Abstract
Temperate forest soils are usually efficient sinks for the greenhouse gas methane, at least in the absence of significant amounts of methanogens. We demonstrate here that trafficking with heavy harvesting machines caused a large reduction in CH4 consumption and even turned well-aerated forest soils into net methane sources. In addition to studying methane fluxes, we investigated the responses of methanogens after trafficking in two different forest sites. Trafficking generated wheel tracks with different impact (low, moderate, severe, and unaffected). We found that machine passes decreased the soils' macropore space and lowered hydraulic conductivities in wheel tracks. Severely compacted soils yielded high methanogenic abundance, as demonstrated by quantitative PCR analyses of methyl coenzyme M reductase (mcrA) genes, whereas these sequences were undetectable in unaffected soils. Even after a year after traffic compression, methanogen abundance in compacted soils did not decline, indicating a stability of methanogens here over time. Compacted wheel tracks exhibited a relatively constant community structure, since we found several persisting mcrA sequence types continuously present at all sampling times. Phylogenetic analysis revealed a rather large methanogen diversity in the compacted soil, and most mcrA gene sequences were mostly similar to known sequences from wetlands. The majority of mcrA gene sequences belonged either to the order Methanosarcinales or Methanomicrobiales, whereas both sites were dominated by members of the families Methanomicrobiaceae Fencluster, with similar sequences obtained from peatland environments. The results show that compacting wet forest soils by heavy machinery causes increases in methane production and release.
INTRODUCTION
Well-aerated temperate forest soils generally act as a sink for atmospheric methane via methanotrophic oxidation (4, 44, 48). When soil physical conditions severely restrict soil aeration, the likelihood of methane emissions is high (1, 28, 46). It was recently shown that the net uptake/oxidation of methane by forest soils can be affected by forest operations (3, 5, 49, 53). Several studies exist documenting the effects of soil compaction on methane fluxes, and these have reported reductions in atmospheric CH4 consumption by 30 to 90% (arable soil [24, 30, 41], grassland soil compaction [40, 45], and forest soil [51]). Soils can even turn from a CH4 sink into a CH4 source after severe compaction (41, 51).
Methane is formed in soils by the microbial breakdown of organic compounds under anaerobic conditions (1). The production of CH4 does not begin until the reduction of molecular oxygen, nitrate, iron(III), manganese(IV), and sulfate (all of which maintain a higher potential) is complete (30). Methanogenic Archaea play an important role in the decomposition of organic matter in anaerobic environments and are responsible for virtually all biogenically produced methane from a variety of habitats, including natural wetlands and tundra, rice paddies, ruminants, landfills, and sediments (30, 55). Methanogens have only been sporadically detected from the upper well-aerated parts of agricultural, forest, and grassland soil environments (39), and the factors promoting methanogens in these environments are still not well understood. Pore size distribution, gas permeability, and hydraulic conductivity govern the distribution and activity of methanogens (33, 40, 54). Analysis of phospholipids fatty acids of compacted forest soil samples demonstrated that compaction resulted in an alteration of the microbial community structure (42). These authors also found a nearly 100-fold increase in cultivable methanogens in the compacted compared to the uncompacted soils. Terminal restriction fragment length polymorphism (T-RFLP) genetic profiling of DNA directly extracted from severely compacted wheel tracks in field experiments showed that total microbial community structures in compacted wheel tracks were changed compared to those of the unaffected soils (15). However, little is known about the structure and function of methanogenic communities in these well-aerated or compacted forest soils.
We report here on the impact of trafficking with heavy harvesting machines on net forest soil methane emissions and relate these effects to the abundance and diversity of soil methanogens. These effects were studied in controlled field experiments, replicated at two sites, in which wheel tracks differing in severity resulting from soil compaction were generated. We hypothesized that machine track formation would alter soil pore structure so as to reduce soil aeration, thereby favoring methanogens. Methanogenic archaea were characterized by PCR cloning and T-RFLP analyses of the functional mcrA gene, which encodes the subunit of methyl coenzyme M reductase, a key enzyme in methanogenesis (7, 12, 18, 31, 32, 47).
MATERIALS AND METHODS
Study sites and traffic experiments.
The traffic experiments were conducted in 2008 at two different forest sites: Ermatingen in the Canton of Thurgau and Heiteren in the Canton of Berne. Both sites are located on the Swiss Plateau and are similar in altitude and climate (550 m above sea level; 900 to 1,100-mm mean annual rainfall; 8.4 to 10.7°C mean annual temperature). Forests were dominated by Fagus sylvatica and Picea abies and had only sparse understory vegetation, and the soils varied markedly between the sites (15). The texture was on average loamy, although in the Ermatingen location there was much more clay (see Table S1 in the supplemental material). The organic carbon content was low, i.e., <1.5% C (see Table S1 in the supplemental material), and the organic layer was very thin (<1 cm) or absent.
Before traffic experiments, the soil moisture contents along the planned lanes (in triplicate, each of 20-m distance) were adjusted to a gradient ranging from 0.17 g of H2O per g of soil (plastic limit) to 0.35 g of H2O per g of soil (liquid limit). Trafficking with forestry machines at the different soil water contents caused three levels of compaction with light, moderate, and heavy impacts on a length of at least 10 m (15). The low soil water content was achieved by covering the soil surface (12 m2) of the planned lanes (light impact) with a plastic sheet for 2 days to prevent the infiltration of rain. The average water content of the soil under plastic covers was ∼0.17 g of H2O per g of soil. The remaining area of the planned lanes was irrigated at different intensities prior to trafficking in order to saturate the plasma porosity and also parts of the structural porosity, resulting in ∼0.27 g of H2O per g of soil (moderate impact) or 0.35 g of H2O per g of soil (severe impact). Before trafficking, 2 days were allowed to ascertain a good redistribution of the applied water.
Trafficking at Ermatingen was done with a fully loaded forwarder, model Valmet 840.2, weighing 26 tons (see Table S2 in the supplemental material). The mean (static) ground contact pressure was 238 kPa under the front tires and 320 kPa under the rear tires. At Heiteren, traffic was done with a skidder (model HSM 805 HD) weighing 14 tons. The ground contact pressure was 209 to 260 kPa beneath the front tires and 223 to 280 kPa beneath the rear tires. The forwarder (Ermatingen) trafficked the plots fully loaded twice forward and backward (four passes), while the skidder (Heiteren) trafficked the soils unloaded four times forward (four passes). Unaffected areas in the vicinity of the compacted soils served as controls (no impact). Traffic lanes were fenced off and further traffic was prevented after the treatment was applied. The study comprised three independent wheel tracks (replicates) per forest site with four levels of compaction (zero, light, moderate, and severe), giving 12 plots (∼12 m2 each) per traffic experiment.
Soil sampling.
