Abstract
Numerous applications of conventional and biogenic magnetic nanoparticles (MNPs), such as in diagnostics, immunomagnetic separations, and magnetic cell labeling, require the immobilization of antibodies. This is usually accomplished by chemical conjugation, which, however, has several disadvantages, such as poor efficiency and the need for coupling chemistry. Here, we describe a novel strategy to display a functional camelid antibody fragment (nanobody) from an alpaca (Lama pacos) on the surface of bacterial biogenic magnetic nanoparticles (magnetosomes). Magnetosome-specific expression of a red fluorescent protein (RFP)-binding nanobody (RBP) in vivo was accomplished by genetic fusion of RBP to the magnetosome protein MamC in the magnetite-synthesizing bacterium Magnetospirillum gryphiswaldense. We demonstrate that isolated magnetosomes expressing MamC-RBP efficiently recognize and bind their antigen in vitro and can be used for immunoprecipitation of RFP-tagged proteins and their interaction partners from cell extracts. In addition, we show that coexpression of monomeric RFP (mRFP or its variant mCherry) and MamC-RBP results in intracellular recognition and magnetosome recruitment of RFP within living bacteria. The intracellular expression of a functional nanobody targeted to a specific bacterial compartment opens new possibilities for in vivo synthesis of MNP-immobilized nanobodies. Moreover, intracellular nanotraps can be generated to manipulate bacterial structures in live cells.
INTRODUCTION
Magnetic nanoparticles (MNPs) are used in a wide range of biomedical in vivo and in vitro applications (6, 31). A novel class of biogenic MNPs with unique characteristics is represented by the magnetosome particles of magnetotactic bacteria (MTB). Magnetosomes are organelles for magnetic orientation and consist of membrane-enveloped magnetite (Fe3O4) particles aligned in well-ordered intracellular chains (14). Magnetite biomineralization occurs within dedicated vesicles formed by the magnetosome membrane (MM), which invaginates from the cytoplasmic membrane and contains a number of specific proteins that are involved in the synthesis of functional magnetosome particles (7, 14, 15, 17). Due to the strict biological control over their biomineralization, magnetosomes have a number of unusual attributes, such as high crystallinity, strong magnetization, and uniform shapes and sizes (typically between 30 and 120 nm), which are difficult to achieve by artificial synthetic approaches (4). In addition, crystal morphologies and the composition of the enveloping MM can be manipulated at the genetic level (4, 21, 22). These characteristics have attracted considerable interest in using magnetosomes as biogenic MNPs in a number of potential applications, such as magnetic separation and detection of analytes, as contrast agents in magnetic resonance imaging, and to generate heat in magnetic hyperthermia (12, 26, 41, 44). Many of these applications depend on the functionalization of isolated magnetosome particles, for instance by the magnetosome-specific display of functional moieties, such as enzymes, coupling groups, gold particles, or oligonucleotides (3, 21, 22, 24, 25, 44).
Applications of conventional and biogenic MNPs in diagnostics, immunomagnetic separations, and magnetic cell labeling require the immobilization of antibodies to the particles (2, 11, 37). For bacterial magnetosomes, this has been achieved by chemical coupling of fluorescein isothiocyanate (FITC)-conjugated monoclonal anti-Escherichia coli antibody (29). Alternatively, display of the IgG-binding ZZ domain of protein A fused to the magnetosome protein MamC (Mms13) in Magnetospirillum magneticum (27) and Magnetospirillum gryphiswaldense (20) resulted in magnetosomes that bind IgG molecules after the isolation of particles from bacteria. However, in vitro coupling of antibodies often requires additional chemistry and is not very efficient.
Alternatively, it has been demonstrated that entire foreign proteins, such as GFP (green fluorescent protein) (23), and even multisubunit complexes like RNase P (30) can be expressed directly on the surface of magnetosomes by genetic fusions to magnetosome proteins, which might also provide a synthetic route for antibody immobilization. However, heterologous expression of conventional antibodies in bacterial systems is hampered by impaired disulfide bond formation in the reducing cytoplasm and inefficient assembly of the light and heavy chains, which requires cosecretion of the variable domains into the periplasmatic space, where protein folding occurs correctly (10, 42). An alternative to conventional antibodies are heavy-chain antibodies (HCAbs) that lack the light chains and are formed by camelids, such as camels, dromedaries, and alpacas (8). HCAbs recognize and bind their antigens via a single variable domain (referred to as VHH or nanobody), which comprises the smallest intact antigen binding fragment (∼15 kDa) known (28). Specific nanobodies can be easily selected from large libraries by display technologies. Due to their small size and rigid folding, nanobodies are highly soluble and stable and can be efficiently expressed in microbial systems like yeast or bacteria (5, 32, 33).
It has been already demonstrated that nanobodies are functional in the cytoplasm of eukaryotic cells. In a recent major advance, Rothbauer et al. (35) developed so-called chromobodies comprising an antigen-specific VHH domain linked to a fluorescent protein. Chromobodies can target their antigen and trace the dynamics of cellular components in real time and can be used for protein modulation and intracellular localization within living human (HeLa) (16) and plant cells (38). It has been further shown that a GFP-specific nanobody (GBP, GFP binding protein) is suitable for expression and localization in vivo. Immobilization of this GBP nanobody at distinct cellular structures like the nuclear lamina generates a nanotrap, which enables efficient targeting of GFP fusion proteins to distinct subcellular structures in eukaryotic cells (34).
