Abstract
Three broiler feeding trials were investigated in order to identify gut bacteria consistently linked with improvements in bird performance as measured by feed efficiency. Trials were done in various geographic locations and varied in diet composition, broiler breed, and bird age. Gut microbial communities were investigated using microbial profiling. Eight common performance-linked operational taxonomic units (OTUs) were identified within both the ilea (180, 492, and 564–566) and ceca (140–142, 218–220, 284–286, 312, and 482) across trials. OTU 564–566 was associated with lower performance, while OTUs 140–142, 482, and 492 were associated with improved performance. Targeted cloning and sequencing of these eight OTUs revealed that they represented 26 bacterial species or phylotypes which clustered phylogenetically into seven groups related to Lactobacillus spp., Ruminococcaceae, Clostridiales, Gammaproteobacteria, Bacteroidales, Clostridiales/Lachnospiraceae, and unclassified bacteria/clostridia. Where bacteria were identifiable to the phylum level, they belonged predominantly to the Firmicutes, with Bacteroidetes and Proteobacteria also identified. Some of the potential performance-related phylotypes showed high sequence identity with classified bacteria (Lactobacillus salivarius, Lactobacillus aviarius, Lactobacillus crispatus, Faecalibacterium prausnitzii, Escherichia coli, Gallibacterium anatis, Clostridium lactatifermentans, Ruminococcus torques, Bacteroides vulgatus, and Alistipes finegoldii). The 16S rRNA gene sequence information generated will allow quantitative assays to be developed which will enable elucidations of which of these phylotypes are truly performance related. This information could be used to monitor strategies to improve feed efficiency and feed formulation for optimal gut health.
INTRODUCTION
Because feed constitutes approximately 70% of the cost of raising broiler chickens (1), the most common measures of bird performance have been linked to weight gain and feed efficiency. Broiler performance is closely linked to the genetics, diet, age, and rearing environment of the bird (1, 23, 32, 54). Genetic selection has largely driven the vast improvements observed in weight gain and feed efficiency in meat chickens over the last 50 years, although a small proportion of these improvements have been attributed to nutrition and other management practices (32). The genetic changes associated with improved weight gain and feed efficiency have also resulted in changes to the gut physiology and gut microbial community composition of birds (44). Diet, age, and environmental factors have also been reported to influence the gut microbiota (43, 71, 72). Therefore, there appears to be a clear link between bird performance and gut microbiota composition.
In medicine, much interest has already focused on the influence of the gut microbiota in human health (35, 78) and energy metabolism (73, 74, 83). Interest in the role of gut microbiota in animal health, performance, and product safety has substantially increased since the European Union ban (2006) on the use of in-feed antimicrobials in livestock production (12). In-feed antimicrobials improve feed conversion and animal growth and reduce morbidity and mortality due to clinical and subclinical disease (24) by modulating the gut microbiota and damping the immune response of the host (51, 58). Feed enzymes are currently used widely in commercial poultry production to improve poultry performance by modifying the gut environment and associated microbiota (13, 72).
The primary functions of the gastrointestinal tract are to absorb nutrients from the diet and to excrete waste products. The gut also contains a unique type of microbial ecosystem which is affected by the flow of nutrients from the diet, host-derived substrates such as mucin and bile acids, the immunological responses of the host, and gut anatomy (36, 44, 57, 82). Conversely, the gut microbiota has significant impacts on the host, such as influencing the chick's gastrointestinal development, biochemistry, immunology, gene expression, physiology, and nonspecific resistance to infection (18, 26, 36, 50, 62, 82). The gastrointestinal microbiota has one of the highest cell densities for any ecosystem and in poultry ranges from 107 to 1011 bacteria per gram gut content (3). Furthermore, the collective microbial genome (microbiome) has a coding capacity that vastly exceeds that of the host's genome, encoding biochemical pathways that the host has not evolved.
Facultative and microaerophilic bacteria (lactobacilli and related genera) dominate the ileum of the chicken, while obligate anaerobes (mainly clostridium related) dominate the ceca (4, 7, 28, 43, 82). Within the ceca, culture-dependent techniques have identified over 40 different types of anaerobic Gram-negative and Gram-positive non-spore-forming rods and cocci, including several species of clostridia (4). However, culture-independent analysis of the chicken cecal microbiota has estimated a bacterial population consisting of over 600 species from more than 100 genera, with a large proportion of these bacteria belonging to unclassified species or genera (3). Hence, our knowledge of the gut microbiota composition, metabolic functions, and influence on animal health, welfare, and performance is far from complete.
The greatest determinant of gut microbiota composition is diet, and the microbiota is influenced by the dietary ingredients, nutrient levels of fat, protein, and carbohydrate, physical structure (particle size and processing technique), use of in-feed antimicrobials (antibiotics and coccidiostats), and use of exogenous feed enzymes (33, 57). Improvement in broiler performance may be due to the presence of beneficial bacterial species and/or absence of detrimental bacterial species. The gut microbiota has been reported to have both buffering (reducing energy loss when the host is in a fasted state) and counterproductive (reducing dietary energy utilization) actions on energy metabolism in chickens (47). Although chicks raised in a germfree environment grow faster than those in conventional environments where they are exposed to microbial challenges (16), germfree chicks have physiological abnormalities such as reduced intestinal motility, lower body temperature, and a poorly developed immune system (50). All of these physiological functions were improved after addition of a normal microbiota to the gut (50).
Several studies have been undertaken to investigate the influence of dietary changes on gut microbial community structure by using microbiological culturing techniques (4, 7, 33), molecular culture-independent techniques (7, 43, 72, 82), and indirect measurement of bacterial metabolic products such as short-chain fatty acids (SCFA) (7, 33). However, understanding how these many diet-induced changes in gut bacterial community composition relate to important metabolic changes, and ultimately to broiler health and performance, is less clear. We have previously shown that overall changes in gut microbiota associated with nonstarch polysaccharide (NSP)-degrading enzyme supplementation were correlated with broiler performance (72). However, it was not known whether this was a cause or effect of improved performance. Although knowledge of the ideal gut microbiota is still incomplete, it is apparent that a variety of diets can equally support optimal bird performance and maintain a healthy gut microbial balance. However, diet-related changes in the gut microbiota have not always translated into improved broiler performance (27, 54). Therefore, the aim of this study was to determine if particular gut bacteria could consistently be linked with either improved or decreased performance, as measured by feed efficiency, across three independent broiler feeding trials.
MATERIALS AND METHODS
Poultry feeding trials.
Linkages were established with three independent poultry feeding trials investigating the influence of feed type on broiler performance as measured by the feed conversion ratio (FCR). Trials were approved by the following animal ethics committees: trial I was approved by the Department of Primary Industries and Fisheries Queensland Government Animal Ethics Committee, trial II by the University of New England Animal Ethics Committee, and trial III by the Primary Industries and Resources South Australia Animal Ethics Committee. Birds in all three trials were fed nutritionally balanced diets which met or exceeded 1994 National Research Council (NRC) standards (52). For each trial, pen bird weights and amounts of feed consumed were recorded to enable FCRs to be calculated as the amount of pen weight gain/amount of feed consumed. All FCR values reported corresponded to the bird age at which gut samples were collected for microbial profiling.
(i) Trial I.
Trial I investigated broiler performance on different dietary regimens, with a specific focus on sorghum varieties as detailed by Perez-Maldonado and Rodrigues (55). One-day-old Arbor Acres broiler chicks (n = 1,800) were obtained from a local hatchery (Darwalla Milling Company, Mt. Cotton, Queensland, Australia). Chicks were assigned to 1 of 60 floor pens (n = 30/pen; 15 male and 15 female, with a stocking density of 4 birds/m2) in a poultry shed at the Poultry Research Centre, Department of Primary Industries and Fisheries, Alexandra Hills, Queensland, Australia. Broilers received one of 10 dietary regimens (n = 6 pens/treatment), with birds receiving starter diet from 0 to 21 days and then grower/finisher diet from 22 to 42 days (55). Broiler gut microbiotas were investigated at 42 days of age from 4 of a possible 10 grower/finisher dietary regimens being evaluated in the trial and included a wheat control diet supplemented with xylanase (65.8% wheat, 17.1% soybean meal, 4.4% soybean oil, and 0.03% xylanase), sorghum B diet (63.9% sorghum, 21.4% soybean meal, 2% soybean oil), a commercial sorghum diet, and a commercial sorghum diet supplemented with exogenous phytase. Both sorghum commercial diets contained 65.5% sorghum, 20% soybean meal, and 1.9% soybean oil, but the phytase-supplemented diet also contained 0.015% phytase. All four diets contained 4% canola meal, 4% meat/bone meal, and 3% sunflower meal and had apparent metabolizable energy (AME) of 13 MJ/kg (see Table S1 in the supplemental material). None of the diets contained any in-feed antibiotics. All birds were raised on pine chip litter.