Samples for soil physical measurements (three replicates) were collected within 1 to 4 days after trafficking at sampling locations below the ruts. Nontrafficked soil samples were taken at a 1-m distance from the center line of the track, thus avoiding influences of traffic and at the same time being in the proximity of the trafficked sample to control spatial heterogeneity. After removal of a thin litter layer, if present, the samples were taken from the topsoil at a 3- to 7-cm depth (referred to as 5 cm) and from the subsoil at 13- to 17-cm depth (referred to as 15 cm) with respect to the soil surface of the respective sampling locations after trafficking using sharpened steel cylinders with a volume of ∼100 cm3 (4.2 cm in height and 5.5 cm in diameter). Due to the formation of ruts, it was not possible to take the samples from the trafficked locations at exactly the depths corresponding to the specified sampling depths before displacement by wheeling. However, the topsoil (3 to 7 cm) and subsoil (13 to 17 cm) of the reconstituted profile were quite homogeneous, and the differences in C content between the sampled layers were small (data not shown). A total of 72 samples per experiment were obtained for the soil physical measurements with three replicate lanes, two depths (5 and 15 cm), and four treatments (zero, light, moderate, and severe compaction). These plots were assumed to be independent, since the wheel tracks were at least 20 m from each other.
Samples for microbial analyses were collected immediately prior to trafficking and 7 days, 1 month, 6 months, and 1 year after trafficking from the same locations as for the physical soil analyses. These samples were combined for each plot and soil layer, immediately cooled to 4°C, and transported to the laboratory. In all, 24 samples per harvest and experiment (four compaction levels × two depths × three lanes) were sieved (2-mm mesh size) and stored frozen (−20°C) in DNA extraction buffer.
Soil physical measurements.
Pore-size distribution was determined using the standard pressure-plate procedure for soil moisture retention curve (15, 25). Proportions of pore size classes were calculated based on the measured water desorption characteristics (50). After draining at 300 kPa, samples were dried at 105°C. Total pore volume (porosity) was determined as mass difference between saturated and oven-dried samples. Saturated hydraulic conductivity (kf) was measured using an ICW soil water permeameter (15).
Methane flux measurements.
Methane fluxes were determined using medium-sized (0.076-m2 area and 7.9-liter volume) unventilated manually operated static chambers (36). These chambers were pushed ca. 16 cm into the soil several weeks before the measurement. Similar locations (n = 3) as for microbial analyses at each compaction level were randomly selected for gas flux measurements and used as independent field replicates. Net soil-atmosphere methane fluxes were measured several times after traffic, concomitantly with the molecular analyses. The static chambers were closed with a lid, and headspace samples were obtained with syringes 5, 20, and 35 min after chamber closure. Gas samples were stored in pre-evacuated glass vials and analyzed by gas chromatography (Agilent 6890 gas chromatograph [Agilent, Palo Alto, CA]; methane was quantified using a flame ionization detector, and CO2 was reduced with hydrogen on a nickel catalyst [methanizer] and then measured on the same detector). CH4 concentration measurements were calibrated against standard samples analyzed with every batch of samples. Soil-atmosphere CH4 fluxes were calculated by linear regression of CH4 concentration against time, accounting for air temperature and pressure at the time of sampling. Concomitantly, soil moisture was measured by using a time-domain reflectometry probe (Trime-FM; IMKO Micromodultechnik GmbH, Ettlingen, Germany), and the temperature at a soil depth of 3 cm was measured with a pocket thermometer with stainless steel penetration probe (Checktemp HI98501; Hanna Instruments, Italy).
DNA extraction.
Total nucleic acids were extracted in duplicate from 5- and 15-cm soil depths by a bead-beating procedure (16). DNA concentrations were determined using a fluorometric assay with PicoGreen (Molecular Probes, Eugene, OR). Before PCR amplification, soil DNA was diluted (1:10) and pretreated with bovine serum albumin (BSA) to bind humic acids and other PCR-inhibiting substances (15).
T-RFLP analysis.
Methanogen-specific methyl coenzyme M reductase a-subunit (mcrA) genes were amplified by using the primer set designed by Luton et al. (32), which amplifies an ∼470-bp fragment of mcrA. PCR mixtures (25 μl) contained 20 pmol of the primer pairs. The forward primer was fluorescently labeled with carboxyfluorescein (6-FAM; Microsynth, Switzerland) for T-RFLP analysis. Reactions also contained 1× PCR buffer (Qiagen, Hilden, Germany), 2 mM MgCl2, 0.4 mM deoxynucleoside triphosphate (dNTP; Promega), 0.6 mg of BSA (Fluka, Buchs, Switzerland) ml−1, and 2 U of HotStar Taq polymerase (Qiagen). Thermal cycling included 40 cycles of denaturation at 94°C (15 min), annealing at 53°C (90 s), and elongation at 72°C (120 s), with a final 6 min of elongation at 72°C. PCR products were purified with the Montage PCR purification cleanup kit (Millipore Corp., Billerica, MA). Restriction of mcrA amplicons was performed with either 10 U of enzyme Sau96I or enzyme MspI (Promega, Mannheim, Germany) plus 1 μl of the appropriate incubation buffer, followed by incubation for 3 h at 37°C. Prior to T-RFLP analysis, digests were desalted with Montage SEQ96 sequencing reaction cleanup kit (Millipore) according to the manufacturer's instructions. T-RFLP analyses were performed as described previously (15, 17), and the T-RFLP profiles were analyzed by using Genotyper v3.7 NT (Applied Biosystems, Foster City, CA). Relative amplicon frequencies were determined as the relative signal intensities of terminal restriction fragments (T-RFs) with peak height analysis as described previously (15, 17). Signals with a peak height below 100 relative fluorescence units were regarded as background noise and excluded from the analysis.
Quantification of mcrA gene by quantitative real-time PCR.
Methanogens were quantified in undisturbed and compacted soil samples by using a mcrA-targeting quantitative real-time PCR assay using the same primers (not fluorescently labeled) as before. Before quantification, the DNA extracts were tested for inhibitory effects of coextracted substances, and the lowest dilution that had no inhibitory effect was used for further measurements. Quantitative real-time PCR assays were performed in an ABI 7500 Fast real-time PCR system (Applied Biosystems). Each 25-μl reaction contained a 0.5 μM concentration of each primer, 12.5 μl of SYBR green PCR master mix (including HotStar Taq DNA polymerase, Quanti-Tec SYBR green PCR buffer, dNTP mix, SYBR green I ROX, and 5 mM MgCl2 [QuantiTect SYBR green PCR kit; Qiagen]), 0.2 mg of BSA ml−1, 11 μl of diluted DNA corresponding to 2.5 ng of total soil DNA, and RNase-free water to complete the 25-μl volume. The PCR conditions were first 15 min at 95°C, followed by 40 cycles of 95°C for 60 s, 52°C for 60 s, and 72°C for 60 s, followed in turn by a final data acquisition step of 80°C for 15 s. Each plate included triplicate reactions per DNA sample and the appropriate set of standards. After the DNA amplification cycles, melting-curve analysis confirmed that the signals obtained were caused by the specific amplicon. Cloned mcrA gene fragments (clone HE_B7 and clone ER_C1) were used to create the standard curve as described earlier (16). The selected clones reflect the range of mcrA sequences encountered. The slope of each standard curve (regression lines of the threshold cycle [CT] versus logN, the log of initial DNA concentration in standard templates) was used to estimate amplification efficiency in our quantitative PCR assays. Similar results were obtained for each standard curve, with an average slope of −3.458, an average efficiency of 94.7%, an average R2 of 0.9951, and a lower limit of detection of approximately 2.1 × 104 gene copies g of dry soil−1. The data are presented as the average copy number of targets per gram of soil (dry weight).