Here, we describe the immobilization of a red fluorescent protein (RFP)-binding nanobody (RBP) on magnetosomes of M. gryphiswaldense by fusion of the RBP to the MM protein MamC. We demonstrate that isolated magnetosomes expressing MamC-RBP efficiently recognize their antigen in vitro and can be used for immunoprecipitation of RFP-tagged proteins and their interaction partners from cell extracts. In addition, we show that coexpression of monomeric RFP (mRFP, in its variant mCherry) and MamC-RBP results in intracellular recognition and magnetosome recruitment of RFP in M. gryphiswaldense.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
All M. gryphiswaldense strains used in this study are shown in Table 1. The M. gryphiswaldense strains were grown microaerobically at 30°C in modified FSM medium (13) as described before (23). For plate cultivation, agar was added to 1.5% (wt/vol) to FSM. Conjugation experiments were performed as described previously (23) with E. coli strain BW29427 [thrB1004 pro thi rpsL hsdS lacZΔM15 RP4-1360 Δ(araBAD)567 ΔdapA1341::(erm+ pir+)] (K. Datsenko and B. L. Wanner, unpublished) as the donor. E. coli strain DH5α (9) was used for DNA cloning. All E. coli strains were grown on Luria-Bertani (LB) medium at 37°C supplemented with kanamycin (25 μg ml−1) or ampicillin (50 μg ml−1) and 1.5% (wt/vol) agar, if appropriate (36). For cultivation of E. coli BW29427, growth media were supplemented with dl-α,ε-diaminopimelic acid (1 mM).
Table 1.
M. gryphiswaldense strains and plasmids used in this study
| Strain or vector | Description | Reference or source |
|---|---|---|
| M. gryphiswaldense strains | ||
| MSR-1 R3/S1 | Rifr Smr, spontaneous mutant | 40 |
| MSR-1B | Spontaneous nonmagnetic mutant | 39 |
| Vectors | ||
| pBBR1MCS-2 | Mobilizable broad-host-range vector; Kmr | 18 |
| pJET1.2/blunt | Cloning vector; Apr | Fermentas |
| pBBRPmamDC | egfp expression vector with PmamDC promoter; Kmr | 20 |
| pRBP | Plasmid expressing rbp; Apr | ChromoTek GmbH |
| pJET1.2 mamC N1 | pJET1.2/blunt + mamC flanked with NdeI restriction sites; Apr | C. Lang, unpublished |
| pBBRPtetmcherry | Plasmid containing mcherry gene; Kmr | C. Lang, unpublished |
| pAP 176 | pJET1.2/blunt with NdeI-BamHI rbp fragment; Apr | This study |
| pAP 178 | pBBRPmamDC with rbp from pAP 176 instead of egfp; Kmr | This study |
| pAP 179 | pAP 178 with mamC from pJET1.2 mamC N1; Kmr | This study |
| pAP 182 | pBBR1MCS-2 with SacI-SacI PmamDC-mcherry fragment; Kmr | This study |
| pAP 183 | pAP 179 with SacI-SacI PmamDC-mcherry fragment; Kmr | This study |
Recombinant DNA techniques.
All recombinant DNA techniques were conducted according to standard procedures (36). DNA fragments were amplified for subsequent cloning by PCR using Phusion high-fidelity DNA polymerase (Finnzymes). Primers were purchased from Sigma-Aldrich (Steinheim, Germany). Cloned genes and fusion constructs were sequenced using BigDye Terminator v3.1 chemistry with an in-house ABI 3730 48 capillary sequencer (Applied Biosystems). All sequences were checked and aligned using DNASTAR SeqMan II software (DNASTAR, Inc.).
Construction of pAP179 encoding the MamC-RBP fusion protein.
The rbp gene was PCR amplified from an RBP-encoding plasmid (ChromoTek, unpublished) by using the forward primer 5′-GGAATTCCAT ATGTCGGGCT CGGGCTCGGG CTCGGGCTCG GGCGCTCAGG TGCAGCTGGT GGAG-3′, introducing an NdeI restriction site and a glycine serine linker to the 5′ end, and the reverse primer 5′-CGCGGATCCT CCCTAGCTGG AGACGGTGAC CTGGGTC-3′, introducing a BamHI site to the 3′ end of the rbp gene. The 404-bp PCR product was purified and cloned into the pJET1.2/blunt vector (Fermentas) to create pAP176. Excision of the rbp gene from pAP176 with NdeI and BamHI and cloning this fragment into pBBRPmamDC (20) by replacing egfp from pBBRPmamDC with rbp yielded pAP178. The mamC gene was amplified from M. gryphiswaldense R3/S1 using primers mamCNdeIfw (5′-CATATGAGCT TTCAACTTGC G-3′) and mamCrevNdeI (5′-CATATGGGCC AATTCTTCCC TCAG-3′) and cloned into pJET1.2 (Fermentas). mamC (378 bp) was excised with NdeI and cloned into the NdeI restriction site upstream of the rbp gene in pAP178 to yield pAP179.
