Abstract
A striking feature of Burkholderia contaminans strain MS14 is the production of a glycolipopeptide named occidiofungin. Occidiofungin has a broad range of antifungal activities against plant and animal pathogens. In this study, a complete covalent structure characterization and identification of the whole genomic DNA region for the occidiofungin gene (ocf) cluster are described. Discovery of the presence of 2,4-diaminobutyric acid and 3-chloro-β-hydroxytyrosine and elucidation of the structure of a novel C18 fatty amino acid residue have been achieved. In addition, seven additional putative open reading frames (the genes from ocfI to ocfN [ocfI-N] and ORF16) were identified. Transcription of all the putative genes ocfI-N identified in the region except ORF16 was regulated by both ambR1 and ambR2. Elucidation of the structure and the ocf gene cluster provides insight into the biosynthesis of occidiofungin and promotes future aims at understanding the biosynthetic machinery. This work provides new avenues for optimizing the production and synthesis of structural analogs of occidiofungin.
INTRODUCTION
Burkholderia contaminans strain MS14, showing a broad range of antifungal activities against plant and animal pathogens, was isolated from a disease-suppressive soil (24). A 45.2-kb genomic DNA fragment harboring 11 open reading frames (ORFs), including the biosynthetase genes and two LuxR regulatory genes, ambR1 and ambR2 (13, 14), was identified. Analysis of transcription demonstrated that both ambR1 and ambR2 are essential for the expression of all the ORFs except for ORF1 and the antifungal activity. Production of a glycolipopeptide, named occidiofungin, is responsible for the antifungal activities of strain MS14 (22). Initial structural characterization of occidiofungin revealed two variants, occidiofungin A and occidiofungin B. Both are composed of 8 amino acids (aa), differing by an addition of oxygen to occidiofungin B forming a β-hydroxy asparagine. The two structurally related antifungal compounds have masses of 1,119.5 and 1,215.5 Da.
Nonribosomal peptide synthetase (NRPS) and polyketide synthetase (PKS) are large multimodular enzymes which are involved in natural product synthesis in many microorganisms (5). NRPS, involved in the biosynthesis of oligopeptide, is grouped by active sites termed modules, in which each module is required for catalyzing one single cycle of product length elongation. The order and number of the modules of an NRPS protein are mainly determined by the “collinearity rule” (12). There are three main domains in each module: the adenylation (A) domain, responsible for amino acid recognition; the thiolation (T) domain, which is the carrier of thioesterified amino acid intermediates; and the condensation (C) domain, which catalyzes peptide bond formation between two consecutive amino acids (5). The epimerization (E) domain as one of the modification domains catalyzes the conversion of l-amino acids to their d-isomers (26). Cyclization and release of the peptide product are catalyzed by the C-terminal thioesterase (TE) domain (36). Polyketide synthetases are a family of enzymes or enzyme complexes that produce polyketides, a large class of secondary metabolites, in bacteria, fungi, plants, and a few animal lineages (17). The biosynthesis of polyketides shares striking architectural and organizational similarities with nonribosomal peptide biosynthesis, and their modules can be integrated to produce the hybrid NRPS-PKS products. Some pharmaceutical antimicrobial agents, such as the precursor of penicillin and erythromycin, are synthesized through these mechanisms (6, 7, 32, 40).
Burkholderia bacteria, which are widely distributed in nature, are reported to be important for plant growth promotion (4). However, taxonomic distinctions have not enabled biological control strains to be clearly distinguished from human pathogenic strains, which has led to the reassessment of the risk of the registered Burkholderia strains as biological control agents (27). For example, some strains of B. contaminans were isolated from sputum and blood samples of debilitated patients such as cystic fibrosis patients (42), while other strains have shown significant antifungal activities (13). Analysis of the genetic elements and molecular mechanisms in Burkholderia may benefit the development of biologically based management approaches while potentially providing means to eliminate the deleterious effects of the microorganism. In this paper, we describe a genomic region that contributes to biosynthesis of occidiofungin, as well as the structure of the unique antifungal compound.
The whole genomic sequences of 22 strains of Burkholderia determined to date are available in the NCBI database (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). According to the annotation of the ExPASy Proteomics Server, eight of them are human pathogens, including three strains of Burkholderia cenocepacia, three strains of Burkholderia pseudomallei, and two strains of Burkholderia mallei. B. cenocepacia is an opportunistic pathogen of human cystic fibrosis or chronic granulomatous diseases (25). B. pseudomallei and B. mallei are the causal agents of melioidosis and glanders diseases, respectively (3, 15). The other 12 strains, such as Burkholderia lata strain 383, were not considered to be human pathogens (http://www.expasy.ch/sprot/hamap/BURM1.html).
In this study, the right border of the ocf gene cluster was sequenced, and further sequence analysis revealed the presence of another seven additional ORFs, which include ocfJ, another NRPS gene, and ocfN, a putative thioesterase gene. Nuclear magnetic resonance (NMR), mass spectrometry, and amino analysis data have helped to resolve regions of structural uncertainty. We have clarified the structure of a novel amino acid and identified the presence of a chloro-β-hydroxytyrosine (chloro-BHY) and 2,4-diaminobutyric acid (DABA). Characterization of the covalent structure and elucidation of the gene cluster provide a solid basis for modeling the biosynthesis of the unique antifungal compound, as well as provide avenues for improving the yield of occidiofungin during fermentation.
MATERIALS AND METHODS
NMR spectroscopy.
Occidiofungin was produced and purified as previously described (13). A 3.5-mg sample of occidiofungin was dissolved in 600 μl of dimethyl sulfoxide (DMSO-d6; Cambridge Isotopes). The NMR data were collected on a Bruker Avance DRX spectrometer, equipped with a CryoProbe, operating at a proton frequency of 600 MHz. The 1H resonances were assigned according to standard methods (45) using correlation spectroscopy (COSY), total correlation spectroscopy (TOCSY), and nuclear Overhauser effect spectroscopy (NOESY) experiments. Rotating-frame Overhauser enhancement spectroscopy (ROESY) and 13C-heteronuclear single quantum correlation (HSQC) experiments were used to clarify some areas of ambiguity in the TOCSY and NOESY spectra. NMR experiments were collected at 25°C. The carrier frequency was centered on the residual water resonance (3.333 ppm), which was suppressed minimally using standard presaturation methods. A 2.0-s relaxation delay was used between scans. The TOCSY experiment was acquired with a 60-ms mixing time using the Bruker DIPSI-2 spinlock sequence. The NOESY and ROESY experiments were acquired with 400-ms mixing times. The parameters for collecting the HSQC spectrum were optimized to observe aliphatic and aromatic CH groups. The spectral sweep width for the TOCSY, NOESY, and ROESY was 11.35 ppm in both dimensions. The spectral sweep widths for HSQC were 11.35 ppm in the proton dimensions and 100 and 150 ppm for the carbon dimension. All two-dimensional (2D) data were collected with 2,048 complex points in the acquisition dimension and 256 complex points for the indirect dimensions, except for the HSQC, which was collected with 2,048 and 128 complex points in the direct and indirect dimensions, respectively. Phase-sensitive indirect detection for NOESY, ROESY, TOCSY, and COSY experiments was achieved using the standard Bruker pulse sequences. 1H chemical shifts were referenced to the residual water peak (3.33 ppm). Data were processed with nmrPipe (8) by first removing the residual water signal by deconvolution, multiplying the data in both dimensions by a squared sinebell function with 45- or 60-degree shifts (for the 1H dimension of HSQC), zerofilling once, Fourier transformation, and baseline correction. Data were analyzed with the interactive computer program NMRView (16). The NOE cross-peak intensities were measured in NMRView. Distances were calibrated using the relationship rab6 = rcal6(Vcal/Vab), where rab is the distance between atoms a and b, Vab is the NOESY a to b cross-peak volume, rcal is a known distance, and Vcal is the corresponding volume of the NOESY calibration cross-peak. The distance used for calibrations was the distance between the β-hydroxy Tyr4 Hδ and Hε aromatic protons (2.46 Å).