Cloning and sequencing of mcrA genes.
Portions (50 ng) of DNA were pooled from three replicate extracts of severely compacted soil samples from the two forest sites at 5 and 15 cm depths after 180 days of trafficking and were used as templates for the amplification of mcrA genes. The reaction mixture and amplification cycle were performed as described above. Purified PCR products from the four replicates were pooled and cloned by using a pGEM-T Easy cloning kit (Promega) according to the manufacturer's instructions. To group similar clone types for subsequent sequence analysis, mcrA clones were subjected to T-RFLP analysis using the restriction enzyme Sau96I and to assign T-RFs obtained in environmental samples with T-RFs from cloned sequences. Representative clones from the most frequently occurring restriction patterns from each library were sequenced by using M13 primers. M13 amplicons were cleaned prior to sequencing with Montage SEQ96 sequencing reaction cleanup kit (Millipore). Cycle sequencing was carried out using the BigDye terminator cycle sequencing kit (version 3.1; Perkin-Elmer Applied Biosystems, Foster City, CA) according to the manufacturer's recommendations. After the reaction, excess dye terminator was removed using a Montage SEQ96 sequencing reaction cleanup kit (Millipore) according to the manufacturer's instructions. Nucleotide sequences of mcrA clones were sequenced on both strands using an ABI310 genetic analyzer (Applied Biosystems).
Between 55 and 60 clones were analyzed for each library (in total four libraries: two sites and two soil horizons), depending on the diversity of recovered phylotypes. Obtained nucleotide sequences were translated to deduced amino acid sequences by using the program Expase. Partial mcrA sequences were subjected to a BLAST search, and sequences in the GenBank database sharing the greatest similarities were imported into the BioEdit sequence alignment editor. For phylogenetic placement, alignments against closest relatives identified by a National Center for Biotechnology Information BLAST search and known taxonomic sequences were carried out by using CLUSTAL X. The resulting alignments were edited using BioEdit. Evolutionary distances were calculated by the Jukes-Cantor algorithm and the phylogenetic tree determined by the neighbor-joining method using Treecon for Windows version 1.3b (52). The percent coverage of the clone libraries was calculated by the method of Good (23) as modified by Singleton et al. (43) using ≥97% sequence similarity to indicate identical clones. Diversity of clone libraries was calculated by using the Shannon index.
Statistics.
Statistical analyses of soil physical parameters and methane fluxes were carried out using one-way analysis of variance (ANOVA) using Statistica 5.1 (Statsoft, Inc., Tulsa, OK). Multiple comparisons of significant differences between unaffected and the different compaction levels within one site and depth (for soil physical parameters only) were made using Scheffe's post hoc test (P = 0.05). One-way ANOVA with repeated measures and Scheffe's post hoc test were used for testing significant differences between treatments for quantitative real-time PCR results.
Accession numbers.
Deduced amino acid sequences generated in the present study were in the GenBank database under accession numbers JF288564 to JF288610.
RESULTS
Soil physical conditions.
Varying the levels of soil water content prior to trafficking allowed the generation of wheel tracks differing in level of compaction (light, moderate, and severe). The high porosity and hydraulic conductivities of the unaffected control samples reveal a well-structured pore system at both sites. In contrast, wheel traffic considerably reduced soil porosity and lowered water movement throughout the soil profile in both soil layers analyzed (Tables 1 and 2). Reflected in this overall decline in porosity were large decreases in macropores (≥50-μm pore size), which decreased by 65% (average of the two soil layers) at Ermatingen and by 76% at Heiteren under severe compaction (Table 1). According to the rut classification from light to severe, saturated hydraulic conductivities declined constantly to ca. 10% compared to the original conductivity of unaffected soils at both soil depths (Table 2). As a consequence, higher water contents were found in compacted wheel tracks compared to unaffected soils (data not shown), indicating that water infiltration was very low, thereby increasing the risk of waterlogging in the ruts.
Table 1.
Total pore volume and proportion of wide coarse pores in different compacted wheel track types at two soil depths
| Site and sample depth (cm) | Compaction level | Mean ± SDa |
|||
|---|---|---|---|---|---|
| Total pore vol (%) | Pore size vol (%) (≥50 μm) | ||||
| Ermatingen | |||||
| 5–10 | Undisturbed | 52 ± 4A | 9 ± 2A | ||
| Light | 45 ± 2B | 6 ± 1AB | |||
| Moderate | 43 ± 2B | 5 ± 2BC | |||
| Severe | 42 ± 3B | 3 ± 1C | |||
| 15–20 | Undisturbed | 44 ± 2A | 8 ± 1A | ||
| Light | 41 ± 1AB | 5 ± 1B | |||
| Moderate | 40 ± 1B | 5 ± 1B | |||
| Severe | 40 ± 2B | 3 ± 2B | |||
| Heiteren | |||||
| 5–10 | Undisturbed | 55 ± 1A | 16 ± 2A | ||
| Light | 50 ± 1B | 6 ± 1B | |||
| Moderate | 45 ± 2C | 6 ± 2B | |||
| Severe | 43 ± 1C | 4 ± 2AB | |||
| 15–20 | Undisturbed | 55 ± 1A | 18 ± 2A | ||
| Light | 48 ± 1B | 11 ± 2B | |||
| Moderate | 45 ± 3B | 7 ± 2BC | |||
| Severe | 42 ± 1BC | 4 ± 1C | |||
Values are means (n = 9). Means followed by the same superscript capital letter within one soil depth and forest site are not significantly different at P < 0.05. Proportion of wide coarse pores, −6 kPa.
Table 2.
Saturated water conductivity in different compacted wheel track types at two soil depths from the Ermatingen and Heiteren forest sites
| Compaction level | Mean saturated water conductivity (m day−1) ± SDa |
|||
|---|---|---|---|---|
| Ermatingen |
Heiteren |
|||
| 5–10 cm | 15–20 cm | 5–10 cm | 15–20 cm | |
| Undisturbed | 0.63 ± 0.38A | 0.86 ± 0.47A | 3.2 ± 1.2A | 2.9 ± 0.8A |
| Light | 0.48 ± 0.23A | 0.47 ± 0.14B | 0.7 ± 0.2B | 0.5 ± 0.1B |
| Moderate | 0.23 ± 0.10B | 0.25 ± 0.12B | 0.4 ± 0.1B | 0.3 ± 0.1B |
| Severe | 0.06 ± 0.04B | 0.19 ± 0.11B | 0.1 ± 0.1C | 0.1 ± 0.1C |
Values are means (n = 9). Means followed by the same superscript capital letter within one soil depth and forest site are not significantly different at P < 0.05.
Methane fluxes.