Construction of pAP183 und pAP182.
The rfp (mcherry) gene was PCR amplified from pBBRPtetmcherry (C. Lang, unpublished data) by using the primer pair PdcmcherrySacIfw (5′-GAGCTCCTTT TTCGCTTTAC TAGCTCTTAG TTCTCCAATA AATTCCCTGC GAGGAGATCA GCATATGGCA ACTAGCGGCA TGGT-3′) and mcherrySacIrev (5′-GAGCTCTTAT TTGTATAGTT CATCCATG-3′). The sequence of forward primer Pdcmcherry SacIfw includes the PmamDC promoter and a strong ribosome binding site, generating a PCR product of 777 bp. Both primers were designed to add a SacI restriction site at the 5′ and 3′ ends of the amplified mcherry gene. The PCR product was purified with a nucleoSpin extract II kit (Macherey & Nagel, Düren, Germany) and cloned into the SacI restriction site of the pAP179 plasmid to create the plasmid pAP183. This PCR product was also cloned into the SacI site of the pBBR1MCS-2 plasmid to yield pAP182, which contains the mcherry gene but not the mamC-rbp fusion.
Construction of pCMV-c-Jun-mRFP full length, pCMV-c-Jun 1-145-mRFP, and pCMV-c-Fos-eGFP.
The coding sequences of the human c-Fos and c-Jun genes were excised from pSV-c-Fos-EGFP (BamHI/EcoRI) and pSV-cJun-mRFP1 (HindIII/BamHI) vectors kindly provided by J. Langowski and inserted into pEGFP-C1 (Clontech) or pmRFP-N2 vector, respectively. The sequence coding for the first 145 amino acids of Jun was amplified by PCR using the forward primer 5′-CCCAAGCTTA TGACTGCAAA GATGGAAAC-3′ inserting a HindIII restriction site and the reverse primer 5′-CGGGATCCCG GGGAGCCACC ATGCCTGCC-3′ inserting a BamHI restriction site. The PCR product (451 bp) was cloned by HindIII and BamHI into the pmRFP-N2 vector.
Microscopy.
DNA of M. gryphiswaldense cells was DAPI (4′,6-diamidino-2-phenylindole) stained using 0.5 mM EDTA and DAPI (1 μg/ml). The stained cells were mounted in 5 μl ProLong gold antifade reagent (Invitrogen) and analyzed with an Olympus IX81 fluorescence microscope equipped with an Orca-ER (Hamamatsu) camera. The excitation wavelengths for DAPI and mCherry were 358 and 587 nm, and emission was recorded at 461 and 610 nm, respectively. Digital images were analyzed with cellM software (Olympus).
Biochemical methods.
Magnetosome isolation from M. gryphiswaldense strains was done as described previously (23). Polyacrylamide gels were prepared according to the method of Laemmli (19). Protein concentrations were determined by the bicinchoninic acid protein kit (Sigma-Aldrich), and 15 μg of magnetosome protein was loaded onto 15% (wt/vol) SDS gels and analyzed by immunoblotting to verify expression and stability of the MamC-RBP fusion protein.
Proteins were electroblotted onto nitrocellulose membranes (Protran [Whatman, Germany]). Membranes were stained with Ponceau S solution (0.1% [wt/vol] in 5% [vol/vol] acetic acid) for general protein detection. Ponceau S was removed from the membranes by washing several times with water. Membranes were blocked for 1 h at room temperature. Primary rabbit anti-MamC IgG antibody (1:500 dilution [Pineda, Germany]) was added to the blocking solution and incubated 1 h at room temperature. Membranes were washed with Tween-Tris-buffered saline (TTBS) and Tris-buffered saline (TBS) and incubated with secondary antibody horseradish peroxidase-labeled goat anti-rabbit IgG (1:5,000 dilution [Dianova GmbH]) for 45 min. Membranes were washed with TBS, and immunoreactive proteins were visualized by using an enhanced chemiluminescence kit (Amersham ECL Plus Western blotting detection reagents [GE Healthcare]) and exposing the membranes to X-ray film (Super RX, Fuji medical X-ray film [Fujifilm]).
Affinity measurements.
For affinity measurements, RBP and mRFP were expressed in E. coli and purified according to established protocols (34). Binding kinetics were analyzed with an Attana A100 C-Fast system. RBP was covalently immobilized on a carboxyl chip via N-hydroxysuccinimide (NHS) esterification resulting in a 90-Hz signal increase. Experiments were performed in HBST running buffer (10 mM HEPES, 150 mM NaCl, 0.005% Tween, pH 7.4); the flow rate was set to 25 μl/min. mRFP was injected at four different concentrations (5, 2.5, 1.25, 0.6125 μg/ml); the association time was set to 80 s, and dissociation was recorded for 220 s. Surface regeneration was carried out with 10 mM clycin-Cl (pH 2.5). Data were recorded with Attester 3.0 (Attana, Sweden), and data fitting and rate constant calculation were performed with ClampXP, using a mass transport model.