Mass spectrometry.
Occidiofungin (10 μg) was evaporated to dryness in a Speed Vac concentrator (ThermoScientific, San Jose, CA), and the residue was taken up in 50 μl methanol and analyzed by direct infusion at 3 μl/min into an LCQ DecaXP (ThermoScientific, San Jose, CA) operated with a capillary voltage of 20 V, a spray voltage of 3,000 V, and a capillary temperature of 150°C. Data were acquired for 3 min over a mass range of m/z 200 to 2,000.
Amino acid analysis.
Amino acid analysis was performed at the Molecular Structure Facility, UC Davis. This facility uses a “post-ion-exchange column” ninhydrin reaction detection system. Amino acid analysis was also performed at the Texas A&M Protein Chemistry Lab by using a Hewlett Packard AminoQuant II system. Occidiofungin samples were aliquoted, mixed with internal standards (norvaline [Int1], sarcosine [Int2], and 2,4-diaminobutyrate [DABA]), dried in glass tubes in a vacuum concentrator, and subjected to vapor phase hydrolysis. The samples were subsequently reconstituted in 0.4 N borate buffer to bring the pH to 10 for optimum derivatization. Primary amino acids tagged with o-phthaldialdehyde (OPA) were detected by the diode array (UV) detector at 338/390 nm, and the fluorometric detector monitored the primaries at excitation/emission wavelengths of 340/450 nm. Precolumn derivatized amino acids were eluted from a narrow-bore 5-μm reverse-phase column.
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are described in Table 1. Escherichia coli strain JM109 was grown in Luria-Bertani medium at 37°C (35). Burkholderia strains were cultured at 28°C on nutrient broth-yeast (NBY) extract agar medium (43). When required, antibiotics were added at the following concentrations: ampicillin (100 μg ml−1), trimethoprim (50 μg ml−1), chloramphenicol (12.5 μg ml−1), and kanamycin (100 μg ml−1 for E. coli and 300 μg ml−1 for the MS14 mutants).
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Escherichia coli JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB)/F′ [traD36 proAB+lacIqlacZΔM15] | Promega |
| Burkholderia contaminans | ||
| MS14 | Wild-type strain | 13 |
| MS14MT15 | ambR2::Tn5 derivative of MS14; Kmr | 13 |
| MS14GG44 | ambR1::nptII derivative of MS14; Kmr | 14 |
| MS14MT24 | ocfH::Tn5 derivative of MS14; Kmr | This study |
| MS14GG78 | ocfJ::nptII derivative of MS14; Kmr | This study |
| Plasmid/fosmid | ||
| pSL604 | EZ-Tn5 carrying the ∼1-kb genomic DNA of MS14MT24; Kmr | This study |
| pBluescript II SK | Cloning vector; Apr | Stratagene |
| pBR325 | Cloning vector; Cmr Tcr Apr | 31 |
| pMLS7 | Expression vector of Burkholderia; Tpr | 21 |
| pGEM-T Easy | Cloning vector; Apr | Promega |
| pBSL15 | Kanamycin resistance gene cassette; Kmr | 1 |
| pGG20 | pGEM-T Easy carrying 4-kb PCR product containing the ocfJ gene; Apr | This study |
| pGG21 | pGG20 with the nptII insertion in ocfJ at the BamHI site; Kmr Apr | This study |
| pGG22 | pBR325 carrying the 5-kb EcoRI fragment containing ocfJ gene disrupted by nptII; Cmr Tcr Kmr | This study |
| pGG23 | pGEM-T Easy carrying 4.5-kb PCR product containing the intact ocfJ gene; Apr | This study |
| pGG24 | pMLS7 carrying 4.5-kb HindIII fragment harboring the intact ocfJ gene; Tpr | This study |
| 4G5 | pCC1FOS carrying 40-kb genomic DNA of MS14; Cmr | This study |
Kmr, kanamycin resistance; Apr, ampicillin resistance; Tpr, trimethoprim resistance; Cmr, chloramphenicol resistance; Tcr, tetracycline resistance.
Random mutagenesis, genomic library screening, and sequencing.
To identify more genes associated with production of antifungal compound of strain MS14, the mutant MS14MT24 (Table 1) was generated and plasmid pSL604 was obtained using an EZ-Tn5 <R6Kγori/KAN-2>Tnp Transposome kit (Epicentre Biotechnologies, Madison, WI) as described previously (13). Fosmid 4G5 was identified from the MS14 genomic library using the 1-kb PCR product from pSL604 as described previously (13). The DNA insert of fosmid 4G5 was sequenced using a random shotgun approach (37). At least triple coverage of sequencing reactions was achieved, and the Lasergene software package (DNASTAR, Inc., Madison, WI) was used for generation of a consensus sequence. Open reading frames (ORFs) and genes were subsequently predicted by the Softberry FGENESB program (Softberry, Inc., Mount Kisco, NY), and the identified ORFs and genes were analyzed using Blastx in the NCBI database. Putative promoter sequences were identified by the Softberry BPROM program. The InterProScan program was used for prediction of functional domains of proteins (33). NRPSpredictor (34) and the NRPS-PKS web-based software (2) were used for specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPSs). The program IslandPick was used to analyze the sequence characteristics as genomic islands (19).
Site-directed mutagenesis and complementation of the ocfJ gene.
The wild-type ocfJ gene was disrupted by the insertion of a kanamycin cassette into its open reading frame as described previously (23). To mutate ocfJ, the 4-kb fragment obtained by PCR using primers MoccEF and MoccER (see Table S1 in the supplemental material) was cloned into the pGEM-T Easy Vector System I (Promega Corporation, Madison, WI), resulting in plasmid pGG20. The nptII gene (1) was inserted into pGG20 at BamHI, generating plasmid pGG21. The 5-kb EcoRI fragment of pGG21 harboring the ocfJ gene disrupted by insertion of nptII was cloned into pBR325 (31) at the EcoRI site to generate pGG22. Mutagenesis of the ocfJ gene was conducted via a marker exchange procedure as described previously (23), to generate the mutant MS14GG78 (Table 1). PCR analysis and sequencing were used to verify the double crossover mutations.