(ii) Trial II.
Trial II investigated broiler performance on high- and low-dietary-fiber treatments in combination with different litter treatments (shredded paper versus hardwood sawdust). One-day-old Cobb 500 broiler chickens (n = 720) were obtained from a local hatchery (Inghams' Casula Hatchery, Casula, New South Wales, Australia) and raised for 6 weeks in a temperature-controlled shed at Ingham Enterprises Research Facility in Leppington, New South Wales, Australia. Each pen (n = 30 birds/pen, with a stocking density of 8.9 birds/m2) was randomly assigned to one of four treatments, with six replicate pens per treatment (n = 3 males and 3 females). Chickens in the experiment were raised from hatch until 42 days of age on one of four treatments. At the time gut microbial communities were investigated (day 35), birds were moving from finisher to withdrawal diet (34 to 36 days of age). The low-fiber finisher/withdrawal diet contained 75.6% wheat, 7.6% soybean meal, 6.3% meat meal, 5% canola meal, 3.8% poultry tallow, and 0.03% xylanase). The high-fiber finisher/withdrawal diet contained 69.1% wheat, 7.4% soybean meal, 6.2% meat meal, 5% canola meal, 7% oat hulls, 3.8% poultry tallow, and 0.03% xylanase) (see Table S2 in the supplemental material). The high- and low-fiber diets had AME of 12.4 MJ/kg and 13.1 MJ/kg, respectively. Both the finisher and withdrawal diets contained 50 ppm zinc bacitracin, whereas only the finisher diet contained 60 ppm salinomycin, 125 ppm dinitolmide, and 2 ppm flavophospholipol.
(iii) Trial III.
Trial III investigated broiler performance on different commercial broiler diets. One-day-old Cobb 500 broiler chickens (n = 960) were obtained from a local hatchery (Inghams' Casula Hatchery, Casula, New South Wales, Australia) and raised for 6 weeks in 32 floor pens in a temperature-controlled shed at Ingham Enterprises Research Facility in Leppington, New South Wales, Australia. Each pen (n = 30 birds/pen, with a stocking density of 8.9 birds/m2) was randomly assigned to one of eight dietary treatments, with four replicate pens per treatment (n = 2 males and 2 females). At the time that gut samples were collected for microbial profiling (42 days of age), birds had been receiving withdrawal feed for at least 6 days. Gut samples were collected from birds fed 4 of the 8 dietary treatments and included commercial diet A (16% wheat, 60.3% sorghum, 4.8% meat meal, 15.5% soybean meal, 1.4% tallow/oil, 0.015% phytase, and 15 ppm avilamycin), commercial diet B (15% wheat, 57.7% sorghum, 4.3% meat meal, 10.8% soybean meal, 7% canola meal, 2% sunflower meal, 1.5% tallow/oil, 0.015% xylanase, 15 ppm avilamycin, and 70 ppm zinc bacitracin), commercial diet F (50.0% wheat, 15.1% barley, 7.1% oats, 4.7% meat meal, 7.7% soybean meal, 6% canola meal, 5% lupin meal, 2.6% tallow/oil, 0.025% xylanase, and 15 ppm avilamycin), and commercial diet G (40.0% wheat, 33.2% sorghum, 6.5% meat meal, 7.5% poultry meal, 11% soybean meal, 0.030% xylanase, 93 ppm monensin sodium, and 15 ppm avilamycin) (see Table S3 in the supplemental material). All diets had AME of 3.0 to 3.1 MJ/kg. All birds in the trial were raised on pine shavings.
Microbial profiling. (i) Sample collection and nucleic acid extraction.
Broiler gut samples from trial I were collected at 42 days of age from two male birds per pen on each of four dietary regimens: wheat control diet supplemented with xylanase, sorghum B, commercial sorghum, and commercial sorghum supplemented with phytase (n = 12 birds/treatment). Gut samples from trial II were taken at 35 days of age from four birds per pen on each of the four treatments: paper litter and low-fiber diet, wood litter and low-fiber diet, paper litter and high-fiber diet, and wood litter and high-fiber diet (n = 24 birds/treatment [12 males and 12 females]). Gut samples from trial III were taken at 42 days of age from six birds per pen on each of four dietary treatments: A, B, F, and G (n = 24 birds/treatment [12 males and 12 females]).
All birds were euthanized by cervical dislocation. An approximately 2-cm section of the ileum (tissue and associated digesta), midway between the Meckel's diverticulum and cecal junction, and one cecum were collected from each chicken. Following collection, samples were kept on ice until frozen at −20°C and later freeze-dried. Total nucleic acid was extracted from chicken gut by a modification (72) of a proprietary extraction method developed by the South Australian Research and Development Institute (66).
(ii) T-RFLP.
Terminal restriction fragment length polymorphism (T-RFLP) analysis was done following a previously described technique (70, 72). In brief, bacterial 16S rRNA was amplified with universal 16S bacterial primers 27F (39) and 907R (48). The forward primer (27F) was 5′-end labeled with 6-carboxyfluorescein (FAM) to enable subsequent detection of terminal restriction fragments (T-RFs) following restriction endonuclease digestion of resulting amplicons with MspI. The lengths of fluorescently labeled T-RFs were determined by comparison with an internal size standard (GeneScan 1200 LIZ; Applied Biosystems, Australia) following separation by capillary electrophoresis on an ABI 3730 automated DNA sequencer (Applied Biosystems, Australia). Data were analyzed using GeneMapper v3.7 software (Applied Biosystems, Australia). Data points generated by the GeneMapper software were further analyzed using a custom-built database containing queries to validate data points and generate outputs for statistical analysis (72). T-RFs were defined as peaks with a size of x ± 2 bp within pseudoreplicates of samples and rounded to the nearest even number to produce operational taxonomic units (OTUs).
Statistical analysis.
Trial I performance data were analyzed statistically using analysis of variance (ANOVA), and significant (P < 0.05) differences between treatment means were determined using the least significant difference (LSD) test in GenStat (55). Trial II and III performance data were each analyzed with SAS for Windows, version 9.1 (base SAS software; SAS Institute Inc., Cary, NC). Data were compared by ANOVA using the general linear model (GLM), with differences between treatments determined by Duncan's multiple range test. All values for weight gain and FCR are expressed as means ± standard errors (SE).
OTUs obtained from the ileal and cecal microbial communities of individual broiler chickens from trial I (n = 48), trial II (n = 96), and trial III (n = 96) were each analyzed using multivariate statistical techniques (Primer 6; Primer-E Ltd., Plymouth, United Kingdom). These analyses were used to examine similarities in chicken ileal and cecal bacterial communities associated with treatment and sex where applicable. Bray-Curtis (10) measures of similarity were calculated to examine similarities between gut microbial communities of birds from the T-RFLP data (following standardization and fourth root transformation). One-way or two-way analysis of similarity (ANOSIM) (15) was used to test if gut microbial communities were significantly different between diets or diets and sex, respectively. The R statistic describes the extent of similarity between each pair in the ANOSIM analysis, with values close to unity indicating that the two groups are entirely separate and a zero value indicating that there is no difference between the groups.
Similarity percentage (SIMPER) (15) analyses were done to determine which OTU contributed most to the dissimilarity between treatments. The overall average dissimilarity (δ̄) between gut microbial communities of birds on differing diets was calculated, and the average contribution of the ith OTU (δ̄i) to the overall dissimilarity was determined. The average abundance (ȳ) of important OTUs in each of the groups was determined. OTUs contributing significantly to the dissimilarity between treatments were calculated as δ̄i/SD(δi) > 1. The percent contribution of individual OTUs (δ̄i%) and cumulative percent contribution (∑δ̄i%) to the top 60% of average dissimilarities were also calculated.
Unconstrained ordinations were done to graphically illustrate the relationships between diet and performance level by using nonmetric multidimensional scaling (nMDS) (38, 61). nMDS ordinations attempt to place all samples in an arbitrary two-dimensional space such that their relative distances apart match the corresponding pairwise similarities. Hence, the closer two samples are in the ordination, the more similar are their overall gut bacterial communities. “Stress” values (Kruskal's formula 1) reflect the difficulty involved in compressing the sample relationship into the two-dimensional ordination.
Isolation, PCR amplification, and cloning of OTUs of interest.