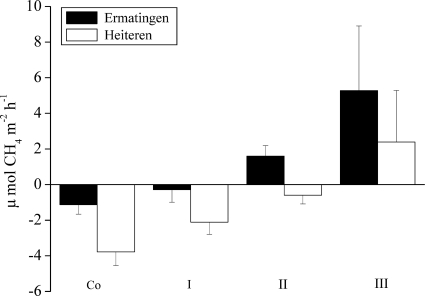
Soil compaction reduced soil-atmosphere CH4 exchange through lower macropore space and hydraulic conductivities. As a consequence, methane fluxes were altered by trafficking, leading to a large reduction in the CH4 consumption rate in the compacted soils (Fig. 1). This decrease was between 2-fold (Ermatingen) and 4-fold (Heiteren) in the lightly compacted soil and 7-fold (Heiteren) in the moderately compacted soil. In the moderately (Ermatingen) and severely compacted wheel tracks at both sites, soils turned from a CH4 sink into a CH4 source (Fig. 1). CH4 emissions were found at both study sites. Averaged over time, CH4 emissions in severely compacted wheel tracks ranged between +5.3 μmol of CH4 m−2 h−1 (Ermatingen) and +2.4 μmol of CH4 m−2 h−1 (Heiteren). In contrast to severely compacted wheel tracks, no CH4 emissions were found in unaffected and lightly compacted soils. Averaged CH4 fluxes in unaffected soils equaled −1.1 μmol of CH4 m−2 h−1 at Ermatingen and −3.8 μmol of CH4 m−2 h−1 at Heiteren. In lightly compacted soils, the corresponding fluxes were −0.3 μmol of CH4 m−2 h−1 and −2.1 μmol of CH4 m−2 h−1, respectively. In the moderately compacted wheel tracks, CH4 fluxes were more variable with CH4 consumption at Heiteren and CH4 emissions at Ermatingen.
Fig. 1.
Averaged CH4 fluxes (mean ± the standard deviation [SD]; n = 3) in unaffected (control) and differently trafficked soils from the two forest sites. Measurements were made four times after traffic when intensive molecular analyses were performed. Co, control (no impact); I, light impact; II, moderate impact; III, severe impact.
Quantitative PCR of mcrA.
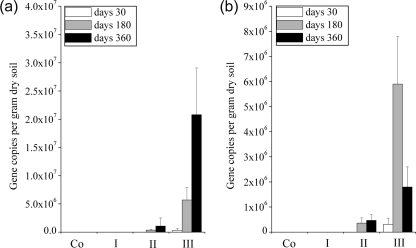
Soil compaction significantly influenced the copy numbers of mcrA genes (P < 0.001; Fig. 2). We could not amplify mcrA genes in unaffected or lightly compacted wheel tracks at any sampling time and in either experiment. Overall, the highest number of mcrA gene copies was found in severely compacted forest soils (Fig. 2) where CH4 emissions had been observed. The number of target molecules ranged from 3.2 × 105 to 2.1 × 107 and from 3.1 × 105 to 5.9 × 106 copies g of soil−1 at Ermatingen and Heiteren, respectively, and were significantly higher than in the moderately compacted wheel tracks (P < 0.001). In the severely compacted soils, mcrA gene copies became detectable 7 days after traffic (in one of three replicates). At 30 days after trafficking, high mcrA gene copy numbers were found at Ermatingen (Fig. 2a), with a steady increase over time. In the more clayey soil at Ermatingen, no decline in mcrA gene abundances was found even 6 and 12 months later. This contrasts with our findings in the more sandy soil at Heiteren, where the methanogenic biomass had declined sharply (Fig. 2b). Factorial analysis of variance indicated that there was no significant difference in the mcrA gene copies between the soil depths at either forest site (P > 0.05 [data not shown]).
Fig. 2.
Numbers of mcrA gene copies (mean ± the SD; n = 3) in unaffected and compacted soils at a 5-cm depth from the Ermatingen (a) and Heiteren (b) sites at different times after trafficking with heavy forestry machinery. Co, control (no impact); I, light impact; II, moderate impact; III, severe impact.
Methanogen community composition and diversity.
We investigated the structure of the methanogenic communities by combining T-RFLP analysis of PCR-amplified mcrA gene fragments with cloning and sequencing of mcrA genes. No PCR products were obtained using environmental DNA extracted from unaffected and lightly compacted soils in both traffic experiments, suggesting that methanogens were not abundant in these soils and confirming the findings of quantitative PCR. Therefore, only mcrA gene fragments from severely compacted wheel tracks, with distinct methanogenic characteristics, were subjected to cloning and sequencing. The four libraries (two sites; two soil horizons) contained between 7 and 10 operational taxonomic units (OTU) (Table 3). There were no depth-characteristic OTU in either soil. The Shannon indices for the mcrA Ermatingen clone library (average of the two soil horizons: H = 1.72) were higher than for the mcrA Heiteren clone library (H = 1.25) (Table 3). Coverage of expected mcrA diversity within each clone library was ascertained by comparison of the observed versus expected phylotypes for each library. Values ranged from 83 to 88% and were highest in the mcrA Heiteren libraries, independent of soil depth (Table 3), indicating that the number of clones was sufficient to cover the diversity of the mcrA OTU.
Table 3.
Diversity indices calculated using OTU distributions from T-RFLP analyses of mcrA clone libraries
| Parameter | Clone librarya |
|||
|---|---|---|---|---|
| ER, 5 cm | ER, 15 cm | HE, 5 cm | HE, 15 cm | |
| No. of clones analyzed | 59 | 58 | 60 | 55 |
| No. of OTU detected | 10 | 9 | 7 | 7 |
| Coverage (%) | 83 | 84 | 88 | 87 |
| Shannon diversity index (H′) | 1.76 | 1.67 | 1.29 | 1.21 |
The libraries originated from the two forest sites (ER, Ermatingen; HE, Heiteren) and two soil depths (5 and 15 cm).