To obtain the intact wild-type gene ocfJ, which was disrupted in the mutant MS14GG78, a 4.5-kb fragment was amplified by PCR using primers EoccEF2 and EoccER2 (see Table S1 in the supplemental material), both of which contain the HindIII site (Table S1), and cloned into the pGEM-T Easy vector, resulting in the plasmid pGG23. The presence of the intact ocfJ gene in the 4.5-kb DNA fragment was verified by sequencing. The 4.5-kb HindIII fragment harboring the intact ocfJ gene was inserted into the Burkholderia gene expression vector pMLS7 (21), to generate the plasmid pGG24. The plasmid pGG24 was electroporated into cells of the mutant MS14GG78, an ocfJ mutant (Table 1). Colonies acquiring trimethoprim resistance were confirmed to contain the plasmid construct pGG24 by plasmid extraction and restriction enzyme digestion. Complementation experiments were conducted using the plate assays to evaluate antifungal activity against Geotrichum candidum as described previously (13).
RNA extraction and quantitative real-time PCR.
Total RNA of strain MS14 and its mutants was extracted using an RNeasy Protect bacteria kit (Qiagen, Valencia, CA) as recommended by the manufacturer. Transcription of the ORFs and genes was analyzed using quantitative real-time PCR (Q-PCR) as described previously (12, 13). Primers for Q-PCR are described in the supplemental data (see Table S1 in the supplemental material). Three replicates of Q-PCR were conducted independently, and statistically significant differences were determined for relative quantity (RQ) values by analysis of variance (ANOVA; P < 0.05) followed by the Bonferroni and Dunnett post hoc multiple comparisons (SAS Institute, Inc., NC).
Nucleotide sequence accession number.
A 58.1-kb genomic DNA fragment, named the ocf gene cluster, was obtained and deposited into GenBank with the accession number EU938698.
RESULTS AND DISCUSSION
Occidiofungin contains nonproteinogenic amino acids.
The covalent structure of occidiofungin was determined to contain a β-hydroxy modification of asparagine (BHN), a β-hydroxy modification of tyrosine (BHY), and a 3-chloro addition to β-hydroxytyrosine (chloro-BHY), as well as a novel amino acid (NAA) derived from fatty acid synthesis (Fig. 1). Expansion of the TOCSY 2D NMR spectra showing the amide to alpha and the amide to side chain spin systems for each assigned residue in occidiofungin is shown in Fig. 2. Chemical shift values are provided in the supplemental data (see Table S2 in the supplemental material). Observed interresidue NOEs are summarized in Fig. S1 in the supplemental material, and an expansion of the NOESY spectra showing interresidue NOEs is also shown in the supplemental data (see Fig. S2 and S3).
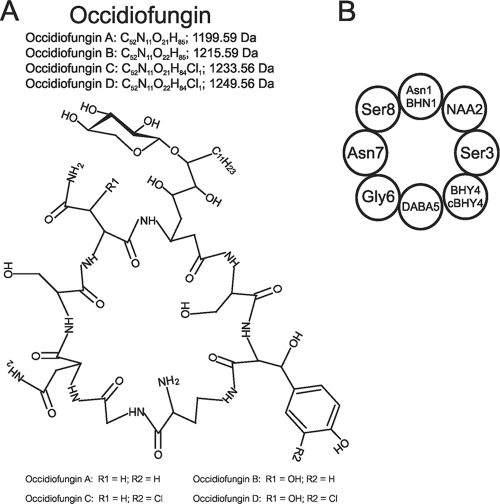
Fig. 1.
Covalent structure of occidiofungin. (A) Monoisotopic masses for each variant, occidiofungins A to D, are provided, along with the location of each variance, designated by R1 and R2. (B) Representative circle diagram of the covalent structure of occidiofungin.
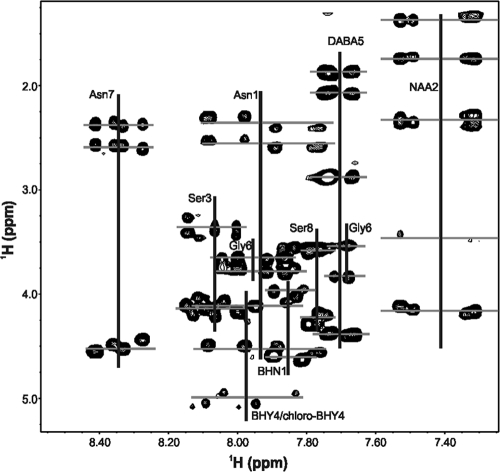
Fig. 2.
Expansion of the TOCSY 2D NMR spectra showing the amide to alpha and amide to side chain spin systems for each assigned residue in occidiofungin. Horizontal lines show the multiple amide proton chemical shift values for each amino acid.
Initial structural characterization was done using an aqueous solvent (50% acetonitrile-d3 [ACN]-50% water) and limited NMR experiments that provided a preliminary understanding of the antifungal compound structure (22). In these analyses, the antifungal peptide was determined to be an eight-residue cyclic glycolipopeptide via amide to alpha and amide to amide proton sequential walk along the backbone atoms. Furthermore, the compound was determined to contain a xylose attached to a novel amino acid. Further analyses using a nonaqueous solvent, along with additional NMR experiments, provided means to refine the structural predictions. In this study, we have determined that position 2 is a C18 glyco-amino acid, position 3 is a serine, position 4 is a mixture of BHY and chloro-BHY, and position 5 is a 2,4-diaminobutyric acid (DABA).
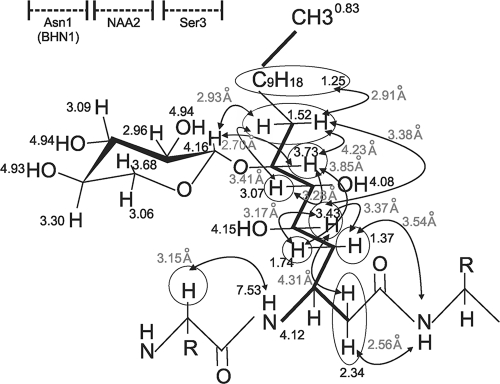
To help elucidate the structure of NAA2, COSY NMR spectra were collected to sequentially assign proton couplings within this residue. An almost complete sequential walk for the proton couplings could be observed in the COSY data set for NAA2 (see Fig. S4 in the supplemental material), providing evidence that the novel amino acid is a C18 fatty acid residue, similar to what has been reported for cepacidine (20). The only break in the proton coupling occurs between C-8 and C-9, which is visible in the TOCSY data set. The attachment site for the xylose sugar was determined to be at the C-7 position (Fig. 3; see Fig. S5 in the supplemental material). C-5, C-6, and C-7 all have chemical shift values supporting the presence of an oxygen in their vicinity and could support the attachment of the xylose. However, C-5 and C-6 have a correlation to a hydroxyl proton, while C-7 lacks a correlation to a proton on a hydroxyl, thus supporting an ether linkage to the sugar. In addition, NOEs are observed between the proton on C-7 and C-8 of NAA2 and the proton on C-1 of the xylose sugar.