IP of recombinant mCherry.
Magnetosomes equivalent to 2 mg iron determined as described before (23) were washed with immunoprecipitation (IP) buffer (10 mM Tris-HCL, 150 mM NaOH, pH 7.5), resuspended in 300 μl IP buffer containing 6 μg mCherry (input), and incubated for 4 h at 4°C at constant rotation. After sedimentation of the magnetosomes by centrifugation or magnetic separation, 10 μl of the supernatant (flowthrough) and 10 μl of the input were loaded onto SDS gels and analyzed by immunoblotting. Magnetosomes were washed three times in 500 μl IP buffer and resuspended in 30 μl IP buffer. An aliquot (20 μl) was loaded onto the SDS gel for Western blot analysis, which was carried out as described using rat anti-mCherry IgG antibody (1:250 dilution [ChromoTek, Germany]) as the primary antibody and horseradish peroxidase-labeled goat anti-rat IgG (1:5,000 dilution [Dianova GmbH]) as the secondary antibody.
Immunoprecipitation of c-Jun-RFP.
BHK cells transfected with pCMV-c-Fos-EGFP and pCMV-c-Jun-mRFP or pCMV-c-Jun1-145-mRFP were lysed in 200 μl lysis buffer (10 mM Tris-Cl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1× mammalian protease inhibitor [Serva]). After sonification, cell lysates were centrifuged for 15 min, at 4°C and 13,000 rpm. Supernatant was diluted in 500 μl dilution buffer (10 mM Tris-Cl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 1 mM PMSF, 1× mammalian protease inhibitor [Serva]). Diluted lysate was added to magnetosomes equivalent to 1.3 mg iron equilibrated in dilution buffer. Pulldown was performed for 2 h at 4°C. As an alternative to magnetic separation, magnetosomes were pelleted by centrifugation at 2,000 rpm for 2 min at 4°C. After removal of the flowthrough, magnetosomes were washed two times in 500 μl of dilution buffer. Finally, magnetosomes were resuspended in SDS sample buffer and pulldown was analyzed by immunoblotting using rat anti-mCherry IgG antibody, rat anti-3H9 GFP antibody (Chromotek, Germany), and horseradish peroxidase-labeled goat anti-rat IgG (1:5,000 dilution [Dianova GmbH]) as the secondary antibody.
RESULTS
RBP is functionally expressed on magnetosomes by fusion to MamC.
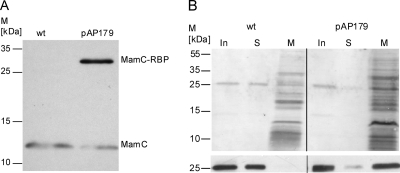
As described before, heterologous fusion of a protein to the abundant MM protein MamC results in targeting of the fusion protein to the MM. For magnetosome display, we translationally fused an RFP-binding nanobody (RBP) to the abundant MM protein MamC as a targeting magnetosome anchor. The mamC-rbp fusion was placed under the control of the PmamDC promoter (23) on pAP179 and conjugated into the M. gryphiswaldense wild-type (WT) strain for magnetosome expression (Fig. 1 A). After isolation of magnetosomes from a strain harboring pAP179, expression of the MamC-RBP fusion protein was analyzed by Western blotting. Whereas only one single band corresponding to the native (12.5-kDa) MamC protein was detected in Western blots of WT magnetosome proteins, an additional band at 27 kDa, reflecting the predicted size of the MamC-RBP fusion protein, was recognized in M. gryphiswaldense containing pAP179 (Fig. 2 A). This result indicated that the full-length MamC-RBP protein was highly expressed in M. gryphiswaldense and incorporated into the MM in addition to the unfused native MamC protein.
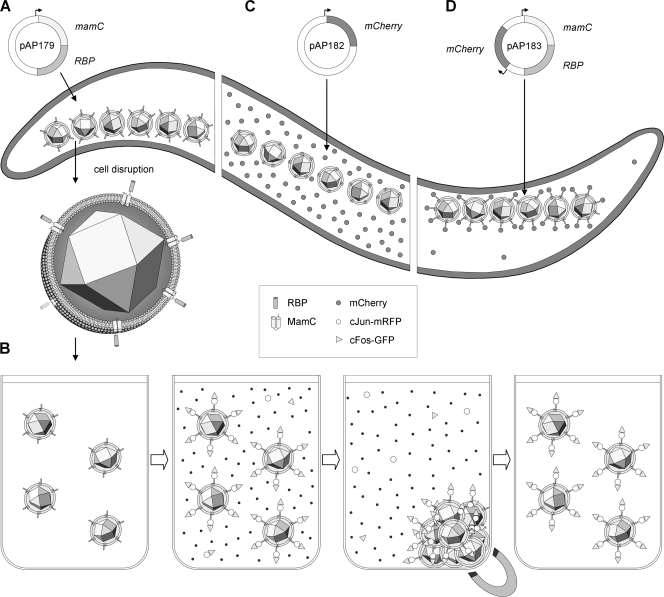
Fig. 1.