T-RFs were isolated from OTUs significantly associated with performance differences. Multiple samples containing OTUs of interest were targeted. When possible, samples were chosen which lacked other OTUs within ±10 bp. A combination of adapter ligation, fragment size selection, and reamplification with adapter-specific PCR was used to isolate T-RFs of interest as previously described (70, 80). Double-stranded MspI-adapter was prepared and ligated to restriction fragments as described by Widmer et al. (80). Size selection of T-RFs of interest was done by gel electrophoresis in a SEA 2000 electrophoresis apparatus (Elchrom Scientific Inc., Switzerland), using precast Spreadex gels (EL 400, 600, 800, or 1200; Elchrom Scientific Inc., Switzerland) (70). Following gel electrophoresis, a size range of approximately ±50 bp of the T-RFs of interest was excised from the gel. The gel slice was cut into equally sized pieces, each corresponding to a size range of approximately 12 bp. DNA was eluted from the gels as described by Widmer et al. (80). Eluted DNA was used as a template for PCR amplification with primers 27F and MspI-adapter-primer (80) as described by Torok et al. (70). PCR products were analyzed in a 2% agarose gel and visualized following staining with ethidium bromide. Single amplification products within the expected size range were excised and purified using a Macherey-Nagel NucleoSpin Extract II kit (Scientifix, Clayton, Australia) according to the manufacturer's instructions. Purified products were cloned and recombinant plasmids purified as previously described (70).
16S rRNA gene sequence analysis.
Plasmids were sequenced by Macrogen Inc., Seoul, South Korea. Vector sequence was removed using Staden Package Pregap4, version 1.5 (9). Sizes of T-RFs were predicted in silico by using WatCut (http://watcut.uwaterloo.ca/watcut/watcut/template.php?act=restriction_new; Michael Palmer, University of Waterloo, Canada). 16S rRNA gene sequence data were assigned to a bacterial taxonomic hierarchy using the Ribosomal Database Project (RDP) Release 10 classifier (79). The classifier estimates the classification reliability by using bootstrapping. For sequences shorter than 250 bp, a bootstrap cutoff threshold of 50% was used, while for longer sequences, the default cutoff of 80% was used (14). T-RFs were assigned to OTUs based on the closest nucleotide length match or within ±8 bp. BLASTn searches with nucleotide collection (nr/nt) databases and the Megablast algorithm (National Centre for Biotechnology Information [NCBI]) were used to identify the similarity of T-RFs to other sequences available in public genome sequence databases (2). Unrooted neighbor-joining trees of 16S rRNA gene sequences from T-RFs of interest and related sequences identified in NCBI databases were constructed. Sequences were aligned with ClustalW (69), and a bootstrapped (n = 500) consensus tree was created with MEGA 4 (67). The evolutionary distances were computed using the maximum composite likelihood method (68).
Nucleotide sequence accession numbers.
Representative 16S rRNA gene sequences from cloning experiments were deposited in GenBank with accession numbers JF797629 to JF798255.
RESULTS
Broiler performance. (i) Trial I.
In trial 1, live weight gain at 0 to 42 days was significantly (P < 0.05) decreased for birds on the wheat control diet supplemented with xylanase (2,595 ± 22 g/bird) compared to those on the sorghum B (2,679 ± 18 g/bird), commercial sorghum (2,675 ± 23 g/bird), and commercial sorghum with phytase (2,674 ± 21 g/bird) diets (55). Feed conversion efficiency was significantly (P < 0.05) decreased for birds at 0 to 42 days on the wheat control (1.788 ± 0.010 g/g) diet compared to those on the sorghum B (1.721 ± 0.013 g/g), commercial sorghum (1.728 ± 0.006 g/g), and commercial sorghum with phytase (1.739 ± 0.004 g/g) diets (55).
(ii) Trial II.
In trial II, the litter type in combination with diet composition significantly (P < 0.05) influenced the FCR at 0 to 35 days of age. Birds were more feed efficient on a low-fiber diet if they were housed on wood litter (1.481 ± 0.018 g/g) than if they were housed on paper litter (1.538 ± 0.023 g/g). The sex of birds also significantly (P < 0.001) influenced the FCR at 35 days of age, with males having a higher feed efficiency (1.482 ± 0.009 g/g) than females (1.551 ± 0.010 g/g). The body weight of male chickens at 35 days of age (2,285 ± 14 g/bird) was significantly (P < 0.001) higher than that of female chickens (1,993 ± 13 g/bird). Litter type in combination with sex was found to significantly (P < 0.05) alter body weight at 35 days of age, with females growing slower on paper litter (1,960 ± 17 g/bird) than on wood litter (2,026 ± 6 g/bird).
(iii) Trial III.
In trial III, there were significant differences (P < 0.001) in FCR among dietary treatments at 42 days of age. Birds on the commercial G (1.501 ± 0.050 g/g) and commercial F (1.554 ± 0.068 g/g) diets had significantly improved feed efficiency compared with birds on the commercial A (1.583 ± 0.052 g/g) and commercial B (1.603 ± 0.061 g/g) diets. Birds fed the commercial G diet had the most significantly improved feed efficiency of birds on any of the diets investigated. The sex of birds also significantly (P < 0.001) influenced the FCR, with males having a higher feed efficiency (1.465 ± 0.008 g/g) than females (1.666 ± 0.009 g/g).
Microbial profiling. (i) Trial I.
In feeding trial I, diet had a significant influence on ileal microbial community composition (global R = 0.079; P = 0.015), with significant pairwise differences detected between birds on the wheat control diet and those on the sorghum commercial diet (R = 0.221; P = 0.007) and between the wheat control diet and the sorghum commercial diet supplemented with phytase (R = 0.128; P = 0.042). Diet did not have a significant influence on the cecal microbial community composition (global R = 0.048; P = 0.075). Seven and six OTUs were identified by SIMPER analysis as significantly contributing to the dissimilarity in ileal bacterial community composition between the wheat control and sorghum commercial diets (see Table S4 in the supplemental material) and between the wheat control diet and sorghum commercial diet with added phytase (see Table S5), respectively. Six OTUs were identified as being common between the control wheat diet and either of the commercial sorghum diets, with OTUs 76, 180, 468, 492, and 936 being more abundant in the better-performing sorghum commercial diets and OTU 564 being more abundant in the poorer-performing wheat control diet (Table 1).
Table 1.
Summary of potential performance-related OTUs identified within the ilea and ceca in each of three broiler performance trials
| Gut section | Trial no. | Potential performance-related OTU(s)a |
|
|---|---|---|---|
| Improved performance | Poorer performance | ||
| Ileum | 1 | 76, 180, 468, 492, 936 | 564 |
| 2 | NS | NS | |
| 3 | 454, 492 | 180, 188, 506, 566, 938 | |
| Cecum | 1 | NS | NS |
| 2 | 92–94, 142, 198, 206–208, 482, 542 | 218, 284–286, 312 | |
| 3 | 140–142, 218–220, 284, 312, 482, 488, 536 | 212 | |
NS, no significant difference in microbial community composition detected between dietary treatments for which performance differences were detected.
Similarities in ileal microbial community composition for birds on the same diet ranged from 32 to 48% and were higher than the similarities in cecal microbial communities for birds on the same diet (24 to 37%). Birds fed the wheat control diet showed the most variability in both ileal and cecal microbial community composition, while birds fed the commercial sorghum diet and the phytase-supplemented commercial sorghum diet showed the least variability in cecal and ileal microbial communities, respectively.
(ii) Trial II.
Multivariate statistical analysis was used to investigate differences in gut microbial communities from either the ilea or ceca of birds. Factors investigated were litter/dietary fiber composition and sex of birds. No significant differences were detected in the ileal microbial community composition among litter-diet combinations (global R = 0.025; P = 0.110) across both sexes or between sexes of birds (global R = 0.033; P = 0.095) across all litter-diet combinations. However, significant differences were detected in the cecal microbial community composition among litter-diet combinations (global R = 0.089; P = 0.001) across both sexes and between sexes of birds (global R = 0.046; P = 0.034) across all litter-diet combinations. Therefore, cecal microbial communities from males and females were further analyzed separately. Significant differences in cecal microbial communities among litter-diet combinations were detected in both males (global R = 0.084; P = 0.006) and females (global R = 0.094; P = 0.001). For both sexes, significant (P < 0.05) pairwise differences were detected between birds fed a low-fiber diet and raised on wood versus paper litter and birds raised on paper and fed a high-fiber diet versus birds raised on wood and fed a high-fiber diet. SIMPER analysis showed that similarities in cecal microbial communities of birds on a particular treatment ranged from 45 to 61% and 33 to 51% for males and females, respectively, while ileal microbial community similarities for birds on the same treatment ranged from 35 to 45%. Birds reared on wood litter showed less variability in both their ileal and cecal microbial communities, regardless of dietary treatment.