Fig. 3.
Covalent structure of novel amino acid 2. The proton chemical shift values (in ppm) are shown next to their respective atoms. NOE values (in gray) are written next to the circles, and arrows designate the proton interactions. The thick lines between the atoms represent proton couplings observed in the COSY 2D NMR data set.
The chemical shift values that were found along the amide frequency for the amino acid at position 3 could account for only a few amino acids, many of which could be ruled out based on their absence in the amino acid analysis (AAA) data. AAA revealed the presence of asparagine, glycine, serine, and lysine in the molecule (see Fig. S6 in the supplemental material). The beta proton chemical shift values (∼3.27 ppm) for the residue at position 3 are out of the normal range for a serine (∼3.88 ppm). The proximity of the tyrosine ring presumably shields the beta proton on Ser3, causing a lower-than-predicted chemical shift (∼3.88 ppm), which accounts for the Ser β-proton chemical shifts of 3.27 ppm (see Table S2 in the supplemental material). Further examination of the NMR data sets revealed that coupling between the beta protons to the hydroxyl proton of Ser3 could be seen (see Fig. S7 in the supplemental material), further supporting the assignment of a serine at this position.
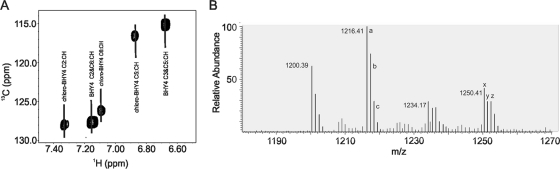
Indole-like resonances in the NMR TOCSY and NOESY data sets were identified. A peak at 10.03 ppm (see Fig. S8 in the supplemental material), which corresponds to the frequency of an indole epsilon proton, was observed. In addition, the characteristic indole proton frequencies were also observed in the aromatic region of the TOCSY spectra. These resonances were characteristic of a tryptophan residue. Additional NMR data were collected to determine the location and structure of the indole-like residue in the compound. Aromatic HSQC data revealed the presence of three additional proton carbon couplings, which is one less than expected for an indole ring (Fig. 4 A). Mass spectrometry data helped to characterize the nature of this residue. Electrospray ionization (ESI) mass spectrometry data revealed the existence of four structural variants of the antifungal peptide, one with a mass [M + 1 (H)] of 1,200.39 Da, one with a mass of 1,216.41 Da, one with a mass of 1,234.17 Da, and the fourth having a mass of 1,250.41 Da, which corresponds to the addition of oxygen and/or chlorine to the first compound (Fig. 4B). These masses are in accordance with the elucidated structure (calculated monoisotopic masses of 1,199.592, 1,215.587, 1,233.553, and 1,249.548 Da) shown in Fig. 1. Natural abundance of chlorine isotope 35 and 37 exists in a 3:1 ratio, which helps to confirm chlorination. As shown in Fig. 4B (peaks labeled with a, b, and c), the relative abundance for each isotope of a nonhalogenated compound will have a typical stair step pattern for M + 1, M + 2, and M + 3, while the chlorinated compound (peaks labeled with x, y, and z) will have an increase in the M + 3 isotope. The increase in the M + 3 isotope is due to the addition of 37Cl. The identification of a chlorination taking place in the molecule helped clarify the nature of the other aromatic residue observed in the NMR data. The three additional proton carbon couplings observed in the aromatic HSQC data are actually resonances for C-2, C-5, and C-6 of the BHY ring, and the chlorination of the tyrosine ring occurs at the C-3 position. The addition of the chlorine to the C-3 ring position is responsible for the observed downfield shift of the hydroxyl proton of Tyr to 10.03 ppm and is responsible for the downfield shift of the other aromatic protons. The presence of three distinct proton resonances on the aromatic ring for chloro-BHY4, instead of two proton resonances for BHY4, arises from the chlorination at the C-3 position, causing the environments for the protons at position C-2, C-5, and C-6 to be chemically distinct. The presence of these four structural variances is similar to what has been reported for the nonribosomally synthesized antibiotics balhimycin and vancomycin (6, 44). Furthermore, the presence of four structural variants of occidiofungin explains the multiple amide proton chemical shift values observed for each amino acid residue (Fig. 2).
Fig. 4.
Identification of a chloro-BHY. (A) Expansion of the HSQC 2D NMR spectra shows the proton and carbon chemical shift values for the chloro-BHY and BHY. (B) ESI mass spectrometry data show the presence of four variants differing in mass by the addition of an oxygen and chlorine. a, b, and c show a typical stair step pattern for each isotope, while x, y, and z show an increase in the M + 3 isotope corresponding to the addition of 37Cl.
Further analysis of the COSY data set revealed that the proton couplings of the predicted lysine at position 5 (prediction based on AAA data) did not match this assignment. Alternatively, the residue appeared to be a 2,4-diaminobutyric acid. This was unexpected given the identification of Lys by amino acid analysis (see Fig. S6A in the supplemental material). Possibly, as was previously shown, lysine and 2,4-diaminobutyric acid may have the same retention time when ion-exchange chromatography is used to separate amino acids using a “post-column” ninhydrin reaction detection system. Therefore, an alternative method was used along with a DABA standard. Following acid hydrolysis, high-performance liquid chromatography (HPLC) separation followed by UV detection (UV) and fluorescent detection (FLD) was used to see if we could distinguish a DABA from a lysine. Using this method, we did not see a lysine, but we did observe a concomitant increase in the glycine-to-serine volume ratio (Fig. S6B). An internally loaded standard of DABA (5 nm) was shown to coelute with glycine using this procedure (Fig. S6C). The amino acid analysis data are in agreement with the NMR data supporting the assignment of a DABA at position 5.
As described above, an extensive amount of new data has been analyzed providing a definitive assignment of the structure of occidiofungin. The assignment of the fatty glyco-amino acid at position 2 and the serine at position 3 and the presence of a β-hydroxytyrosine and a 3-chloro-β-hydroxytyrosine at position 4 and 2,4-diaminobutyric acid at position 5 satisfy all chemical shifts present in the NMR data sets, ESI mass spectrometry data, and amino acid analysis data. Furthermore, our structure is in agreement with the genetic data in the ocf gene cluster.
The ocf gene cluster.
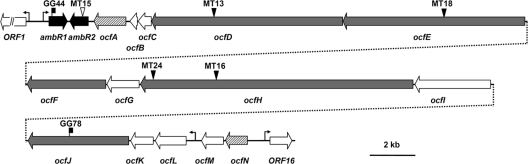
Seven new ORFs (the genes from ocfI to ocfN [ocfI-N] and ORF16) were identified (Fig. 5; Table 2) upstream of the 45.2-kb genomic fragment that was previously reported (14). This region was sequenced through the sequencing of the fosmid 4G5. A 58.1-kb genomic DNA fragment, named the ocf gene cluster, was obtained and deposited into GenBank.
Fig. 5.