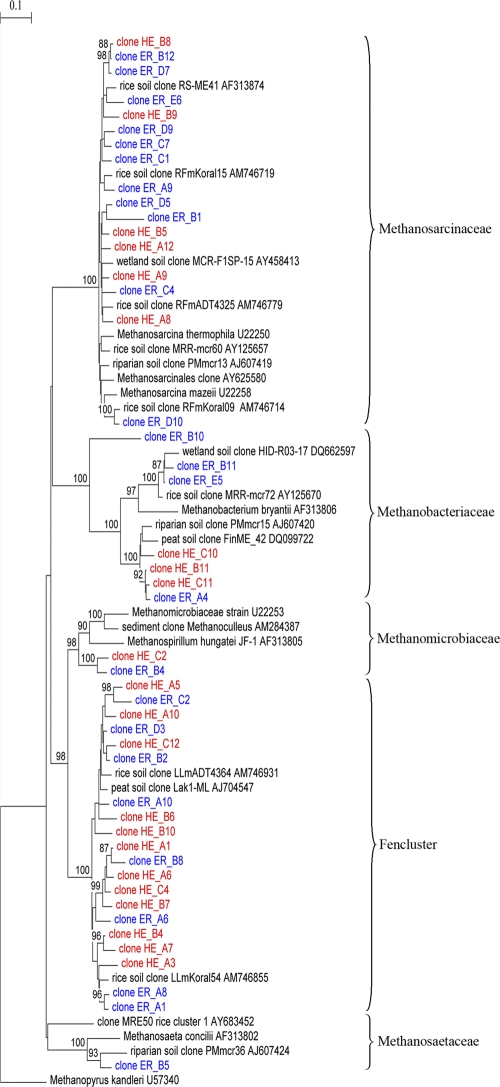
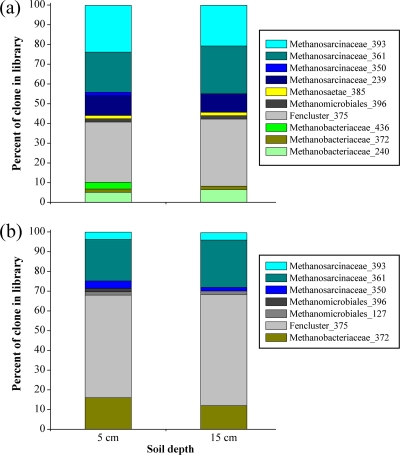
Phylogenetic analysis of mcrA clones revealed that almost all retrieved mcrA sequences were related to uncultivated methanogenic archaean phylotypes isolated from a variety of environments, including peat (19, 26), rice roots (9), and riparian soils (8, 27). The mcrA gene libraries were dominated by members of the families Methanosarcinaceae and Methanomicrobiaceae Fencluster (Fig. 3). T-RFLP analysis of representative mcrA clones (n = 232) produced terminally labeled fragments with characteristic lengths of mostly 361, 375, and 393 bp with Sau961 (Fig. 4) and 271, 292, and 310 bp with MspI (data not shown). Sequence analysis allowed the determination of the phylogenetic affiliation of the individual clones with their characteristic T-RFs. The four mcrA gene libraries were dominated by members of the families Methanosarcinaceae (239, 361, and 393 bp) and Methanomicrobiaceae Fencluster (375 bp). More than 30% of all mcrA gene OTU at Ermatingen (Fig. 4a) and more than 50% of the mcrA gene OTUs at Heiteren (Fig. 4b) belonged to the order Methanomicrobiales. Methanosarcinales were also abundant mcrA gene OTU at Heiteren (ca. 25%) and Ermatingen (>50%). The T-RF of 385 bp was found only in the mcrA Ermatingen clone library and represented 2% of all mcrA clones (Fig. 4a). Methanobacteriaceae-like sequences exhibited T-RFs of 240, 372, and 436 bp, constituting 9 and 14% of all mcrA clones at Ermatingen and Heiteren, respectively.
Fig. 3.
Phylogram showing phylogenetic relationships of mcrA sequences of forest soils from Ermatingen (ER, red type) and Heiteren (HE, blue type) collected 180 days after traffic at the severe impacted wheel tracks and reference sequences of described methanogen species and environmental clones. The tree was constructed from inferred amino acid sequences (141 positions) by the neighbor-joining method. Numbers at the nodes (>75%) represent the percentages of bootstrap resamplings based on 500 replicates. The scale bar represents 0.1 nucleotide substitutions per site. Methanopyrus kandleri was used as an outgroup reference.
Fig. 4.
Relative proportions of mcrA clones in libraries of the Ermatingen (a) and Heiteren (b) samples from two soil depths (5 and 15 cm) after 6 months of traffic. Only mcrA gene fragments from severely compacted wheel tracks were subjected to cloning and sequencing. Sau96I was used as a restriction enzyme to screen mcrA clones with T-RFLP analyses. T-RFs of 239, 350, 361, and 393 bp are representative of Methanosarcinaceae; T-RFs of 127 and 396 bp are representative of Methanomicrobiales; a T-RF of 375 bp is representative of the Fencluster within Methanomicrobiales; T-RFs of 240, 372, and 436 bp are representative of Methanobacteriaceae; and a T-RF of 385 bp is representative of Methanosaetae.
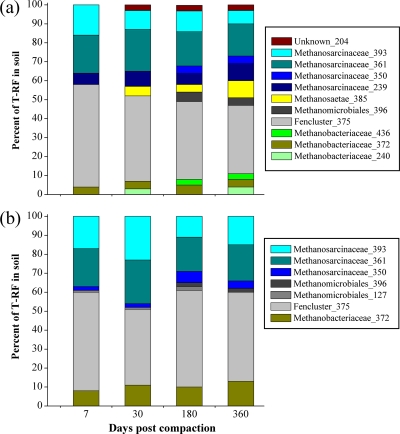
All restriction fragments detected in the clone libraries were also found when the soil DNA was analyzed directly by T-RFLP (Fig. 5). T-RFLP patterns were able to resolve the dominant methanogen-affiliated taxa and generally supported the proportions recovered in the clone libraries. A T-RF peak at 204 bp was exclusively found in environmental DNA of Ermatingen but could not be assigned, since clones with this T-RF were not found. In general, no significant temporal changes in the relative abundance of identified T-RFs were observed at Heiteren, whereas the T-RFLP patterns at Ermatingen were more dynamic over time (Fig. 5). Also, there was no depth dependency of the T-RFs observed in both soils (data not shown). Eleven T-RFs were found in T-RFLP profiles in the soil samples of Ermatingen (Fig. 5a) and seven T-RFs in the Heiteren soil samples (Fig. 5b). Six T-RFs (350, 361, 372, 375, 393, and 396 bp) were detected in all soil samples. The dominant T-RF group of 375 bp in the compacted wheel tracks of both soils comprising between 36 and 54% of the total abundance in the profiles may correspond to the sequence assigned to the Methanomicrobiales Fencluster. Four T-RFs (72, 127, 204, and 436 bp) were detected only occasionally and at low abundance. The following T-RFs assigned were exclusively found in soil samples from Ermatingen: 204, 239, 240, and 436 bp.
Fig. 5.
Changes over time in the abundance of methanogen subgroups recovered from severely compacted forest soils (5-cm soil depth) from the Ermatingen (a) and Heiteren (b) sites at four different time points (7, 30, 180, and 360 days after traffic). The relative abundance of T-RFs was used as a measure of the composition of the mcrA community. mcrA gene-based T-RFLP profiles were created with Sau96I as a restriction enzyme. T-RFs of 239, 350, 361, and 393 bp are representative of Methanosarcinaceae; T-RFs of 127 and 396 bp are representative of Methanomicrobiales; a T-RF of 375 bp is representative of Fencluster within Methanomicrobiales; T-RFs of 240, 372, and 436 bp are representative of Methanobacteriaceae; and a T-RF of 385 bp is representative of Methanosaetae. A T-RF of 204 bp could not be affiliated with any clone sequences.