SIMPER analysis identified 22 OTUs as being significantly different between birds fed the low-fiber diet and raised on shredded paper versus hardwood litter for males (see Table S6 in the supplemental material) and for females (see Table S7), although only 16 OTUs were common to both sexes. In both sexes, OTUs 218, 284 to 286, and 312 were more abundant in the lower-performing birds on low dietary fiber and paper litter treatment, while OTUs 92 to 94, 142, 198, 206 to 208, 482, and 542 were more abundant in the higher-performing birds on the low-fiber diet and wood litter treatment (Table 1).
(iii) Trial III.
Multivariate statistical analysis was used to investigate differences in gut microbial communities from either the ilea or ceca of birds. Factors investigated were diet and sex of birds. No significant differences were detected in the ileal microbial community composition between sexes of birds (global R = 0.031; P = 0.127); however, significant differences were detected among diets (global R = 0.331; P = 0.001) across both sexes. All ileal dietary pairwise comparisons were significant (P < 0.05), with the exception of that between diet A and diet B. Significant differences (global R = 0.253; P = 0.001) in ileal microbial communities were also observed between higher-performing birds (fed diets F and G) and poorer-performing birds (fed diets A and B). Seven OTUs were identified as significantly contributing to dissimilarity in ileal bacterial community composition between birds with improved performance and poorer-performing birds (see Table S8 in the supplemental material). OTUs 454 and 492 were more abundant in the groups with improved performance, while OTUs 180, 188, 506, 566, and 938 were more abundant in the lower-performing groups (Table 1).
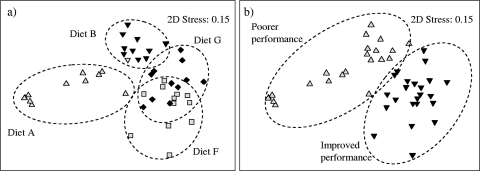
Within the ceca, significant differences in microbial community composition were detected between the sexes across all diets (global R = 0.281; P = 0.001) and among diets across both sexes (global R = 0.541; P = 0.001), with all dietary pairwise comparisons being significantly different. Therefore, cecal microbial communities from male and female birds were analyzed separately. Significant differences were also observed in cecal microbial communities between birds with improved performance (diets F and G) and poorer-performing birds (diets A and B) for both males (global R = 0.535; P = 0.001) and females (global R = 0.297; P = 0.001). Differences in cecal microbial communities of male birds fed the four diets are shown in Fig. 1a, with separation into birds with improved performance and poorer-performing birds shown in Fig. 1b. Eleven and 16 OTUs were identified as contributing significantly to the dissimilarity in cecal bacterial community composition between birds with improved performance and poorer-performing birds for females (see Table S9 in the supplemental material) and males (see Table S10), respectively, with 11 common OTUs identified in both sexes. OTUs 140 to 142, 218 to 220, 284, 312, 482, 488, and 536 were more abundant in the better-performing groups, while OTU 212 was more abundant in the lower-performing groups (Table 1). Similarities in ileal microbial communities ranged from 41 to 54%, while cecal microbial communities ranged from 35 to 49% (females) and 45 to 51% (males) for birds on the same dietary treatment. Similarities in gut microbial communities were generally higher for birds on diets resulting in improved performance.
Fig. 1.
nMDS ordination of cecal microbial communities from male broilers in feeding trial III. (a) Cecal microbial communities by diet. ▴, diet A; ▾, diet B; ▪, diet F; ⧫, diet G. (b) Same ordination as in panel a, but microbial communities are identified as being from either birds with improved performance (▾) or poorer-performing birds (▴). The ordination is based on Bray-Curtis similarities calculated from standardized and 4th-root-transformed OTU abundances. nMDS ordinations attempt to place all samples in an arbitrary two-dimensional space such that their relative distances apart match the corresponding pairwise similarities. Hence, the closer two samples are in the ordination, the more similar are their overall gut bacterial communities. “Stress” values (Kruskal's formula 1) reflect the difficulty involved in compressing the sample relationship into the two-dimensional ordination.
Results from all three performance trials are summarized in Table 1. OTUs are listed if changes in ileal and/or cecal microbial communities were detected in response to dietary treatment and these could also be linked to broiler performance as determined by FCR. Within the ilea, three common OTUs were identified across trials (180, 492, and 564–566). Of these, OTU 492 was consistently associated with improved performance, and OTU 564–566 was associated with poorer performance. Within the ceca, five common OTUs were identified (140–142, 218–220, 284–286, 312, and 482), with OTUs 140–142 and 482 consistently associated with improved performance.
Sequence information on OTUs.
Targeted cloning and sequencing of OTUs generated 16S rRNA T-RF sequence information from 627 clones. Not all bacterial sequences identified represented significant performance-related OTUs, because size estimation was based on gel electrophoresis. Some of the 16S rRNA gene sequences could be identified only as unclassified bacteria. In cases where 16S rRNA gene sequences representing the commensal gut microbiota which included potential performance-related OTUs could be classified further, they belonged to the phyla Firmicutes, Bacteroidetes, Proteobacteria, and Verrucomicrobia. Many sequences could be classified further, using the RDP classifier, to the class (Bacilli and Clostridia), order (Clostridiales and Lactobacillales), family (Enterobacteriaceae, Lachnospiraceae, and Ruminococcaceae), or genus (Akkermansia, Alistipes, Anaerotruncus, Bacteroides, Campylobacter, Gallibacterium, Lachnospiraceae incertae sedis, Lactobacillus, Peptostreptococcaceae incertae sedis, Shigella, and Subdoligranulum) level.
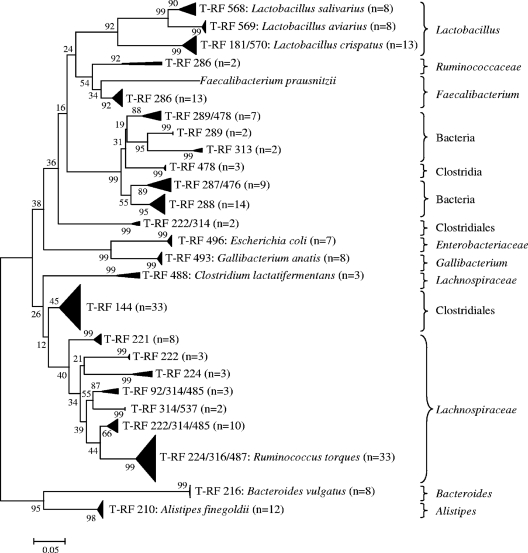
Only the 16S rRNA T-RF sequence data generated from OTUs significantly associated with performance were used to construct a phylogenetic tree (Fig. 2). Some T-RFs were shown to form several clades on the phylogenetic tree: T-RF 286 formed two clades, related to Ruminococcaceae and Faecalibacterium. Significant T-RFLP-generated OTUs and the 16S rRNA T-RF sequence information they are likely to represent are indicated in Table 2. Discrepancies between sequence-determined T-RFs and observed fragment sizes (T-RFLP-generated OTUs) were evident. Furthermore, some T-RFLP-generated OTUs could represent several bacterial groups, such as T-RFLP-generated OTUs 492, 564–566, 312, 218–222, 284–286, and 482 (Table 2). The 16S rRNA genome sequences of T-RFs indicate that some of the T-RFLP-generated OTUs may have resulted from incomplete restriction enzyme digestion, e.g., T-RF 181 (OTU 180) and T-RF 181/570 (OTU 564–566) were identical (Lactobacillus crispatus) (Fig. 2). However, similar in silico-predicted endonuclease restriction sites did not always result in identifying the same bacterial species, e.g., T-RF 222/314 (Clostridiales) was not a partial digest of T-RF 222 (Lachnospiraceae) (Fig. 2). The phylogenetic tree clustered phylotypes into 7 major groups according to classification: T-RFs 568, 569, and 570 (Lactobacillus spp.); T-RF 286 (Ruminococcaceae); T-RFs 289/478, 289, 313, 478, 287/476, and 288 (unclassified bacteria and clostridia); T-RF 222/314 (Clostridiales); T-RFs 488, 144, 221, 222, 224, 92/314/485, 314/537, 222/314/485, and 224/316/487 (Lachnospiraceae and Clostridiales); T-RFs 216 and 210 (Bacteroidales); and T-RFs 496 and 493 (Gammaproteobacteria). Forty-six percent of the potential performance-related T-RFs showed >90% nucleotide sequence identity with culturable bacterial type strains (Table 2). These included T-RF 210 (Alistipes finegoldii), T-RF 216 (Bacteroides vulgatus), T-RF 224/316/487 (Ruminococcus torques), T-RF 286 (Faecalibacterium prausnitzii), T-RF 488 (Clostridium lactatifermentans), T-RF 493 (Gallibacterium anatis), T-RF 496 (Escherichia coli), T-RF 568 (Lactobacillus salivarius), T-RF 569 (Lactobacillus aviarius), and T-RFs 181 and 181/570 (Lactobacillus crispatus) (Table 2).