Map of the 58.1-kb occidiofungin gene cluster region of Burkholderia contaminans strain MS14. The positions and orientations of the known genes and potential ORFs are shown as horizontal arrows. Vertical arrows and flags indicate the insertion positions of the Tn5 transposon and the nptII gene cassette, respectively. The open and solid vertical arrows/flags represent reduction and elimination of antifungal activity, respectively. The arrows indicate putative promoter sequences. The genotypes of the MS14 mutants are GG44(ambR1::nptII), MT15(ambR2::Tn5), MT13(ocfD::Tn5), MT18(ocfE::Tn5), MT14(ocfH::Tn5), MT16(ocfH::Tn5), and GG78(ocfJ::nptII).
Table 2.
Putative genes identifieda
| Gene or ORF | Size (bp) | Homologb | Identity (%) | Predicted functionc |
|---|---|---|---|---|
| ORF1 (partial) | 1,175 | bamb_6465 | 93 | FAD-linked oxidase domain protein |
| ambR1 | 822 | bamb_6466 | 89 | LuxR-type regulator |
| ambR2 | 891 | bamb_6468 | 77 | LuxR-type regulator |
| ocfA | 1,704 | bamb_6469 | 90 | Cyclic peptide transporter |
| ocfB | 483 | bamb_6470 | 82 | Hypothetic protein |
| ocfC | 657 | bamb_6471 | 94 | Glycosyl transferase |
| ocfD | 9,495 | bamb_6472 | 88 | NRPS |
| ocfE | 9,066 | bamb_6473 | 89 | NRPS |
| ocfF | 3,921 | bamb_6474 | 90 | NRPS |
| ocfG | 1,617 | bamb_6475 | 93 | Hydroxylase |
| ocfH | 13,410 | bamb_6476 | 91 | Hybrid NRPS-PKS |
| ocfI | 3,324 | bamb_6477 | 92 | Flavin-dependent monooxygenase |
| ocfJ | 4,428 | bamb_6478 | 91 | NRPS |
| ocfK | 987 | bamb_6479 | 91 | Halogenase |
| ocfL | 1,371 | bamb_6480 | 91 | Transaminase |
| ocfM | 951 | bamb_6481 | 94 | Epimerase |
| ocfN | 720 | bamb_6482 | 90 | Thioesterase |
| ORF16 | 288 | C7511 | 93 | Conserved hypothetical proteind |
The shaded gray region represents previously published work (13, 14). Annotations of a 12.9-kb genomic DNA fragment upstream of ocfH are not shaded.
Homolog to the putative proteins of Burkholderia ambifaria AMMD (GenBank accession no. NC_008392).
Predicted functions are based on annotation of strain AMMD and strain 383.
Homolog to C7511 of B. lata 383 (GenBank accession no. NC_007509).
Sequence analysis revealed that the ocf gene cluster, excluding ORF1 and ORF16, shares high similarity (99% nucleotide coverage and 91% identity) to an uncharacterized DNA region of Burkholderia ambifaria strain AMMD, which was used for biological control of plant diseases (28). The gene cluster was not found in other Burkholderia strains or any other strains available in GenBank (nucleotide coverage, <24%), including some identified opportunistic human pathogens: B. cenocepacia strains J2315, AU1054, and HI2424, B. pseudomallei strains 668, 1710b, and 1106a, and B. mallei strains NCTC 10247 and NCTC 10229. The sequences of the flanking regions of the ocf gene cluster in strain MS14 share highest identities (89% nucleotide identity) with the homologs C7522 (left) and C7511 (right) in chromosome 3 of B. lata 383. The average G+C content of the ocf gene cluster is 67.74%, which is closer to those of flanking region sequences in strain AMMD (left: bam_6475, 68.5%; right: bam_6483, 65.89%) but different from those of strain MS14 (55.16% left and 61.81% right). Besides, sequence analysis using the program IslandPick indicated that the uncharacterized exclusive homolog of the ocf gene cluster in strain AMMD possesses the characteristics of a genomic island. These data suggest that the ocf gene cluster may have been horizontally transferred from a strain similar to B. ambifaria AMMD and had integrated into a strain similar to B. lata 383. More importantly, the absence of the gene cluster in clinical strains of Burkholderia suggests that the gene cluster is not required for potential human pathogenesis and presumably is not involved in pathogenesis. This finding has provided insights for the usage of the gene cluster for engineering biocontrol agents for agricultural use. Furthermore, a detailed understanding of the gene cluster provides avenues for improving the production of occidiofungin, thus promoting studies aimed at understanding the compound's therapeutic potential.
Organization and orientations of the genes ocfI-N are the same as those of the B. ambifaria strain AMMD genome, with an average amino acid identity of 91%, and ORF16 has the highest similarity (93%) to its homolog C7511 of B. lata strain 383 (Fig. 5; Table 2), while its homolog was not found in the genome of the strain AMMD with BLAST. The putative protein (1,107 aa) from ocfI was predicted to encode a hybrid monooxygenase-PKS. The gene ocfJ was predicted to encode an NRPS (1,475 aa). The genes ocfK, ocfL, and ocfM were predicted to encode oxygenase or halogenase, transaminase, and an epimerase or dehydratase, respectively (Table 2). The putative protein sequence of ocfN is 219 aa in length and carries a TE domain. Putative promoters were identified upstream of ORF1, ambR1, ocfL, and ORF16, and their locations are shown in Fig. 5.
Disruption of the NRPS gene ocfJ eliminated the antifungal activity of strain MS14.
A nonpolar mutation was constructed by insertion of an nptII cassette into BamHI of ocfJ, and the mutant MS14GG78 was generated by marker exchange mutagenesis. PCR analysis demonstrated that ocfJ was disrupted by insertion of nptII in the genome of the mutant. The mutant exhibited negligible antifungal activity toward G. candidum (inhibitory zone radius ± standard error of the mean [SEM], 0.33 ± 0.33 mm), which is similar to results for the mutants MS14MT13, MS14MT18 (13), MS14MT16 (14), and MS14MT24 (see Fig. S9 in the supplemental material). As expected, the wild-type strain MS14 showed a strong inhibition against the fungus (13.00 ± 0.58 mm). The wild-type level of antifungal activities against G. candidum was observed for MS14GG78 complemented in trans with plasmid pGG24 (12.67 ± 0.33 mm). As expected, the presence of the empty vector pMLS7 had no effect on antifungal activities of either the wild-type strain MS14 or the mutant MS14GG78 (data not shown). These results further confirmed the essential role of the ocf gene cluster in the production of antifungal activity of strain MS14.
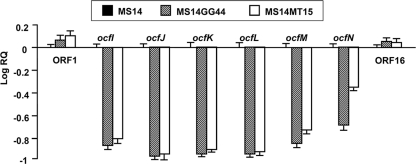
Transcription of ocfI-N but not ORF16 was regulated by both ambR1 and ambR2, the LuxR-type regulatory genes.