DISCUSSION
We investigated the long-term (1 year) responses of methanogenic communities in well-aerated forest soils to traffic with heavy forestry machinery. Traffic caused a profound change in soil physical properties, which in turn led to a significant increase in the abundance of methanogens. High methanogenic abundances, as demonstrated by quantitative PCR of the mcrA gene, were found only in severely compacted soils. In these soils, macropore volume was strongly reduced and water infiltration decreased, leading to severely hindered soil aeration, net CH4 emissions, and the development of distinct methanogenic communities. Reduced gas exchange between the soil and the atmosphere has also been observed through machine impact (1, 15, 45), leading to increasingly anaerobic conditions. Under these conditions, methanogenesis is the dominant electron-accepting process during the oxidation of organic matter (10).
Temperate forest soils are usually efficient sinks for the greenhouse gas CH4 (48). The methane uptake rates of our undisturbed soils were comparable to those reported for European and North American temperate forest soils (4, 35, 48, 49). Here, traffic with heavy harvesting machines caused a large reduction (between 2 and 80%) in CH4 consumption rate. A similarly large reduction in CH4 consumption rates after compaction has also been found in agricultural (24, 45) and forest soils (51). The net soil-atmosphere CH4 flux is the result of the dynamic equilibrium between production (under anoxic conditions, by methanogens [methanogenesis]) and oxidation of CH4 (under oxic conditions, by methanotrophs [methanotrophy]) (11). The CH4 emissions in severely compacted wheel tracks that we observed were in accordance with the increased CH4 emissions measured in compacted beech forest soils in Germany (51). The authors of that study observed that soils turned from a sink to a source of CH4 (∼1.4 kg of CH4-C ha−1 year−1) when water-filled pore space increased to ca. 100% in compacted wheel tracks. Therefore, forest operations with heavy machinery appear to contribute to accelerated global warming. In addition to methane, we have also measured CO2 emitted from the soils in the traffic lanes (see Materials and Methods). We observed a small but nonsignificant decrease in soil respiration in the heavily compacted traffic lanes (data not shown). Despite this weak decrease in CO2 emissions, an overall effect on the global warming potential due to the strong methane emissions is evident. Assuming that severe soil compaction during forest operations is restricted to 3% of the harvested area (6), CH4 emissions amounted to 45 g of CH4 ha−1 year−1. Extrapolated to the area of the forested area of the Swiss Plateau, forestry machine traffic amounts to ∼55 tons of CH4 year−1 (corresponding to 1,265 tons of CO2 eq) for the Swiss Plateau.
Methanogens became abundant in traffic lanes that were severely compacted, which is consistent with the observed reduction in CH4 consumption, or CH4 emissions. Undisturbed and lightly compacted soils in the two forest sites apparently contained only low numbers of methanogens, from which the mcrA gene could not be amplified. Interestingly, there was a lag phase in the mcrA gene detection. After 7 days of traffic, mcrA gene copies could be detected for the first time in severely compacted soils. Only after 30 days of traffic were high mcrA gene copy numbers found, and these remained stable for many months. The generally slow growth rate of methanogens may explain the observed lag phase, as was previously pointed out (39). In contrast to the well-studied rice paddy soils and peat bogs, methanogens have only been detected sporadically in the upper well-aerated parts of soil environments (21, 38, 39, 40). Aggregate interiors with low oxygen concentrations may provide a good habitat for methanogenic microbes to survive the otherwise unfavorable conditions in oxic soils (42). The survival and rapid reactivation of methanogens after prolonged drying have been reported in oxic soils (14, 39) and floodplain wetlands (2).
Since until now no quantitative data on methanogens by targeting mcrA in temperate oxic forest soils have been reported, comparisons in this respect can hardly be made. However, the mcrA gene copy numbers found were comparable to the ones reported in arable soils compacted by cattle (animal treading impacts) (40). Twelve months after traffic, methanogenic abundances did not decline at Ermatingen, and mcrA genes were still detectable in high gene copy numbers. The present study demonstrates large site differences in the resilience of soil methane fluxes. We argue that the clayey soil at Ermatingen led to a more complete and more persistent inhibition of soil aeration, whereas this effect was only of short-term nature in the more sandy soil at Heiteren. The importance of soil texture in methane production has been shown as pointed out by Wagner et al. (54). In another forest site (Messen, canton of Solothurn, with a clay content of 15%) on the Swiss Plateau that was severely affected by traffic of heavy harvesting machines 6 years ago (15), we still found a high abundance of mcrA gene copies and methane emissions (data not shown). Therefore, we assume that several years after traffic without the artificial regeneration of soils, the abundance of mcrA gene copies remains high, indicating no disappearance of methanogens with time. Strict anaerobes would be sensitive to mixing events that introduce traces of oxygen into compacted soil, but that was not the case here.
In our compacted forest soils we found several persisting mcrA sequence types continuously present at all sampling times, indicating that forest methanogenic community composition is relatively stable, in line with findings in rice fields (9) and lake sediments (22). In contrast, methanogenic communities in ephemeral floodplain wetlands were found to be rather dynamic, dominated by changing methanogenic families (27). Phylogenetic analysis of our retrieved mcrA clone sequences revealed a rather large methanogen diversity in the compacted forest soils. The majority of methanogen sequences from our permanently compacted forest soils belonged to either the Methanosarcinales order or the Methanomicrobiales order. Members of the families Methanosaetaceae and Methanobacteriaceae were only rarely detected in these soils. A substantial part of the retrieved clones can be considered as colonizers of wetlands, so most of the detected methanogenic taxa were also found in peatland (7, 19, 26), rice fields (9), and riparian soil (27). Between 36 and 54% of the sequences formed a separate cluster within the Methanomicrobiales and showed close relationships to the Fen cluster (26).
Interpretation of physiology from Ermatingen and Heiteren phylotypes suggested the presence of hydrogenotrophic (7) and acetotrophic methanogens in the forest soils investigated (20), indicating that populations include generalists, species that are metabolically diverse in terms of their substrate utilization for methane production (e.g., Methanosarcina). Substrate quality appears to modify or determine methanogenic community composition (7). Acetotrophic methanogenesis has been connected to labile carbon in surface peat, whereas hydrogenotrophic methanogenesis has been suggested to prevail in environments with less reactive, further decomposed, carbon compounds (29).
Although there appears to be potential for the population to utilize a range of substrates, the Methanomicrobiales-related sequences in the methanogen-specific gene libraries and in the T-RFLP profile suggest a predominantly H2/CO2-utilizing population (reducing CO2 to methane in their energy metabolism). The methanogenic pathways of the Methanomicrobiales-associated Fen cluster dominating our compacted forest soils are unknown but could be hydrogenotrophic, since the closest cultured relatives are hydrogenotrophs (order Methanomicrobiales) (26). These hydrogenotrophic methanogens in soil may be growing symbiotically through interspecies hydrogen transfer with fermentative bacteria (34). These syntrophic bacteria living in close relationship with methanogens and providing the substrate for hydrogenotrophic CH4 production often have much broader substrate specificities than any methanogen would have. Acetate utilization is restricted to two genera in the order Methanosarcinales, Methanosaeta and Methanosarcina; all other species of methanogens use H2. Methanosaeta sequences were observed in small proportions, which may indicate soil niches where acetate concentrations are locally high. The genus Methanosaeta belongs to the family Methanosaetaceae and includes exclusively acetotrophic species that utilize acetate as sole substrate for energy metabolism (13). On the other hand, Methanosarcinaceae are generally considered as somewhat ubiquitous methanogens occurring in oxic and anoxic soils of the temperate zone (31). Members of the family Methanobacteriaceae, as shown in other studies on archaeal diversity in northern peatlands (26) were underrepresented here. Methanobacteriales mcrA gene sequences have, however, been recovered from a wider range of environments, such as from deep marine sediments (37), rice field soil (31), and landfill leachate (32).