Fig. 2.
Unrooted neighbor-joining phylogenetic tree of 16S rRNA gene sequences (T-RFs) obtained by targeted cloning and sequencing of OTUs identified as significantly affected by broiler performance. The tree is drawn to scale, with branch lengths in the same units as those for the evolutionary distances used to infer the phylogenetic tree. The scale bar indicates the number of base substitutions per site. The values along branches indicate percent confidence. The numbers in parentheses indicate the numbers of sequences analyzed from multiple samples. Closest identities of T-RFs to bacterial type strains (>90% identity) are indicated in the tree. Classification of T-RF sequences according to the RDP classifier is indicated to the right of the tree.
Table 2.
Bacterial classification based on 16S rRNA gene sequence information on potential broiler performance-related OTUs
| T-RF (bp) | Assigned OTU (gut section) | Phylogenetic classificationa | Organism with closest bacterial identity (NCBIn BLAST % identity)b | Reference |
|---|---|---|---|---|
| 181 | 180 (ileum) | Lactobacillus | Lactobacillus crispatus ATCC 33820 (100) | |
| 493 | 492 (ileum) | Gallibacterium | Gallibacterium anatis CCM 5995 (99) | |
| 496 | 492 (ileum) | Enterobacteriaceae | Escherichia coli DSM 4235 (99) | |
| 568 | 564–566 (ileum) | Lactobacillus | Lactobacillus salivarius CECT 5713 (100) | |
| 569 | 564–566 (ileum) | Lactobacillus | Lactobacillus aviarius JCM 5666 (99) | |
| 181/570 | 564–566 (ileum) | Lactobacillus | Lactobacillus crispatus ATCC 33820 (100) | |
| 144 | 140–142 (cecum) | Clostridiales | Uncultured rat fecal bacterium EU622670 (96) | |
| 313 | 312 (cecum) | Unclassified bacterium | Uncultured human fecal bacterium FJ365262 (98) | 73 |
| 222/314 | 312 (cecum) | Clostridiales | Uncultured human fecal bacterium AM277037 (98) | 35 |
| 314/537 | 312 (cecum) | Lachnospiraceae | Uncultured human fecal bacterium GQ896909 (94) | |
| 210 | 218–220 (cecum) | Alistipes | Alistipes finegoldii JCM 16770 (91) | |
| 216 | 218–220 (cecum) | Bacteroides | Bacteroides vulgatus JCM 5826 (99) | |
| 221 | 218–220 (cecum) | Lachnospiraceae | Uncultured mouse colon bacterium EF407360 (94) | |
| 222 | 218–220 (cecum) | Lachnospiraceae | Uncultured human colonic mucosal Firmicutes sequence EF071375 (100) | |
| 224 | 218–220 (cecum) | Lachnospiraceae | Uncultured cheetah fecal bacterium EU773752 (94) | 40 |
| 286 | 284–286 (cecum) | Ruminococcaceae | Uncultured human fecal bacterium GU105306 (99) | |
| 286 | 284–286 (cecum) | Faecalibacterium | Faecalibacterium prausnitzii ATCC 27768 (93) | |
| 288 | 284–286 (cecum) | Unclassified bacterium | Uncultured rat fecal Firmicutes sequence GU958367 (97) | |
| 289 | 284–286 (cecum) | Unclassified bacterium | Uncultured turkey fecal bacterium EU873594 (99) | 42 |
| 287/476 | 482 (cecum) | Unclassified bacterium | Uncultured bovine fecal bacterium GQ448052 (96) | |
| 478 | 482 (cecum) | Clostridia | Uncultured chicken cecal bacterium GQ175453 (99) | |
| 92/314/485 | 482 (cecum) | Lachnospiraceae | Uncultured capercaillie cecal bacterium DQ071480 (98) | |
| 222/314/485 | 482 (cecum) | Lachnospiraceae | Uncultured human fecal bacterium DQ80034 (99) | 41 |
| 224/316/487 | 482 (cecum) | Lachnospiraceae | Ruminococcus torques ATCC 27756 (94) | |
| 289/478 | 482 (cecum) | Unclassified bacterium | Uncultured human fecal bacterium FJ370890 (98) | 73 |
| 488 | 482 (cecum) | Lachnospiraceae | Clostridium lactatifermentans DSM 14214 (95) |
Broad bacterial classification was determined using the RDP classifier function.
Closest bacterial identities are listed for culturable bacteria for which a nucleotide identity of >90% was detected; otherwise, the closest uncultured bacterial identity is indicated.
DISCUSSION
Three broiler performance trials were investigated in an attempt to identify gut bacteria which could be linked with bird performance. Although there are several ways of assessing broiler performance, such as weight gain, feed efficiency, and energy utilization, FCR (amount of feed consumed per unit of body weight gain) was used in this study because it is widely used in the poultry industry (1). Trials were done in various geographic locations and varied in composition of raw dietary ingredients, dietary supplementation with antimicrobials (antibiotics and coccidiostats) and exogenous feed enzymes, broiler breed, bird age, and rearing environment. All of these factors have been shown to influence broiler performance (1, 23, 32, 54) and/or to alter gut microbiota composition (43, 44, 71, 72). Within trials, different consortia of gut microorganisms were identified as being able to equally maintain and promote optimal broiler performance, yet we were able to identify eight common OTUs linked to broiler performance across the three feeding trials investigated. These included OTUs 180, 492, and 564–566, identified within the ileum, and OTUs 140–142, 218–220, 284–286, 312, and 482, identified within the ceca. OTU 564–566 was associated predominantly with lower performance, while OTUs 492, 140–142, and 482 were associated predominantly with improved performance. In this study, birds on diets exhibiting significantly lower performances generally showed greater between-bird variation in gut microbial communities. Large variations in chick performance have also been reported between birds on the same treatment (16) and may be linked to the variability in the gut microbiota. Furthermore, maternal nutrition has been shown to alter chick gut development (56), which has been linked indirectly to microbiota development and biological variation between birds (44).
Targeted cloning and sequencing of the eight performance-related OTUs revealed that they may represent 26 different bacterial species or phylotypes. Therefore, many of the OTUs identified by T-RFLP analysis contained several bacterial phylotypes which could contribute to the observed changes in microbial community composition. T-RFLP analysis has been shown to significantly underestimate bacterial richness, but it does detect the same dominant species within a sample as that detected by clone libraries (53). Furthermore, similar levels of relative diversity have been predicted by both methods (53). Five of the 26 potential performance-related phylotypes identified could be classified only as unknown bacteria, and the remaining 21 phylotypes could be classified to the class, order, family, genus, or even species level. Twelve of the phylotypes identified belonged to the order Clostridiales and represented sequences obtained from the ceca. The chicken cecum and its mucosal tissue are dominated by clostridium-related species (7, 28, 45). Clostridium-related bacteria within the gut have been shown to produce SCFA, with clostridium cluster IX producing predominantly propionate and clusters IV (Clostridium leptum group) and XIVa (Clostridium coccoides group) producing primarily butyrate (75). The C. coccoides group is likely to be important in colonization resistance, acting as a barrier against invasion by other potentially pathogenic microbes (75).
The potential performance-related bacteria identified in this study belonged predominantly to the phylum Firmicutes, with Bacteroidetes and Proteobacteria also detected. In humans, changes in abundance of Firmicutes and Bacteroidetes have been implicated in differences in the ability to harvest energy, with the latter being more dominant in obese individuals (74, 83). Although some of the potential performance-related bacteria could be classified only as unknown bacteria, they did show high sequence identities (90 to 100%) with other unclassified bacteria in public genome sequence databases. Many of these similar sequences were identified from studies investigating the relationships between the gut microbiome and host metabolic phenotypes, gut microbiotas in various host species, including poultry, and the role of the gut microbiota in gut health (35, 40–42, 73). Nearly half of the potential performance-related T-RFs identified in this study showed high nucleotide sequence identity with culturable bacterial type strains in public genome databases.