To test the relationship between the newly identified genes or ORFs with the LuxR regulatory genes ambR1 and ambR2, Q-PCR analyses were performed with RNAs isolated from strain MS14 and the mutants MS14GG44 and MS14MT15. Previous studies demonstrated that both ambR1 and ambR2 positively regulated transcription of ocfA-H (14). As expected, significant differences (P < 0.05) of expression levels of ocfI-N in the mutants MS14GG44 and MS14MT15 were observed compared with those in the wild-type strain MS14. Transcript levels of the genes in mutants MS14GG44 and MS14MT15, with mutations in LuxR-type regulatory genes, were reduced 13.2- and 12.0-fold, respectively (Fig. 6). These data suggest that transcription of ocfI-N is promoted by both the ambR1 and ambR2 genes.
Fig. 6.
Expression of the ORFs identified in the ocf gene cluster in strain MS14 and its mutants MS14MT15 and MS14GG44 (Table 1). Transcript levels of the tested genes and ORFs are presented relative to the transcript levels in the wild-type MS14. Mean values for three biological replicates are given, and error bars represent the standard errors of the means.
Transcription of ORF16 was not significantly affected by the mutations in either ambR1 or ambR2, indicating that the ORF16 is not regulated by either of them. In addition, the ORF16 gene, which codes for a hypothetically conserved protein, has highest identity (93%) to the protein encoded by ORF C7511 of B. lata strain 383; however, no significant homolog was found in the genome of strain AMMD, which has an uncharacterized genomic region sharing the highest similarity to the ocf gene cluster (ambR1 to ocfN). These data imply that ORF16, similar to ORF1 (14), may not be part of the gene cluster required for the antifungal activity of strain MS14. Therefore, ocfN is most likely the last gene at the right border of the gene cluster, while ambR1 is the first gene at the left border of the gene cluster. These results indicate that the whole length of the ocf gene cluster responsible for the production of occidiofungin has been identified (Fig. 5).
In summary, the whole length of the ocf gene cluster has been characterized, which is composed of 16 ORFs. Among the 16 members of this cluster, ocfD, ocfE, ocfF, ocfH, and ocfJ were predicted to encode NRPS or NRPS-PKS and are directly related to the biosynthesis of the antifungal compound occidiofungin. The genes ocfC, ocfK, ocfL, ocfM, and ocfN were predicted to be involved in the modification of occidiofungin or its components. AmbR1 and AmbR2 are the regulators controlling the transcription and expression of occidiofungin. The predicted functions of the ocf gene cluster are basically in agreement with the covalent structure.
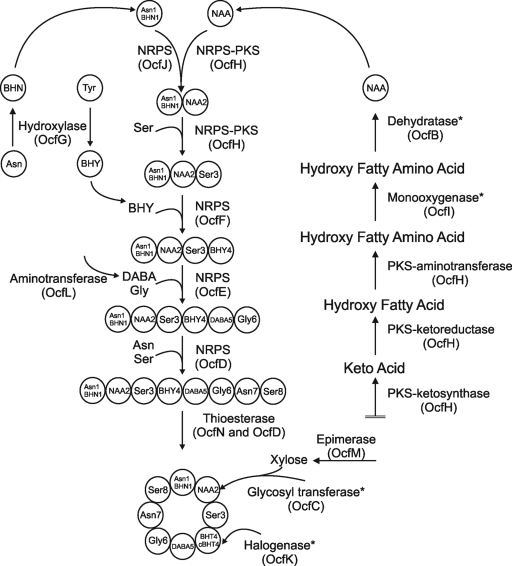
Occidiofungin biosynthesis.
Completion of the sequencing of the ocf gene cluster and the completion of the detailed structural characterization of occidiofungin provides an excellent opportunity to model the biosynthesis of occidiofungin (Fig. 7). Gene products required for the synthesis of the novel amino acid are all located in the ocf gene cluster (Fig. 5). The order in which the modules function, so that they are in agreement with the structure, is as follows: OcfJ-OcfI-OcfH-OcfF-OcfE-OcfD. Through NRPS-PKS web-based software and interProScan software in EMBL-EBI, the predicted domains in these modules are A-T-KS (ketosynthase; OcfJ) FDM (flavin-dependent monooxygenase)-KS-T (OcfI) KS-AT (acyl transferase)-KR (ketoreductase)-T-KS-AT-T-AmT (aminotransferase)-C-A-T-C (OcfH) A-T-E (OcfF) C-A-T-E-C-A-T-C (OcfE) A-T-C-A-T-E-C-T-Te (OcfD). These predictions are in agreement with the structural analyses of occidiofungin.
Fig. 7.
Representative diagram for the biosynthesis of occidiofungin. Some steps are demarcated with an asterisk, indicating the high potential of alternative routes of synthesis in the model. For instance, it is conceivable that glycosylation can occur before the synthesis of the peptide. In addition, no homology exists in the database for the proposed dehydratase activity of OcfB, and this activity may be attributed to another gene outside the gene cluster.
The ocfJ gene encodes a hybrid NRPS-PKS peptide containing domains characteristic of both NRPSs and PKSs. As described above, the potential domains are A-T-KS. There is no AT domain to accompany the KS domain in ORF11. Therefore, it is possible that OcfJ functions solely as an NRPS. This NRPS domain is presumably important for the addition of Asn1/BHN1 to NAA2. Evidence for this assumption comes from the lack of a C domain, which can occur in the first module of NRPS (9). A condensation module is present in OcfH, which can join Asn1/BHN1 to NAA2.
Based on interProScan and NRPS-PKS web-based software predictions described above, the ocfH gene has the following domains: KS-KR-T-AT-T-AmT-C-A-T-C. OcfH has the characteristics of a hybrid PKS-NRPS and is presumably responsible for the partial synthesis of NAA2. The ketoreductase domain is likely to be involved in the oxidation of the carbonyl ketones on the fatty amino acid-forming alcohols, while the aminotransferase domain is likely to be involved in the transfers of an amino group to C-3 of the fatty acid forming a fatty amino acid. ocfC, ocfI, and ocfM are also predicted to be involved in the synthesis of NAA2. C-5, C-6, and C-7 all carry a hydroxyl group. The presence of a hydroxyl group on C-5 and C-7 could arise from the reduction of the ketone by the ketoreductase present in OcfH during fatty acid synthesis. The hydroxyl group on C-6 requires an independent hydroxylation. OcfI has a flavin-dependent monooxygenase domain which may be responsible for the addition of the hydroxyl group to C-6 (29). OcfC and OcfM are presumably important for the addition of xylose to C-7. OcfM codes for a homolog of a NAD-dependent epimerase/dehydratase called UDP-d-glucuronate 4-epimerase (39). This enzyme is involved in converting UDP-glucuronate to UDP-xylose. The ocfC gene codes for the putative glycosyl transferase and is predicted to catalyze the transfer of a xylose to C-7, forming the glyco-amino acid (Fig. 3; see Fig. S5 in the supplemental material). There was no predicted dehydratase domain, which is needed for the subsequent removal of the hydroxyl groups from the polyketide chain (from C8 to C18 carbons) forming the fatty acid chain observed in the NMR data. Presumably, this activity is being harnessed from fatty acid biosynthesis or possibly OcfB, a hypothetical protein. However, gene knockout and complementation experiments should provide confirmation of the function of this gene product.