Schematic overview of the approach for MamC-RBP and RFP/mCherry expression in M. gryphiswaldense and the application of modified magnetosomes for immunoprecipitation. (A) Transformation of bacteria with pAP179 resulted in expression and magnetosome targeting of MamC-RBP fusion protein due to MamC serving as an MM anchor. (B) Application of MamC-RBP-decorated magnetosomes for coimmunoprecipitation (IP). The modified magnetosomes were isolated from disrupted cells, equilibrated in IP buffer, and incubated with RFP-tagged c-Jun protein. Interacting c-Fos-GFP protein was pulled down after incubation with cell lysate (Co-IP). The bound proteins were separated magnetically or by centrifugation, whereas residual noninteracting proteins were removed by washing. (C) mCherry alone is expressed from pAP182 and disperses in the cytoplasm in the absence of MamC-RBP. (D) Coexpression of MamC-RBP and mCherry from pAP183 results in efficient recruitment of the soluble mCherry protein to the magnetosomes expressing MamC-RBP and its depletion from cytoplasm.
Fig. 2.
Expression of MamC-RBP on isolated magnetosomes. (A) Immunoblot analysis of proteins solubilized from isolated magnetosomes of WT cells and cells harboring pAP179 (MamC-RBP). Blots were probed with an anti-MamC antibody. (B) Binding of mCherry to RBP-expressing magnetosomes. An mCherry-containing solution was incubated with isolated magnetosomes of WT cells and cells harboring pAP179 (MamC-RBP). Protein fractions from the input (In), supernatant (S), and bound magnetosome particles (M) were resolved by SDS-PAGE, blotted to a nitrocellulose membrane, Ponceau stained (top), and probed with an anti-mCherry antibody (bottom). M, molecular mass.
For functional analysis, we tested whether isolated magnetosomes expressing MamC-RBP are able to pull down recombinantly expressed mCherry (mRFP). Magnetosomes from untransformed WT strain and WT strain harboring pAP179 were incubated with purified mCherry. After incubation, separation, and washing of magnetosome particles, the input, supernatant, and magnetosome fractions were analyzed by Western blotting. Bands of about 28 kDa, corresponding to the molecular mass of purified mCherry of identical intensities, were found in the input and supernatant before and after incubation with WT magnetosomes, respectively, but not in the magnetosome fraction from the WT strain. In contrast, only minor amounts of mCherry could be detected in the flowthrough fraction after incubation with magnetosomes expressing MamC-RBP (M. gryphiswaldense containing pAP179), whereas a strong band was present in the magnetosome fraction from this strain after incubation (Fig. 2B, bottom). mCherry protein was also detectable on the Ponceau S-stained membrane (Fig. 2B, top). These results clearly indicated that the mCherry protein was largely depleted from the input fraction and had bound to the magnetosomes expressing MamC-RBP. Due to bound mCherry, these magnetosomes displayed strong red fluorescence if analyzed under the microscope (see Fig. S1 in the supplemental material).
Immunoprecipitation of an RFP-tagged protein and its interaction partner from cell extracts by RBP-expressing magnetosomes.
We investigated whether magnetosome-bound RBP can recognize and specifically bind its antigen from complex protein mixtures. First, to analyze the binding kinetics of the RBP-mRFP interaction in vitro, we first subcloned the RBP cDNA into expression vector pHEN6, thereby adding a 6×His tag for immobilized metal ion adsorption chromatography (IMAC) purification. Affinity measurements with purified proteins were performed by quartz crystal microbalance (QCM), revealing association rate constant ka (M−1 s−1) and dissociation rate constant kd (s−1) values of 1.5 × 105 ± 1,477 and 6.6 × 10−4 ± 5.6 × 10−6, respectively, which results in a binding affinity (equilibrium dissociation constant [KD]) of 4.4 nM (for ligand RBP and analyte mRFP) (see Fig. S2 in the supplemental material).
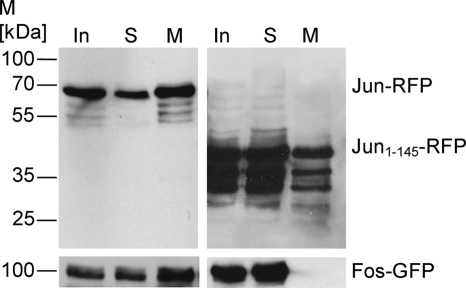
Next, we performed immunoprecipitation of a native nuclear protein from soluble mammalian cell extracts (Fig. 1B). For initial experiments, we chose the RFP-labeled Jun protein, which is a transcription factor that is known to form a heterodimer with another transcription factor, Fos (43). Incubation of magnetosomes expressing MamC-RBP with cell lysates of BHK cells coexpressing c-Jun-RFP and c-Fos-GFP resulted in an efficient pulldown of c-Jun-RFP and a coprecipitation of c-Fos-GFP (Fig. 3, left). As expected, no coprecipitation of c-Fos-GFP was observed when c-Jun1-145-RFP, a mutant lacking the DNA-binding and dimerization domain (1), was pulled down using magnetosomes expressing MamC-RBP (Fig. 3, right). These results showed that RBP magnetosomes specifically precipitated an RFP-tagged protein and its interaction partner from complex cell extracts.