In this study, OTU 564–566 was consistently identified within the ileum as being associated with decreased performance across trials and potentially represents three Lactobacillus spp. (L. salivarius [T-RF 568], L. aviarius [T-RF 569], and L. crispatus [T-RF 181/570]). Some lactobacilli, including L. salivarius, have been reported to have bile-deconjugating activity (31, 37). This bacterial activity lowers the detergent properties of bile acids in the emulsification of fat and leads to growth depression in chickens (31, 37). Lactobacillus communities are influenced by the presence of in-feed antimicrobials; however, this is dependent on the antimicrobial used and the bacterial species. L. salivarius has been shown to be decreased with antibiotic treatment (21, 29), while L. crispatus has been shown to increase in prevalence in chicks fed various in-feed antimicrobials posthatch (21, 70).
OTU 482, which was associated predominantly with improved performance in the ceca, may represent six phylotypes. Two of these phylotypes could be related to C. lactatifermentans (T-RF 488) or R. torques (T-RF 224/316/487). C. lactatifermentans is a Gram-positive lactate-fermenting obligate anaerobe belonging to clostridium cluster XIVb and has been isolated from the ceca of chickens (77). C. lactatifermentans grows on a variety of organic compounds which are converted to acetate, propionate, and traces of butyrate and isovalerate. C. lactatifermentans in combination with L. crispatus has been shown to inhibit Salmonella enterica serovar Enteritidis in vitro (76). R. torques belongs to the C. coccoides (XIVa) group and is known to degrade gastrointestinal mucin (81). Mucolytic potential is widespread among the intestinal bacteria (20, 75). Mucin production has been shown to change in chicks as a result of gut bacterial colonization, suggesting altered host gene expression in response to the gut microbiota (26). Withholding of enteral nutrients results in commensal and pathogenic bacteria using mucus as a substrate, hence weakening the protection provided by the intestinal mucous layer (19). In poultry, probiotics, in-feed antibiotics, feed enzymes, and feed withdrawal have all been shown to alter mucin biosynthesis, composition, and/or degradation via changes to the intestinal bacterial populations (60, 63, 64).
OTU 492 was associated predominantly with improved broiler performance and may represent two phylotypes. These are bacteria related to G. anatis (T-RF 493) and E. coli (T-RF 496). This was somewhat surprising, as both of these bacteria have the potential to be pathogenic. G. anatis is capable of causing serious systemic infections affecting multiple organ systems, although the mechanism of pathogenesis remains obscure (8). Gallibacterium spp. have been found to be present in the upper respiratory tract as well as the lower genital tract of chickens (8). The 16S rRNA gene sequences related to G. anatis were isolated from the guts of healthy male and female birds across trials. E. coli is also viewed as a pathogen in both livestock and humans. E. coli has been shown to produce colitis in mice, but not all E. coli strains are capable of inducing colitis, such as E. coli Nissle (78). Indeed, E. coli Nissle has been used widely as a probiotic in both humans and livestock (5). Hence, the metabolic activity of bacterial strains of a species can vary greatly, and bacterial strains can dramatically differ in their probiotic potential (78), often displaying host specificity (4).
The fact that several phylotypes were represented by a single OTU may explain why some of the potential performance-related OTUs identified were not consistently associated with either improved or poorer performance across trials. For example, OTU 218–220 was more abundant in the ceca of poorer-performing birds in trial II, while it was more closely associated with improved performance in trial III. T-RFLP analysis determined the OTU 218–220 represents five phylotypes (T-RFs 210, 216, 221, 222, and 224). If one of these phylotypes was performance related and hypothetically decreased in abundance in birds with improved performance, its response may have been masked by another phylotype in the same OTU cluster, which could have been increased in abundance due to some other, non-performance-related factors. Hence, if a particular OTU represents several species, the relative abundance of an individual phylotype within the group cannot be established. Further investigation of these phylotypes by use of quantitative assays is required to determine if they are truly performance related. This could be resolved by designing quantitative PCR (qPCR) assays to obtained 16S rRNA gene sequence information, but this was beyond the scope of the current work.
OTU 218–220 may represent five phylotypes and was not consistently associated with either improved or poorer performance across trials. Two of these phylotypes are related to A. finegoldii (T-RF 210) and B. vulgatus (T-RF 216). Although Alistipes spp. (anaerobic, non-spore-forming, nonmotile Gram-positive bacteria) have been isolated from feces of humans (49), no putative function has been reported. Alternatively, B. vulgatus, a Gram-negative anaerobe, is one of the most numerous bacterial species in the human colon, accounting for approximately 10 to 12% of the bacterial population (46). An inverse relationship between B. vulgatus and salmonella has been observed in the chicken gut (4). Although the possible relationship is not understood, it is believed to be one of competition for limiting nutrients rather than antibacterial activity (4). A strain of B. vulgatus has also been shown to have the potential to ameliorate E. coli-induced colitis development in mice (78).
OTU 284–286 may represent six phylotypes and also was not consistently associated with either improved or poorer performance across trials. Two of these phylotypes (T-RF 286) may be related to F. prausnitzii, a Gram-negative obligate anaerobe belonging to the C. leptum group (cluster IV), which is dominant in the ceca of chickens (45). F. prausnitzii is urolytic, has a requirement for acetate, and produces SCFA such as butyrate, formate, and lactate (59). Butyric acid has been shown to have an important function in protection against pathogens in poultry (25).
Weaknesses noted in the techniques used to identify OTUs within this study were the potential for incomplete restriction enzyme digestion of PCR amplicons and discrepancies between T-RFLP-generated OTUs and actual T-RF lengths, both of which have been reported previously (11, 22, 34). Despite these shortcomings, the techniques employed in this study did successfully identify treatment (dietary and performance) differences. Fingerprinting techniques such as T-RFLP analysis are still orders of magnitude cheaper and faster to perform than metagenomic pyrosequencing and can be used successfully to analyze large numbers of samples while maintaining interindividual variability (30). Although metagenomic pyrosequencing would have produced a greater depth of bacterial phylogenetic sequence information (including the non-treatment-related microbiota), this high-resolution analysis is not always warranted (14).
Despite the diversity in gut bacterial assemblages among human individuals, it has been shown that they yield a core microbiome at the functional level and that deviations from this core are associated with differences in the host physiological state (73). Gut bacterial function was not investigated in our study but may explain why a variety of gut microbiota were able to maintain optimal performance. Indeed, several gut bacteria isolated from chickens have already been shown to have various important biochemical properties. Recently, Clostridium perfringens, Enterococcus faecium, Streptococcus bovis, and Bacteroides spp. were shown to have polysaccharide-degrading activity against NSPs found in grain (6). Furthermore, bacterial species or genera may have multiple functions (6, 17, 37). It appears that it is both the bacterial genes and the bacterial species present within the gut that are important. Turnbaugh et al. (73) found that obese individuals had a larger proportion of genes for digesting fat, protein, and carbohydrates, which might make them better at extracting and storing energy from food.
The 16S rRNA gene sequences generated in this study have established the basis for developing quantitative assays for these organisms. This will ultimately allow us to validate the presence of performance-related gut bacteria and potentially use these assays to monitor strategies to improve feed efficiency. Despite some of the 16S rRNA gene sequences being identified as unclassified bacteria, the sequence information can be used to PCR screen and potentially isolate these bacteria from culture (65). It is also promising that so many 16S rRNA gene sequences did show similarity with classified bacteria, which could lead to the development of poultry-specific probiotics. Overall, our results are promising in our quest to identify potential performance-related gut bacteria in poultry. Understanding the dynamics of the gut microbial community, or microbial balance, is necessary to establish or develop strategies to improve feed efficiency and growth rate, avoid intestinal diseases and proliferation of food-borne pathogens, and identify better feed additives and nutrient levels that influence beneficial microbial communities. Nutritional strategies to manage the composition of the intestinal microbiota, and thus the detrimental or beneficial outcomes, will have practical value in the future.
Supplementary Material
ACKNOWLEDGMENTS
This research was partly conducted within the Poultry CRC, established and supported under the Australian Government's Cooperative Research Centres program.
We thank Mark Geier, Hugh Rodrigues, Greg McDonald, Valetta Taylor-Graig, and Julie Bleeker for help with sample collection. We also thank Teresa Mammone and Rohit Philip for technical assistance.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 8 July 2011.
