martini M. 1991. Sampling and analysis of light hydrocarbons in volcanic gases. Chromatographia 32:441–444 [Google Scholar]
- 48. Miller T. L., Wolin M. J. 1974. A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl. Microbiol. 27:985–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Musat F., Widdel F. 2008. Anaerobic degradation of benzene by a marine sulfate-reducing enrichment culture, and cell hybridization of the dominant phylotype. Environ. Microbiol. 10:10–19 [DOI] [PubMed] [Google Scholar]
- 50. Oka A. R., et al. 2008. Identification of critical members in a sulfidogenic benzene-degrading consortium by DNA stable isotope probing. Appl. Environ. Microbiol. 74:6476–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Phelps C. D., Kazumi J., Young L. Y. 1996. Anaerobic degradation of benzene in BTX mixtures dependent on sulfate reduction. FEMS Microbiol. Lett. 145:433–437 [DOI] [PubMed] [Google Scholar]
- 52. Phelps C. D., Kerkhof L. J., Young L. Y. 1998. Molecular characterization of a sulfate-reducing consortium which mineralizes benzene. FEMS Microbiol. Ecol. 27:269–279 [Google Scholar]
- 53. Rooney-Varga J. N., Anderson R. T., Fraga J. L., Ringelberg D., Lovley D. R. 1999. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sakai N., Kurisu F., Yagi O., Nakajima F., Yamamoto K. 2009. Identification of putative benzene-degrading bacteria in methanogenic enrichment cultures. J. Biosci. Bioeng. 108:501–507 [DOI] [PubMed] [Google Scholar]
- 55. Salinero K. K., et al. 2009. Metabolic analysis of the soil microbe Dechloromonas aromatica str. RCB: indications of a surprisingly complex life-style and cryptic anaerobic pathways for aromatic degradation. BMC Genomics 10:351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Theis T., Skurray R. A., Brown M. H. 2007. Identification of suitable internal controls to study expression of a Staphylococcus aureus multidrug resistance system by quantitative real-time PCR. J. Microbiol. Methods 70:355–362 [DOI] [PubMed] [Google Scholar]
- 57. Tor J. M., Kashefi K., Lovley D. R. 2001. Acetate oxidation coupled to Fe(III) reduction in hyperthermophilic microorganisms. Appl. Environ. Microbiol. 67:1363–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tor J. M., Lovley D. R. 2001. Anaerobic degradation of aromatic compounds coupled to Fe(III) reduction by Ferroglobus placidus. Environ. Microbiol. 3:281–287 [DOI] [PubMed] [Google Scholar]
- 59. Ulrich A. C., Beller H. R., Edwards E. A. 2005. Metabolites detected during biodegradation of 13C6-benzene in nitrate-reducing and methanogenic enrichment cultures. Environ. Sci. Technol. 39:6681–6691 [DOI] [PubMed] [Google Scholar]
- 60. Ulrich A. C., Edwards E. A. 2003. Physiological and molecular characterization of anaerobic benzene-degrading mixed cultures. Environ. Microbiol. 5:92–102 [DOI] [PubMed] [Google Scholar]
- 61. Villatoro-Monzon W. R., Mesta-Howard A. M., Razo-Flores E. 2003. Anaerobic biodegradation of BTEX using Mn(IV) and Fe(III) as alternative electron acceptors. Water Sci. Technol. 48:125–131 [PubMed] [Google Scholar]
- 62. Weelink S. A. B., et al. 2008. Isolation and characterization of Alicycliphilus denitrificans strain BC, which grows on benzene with chlorate as the electron acceptor. Appl. Environ. Microbiol. 74:6672–6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Weelink S. A. B., van Eekert M. H. A., Stams A. J. M. 2010. Degradation of BTEX by anaerobic bacteria: physiology and application. Rev. Environ. Sci. Biotechnol. 9:359–385 [Google Scholar]
- 64. Weiner J. M., Lovley D. R. 1998. Anaerobic benzene degradation in petroleum-contaminated aquifer sediments after inoculation with a benzene-oxidizing enrichment. Appl. Environ. Microbiol. 64:775–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Weiner J. M., Lovley D. R. 1998. Rapid benzene degradation in methanogenic sediments from a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 64:1937–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Widdel F., Rabus R. 2001. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr. Opin. Biotechnol. 12:259–276 [DOI] [PubMed] [Google Scholar]
- 67. Zhang T., Gannon S. M., Nevin K. P., Franks A. E., Lovley D. R. 2010. Stimulating the anaerobic degradation of aromatic hydrocarbons in contaminated sediments by providing an electrode as the electron acceptor. Environ. Microbiol. 12:1011–1020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.