Conclusions.
Temperate forest soils with low numbers of methanogens are usually efficient sinks for the greenhouse gas methane. However, these methanogenic populations became active and proliferated after traffic with heavy forestry machines, leading to a dramatic increase in the abundance of methanogens, and therefore substantially reduced the strength of the soil methane sink. The compacted forest soils were dominated by members of the families Methanomicrobiaceae Fencluster with similar mcrA sequences obtained from peatland environments. This leads to the assumption that a substantial part of the methanogenic community in our compacted forest soils were identical to cloned sequences from acidic, oligotrophic bogs. When soil physical properties became completely unfavorable for adequate soil aeration, the risk of methane emission was likely. Therefore, intensive logging practices may turn oxic forest soils into a source of methane and increase radiative forcing.
Supplementary Material
ACKNOWLEDGMENTS
We thank the forest services in Ermatingen ct. TG (Werner Kreis) and in Heiteren ct. BE (Roland Ruppli), who agreed to collaborate with trafficking experiments in the forests. The technical assistance of Daniela Steiner and Andreas Rüdt in the laboratory is gratefully acknowledged. We also thank Peter Christie (Queen's University Belfast) for reviewing earlier versions of the manuscript.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 8 July 2011.
REFERENCES
- 1. Ball B. C., Dobbie K. E., Parker J. P., Smith K. A. 1997. The influence of gas transport and porosity on methane oxidation in soils. J. Geophys. Res. Atmos. 102:23301–23308 [Google Scholar]
- 2. Boon P. I., Mitchell A., Lee K. 1997. Effects of wetting and drying on methane emissions from ephemeral floodplain wetlands in southeastern Australia. Hydrobiol. 357:73–87 [Google Scholar]
- 3. Borken W., Xu Y. J., Beese F. 2003. Conversion of hardwood forests to spruce and pine plantations strongly reduced soil methane sink in Germany. Glob. Chang. Biol. 9:956–966 [Google Scholar]
- 4. Borken W., Beese F. 2006. Methane and nitrous oxide fluxes of soils in pure and mixed stands of European beech and Norway spruce. Eur. J. Soil Sci. 57:617–625 [Google Scholar]
- 5. Bradford M. A., Ineson P., Wookey P. A., Lappin-Scott H. M. 2000. Soil CH4 oxidation, response to forest clearcutting and thinning. Soil Biol. Biochem. 32:1035–1038 [Google Scholar]
- 6. Brändli U. B. 2010. The Swiss National Forest Inventory: results of the third survey 2004–2006. Swiss Federal Research Institute WSL, Federal Office for the Environment, Geneva, Switzerland [Google Scholar]
- 7. Cadillo-Quiroz H., et al. 2006. Vertical profiles of methanogenesis and methanogens in two contrasting acidic peatlands in central New York State, U.S.A. Environ. Microbiol. 8:1428–1440 [DOI] [PubMed] [Google Scholar]
- 8. Castro H. F., Reddy K. R., Ogram A. V. 2004. Phylogenetic characterization of methanogenic assemblages in eutrophic and oligotrophic areas of the Florida Everglades. Appl. Environ. Microbiol. 70:6559–6568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Conrad R., Klose M., Noll M., Kemnitz D., Bodelier P. 2008. Soil type links microbial colonization of rice roots to methane emission. Glob. Chang. Biol. 14:657–669 [Google Scholar]
- 10. Conrad R. 2007. Microbial ecology of methanogens and methanotrophs. Adv. Agron. 96:1–63 [Google Scholar]
- 11. Dalal R. C., Allen D. E., Livesley S. J., Richards G. 2008. Magnitude and biophysical regulators of methane emission and consumption in the Australian agricultural, forest, and submerged landscapes: a review. Plant Soil 309:43–76 [Google Scholar]
- 12. Ermler U., Grabarse W., Shima S., Goubeaud M., Thauer R. K. 1997. Crystal structure of methyl coenzyme M reductase: the key enzyme of biological methane formation. Science 278:1457–1462 [DOI] [PubMed] [Google Scholar]
- 13. Ferry J. G. 2010. Biochemistry of acetotrophic methanogenesis, p. 357–367. In Timmis K. N. (ed.), Handbook of hydrocarbon and lipid microbiology. Springer Verlag, Berlin, Germany [Google Scholar]
- 14. Fetzer S., Bak F., Conrad R. 1993. Sensitivity of methanogenic bacteria from paddy soil to oxygen and desiccation. FEMS Microbiol. Ecol. 12:107–115 [Google Scholar]
- 15. Frey B., et al. 2009. Compaction of forest soil with heavy logging machinery affects soil bacterial community structure. Eur. J. Soil Biol. 45:312–320 [Google Scholar]
- 16. Frey B., Pesaro M., Rüdt A., Widmer F. 2008. Dynamics of bacterial communities in bulk and poplar rhizosphere soil contaminated with heavy metals. Environ. Microbiol. 10:1433–1449 [DOI] [PubMed] [Google Scholar]
- 17. Frey B., Stemmer M., Widmer F., Luster J., Sperisen C. 2006. Microbial characterization of a heavy metal-contaminated soil in a model forest ecosystem. Soil Biol. Biochem. 38:1745–1756 [Google Scholar]
- 18. Friedrich M. W. 2005. Methyl-coenzyme M reductase genes: unique functional markers for methanogenic and anaerobic methane-oxidizing Archaea. Methods Enzymol. 397:428–442 [DOI] [PubMed] [Google Scholar]
- 19. Galand P. E., Fritze H., Conrad R., Yrjälä K. 2005. Pathways for methanogenesis and diversity of methanogenic archaea in three boreal peatland ecosystems. Appl. Environ. Microbiol. 71:2195–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garcia J. L., Patel B. K. C., Ollivier B. 2000. Taxonomic, phylogenetic, and ecological diversity of methanogenic Archaea. Anaerobe 6:205–226 [DOI] [PubMed] [Google Scholar]
- 21. Gattinger A., et al. 2007. Traditional cattle manure application determines abundance, diversity and activity of methanogenic Archaea in arable European soils. Environ. Microbiol. 9:612–624 [DOI] [PubMed] [Google Scholar]
- 22. Glissmann K., Chin K. J., Casper P., Conrad R. 2004. Methanogenic pathway and archaeal community structure in the sediment of eutrophic Lake Dagow: effect of temperature. Microb. Ecol. 48:389–399 [DOI] [PubMed] [Google Scholar]
- 23. Good I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264 [Google Scholar]
- 24. Hansen S., Mæhlum J. E., Bakken L. R. 1993. N2O and CH4 fluxes in soil influenced by fertilization and tractor traffic. Soil Biol. Biochem. 5:621–630 [Google Scholar]