REFERENCES
- 1. Aggrey S. E., Karnuah A. B., Sebastian B., Anthony N. B. 2010. Genetic properties of feed efficiency parameters in meat-type chickens. Genet. Sel. Evol. 42:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Apajalahti J., Kettunen A., Graham H. 2004. Characteristics of the gastrointestinal microbial communities, with special reference to the chicken. Worlds Poult. Sci. J. 60:223–232 [Google Scholar]
- 4. Barnes E. M. 1979. The intestinal microflora of poultry and game birds during life and after storage. J. Appl. Bacteriol. 46:407–419 [DOI] [PubMed] [Google Scholar]
- 5. Barth S., et al. 2009. Escherichia coli Nissle 1917 for probiotic use in piglets: evidence for intestinal colonization. J. Appl. Microbiol. 107:1697–1710 [DOI] [PubMed] [Google Scholar]
- 6. Beckmann L., Simon O., Vahjen W. 2006. Isolation and identification of mixed linked β-glucan degrading bacteria in the intestine of broiler chickens and partial characterization of respective 1,3-1,4-β-glucanase activities. J. Basic Microbiol. 46:175–185 [DOI] [PubMed] [Google Scholar]
- 7. Bjerrum L., et al. 2006. Microbial community composition of the ileum and cecum of broiler chickens as revealed by molecular and cellular-based techniques. Poult. Sci. 85:1151–1164 [DOI] [PubMed] [Google Scholar]
- 8. Bojesen A. M., Christensen H., Nielsen O. L., Olsen J. E., Bisgaard M. 2003. Detection of Gallibacterium spp. in chickens by fluorescent 16S rRNA in situ hybridization. J. Clin. Microbiol. 41:5167–5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bonfield J. K., Staden R. 1996. Experiment files and their application during large-scale sequencing projects. DNA Seq. 6:109–117 [DOI] [PubMed] [Google Scholar]
- 10. Bray J. R., Curtis K. R. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27:325–349 [Google Scholar]
- 11. Bukovska P., Jelinkova M., Hrselova H., Sykorova Z., Gryndler M. 2010. Terminal restriction fragment length measurement errors are affected mainly by fragment length, G+C nucleotide content and secondary structure melting point. J. Microbiol. Methods 82:223–228 [DOI] [PubMed] [Google Scholar]
- 12. Castanon J. I. R. 2007. History of the use of antibiotic as growth promoters in European poultry feeds. Poult. Sci. 86:2466–2471 [DOI] [PubMed] [Google Scholar]
- 13. Choct M., Sinlae M., Al-Jassim R. A. M., Pettersson D. 2006. Effects of xylanase supplementation on between-bird variation in energy metabolism and the number of Clostridium perfringens in broilers fed a wheat-based diet. Aust. J. Agric. Res. 57:1017–1021 [Google Scholar]
- 14. Claesson M. J., et al. 2009. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669 doi:10.1371/journal.pone.0006669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clarke K. R. 1993. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18:117–143 [Google Scholar]
- 16. Coates M. E., Fuller R., Harrison G. F., Lev M., Suffolk S. F. 1963. A comparison of the growth of chicks in the Gustafsson germ-free apparatus and in a conventional environment, with and without dietary supplements of penicillin. Br. J. Nutr. 17:141–150 [DOI] [PubMed] [Google Scholar]
- 17. Collier C. T., van der Klis J. D., Deplancke B., Anderson D. B., Gaskins H. R. 2003. Effects of tylosin on bacterial mucolysis, Clostridium perfringens colonization, and intestinal barrier function in a chick model of necrotic enteritis. Antimicrob. Agents Chemother. 47:3311–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Angelis M., et al. 2006. Selection of potential probiotic lactobacilli from pig feces to be used as additives in pelleted feeding. Res. Microbiol. 157:792–801 [DOI] [PubMed] [Google Scholar]
- 19. Deplancke B., et al. 2006. Selective growth of mucolytic bacteria including Clostridium perfringens in a neonatal piglet model of total parenteral nutrition. Am. J. Clin. Nutr. 76:1117–1125 [DOI] [PubMed] [Google Scholar]
- 20. Derrien M., Collado M. C., Ben-Amor K., Salminen S., De Vos W. M. 2008. The mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl. Environ. Microbiol. 74:1646–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dumonceaux T. J., Hill J. E., Hemmingsen S. M., Van Kessel A. G. 2006. Characterization of intestinal microbiota and response to dietary virginiamycin supplementation in the broiler chicken. Appl. Environ. Microbiol. 72:2815–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Egert M., Friedrich W. 2003. Formation of pseudoterminal restriction fragments, a PCR related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structures. Appl. Environ. Microbiol. 69:2555–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Engberg R. M., Hedemann M. S., Steenfeldt S., Jensen B. B. 2004. Influence of whole wheat and xylanase on broiler performance and microbial composition and activity in the digestive tract. Poult. Sci. 83:925–938 [DOI] [PubMed] [Google Scholar]
- 24. Feighner S. D., Dashkevicz M. P. 1987. Subtherapeutic levels of antibiotics in poultry feed and their effects on weight gain, feed efficiency, and bacterial cholyltaurine hydrolase activity. Appl. Environ. Microbiol. 53:331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fernández-Rubio C., et al. 2009. Butyric acid-based feed additives help protect broiler chickens from Salmonella Enteritidis infection. Poult. Sci. 88:943–948 [DOI] [PubMed] [Google Scholar]
- 26. Forder R. E. A., Howarth G. S., Tivey D. R., Hughes R. J. 2007. Bacterial modulation of small intestinal goblet cells and mucin composition during early posthatch development of poultry. Poult. Sci. 86:2396–2403 [DOI] [PubMed] [Google Scholar]
- 27. Geier M. S., Torok V. A., Allison G. E., Ophel-Keller K., Hughes R. J. 2009. Indigestible carbohydrates alter the intestinal microbiota but do not influence the performance of broiler chickens. J. Appl. Microbiol. 106:1540–1548 [DOI] [PubMed] [Google Scholar]
- 28. Gong J., et al. 2007. 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts; from crops to ceca. FEMS Microbiol. Ecol. 59:147–157 [DOI] [PubMed] [Google Scholar]
- 29. Guban J., Korver D., Allison G., Tannock G. 2006. Relationship of dietary antimicrobial drug administration with broiler performance, decreased population levels of Lactobacillus salivarius, and reduced bile salt deconjugation in the ileum of broiler chickens. Poult. Sci. 85:2186–2194 [DOI] [PubMed] [Google Scholar]
- 30. Hamady M., Knight R. 2009. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. 19:1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harrow S., Ravindran V., Butler R., Marshall J., Tannock G. 2007. Real-time quantitative PCR measurement of ileal Lactobacillus salivarius populations from broiler chickens to determine the influence of farming practices. Appl. Environ. Microbiol. 73:7123–7127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Havenstein G. B., Ferket P. R., Qureshi M. A. 2003. Growth, livability, and feed conversion of 1957 versus 2001 broilers when fed representative 1957 and 2001 broiler diets. Poult. Sci. 82:1500–1508 [DOI] [PubMed] [Google Scholar]
- 33. Hubener K., Jahjen W., Simon O. 2002. Bacterial responses to different dietary cereal types and xylanase supplementation in the intestine of broiler chicken. Arch. Anim. Nutr. 56:167–187 [DOI] [PubMed] [Google Scholar]
- 34. Kaplan C. W., Kitts C. L. 2003. Variation between observed and true terminal restriction fragment length is dependent on true TRF length and purine content. J. Microbiol. Methods 54:121–125 [DOI] [PubMed] [Google Scholar]
- 35. Kassinen A., et al. 2007. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology 133:24–33 [DOI] [PubMed] [Google Scholar]
- 36. Klasing K. C. 2007. Nutrition and the immune system. Br. Poult. Sci. 48:525–537 [DOI] [PubMed] [Google Scholar]
- 37. Knarreborg A., Engberg R. M., Jensen S. K., Jensen B. B. 2002. Quantitative determination of bile salt hydrolase activity in bacteria isolated from the small intestine of chickens. Appl. Environ. Microbiol. 68:6425–6428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kruskal J. B. 1964. Multidimensional scaling by optimising a goodness of fit to a nonmetric hypothesis. Psychometrika 29:1–28 [Google Scholar]
- 39. Lane D. J. 1991. 16S/23S rRNA sequencing, p. 115–175 In Stackebrandt E., Goodfellow M. (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 40. Ley R., et al. 2008. Evolution of mammals and their gut microbes. Science 320:1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022–1023 [DOI] [PubMed] [Google Scholar]