Fig. 3.
Immunoblots of immunoprecipitation fractions of BHK cell extracts. Immunoblot analysis of Jun-RFP probed with an anti-RFP antibody show signals in all three fractions of immunoprecipitation (top left). Coprecipitated Fos-GFP is also detectable in each fraction due to the interaction of Jun and Fos (bottom left). Immunoblot of truncated Jun1-145-RFP shows signals in each fraction of the immunoprecipitation (top right), whereas Fos-GFP is not detectable in the magnetosome fraction due to the loss of the Fos binding site in truncated Jun1-145 (bottom right). M, molecular mass.
Intracellular binding and antigen recognition of mCherry within M. gryphiswaldense cells.
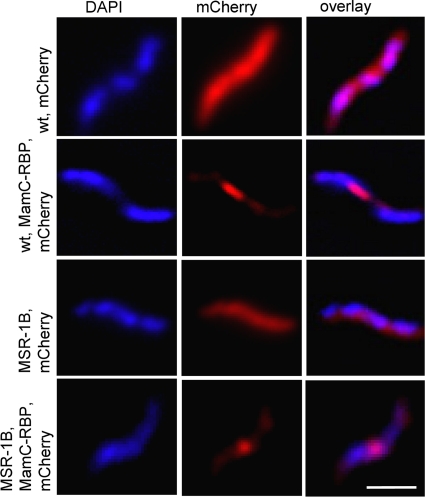
Next, we investigated the ability of magnetosome-bound RBP to access and bind its antigen in vivo if coexpressed within cells of M. gryphiswaldense. To this end, we generated two constructs: pAP182 expressing mCherry alone under the control of a PmamDC promoter construct (Fig. 1C), and pAP183, in which in addition, the mamC-rbp fusion is transcribed from PmamDC promoter (20) and inserted in tandem with PmamDC::rfp (Fig. 1D). If the fusion protein MamC-RBP is functional in vivo, the mCherry protein should be recruited onto the surface of the magnetosomes due to RBP::mCherry binding. Fluorescence microscopy of M. gryphiswaldense cells expressing mCherry alone from pAP182 revealed a strong red fluorescence of the entire cytoplasm (Fig. 4), which is consistent with the expected cytoplasmic expression of the soluble mCherry protein. In striking contrast, M. gryphiswaldense cells that harbored pAP183 coexpressing MamC-RBP and mCherry showed a linear fluorescence signal, which was confined to the characteristic intracellular position of magnetosome chains, whereas the cytoplasm no longer displayed detectable fluorescence (Fig. 4). This suggested that intracellularly expressed mCherry protein became depleted from the cytoplasm and bound to the magnetosomes upon coexpression of MamC-RBP in the MM. In contrast, coexpression of MamC-RBP and mCherry in a magnetosome-free background, i.e., in strain MSR-1B, which is nonmagnetic due to a large chromosomal deletion within the magnetosome island (39), showed an irregular, spotted fluorescence pattern (Fig. 4) that was very similar to the localization of a MamC-GFP fusion in MSR-1B (Lang, unpublished).
Fig. 4.
Fluorescence micrographs of representative cells of M. gryphiswaldense WT expressing pAP182 and pAP183 and M. gryphiswaldense strain MSR-1B expressing pAP182 and pAP183. Left: DAPI fluorescence signals. Middle: mCherry signal from the same cell. Right: overlay of DAPI (blue) and mCherry (red) signals. Bar, 3 μm.
DISCUSSION
In this work, we accomplished the functional magnetosome display of a 14.6-kDa RFP binding fragment derived from an alpaca single-chain antibody (RBP). RBP binds monomeric red fluorescent proteins with nanomolar affinity, making it a potent binding entity for biotechnological applications. Translational fusion of RBP to the magnetosome protein MamC resulted in RBP magnetosomes, which exhibited binding activity to its cognate antigen in vitro and in vivo. In situ expression and immobilization of nanobodies as fusions to specific magnetosome anchors has several advantages over previous approaches, which attempted antibody immobilization on isolated magnetosomes in vitro by either chemical coupling (29) or coupling to the magnetosome-expressed IgG-binding ZZ domain of protein A in M. magneticum (45) and M. gryphiswaldense (20). First, in vivo genetic coupling is independent of chemical reagents, additional connectors, and purified antibodies. Second, it provides tight covalent binding to abundant, autochthonous magnetosome proteins. RBP magnetosomes allowed the specific pulldown of an RFP-tagged Jun protein, which could be selectively precipitated together with its interacting partner from BHK cell extracts. In future approaches this might be used for the production and application of magnetosome-immobilized nanobodies for immunoprecipitation of antigens from complex samples such as cell lysates or blood sera. This may also provide easy magnetic manipulation of a nanotrap for in vitro binding of target proteins and identification of their interactors.