- 25. Hartge K. H., Horn R. 1992. Die Physikalische Untersuchung von Böden. Enke Verlag, Stuttgart, Germany [Google Scholar]
- 26. Juottonen H., et al. 2005. Methanogen communities and bacteria along an ecohydrological gradient in a northern raised bog complex, Environ. Microbiol. 7:1547–1557 [DOI] [PubMed] [Google Scholar]
- 27. Kemnitz D., Chin K. J., Bodelier P., Conrad R. 2004. Community analysis of methanogenic archaea within a riparian flooding gradient. Environ. Microbiol. 6:449–462 [DOI] [PubMed] [Google Scholar]
- 28. Kettunen A., et al. 1999. Methane production and oxidation potentials in relation to water table fluctuations in two boreal mires. Soil Biol. Biochem. 31:1741–1749 [Google Scholar]
- 29. Kotsyurbenko O. R., et al. 2004. Acetoclastic and hydrogenotrophic methane production and methanogenic populations in an acidic West-Siberian peat bog. Environ. Microbiol. 6:1159–1173 [DOI] [PubMed] [Google Scholar]
- 30. Le Mer J., Roger P. 2001. Production, oxidation, emission and consumption of methane by soils: a review. Eur. J. Soil Biol. 37:25–50 [Google Scholar]
- 31. Lueders T., Chin K. J., Conrad R., Friedrich M. 2001. Molecular analyses of methyl-coenzyme M reductase alpha-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3:194–204 [DOI] [PubMed] [Google Scholar]
- 32. Luton P. E., Wayne J. M., Sharp R. J., Riley P. W. 2002. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148:3521–3530 [DOI] [PubMed] [Google Scholar]
- 33. Mayer H. P., Conrad R. 1990. Factors influencing the population of methanogenic bacteria and the initiation of methane production upon flooding of paddy soil. FEMS Microbiol. Ecol. 73:103–112 [Google Scholar]
- 34. McInerney M. J., et al. 2008. Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann. N. Y. Acad. Sci. 1125:58–72 [DOI] [PubMed] [Google Scholar]
- 35. Menyailo O. V., Hungate B. A., Abraham W. R., Conrad R. 2008. Changing land use reduces soil CH4 uptake by altering biomass and activity but not composition of high-affinity methanotrophs. Glob. Chang. Biol. 14:2405–2419 [Google Scholar]
- 36. Mosier A. R., Schimel D., Valentine D., Bronson K., Parton W. 1991. Methane and nitrous oxide fluxes in native, fertilized and cultivated grasslands. Nature 350:330–332 [Google Scholar]
- 37. Newberry C. J., et al. 2004. Diversity of prokaryotes and methanogenesis in deep subsurface sediments from the Nankai Trough, Ocean Drilling Program Leg 190. Environ. Microbiol. 6:274–287 [DOI] [PubMed] [Google Scholar]
- 38. Pesaro M., Widmer F. 2002. Identification of novel Crenarchaeota and Euryarchaeota clusters associated with different depth layers of a forest soil. FEMS Microbiol. Ecol. 42:89–98 [DOI] [PubMed] [Google Scholar]
- 39. Peters V., Conrad R. 1995. Methanogenic and other strictly anaerobic bacteria in desert soil and other oxic soils. Appl. Environ. Microbiol. 61:1673–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Radl V., et al. 2007. Effects of cattle husbandry on abundance and activity of methanogenic archaea in upland soils. ISME J. 1:443–452 [DOI] [PubMed] [Google Scholar]
- 41. Ruser R., Flessa H., Schilling R., Steindl R., Beese F. 1998. Soil compaction and fertilization effects on nitrous oxide and methane fluxes in potato fields. Soil Sci. Soc. Am. J. 62:1587–1595 [Google Scholar]
- 42. Schnürr-Putz S., Baath E., Guggenberger G., Drake H. L., Küsel K. 2006. Compaction of forest soil by logging machinery favors occurrence of prokaryotes. FEMS Microbiol. Ecol. 58:503–516 [DOI] [PubMed] [Google Scholar]
- 43. Singleton D. R., Furlong M. A., Rathbun S. L., Whitman W. B. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Singh B. K., et al. 2007. Effect of afforestation and reforestation of pastures on the activity and population dynamics of methanotrophic bacteria. Appl. Environ. Microbiol. 73:5153–5161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sitaula B. K., Hansen S., Sitaula I. B., Bakken L. R. 2000. Methane oxidation and fluxes in agricultural soil: effects of fertilisation and soil compaction. Biogeochemistry 48:323–339 [Google Scholar]
- 46. Smith K. A., et al. 2003. Exchange of greenhouse gases between soil and atmosphere: interactions of soil physical factors and biological processes. Eur. J. Soil Sci. 54:779–791 [Google Scholar]
- 47. Smith J. M., Castro H., Ogram A. 2007. Structure and function of methanogens along a short-term restoration chronosequence in the Florida Everglades. Appl. Environ. Microbiol. 73:4135–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smith K. A., et al. 2000. Oxidation of atmospheric methane in Northern European soils, comparison with other ecosystems, and uncertainties in the global terrestrial sink. Glob. Chang. Biol. 6:791–803 [Google Scholar]
- 49. Tate K. R., et al. 2007. Methane uptake in soils from Pinus radiata plantations, a reverting shrubland and adjacent pastures: effects of land-use change, and soil texture, water, and mineral nitrogen. Soil Biol. Biochem. 39:1437–1449 [Google Scholar]
- 50. Tebrügge F., Düring R. A. 1999. Reduced tillage intensity: a review of results from a long-term study in Germany. Soil Till. Res. 53:15–28 [Google Scholar]
- 51. Teepe R., Brumme R., Beese F., Ludwig B. 2004. Nitrous oxide emission and methane consumption following compaction of forest soils. Soil Sci. Soc. Am. J. 68:605–611 [Google Scholar]
- 52. Van de Peer Y., De Wachter R. 1994. Treecon for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569–570 [DOI] [PubMed] [Google Scholar]
- 53. Yashiro Y., Rashidah Kadir W., Okuda T., Koizumi H. 2008. The effects of logging on soil greenhouse gas (CO2, CH4, N2O) flux in a tropical rain forest, Peninsular Malaysia. Agric. For. Meteorol. 148:799–806 [Google Scholar]
- 54. Wagner D., Pfeiffer E. M., Bock E. 1999. Methane production in aerated marshland and model soils: effects of microflora and soil texture. Soil Biol. Biochem. 31:999–1006 [Google Scholar]
- 55. Whitman W. B., Bowen T. L., Boone D. R. 2006. The methanogenic bacteria, p. 165–207. In Dworkin M., et al. (ed.), The prokaryotes, vol. 3 Springer-Verlag, New York, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.