- 42. Lu J., Domingo J. 2008. Turkey fecal microbial community structure and functional gene diversity revealed by 16S rRNA gene and metagenomic sequences. J. Microbiol. 46:469–477 [DOI] [PubMed] [Google Scholar]
- 43. Lu J., et al. 2003. Diversity and succession of the intestinal bacterial communities of the maturing boiler chicken. Appl. Environ. Microbiol. 69:6816–6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lumpkins B. S., Batal A. B., Lee M. D. 2010. Evaluation of the bacterial community and intestinal development of different genetic lines of chickens. Poult. Sci. 89:1614–1621 [DOI] [PubMed] [Google Scholar]
- 45. Lund M., Bjerrum L., Pedersen K. 2010. Quantification of Faecalibacterium prausnitzii- and Subdoligranulum variabile-like bacteria in the cecum of chickens by real-time PCR. Poult. Sci. 89:1217–1224 [DOI] [PubMed] [Google Scholar]
- 46. McCarthy R. E., Pajeau M., Salyers A. A. 1988. Role of starch as a substrate for Bacteroides vulgatus growing in the human colon. Appl. Environ. Microbiol. 54:1911–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Muramatsu T., Nakajima S., Okumura J. 1994. Modification of energy metabolism by the presence of the gut microflora in the chicken. Br. J. Nutr. 71:709–717 [DOI] [PubMed] [Google Scholar]
- 48. Muyzer G., Teske A., Wirsen C. O., Jannasch H. W. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165–172 [DOI] [PubMed] [Google Scholar]
- 49. Nagai F., Morotomi M., Watanabe Y., Sakon H., Tanaka R. 2010. Alistipes indistinctus sp. nov. and Odoribacter laneus sp. nov., common members of the human intestinal microbiota isolated from faeces. Int. J. Syst. Evol. Microbiol. 60:1296–1302 [DOI] [PubMed] [Google Scholar]
- 50. Niba A. T., Beal J. D., Kudi A. C., Brooks P. H. 2009. Bacterial fermentation in the gastrointestinal tract of non-ruminants: influence of fermented feeds and fermentable carbohydrates. Trop. Anim. Health Prod. 41:1393–1407 [DOI] [PubMed] [Google Scholar]
- 51. Niewold T. 2007. The nonantibiotic anti-inflammatory effect of antimicrobial growth promoters, the real mode of action? A hypothesis. Poult. Sci. 86:605–609 [DOI] [PubMed] [Google Scholar]
- 52. NRC 1994. Nutrient requirements of poultry, 9th ed. National Academy Press, Washington, DC [Google Scholar]
- 53. Orcutt B., Bailey B., Staudigel H., Tebo B. M., Edwards K. J. 2009. An interlaboratory comparison of 16S rRNA gene-based terminal restriction fragment length polymorphism and sequencing methods for assessing microbial diversity of seafloor basalts. Environ. Microbiol. 11:1728–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pedroso A. A., et al. 2006. Intestinal bacterial community and growth performance of chickens fed diets containing antibiotics. Poult. Sci. 85:747–752 [DOI] [PubMed] [Google Scholar]
- 55. Perez-Maldonado R. A., Rodrigues H. D. 2009. Nutritional characteristics of sorghums from Queensland and New South Wales for chicken meat production. RIRDC publication no. 09/170 Union Offset Printing, Canberra, Australian Capital Territory, Australia [Google Scholar]
- 56. Rebel J. M. J., et al. 2006. Maternal diet influences gene expression in intestine of offspring in chicken (Gallus gallus). Comp. Biochem. Physiol. Part A 145:502–508 [DOI] [PubMed] [Google Scholar]
- 57. Rehman H. U., Vanhjen W., Awad W. A., Zentek J. 2007. Indigenous bacteria and bacterial metabolic products in the gastrointestinal tract of broiler chickens. Arch. Anim. Nutr. 61:319–335 [DOI] [PubMed] [Google Scholar]
- 58. Roura E., Homedes J., Klasing K. C. 1992. Prevention of immunologic stress contributes to the growth-permitting ability of dietary antibiotics in chicks. J. Nutr. 122:2383–2390 [DOI] [PubMed] [Google Scholar]
- 59. Scupham A. J. 2007. Succession in the intestinal microbiota of preadolescent turkeys. FEMS Microbiol. Ecol. 60:136–147 [DOI] [PubMed] [Google Scholar]
- 60. Sharma R., Fernandez F., Hinton M., Schumacher U. 1997. The influence of diet on the mucin carbohydrates in the chick intestinal tract. Cell. Mol. Life Sci. 53:935–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shepard R. N. 1962. The analysis of proximities: multidimensional scaling with an unknown distance function. Psychometrika 27:125–140, 219–246 [Google Scholar]
- 62. Shira E. B., Sklan D., Friedman A. 2005. Impaired immune responses in broiler hatchling hindgut following delayed access to feed. Vet. Immunol. Immunopathol. 105:33–45 [DOI] [PubMed] [Google Scholar]
- 63. Smirnov A., Perez R., Amit-Romach E., Sklan D., Uni Z. 2005. Mucin dynamics and microbial populations in chicken small intestine are changed by dietary probiotic and antibiotic growth promoter supplementation. J. Nutr. 135:187–192 [DOI] [PubMed] [Google Scholar]
- 64. Smirnov A., Sklan D., Uni Z. 2004. Mucin dynamics in the chick small intestine are altered by starvation. J. Nutr. 134:736–742 [DOI] [PubMed] [Google Scholar]
- 65. Stevenson B. S., Eichorst S. A., Wertz J. T., Schmidt T. M., Breznak J. A. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748–4755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stirling G. R., et al. 2004. Combining an initial risk assessment process with DNA assays to improve prediction of soilborne diseases caused by root-knot nematode (Meloidogyne spp.) and Fusarium oxysporum f. sp. lycopersici in the Queensland tomato industry. Aust. Plant Pathol. 33:285–293 [Google Scholar]
- 67. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 68. Tamura K., Nei M., Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030–11035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Torok V. A., Allison G. E., Percy N. J., Ophel-Keller K., Hughes R. J. 2011. Influence of antimicrobial feed additives on broiler commensal posthatch gut microbiota development and performance. Appl. Environ. Microbiol. 77:3380–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Torok V. A., Hughes R. J., Ophel-Keller K., Ali M., MacAlpine R. 2009. Influence of different litter materials on cecal microbiota colonization in broiler chickens. Poult. Sci. 88:2474–2481 [DOI] [PubMed] [Google Scholar]
- 72. Torok V. A., Ophel-Keller K., Loo M., Hughes R. J. 2008. Application of methods for identifying broiler chicken gut bacterial species linked with increased energy metabolism. Appl. Environ. Microbiol. 74:783–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Turnbaugh P., et al. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Turnbaugh P. J., et al. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031 [DOI] [PubMed] [Google Scholar]
- 75. Van den Abbeele P., et al. 2010. Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for Bacteroidetes and Clostridium cluster IX. Appl. Environ. Microbiol. 76:5237–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. van der Wielen P. W. J. J., Lipman L. J. A., van Knapen F., Biesterveld S. 2002. Competitive exclusion of Salmonella enterica serovar Enteritidis by Lactobacillus crispatus and Clostridium lactatifermentans in a sequencing fed-batch culture. Appl. Environ. Microbiol. 68:555–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. van der Wielen P. W. J. J., Rovers G. M. L. L., Scheepens J. M. A., Biesterveld S. 2002. Clostridium lactatifermentans sp. nov., a lactate-fermenting anaerobe isolated from the caeca of a chicken. Int. J. Syst. Evol. Microbiol. 52:921–925 [DOI] [PubMed] [Google Scholar]
- 78. Waidmann M., et al. 2003. Bacteroides vulgatus protects against Escherichia coli-induced colitis in gnotobiotic interleukin-2-deficient mice. Gastroenterology 125:162–177 [DOI] [PubMed] [Google Scholar]
- 79. Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Widmer F., Hartmann M., Frey B., Kolliker R. 2006. A novel strategy to extract specific phylogenetic sequence information from community T-RFLP. J. Microbiol. Methods 66:512–520 [DOI] [PubMed] [Google Scholar]
- 81. Wilson K. H., Ikeda J. S., Blitchington R. B. 1997. Phylogenetic placement of community members of human colonic biota. Clin. Infect. Dis. 25:S114–S116 [DOI] [PubMed] [Google Scholar]
- 82. Yin Y., et al. 2010. Exposure of different bacterial inocula to newborn chicken affects gut microbiota development and ileum gene expression. ISME J. 4:367–376 [DOI] [PubMed] [Google Scholar]
- 83. Zhang H., et al. 2009. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. U. S. A. 106:2365–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.