A third advantage of in vivo coupling of nanobodies is that it can also be used for application within living bacterial cells. Magnetosome expression of RBP resulted in intracellular depletion of cytoplasmic mCherry and recruitment to the magnetosome membrane as indicated by the altered localization from diffuse cytoplasmic to linear (magnetosome-bound) localization upon coexpression of mCherry and RBP. This indicates that nanobodies can be expressed in the reducing environment of the bacterial cytoplasm in a fully functional form.
In eukaryotic systems, it has also already been shown that GFP-binding nanobodies can selectively alter phenotypes mediated by the targeted proteins. For example, protein properties such as intracellular localization, conformation, and spectral properties could be modulated in living human (HeLa) cells (16, 34). In plants, GBP-RFP could be applied to interfere with the function of a GFP fusion protein and to mislocalize (trap) GFP fusions to ectopic intracellular localizations (38). While overexpression of recombinant nanobodies in E. coli was described previously (32), this is the first report of the functional expression of a nanobody targeted to a particular subcellular compartment in living bacteria. The successful in vivo expression of functional nanobodies thus may extend the use of chromobody technology to bacterial cells. By binding nanobodies to bacterial organelles, specific compartments or other spatial determinants within bacterial cells, intracellular nanotraps that allow functional studies and manipulation of bacterial intracellular structures might be constructed.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (grant DFG Schu1080/12-1 to D.S.) and the GO-Bio program (BMBF) for U.R., T.R., and J.H. U.R. and H.L. are shareholders of ChromoTek GmbH, which currently has the exclusive right to commercialize and distribute RFP-binding proteins derived from single-domain antibodies. The use of such molecules for diverse applications is within the scope of interest of ChromoTek GmbH.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Baudendistel N., Muller G., Waldeck W., Angel P., Langowski J. 2005. Two-hybrid fluorescence cross-correlation spectroscopy detects protein-protein interactions in vivo. Chemphyschem 6:984–990 [DOI] [PubMed] [Google Scholar]
- 2. Bhirde A., Xie J., Swierczewska M., Chen X. 2011. Nanoparticles for cell labeling. Nanoscale 3:142–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ceyhan B., Alhorn P., Lang C., Schüler D., Niemeyer C. M. 2006. Semisynthetic biogenic magnetosome nanoparticles for the detection of proteins and nucleic acids. Small 2:1251–1255 [DOI] [PubMed] [Google Scholar]
- 4. Faivre D., Schüler D. 2008. Magnetotactic bacteria and magnetosomes. Chem. Rev. 108:4875–4898 [DOI] [PubMed] [Google Scholar]
- 5. Frenken L. G., et al. 2000. Isolation of antigen specific llama VHH antibody fragments and their high level secretion by Saccharomyces cerevisiae. J. Biotechnol. 78:11–21 [DOI] [PubMed] [Google Scholar]
- 6. Frimpong R. A., Hilt J. Z. 2010. Magnetic nanoparticles in biomedicine: synthesis, functionalization and applications. Nanomedicine (Lond.) 5:1401–1414 [DOI] [PubMed] [Google Scholar]
- 7. Grünberg K., et al. 2004. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 70:1040–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hamers-Casterman C., et al. 1993. Naturally occurring antibodies devoid of light chains. Nature 363:446–448 [DOI] [PubMed] [Google Scholar]
- 9. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 10. Harmsen M. M., De Haard H. J. 2007. Properties, production, and applications of camelid single-domain antibody fragments. Appl. Microbiol. Biotechnol. 77:13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haun J. B., Yoon T. J., Lee H., Weissleder R. 2010. Magnetic nanoparticle biosensors. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2:291–304 [DOI] [PubMed] [Google Scholar]
- 12. Hergt R., et al. 2005. Magnetic properties of bacterial magnetosomes as diagnostic and therapeutic tools. J. Magn. Magn. Mater. 293:80–86 [Google Scholar]
- 13. Heyen U., Schüler D. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl. Microbiol. Biotechnol. 61:536–544 [DOI] [PubMed] [Google Scholar]
- 14. Jogler C., Schüler D. 2009. Genetics, genomics, and cell biology of magnetosome formation in magnetotactic bacteria. Annu. Rev. Microbiol. 63:501–521 [DOI] [PubMed] [Google Scholar]
- 15. Jogler C., et al. 2011. Conservation of proteobacterial magnetosome genes and structures in an uncultivated member of the deep-branching Nitrospira phylum. Proc. Natl. Acad. Sci. U. S. A. 108:1134–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kirchhofer A., et al. 2010. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17:133–138 [DOI] [PubMed] [Google Scholar]
- 17. Komeili A. 2007. Molecular mechanisms of magnetosome formation. Annu. Rev. Biochem. 76:351–366 [DOI] [PubMed] [Google Scholar]