By use of the NRPS-PKS web-based software, the C-terminal part of OcfH is predicted to catalyze the addition of Ser, and it has an identity of 54% and similarity of 67% to the other NRPS domains that catalyze addition of Ser. Therefore, it is predicted that the NRPS adds a serine to the Asn-NAA2 product. Subsequently, OcfF is predicted to be responsible for the addition of the BHY to the Asn/BHN1-NAA2-Ser3 product. The NRPS-PKS web-based software predicts OcfF to catalyze the addition of Tyr, and this protein has 38% identity and 55% similarity to those responsible for Tyr addition.
There were no confirmed modular predictions for OcfE using the NRPS-PKS web-based software. The two modules are predicted to catalyze the addition of DABA5 to the Asn/BHN1-NAA2-Ser3-BHY4 product and subsequently add Gly6 to the Asn/BHN1-NAA2-Ser3-BHY4-DABA5 product. The NRPS-PKS web-based software predicts that OcfD catalyzes the addition of Asn and Ser. Therefore, the first module would add an Asn to the Asn1/BHN1-NAA2-Ser3-BHY4-DABA5-Gly6 product, and the second module would add a Ser to the Asn/BHN1-NAA2-Ser3-BHY4-DABA5-Gly6-Asn7 product.
The gene ocfL, predicted to code an α-ketoglutarate 4-aminotransferase, is presumably responsible for the transfers of an amino group during the biosynthesis of DABA (41). Substrates are predicted to be l-glutamate and l-aspartate, and the resulting products would be l-2,4-diaminobutyric acid and l-α-ketoglutarate. The ocfG gene encodes a homolog of an enzyme known to be involved in the formation of β-hydroxytyrosine, StaM of the A47934 cluster (30). This enzyme oxidizes the beta carbon, while Tyr is tethered to the T domain. It is likely that the enzyme is responsible for the beta-hydroxylations of Tyr4 and Asn1. Given the presence of Asn and BHN at position 1, it is likely that the hydoxylation of Asn on the T domain of module 1 is inefficient. Based on the structural predictions presented above, three of the eight modules in the ocf gene cluster harbor an internal E-domain for the conversion of L to D isomers. Therefore, it is likely that BHY4, DABA5, and Ser8 residues in occidiofungin are D form.
The C-terminal region of OcfD is predicted to have a TE domain, which is presumably important for the condensation reaction of Ser8 to Asn/BHN1, which terminates synthesis and forms the cyclic peptide. The ocfN gene also codes for a thioesterase. Given that the N-terminal end of the linear peptide is an Asn or BHN, it is possible that two thioesterases are required to form the Asn and BHN structural variants of the cyclic peptide.
The gene ocfA is predicted to encode an ATP-binding cassette (ABC) transporter. This large superfamily of integral membrane proteins are known to carry various substrates across cellular membranes. Given that it is regulated by ambR1 and ambR2, this is presumably the transporter for effective secretion of the antifungal compound. The ocfK gene codes for a homolog of nonheme iron dioxygenases that can catalyze oxidations, as well as chlorinations (30). Potentially this enzyme catalyzes the chlorination that is observed on BHY. Another possibility is that the halogenase activity is attributed to another gene outside the ocf gene cluster. In Fig. 7, the chlorination event is arbitrarily shown to occur at the end. Previous studies have shown that chlorination takes place sometime during peptide synthesis (32).
The genetic information of the ocf gene cluster, along with the covalent structure, has provided a solid foundation to engineer strains that will enhance the production of occidiofungin and to engineer new chemical variants with increased antifungal activity, while minimizing possible toxicity to plants and animals. For example, overexpression of ocfA and/or ambR1 (regulator) may increase occidiofungin production by strain MS14. The study of hybrid NRPS-PK natural products, which further expands the perspective of combinatorial biosynthesis, is attracting more attention (11). Some hybrid products with medicinal importance have been reported, such as rapamycin, bleomycin, and leinamycin (10, 18, 38). Understanding the genetic structures for the production of occidiofungin, especially the biosynthesis of the novel amino acid, will benefit the optimization and production of novel products.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Jinny Johnson and Lawrence Dangott at Texas A&M Protein Core for their proteomic assistance. We are also grateful to Kuan-Chih Chen and Sonya M. Baird for laboratory assistance. Lastly, we thank Ron Shin and Rama Krishna at the UAB High-Field NMR Facility for NMR data collection.
This research was supported by the Special Research Initiative Program, Mississippi Agricultural and Forestry Experiment Station (S.-E.L.) and funds from Texas A&M University.
This work was approved for publication as journal article no. J-11999 of the Mississippi Agricultural and Forestry Experiment Station (S.-E.L.).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 8 July 2011.
REFERENCES
- 1. Alexeyev M. F. 1995. Three kanamycin resistance gene cassettes with different polylinkers. Biotechniques 18:52–56 [PubMed] [Google Scholar]
- 2. Ansari M. Z., Yadav G., Gokhale R. S., Mohanty D. 2004. NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 32:W405–W413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bondi S. K., Goldberg J. B. 2008. Strategies toward vaccines against Burkholderia mallei and Burkholderia pseudomallei. Expert Rev. Vaccines 7:1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cain C. C., Henry A. T., Waldo R. H., Casida L. J., Falkinham J. O. 2000. Identification and characteristics of a novel Burkholderia strain with broad-spectrum antimicrobial activity. Appl. Environ. Microbiol. 66:4139–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cane D. E., Walsh C. T. 1999. The parallel and convergent universes of polyketide synthases and nonribosomal peptide synthetases. Chem. Biol. 6:R319–R325 [DOI] [PubMed] [Google Scholar]
- 6. Choroba O. W., Williams D. H., Spencer J. B. 2000. Biosynthesis of the vancomycin group of antibiotics: involvement of an unusual dioxygenase in the pathway to (S)-4-hydroxyphenylglycine. J. Am. Chem. Soc. 122:5389–5390 [Google Scholar]
- 7. Cortes J., Haydock S. F., Roberts G. A., Bevitt D. J., Leadlay P. F. 1990. An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea. Nature 348:176–178 [DOI] [PubMed] [Google Scholar]
- 8. Delaglio F., et al. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277–293 [DOI] [PubMed] [Google Scholar]
- 9. Donadio S., Monciardini P., Sosio M. 2007. Polyketide synthases and nonribosomal peptide synthetases: the emerging view from bacterial genomics. Nat. Prod. Rep. 24:1073–1109 [DOI] [PubMed] [Google Scholar]
- 10. Du L. C., Sanchez C., Chen M., Edwards D. J., Shen B. 2000. The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase. Chem. Biol. 7:623–642 [DOI] [PubMed] [Google Scholar]
- 11. Du L. H., Sanchez C., Shen B. 2001. Hybrid peptide-polyketide natural products: biosynthesis and prospects toward engineering novel molecules. Metab. Eng. 3:78–95 [DOI] [PubMed] [Google Scholar]
- 12. Dubern J. F., Coppoolse E. R., Stiekema W. J., Bloemberg G. V. 2008. Genetic and functional characterization of the gene cluster directing the biosynthesis of putisolvin I and II in Pseudomonas putida strain PCL1445. Microbiology 154:2070–2083 [DOI] [PubMed] [Google Scholar]
- 13. Gu G., Smith L., Wang N., Wang H., Lu S.-E. 2009. Biosynthesis of an antifungal oligopeptide in Burkholderia contaminans strain MS14. Biochem. Biophys. Res. Commun. 380:328–332 [DOI] [PubMed] [Google Scholar]
- 14. Gu G., Wang N., Chaney N., Smith L., Lu S.-E. 2009. AmbR1 is a key transcriptional regulator for production of antifungal activity of Burkholderia contaminans strain MS14. FEMS Microbiol. Lett. 297:54–60 [DOI] [PubMed] [Google Scholar]
- 15. Inglis T. J. J., Merritt A., Chidlow G., Aravena-Roman M., Harnett G. 2005. Comparison of diagnostic laboratory methods for identification of Burkholderia pseudomallei. J. Clin. Microbiol. 43:2201–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson B. A., Blevins R. A. 1994. NMR View—a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4:603–614 [DOI] [PubMed] [Google Scholar]
- 17. Khosla C., Gokhale R. S., Jacobsen J. R., Cane D. E. 1999. Tolerance and specificity of polyketide synthases. Annu. Rev. Biochem. 68:219–253 [DOI] [PubMed] [Google Scholar]
- 18. König A., et al. 1997. The pipecolate-incorporating enzyme for the biosynthesis of the immunosuppressant rapamycin—nucleotide sequence analysis, disruption and heterologous expression of rapP from Streptomyces hygroscopicus. Eur. J. Biochem. 247:526–534 [DOI] [PubMed] [Google Scholar]
- 19. Langille M. G., Hsiao W. W., Brinkman F. S. 2008. Evaluation of genomic island predictors using a comparative genomics approach. BMC Bioinformatics 9:329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee C. H., Kempf H. J., Lim Y., Cho Y. H. 2000. Biocontrol activity of Pseudomonas cepacia AF2001 and anthelmintic activity of its novel metabolite, cepacidine A. J. Microbiol. Biotechnol. 10:568–571 [Google Scholar]
- 21. Lefebre M. D., Valvano M. A. 2002. Construction and evaluation of plasmid vectors optimized for constitutive and regulated gene expression in Burkholderia cepacia complex isolates. Appl. Environ. Microbiol. 68:5956–5964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu S.-E., et al. 2009. Occidiofungin, a unique antifungal glycopeptide produced by a strain of Burkholderia contaminans. Biochemistry 48:8312–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu S.-E., Scholz-Schroeder B. K., Gross D. C. 2002. Characterization of the salA, syrF, and syrG regulatory genes located at the right border of the syringomycin gene cluster of Pseudomonas syringae pv. syringae. Mol. Plant Microbe Interact. 15:43–53 [DOI] [PubMed] [Google Scholar]
- 24. Lu S.-E., Woolfolk S., Caceres J. 2005. Isolation and identification and genetic analysis of rhizobacteria antagonistic to plant soilborne fungal pathogens. Phytopathology 95:S62–S63 [Google Scholar]
- 25. Mahenthiralingam E., Urban T. A., Goldberg J. B. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144–156 [DOI] [PubMed] [Google Scholar]
- 26. Marahiel M. A., Stachelhaus T., Mootz H. D. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97:2651–2673 [DOI] [PubMed] [Google Scholar]
- 27. Parke J. L., Gurian-Sherman D. 2001. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol. 39:225–258 [DOI] [PubMed] [Google Scholar]
- 28. Parke J. L., Rand R. E., Joy A. E., King E. B. 1991. Biological control of Pythium damping-off and Aphanomyces root rot of peas by application of Pseudomonas cepacia to seed. Plant Dis. 75:987–992 [Google Scholar]
- 29. Pohle S., Appelt C., Roux M., Fiedler H. P., Sussmuth R. D. 2011. Biosynthetic gene cluster of the non-ribosomally synthesized cyclodepsipeptide skyllamycin: deciphering unprecedented ways of unusual hydroxylation reactions. J. Am. Chem. Soc. 133:6194–6205 [DOI] [PubMed] [Google Scholar]
- 30. Pootoolal J., et al. 2002. Assembling the glycopeptide antibiotic scaffold: the biosynthesis of A47934 from Streptomyces toyocaensis NRRL15009. Proc. Natl. Acad. Sci. U. S. A. 99:8962–8967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Prentki P., Karch F., Iida S., Meyer J. 1981. The plasmid cloning vector pBR325 contains a 482 base-pair-long inverted duplication. Gene 14:289–299 [DOI] [PubMed] [Google Scholar]
- 32. Puk O., et al. 2004. Biosynthesis of chloro-beta-hydroxytyrosine, a nonproteinogenic amino acid of the peptidic backbone of glycopeptide antibiotics. J. Bacteriol. 186:6093–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quevillon E., et al. 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33(Suppl 2):W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rausch C., Weber T., Kohlbacher O., Wohlleben W., Huson D. H. 2005. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Res. 33:5799–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 36. Schwarzer D., Mootz H. D., Marahiel M. A. 2001. Exploring the impact of different thioesterase domains for the design of hybrid peptide synthetases. Chem. Biol. 8:997–1010 [DOI] [PubMed] [Google Scholar]
- 37. Staden R. 1979. A strategy of DNA sequencing employing computer programs. Nucleic Acids Res. 6:2601–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tang G. L., Cheng Y. Q., Shen B. 2004. Leinamycin biosynthesis revealing unprecedented architectural complexity for a hybrid polyketide synthase and nonribosomal peptide synthetase. Chem. Biol. 11:33–45 [DOI] [PubMed] [Google Scholar]
- 39. Thoden J. B., et al. 1997. Structural analysis of UDP-sugar binding to UDP-galactose 4-epimerase from Escherichia coli. Biochemistry 36:6294–6304 [DOI] [PubMed] [Google Scholar]
- 40. Tobin M. B., et al. 1991. Localization of the lysine ε-aminotransferase (lat) and δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine synthetase (pcbAB) genes from Streptomyces clavuligerus and production of lysine ε-aminotransferase activity in Escherichia coli. J. Bacteriol. 173:6223–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vandenende C. S., Vlasschaert M., Seah S. Y. 2004. Functional characterization of an aminotransferase required for pyoverdine siderophore biosynthesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 186:5596–5602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vanlaere E., et al. 2009. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int. J. Syst. Evol. Microbiol. 59:102–111 [DOI] [PubMed] [Google Scholar]
- 43. Vidaver A. K. 1967. Synthetic and complex media for the rapid detection of fluorescence of phytopathogenic pseudomonads: effect of the carbon source. Appl. Microbiol. 15:1523–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wagner C., El Omari M., Konig G. M. 2009. Biohalogenation: nature's way to synthesize halogenated metabolites. J. Nat. Prod. 72:540–553 [DOI] [PubMed] [Google Scholar]
- 45. Wüthrich K. 1986. NMR of proteins and nucleic acids. Wiley, New York, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.