- 18. Kovach M. E., et al. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 19. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 20. Lang C., Pollithy A., Schüler D. 2009. Identification of promoters for efficient gene expression in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 75:4206–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lang C., Schüler D. 2006. Biogenic nanoparticles: production, characterization, and application of bacterial magnetosomes. J. Phys. Condens. Matter 18:2815–2828 [Google Scholar]
- 22. Lang C., Schüler D. 2005. Biomineralization of magnetosomes in bacteria: nanoparticles with potential applications, p. 107–124In Rehm B.(ed.),Microbial bionanotechnology: biological self-assembly systems and biopolymer-based nanostructures. Horizon Scientific Press, Norfolk, United Kingdom [Google Scholar]
- 23. Lang C., Schüler D. 2008. Expression of green fluorescent protein fused to magnetosome proteins in microaerophilic magnetotactic bacteria. Appl. Environ. Microbiol. 74:4944–4953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maeda Y., Yoshino T., Matsunaga T. 2010. In vivo biotinylation of bacterial magnetic particles by a truncated form of Escherichia coli biotin ligase and biotin acceptor peptide. Appl. Environ. Microbiol. 76:5785–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maeda Y., et al. 2008. Noncovalent immobilization of streptavidin on in vitro- and in vivo-biotinylated bacterial magnetic particles. Appl. Environ. Microbiol. 74:5139–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsunaga T., Okamura Y., Tanaka T. 2004. Biotechnological application of nano-scale engineered bacterial magnetic particles. J. Mater. Chem. 14:2099–2105 [Google Scholar]
- 27. Matsunaga T., Takahashi M., Yoshino T., Kuhara M., Takeyama H. 2006. Magnetic separation of CD14+ cells using antibody binding with protein A expressed on bacterial magnetic particles for generating dendritic cells. Biochem. Biophys. Res. Commun. 350:1019–1025 [DOI] [PubMed] [Google Scholar]
- 28. Muyldermans S. 2001. Single domain camel antibodies: current status. J. Biotechnol. 74:277–302 [DOI] [PubMed] [Google Scholar]
- 29. Nakamura N., et al. 1993. Detection and removal of Escherichia coli using fluorescin isothiocyanate conjugated monoclonal antibody immobilized on bacterial magnetic particles. Anal. Chem. 65:2036–2039 [DOI] [PubMed] [Google Scholar]
- 30. Ohuchi S., Schüler D. 2009. In vivo display of a multisubunit enzyme complex on biogenic magnetic nanoparticles. Appl. Environ. Microbiol. 75:7734–7738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pankhurst Q. A., Connolly J., Jones S. K., Dobson J. 2003. Applications of magnetic nanoparticles in biomedicine. J. Phys. D Appl. Phys. 36:R167–R181 [Google Scholar]
- 32. Rahbarizadeh F., Rasaee M. J., Forouzandeh-Moghadam M., Allameh A. A. 2005. High expression and purification of the recombinant camelid anti-MUC1 single domain antibodies in Escherichia coli. Protein Expr. Purif. 44:32–38 [DOI] [PubMed] [Google Scholar]
- 33. Rahbarizadeh F., Rasaee M. J., Forouzandeh M., Allameh A. A. 2006. Overexpression of anti-MUC1 single-domain antibody fragments in the yeast Pichia pastoris. Mol. Immunol. 43:426–435 [DOI] [PubMed] [Google Scholar]
- 34. Rothbauer U., et al. 2008. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics 7:282–289 [DOI] [PubMed] [Google Scholar]
- 35. Rothbauer U., et al. 2006. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 3:887–889 [DOI] [PubMed] [Google Scholar]
- 36. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Sandhu A., Handa H., Abe M. 2010. Synthesis and applications of magnetic nanoparticles for biorecognition and point of care medical diagnostics. Nanotechnology 21:442001. [DOI] [PubMed] [Google Scholar]
- 38. Schornack S., et al. 2009. Protein mislocalization in plant cells using a GFP-binding chromobody. Plant J. 60:744–754 [DOI] [PubMed] [Google Scholar]
- 39. Schübbe S., et al. 2003. Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J. Bacteriol. 185:5779–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schultheiss D., Kube M., Schüler D. 2004. Inactivation of the flagellin gene flaA in Magnetospirillum gryphiswaldense results in non-magnetotactic mutants lacking flagellar filaments. Appl. Environ. Microbiol. 70:3624–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schwarz S., et al. 2009. Synthetic and biogenic magnetite nanoparticles for tracking of stem cells and dendritic cells. J. Magn. Magn. Mater. 312:1533–1538 [Google Scholar]
- 42. Skerra A., Plückthun A. 1988. Assembly of a functional immunoglobulin Fv fragment in Escherichia coli. Science 240:1038–1041 [DOI] [PubMed] [Google Scholar]
- 43. Vamosi G., et al. 2008. Conformation of the c-Fos/c-Jun complex in vivo: a combined FRET, FCCS, and MD-modeling study. Biophys. J. 94:2859–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wacker R., et al. 2007. Magneto immuno-PCR: a novel immunoassay based on biogenic magnetosome nanoparticles. Biochem. Biophys. Res. Commun. 357:391–396 [DOI] [PubMed] [Google Scholar]
- 45. Yoshino T., Matsunaga T. 2006. Efficient and stable display of functional proteins on bacterial magnetic particles using mms13 as a novel anchor molecule. Appl. Environ. Microbiol. 72:465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.